Significance
Therapeutic antibodies have revolutionized the treatment of human disease. Despite these advances, antibody bivalency limits their utility against some targets. Here, we describe the development of a one-armed (monovalent) antibody, onartuzumab, targeting the receptor tyrosine kinase MET. While initial screening of bivalent antibodies produced agonists of MET, engineering them into monovalent antibodies produced antagonists instead. We explain the structural basis of the mechanism of action with the crystal structure of onartuzumab antigen-binding fragment in complex with MET and HGF-β. These discoveries have led to an additional antibody-based therapeutic option and shed light on the underpinnings of HGF/MET signaling.
Keywords: scatter factor, HGFR, MetMAb, OA5D5
Abstract
Binding of hepatocyte growth factor (HGF) to the receptor tyrosine kinase MET is implicated in the malignant process of multiple cancers, making disruption of this interaction a promising therapeutic strategy. However, targeting MET with bivalent antibodies can mimic HGF agonism via receptor dimerization. To address this limitation, we have developed onartuzumab, an Escherichia coli-derived, humanized, and affinity-matured monovalent monoclonal antibody against MET, generated using the knob-into-hole technology that enables the antibody to engage the receptor in a one-to-one fashion. Onartuzumab potently inhibits HGF binding and receptor phosphorylation and signaling and has antibody-like pharmacokinetics and antitumor activity. Biochemical data and a crystal structure of a ternary complex of onartuzumab antigen-binding fragment bound to a MET extracellular domain fragment, consisting of the MET Sema domain fused to the adjacent Plexins, Semaphorins, Integrins domain (MET Sema-PSI), and the HGF β-chain demonstrate that onartuzumab acts specifically by blocking HGF α-chain (but not β-chain) binding to MET. These data suggest a likely binding site of the HGF α-chain on MET, which when dimerized leads to MET signaling. Onartuzumab, therefore, represents the founding member of a class of therapeutic monovalent antibodies that overcomes limitations of antibody bivalency for targets impacted by antibody crosslinking.
Monoclonal antibodies (mAbs) have revolutionized our arsenal of modern medicines (1). They allow target specificity that generally results in negligible off-target side effects. Despite the influx of antibodies into the clinic, limitations still remain with respect to selected targets. In particular, receptor tyrosine kinases (RTKs) are generally activated by binding to their respective ligands that dimerize or oligomerize RTKs, leading to kinase autoactivation (2, 3). This structural interplay between ligand and receptor is challenging for therapeutic antibodies because their bivalent nature can dimerize and agonize rather than antagonize their intended target. We describe a class of therapeutics with the generation of a mAb that is monovalent against its target, the MET RTK. This unique antibody, onartuzumab (MetMAb, or the anti-MET monovalent monoclonal antibody), consists of a single humanized and affinity-matured antigen-binding fragment (Fab) fused to a complete constant domain fragment (Fc) engineered to assemble in Escherichia coli through the use of “knob” and “hole” mutations in the CH3 domain within the Fc (4). This design has enabled large-scale production of a functional monovalent antibody that effectively antagonizes hepatocyte growth factor (HGF)/MET signaling.
MET signaling is initiated by binding to its cognate ligand HGF. HGF is secreted as a single-chain ligand (pro-HGF) that is proteolytically processed to generate a disulfide-linked α/β-heterodimer. The α-chain comprises an N-terminal plasminogen family, apple domain, and nematode protein (PAN) domain, followed by four Kringle domain repeats and the β-chain contains a C-terminal trypsin-like serine protease domain (5). Although both pro-HGF and HGF α/β-heterodimer (mature HGF) bind MET with high affinity (6, 7), signaling is elicited only by cleaved HGF.
MET consists of a seven-bladed β-propeller Semaphorin domain (Sema), a Plexin, Semaphorin, Integrin cysteine-rich domain (PSI), four Ig-like domains, a transmembrane region, a juxtamembrane region, and a kinase domain (3, 8). Binding of HGF to the MET Sema domain leads to receptor oligomerization and initiation of cell signaling that results in invasive growth (8). This enables HGF/MET to orchestrate complex cellular biology during embryogenesis (9), wound healing, and tissue repair (10–13). HGF/MET signaling has also been implicated in the metastatic growth of multiple cancers (8, 14), making it an attractive target for various therapeutic agents (14). Onartuzumab, derived from the 5D5 antibody previously shown to bind the MET Sema domain (15), has shown preclinical activity in glioblastoma (GBM), pancreatic cancer, and non–small-cell lung cancer (NSCLC), among other tumor types (16, 17). More recently, onartuzumab demonstrated significant activity in a phase I study in a gastric cancer patient (18, 19) and in a phase II trial in patients with NSCLC in combination with erlotinib (19).
Although antibodies against MET have been described that induce receptor shedding (20) or dimerization (21), development of therapeutic antibodies against MET has been hindered by bivalent antibody-induced crosslinking and consequent downstream signal activation (21). We describe the development of onartuzumab and reveal the ternary structure of the onartuzumab Fab in complex with Sema-PSI of MET bound to the HGF β-chain. The implications of our findings are discussed with respect to therapeutic development of onartuzumab and the mechanism for HGF-dependent activation of MET signaling.
Results
Identification of Monovalent Anti-MET Antibodies.
A protein consisting of the human MET (huMET) extracellular domain (residues 25–929) fused to an IgG1 (huMET-IgG) (7) was used in BALB/c mice to generate anti-MET antibody-producing hybridomas. Prospective mAb candidates were screened for their ability to bind to huMET, compete with human HGF (huHGF) binding, and inhibit proliferation of the Ba/F3-huMET mouse cell line (22) treated with or without huHGF. Although several of the MET-binding mAbs, including 5D5, had HGF-blocking function, none acted as pure antagonists of HGF-stimulated proliferation of Ba/F3-huMET (Fig. S1). Rather, most acted as weak to strong agonists leading to increasing cell growth. The most potent agonist was 5D5, which maximally stimulated Ba/F3-huMET cells regardless of the presence of HGF.
We hypothesized that the bivalency of these agonistic mAbs led to MET activation via receptor crosslinking. Thus, Fab fragments from the 5D5 antibody were generated and evaluated for MET-binding and HGF-binding competition and Ba/F3-huMET cell growth in the presence or absence of HGF. Whereas the 5D5 Fab retained the ability to inhibit HGF-MET binding (Fig. 1A), agonist activity was eliminated in the absence of HGF (Fig. 1B), resulting in concentration-dependent competitive antagonism of HGF-dependent MET signaling (Fig. 1C).
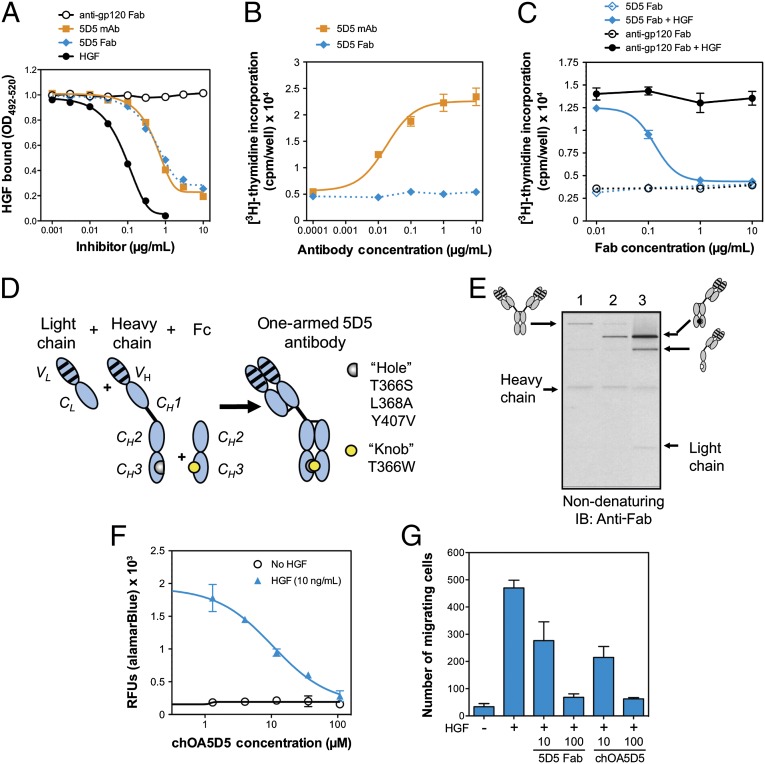
Fig. 1.
Identification of the 5D5 antibody and generation of the one-armed 5D5 antibody. (A) HGF binding to plate-bound MET-IgG is inhibited equally by the bivalent 5D5 mAb and the monovalent 5D5 Fab, relative to the positive control, HGF, and negative control, anti-gp120 Fab. (B) Only the bivalent 5D5 mAb, and not the monovalent 5D5 Fab, agonizes MET-dependent cell proliferation in the absence of HGF in Ba/F3-huMET cells, as measured by [3H]thymidine incorporation. (C) The 5D5 Fab, but not negative control anti-gp120 Fab, blocks HGF-dependent cell proliferation in Ba/F3-huMET in the presence of human HGF, as measured by [3H]thymidine incorporation. (D) Schematic of the design of a one-armed monovalent antibody, produced by coexpression of a light chain, heavy chain, and a truncated Fc domain. The heavy chain incorporates hole mutations, and the truncated Fc domain incorporates a knob mutation, as described previously (4). (E) Anti-Fab immunoblot (IB) analysis of the production of intact monovalent antibodies. E. coli were cotransfected with expression constructs for the following antibody components: (i) full-length light and heavy chains (lane 1); (ii) full-length light and heavy chain plus a nonmutated Fc chain (lane 2); or (iii) a full-length light chain, a full-length heavy chain containing hole mutations (T366S, L368A, and Y407V), and an Fc chain containing the knob mutation (T366W) (lane 3). The expected antibody products are indicated on the side of the blot. (F and G) The chimeric OA5D5 (chOA5D5) demonstrates dose-dependent inhibition of cell proliferation in Ba/F3-huMET cells (F), as well as dose-dependent inhibition of MDA-MB-435 cells (G), in transwell migration assays. All plots reflect group mean plus and minus the SEM (n = 3).
Fab fragments have been used therapeutically (23), but their short half-life in vivo limits their broad application. By constructing, a monovalent antibody (chimeric OA5D5 or chOA5D5), initially consisting of a murine/human chimeric IgG1 with only one 5D5 Fab arm, we sought to overcome the short half-life of a Fab while eliminating the bivalent binding inherent in a full-length IgG (Fig. 1D). Protein was generated in E. coli by coexpression of the chimeric 5D5 light chain, chimeric full-length heavy chain with or without a human Fc chain starting at the hinge region. When only full-length heavy and light chains were coexpressed, the antibody fragments assembled a bivalent full-length antibody of ∼150 kDa (Fig. 1E, lane 1). Inclusion of an expression construct encoding a nonmutated Fc chain with full-length heavy and light chains produced a monovalent antibody of expected size (∼100 kDa), but this did not fully prevent the generation of bivalent antibody (Fig. 1E, lane 2). To generate purely monovalent forms, mutations in the CH3 Fc domains were introduced to favor heterodimerization between two distinct Fc domains, harboring hole mutations (T366S, L368A, Y407V) and a knob mutation (T366W) (24). These mutations promoted heterodimerization of the knob containing Fc and the hole containing heavy chain, improving the assembly of monovalent antibody and reducing the level of bivalent antibody to trace levels (Fig. 1E, lane 3). Like the 5D5 Fab, chOA5D5 acted as a pure MET antagonist in cell-based assays, including dose-dependent inhibition of HGF-dependent cell proliferation of Ba/F3-huMET cells (Fig. 1F) and HGF-dependent migration in MDA-MB-435 cells (Fig. 1G). No agonistic activity was observed in the growth of Ba/F3-huMET cells lacking HGF (Fig. 1F).
Humanization, Affinity Maturation, and Functional Evaluation of Onartuzumab.
A humanized variant of chOA5D5 was generated by grafting the complementarity-determining regions (CDRs) from the 5D5 mAb into a human consensus κI variable light (VL) and subgroup 3 variable heavy (VH3) framework (Fig. S2). Because monovalent display of the Fab CDR graft on phage did not bind to MET, we generated randomized CDR libraries and selected for binding to immobilized MET. Following selection and sequence analysis, a single clone, 5D5 version 1 (5D5.v1), was identified, which contained a single change arising from a mutation (R94S) outside the Kabat (25) definition of CDR-H3 that restored binding to MET comparable to murine 5D5 (Fig. 2A). Notably, the murine 5D5 hybridoma contains a similar residue, threonine, at this position. Although the importance of this change is supported by analyses of antibody and antigen complex crystal structures, which show that heavy chain positions 93 and 94 often contact antigen (26), our complex structure with MET indicates an important structural role for this position (see below).
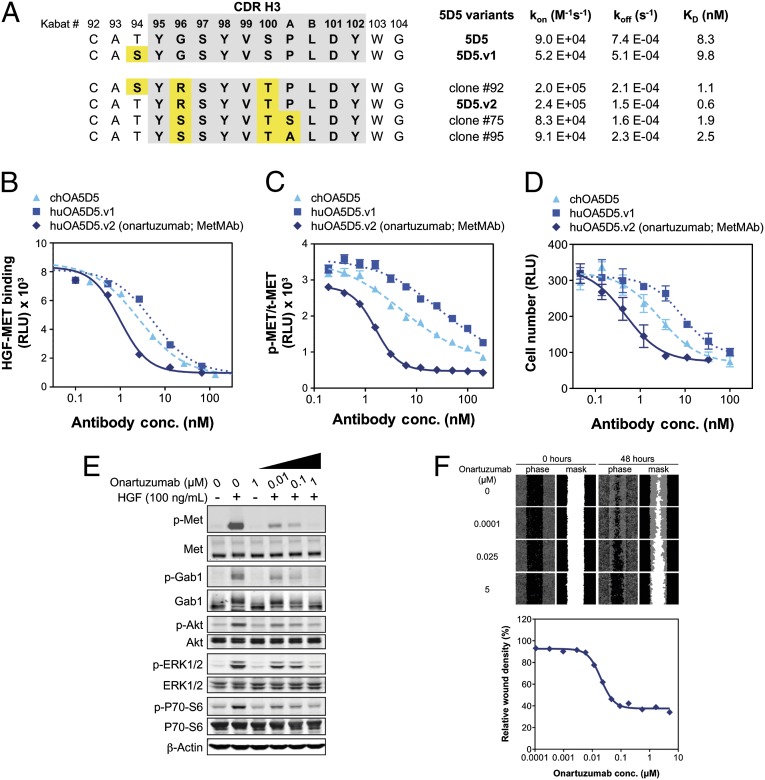
Fig. 2.
Humanization and affinity maturation of mouse 5D5 to generate onartuzumab, which functionally inhibits HGF/MET signaling. (A) CDR-H3 sequences and kinetic binding constants for selected humanized 5D5 variants, including the original chimeric 5D5 (ch5D5), 5D5.v1, and 5D5 clones identified during affinity maturation (5D5 clones 92, 75, and 95), which had slight alterations in the sequence of the CDR-H3 similar to that of 5D5.v2. Positions are numbered according to Kabat (25). The on (kon) and off (koff) rates and affinity measurements (KD) are summarized for each clone. The relative inhibition of HGF binding (B), MET phosphorylation by KIRA in A549 cells treated with HGF (C), and cell proliferation in Ba/F3-huMET cells (D) is shown for chOA5D5, huOA5D5.v1, and huOA5D5.v2 [onartuzumab (MetMAb)]. (E) Onartuzumab inhibits HGF-dependent MET phosphorylation and downstream activation of GAB1, AKT, ERK1/2, and P70-S6 in a dose-dependent fashion but does not induce any MET signaling in the absence of HGF. (F) Onartuzumab inhibits HGF-dependent migration and wound closure in HGF-treated A549 cells with an IC50 of 20 nM. All plots reflect group mean plus and minus the SEM (n = 3).
The affinity of 5D5.v1 (9.8 nM) to MET was essentially identical to the 5D5 Fab (8.3 nM), as determined by surface plasmon resonance (Fig. 2A). Based on 5D5.v1, new randomized CDR libraries were generated and multiple clones selected for improved binding to MET. Clone 92, encoding a serine at position 94, and clones 75 and 95, encoding a serine and alanine, respectively, (instead of a proline) at position 100a of CDR-H3, (Fig. 2A) each demonstrated higher affinity for MET than 5D5 and 5D5.v1, but the highest affinity clone, 5D5.v2, had three changes in CDR-H3 (94T, 96R, and 100T), resulting in a 14-fold higher affinity than 5D5.
The variable heavy and light domains of 5D5.v2 were cloned into constructs enabling expression of humanized OA5D5.v2 [onartuzumab (MetMAb)]. Small-scale fermentations in E. coli (27) confirmed self-assembly of onartuzumab, producing a protein with good stability. Overexpression of the endogenous E. coli periplasmic disulfide bond isomerases dsbA and dsbC genes (28) resulted in >fourfold increases in onartuzumab expression levels and was incorporated into large-scale fermentation and purification processes. The purified antibody is typically ≥95% main peak by analytical size-exclusion chromatography.
In agreement with the surface plasmon resonance data, onartuzumab inhibits HGF binding to MET in a competitive ELISA with a half-maximal inhibition (IC50) of 6.7 nM, which is 5- and 10-fold more potent than chOA5D5 and huOA5D5.v1, respectively (Fig. 2B). This inhibition corresponds with more potent cellular activity where onartuzumab inhibits HGF-dependent MET phosphorylation in A549 cells, as measured by a kinase receptor activation (KIRA) assay (IC50 of 1.6 nM). This IC50 is 3- and 18-fold lower than chOA5D5 and huOA5D5.v1, respectively (Fig. 2C). Furthermore, onartuzumab potently inhibited HGF-dependent cell proliferation in Ba/F3-huMET cells with an IC50 of 0.5 nM, a 6- and 18-fold improvement over chOA5D5 and huOA5D5.v1, respectively (Fig. 2D).
Onartuzumab exhibits dose-dependent suppression of HGF/MET downstream signaling components, as observed in A549 cells for activation of Gab1, Akt, ERK, and 70-S6K (Fig. 2E). It also potently suppresses HGF-dependent cell migration in A549 cells (Fig. 2F) and proliferation of various cancer cell lines with IC50 values in the range of 1–100 nM (Fig. S3).
As a monovalent antibody with an intact Fc domain, onartuzumab binds the neonatal Fc receptor (FcRn), a function important for the long half-life of IgGs in vivo (29, 30), to a comparable level as a full-length IgG1, such as trastuzumab (Fig. S4A). Because onartuzumab is expressed in E. coli, it is aglycosylated and, therefore, incapable of binding to Fcγ receptors or complement factor, C1q (Fig. S4 B and C) (27, 31). This suggests that onartuzumab could have pharmacokinetic (PK) properties similar to other mAbs, while being devoid of antibody-dependent cytotoxicity (ADCC) or complement-dependent cytotoxicity (CDC) against normal MET-expressing cells found on most epithelial and endothelial cell types.
In Vivo PK and Antitumor Efficacy of chOA5D5 and Onartuzumab.
The impact of the monovalent design of chOA5D5 on PK was evaluated in nude mice. The presence of the Fc prolonged the serum elimination half-life (t1/2) of the 5D5 Fab from a few minutes to approximately 6 d, with a clearance of 30 mL⋅d−1⋅kg−1 in mice at a 5 mg/kg i.v. dose (Fig. 3A). The chOA5D5 antibody was stable in mice at day 7, with no evidence of degradation (Fig. 3B), consistent with its PK profile. The mean t1/2 of onartuzumab at doses of 3, 10, and 30 mg/kg in athymic nude mice is approximately 6 d, with a mean clearance of 21 mL⋅d−1⋅kg−1 (32–34). The serum concentration–effect relationship observed in tumor-bearing mice for OA5D5 and onartuzumab supported a once every 1- to 3-wk dosing schedule, providing flexibility for clinical application (18, 32–35).
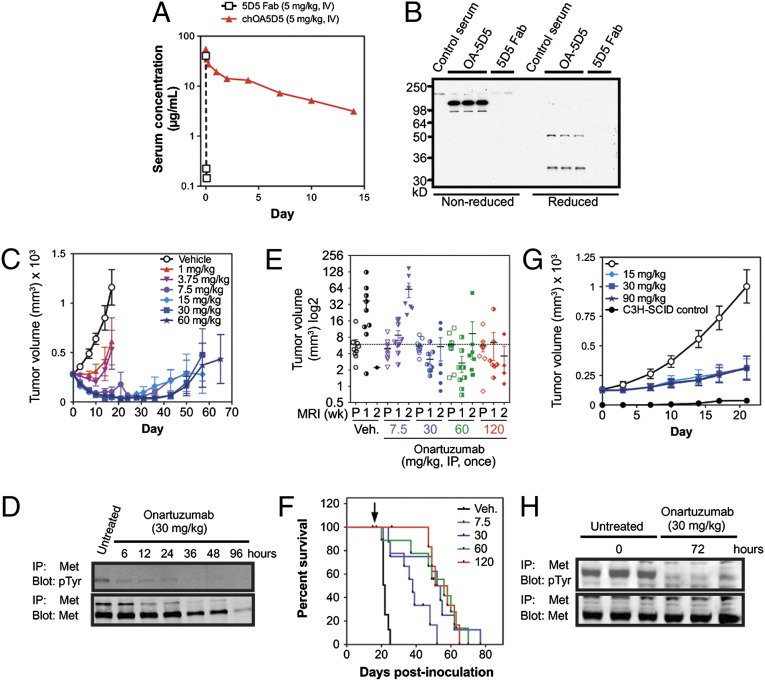
Fig. 3.
The one-armed 5D5 antibody and onartuzumab have prolonged exposure in mice and antitumor activity in HGF-dependent tumor models. (A) The chOA5D5 and 5D5 Fab proteins were dosed at 5 mg/kg i.v. in nude mice (n = 5), and their serum concentrations were measured over a 2-wk period. The serum half-life for the 5D5 Fab was a few minutes versus 5.8 d for the chOA5D5 antibody. (B) Serum drawn from nude mice dosed in A at day 7 posttreatment was assessed via Western blot using anti-Fab antibodies in nonreduced (Left) or reduced (Right) conditions, demonstrating stability of chOA5D5 in vivo relative to the 5D5 Fab. (C) Nude mice (n = 10 per group) bearing s.c. U-87 MG xenograft tumors were treated with onartuzumab at doses between 1–60 mg/kg, given once via i.p. injection. Data are plotted as group mean tumor volumes, with SEM indicated. Overall tumor response rates were calculated based upon the combined number of partial responses (PRs) and complete regressions (CRs), defined as >50% or 100% drop in tumor volumes, respectively, at any time during the study, and were determined for each dose group to be as follows: 1 mg/kg, 5 PRs and 0 CRs; 3.75 mg/kg, 5 PRs and 0 CRs; 7.5 mg/kg, 90% (7 PRs and 2 CRs); 15 mg/kg, 100% (5 PRs and 5 CRs); 30 mg/kg, 70% (2 PRs and 5 CRs); and 60 mg/kg, 100% (6 PRs and 4 CRs). (D) Immunoprecipitation–immunoblot analysis for p-MET (IP-MET, IB-pTyr) and total MET (IP-MET, IB-MET) from U-87 MG xenograft tumors from nude mice following treatment with 30 mg/kg, i.p., once at the indicated time points. (E) MRI evaluation of intracranial U-87 MG xenograft tumors (n = 10 per group) treated with vehicle (black circles), onartuzumab at doses of 7.5 mg/kg (purple triangles), 30 mg/kg (blue circles), 60 mg/kg (green squares), or 120 mg/kg (red diamonds), dosed i.p., once at pretreatment (P), 1 and 2 wk postdose. The mean starting tumor volume for the vehicle group is indicated by the dashed line. Group mean and SEM for each group is shown overlaying the individual animal tumor volumes. (F) Kaplan–Meier analysis for intracranial U-87 MG tumors treated with vehicle (black line) or onartuzumab at 7.5 mg/kg (purple line), 30 mg/kg (blue line), 60 mg/kg (green line), and 120 mg/kg (red line). The median survival for each group was as follows: vehicle, 21 d; 7.5 mg/kg, 37 d; 30 mg/kg, 52 d; 60 mg/kg, 53 d; 120 mg/kg, 51 d. (G) hHGFtg-C3H-SCID mice (n = 10 per group) bearing s.c. NCI-H596 xenograft tumors were treated with either vehicle (white circles) or onartuzumab dosed at 15 mg/kg (light blue diamonds), 30 mg/kg (medium blue squares), or 60 mg/kg mg/kg (dark blue stars). Growth of NCI-H596 in littermate C3H-SCID mice is shown (black circles). Data are shown as group mean tumor volumes, with SEM indicated. (H) Immunoprecipitation–immunoblot analysis for p-MET (IP-MET, IB-pTyr) and total Met (IP-MET, IB-MET) from NCI-H596 xenograft tumors from hHGFtg-C3H-SCID mice following treatment with 30 mg/kg, i.p., once at 0 and 72 h.
As onartuzumab blocks HGF binding to MET, ligand-driven models were used to evaluate antitumor efficacy in vivo. The autocrine U-87 MG (HGF/MET autocrine, PTENnull) GBM orthotopic xenograft tumor model was previously shown to respond to intracranial infused OA5D5 via osmotic pumps (16). To evaluate the dose responsiveness of this model with systemic delivery, U-87 MG tumors were established s.c. and treated with onartuzumab at doses of 1–60 mg/kg given once via i.p. injection. Onartuzumab resulted in dose-dependent inhibition of tumor growth, with doses of 1 and 3.75 mg/kg delaying tumor growth, whereas doses of ≥7.5 mg/kg drove tumor regression (Fig. 3C). There was no significant impact upon body weight. Consistent with these results, in pharmacodynamic (PD) excised tumor samples onartuzumab inhibited MET phosphorylation as early as 6 h postdose, resulting in 85% reduction in phosphorylated MET (p-MET). There was also a decrease in total MET levels starting at 36 h and reaching 92% at 96 h postdose (Fig. 3D). These data demonstrate that the potent antitumor activity of onartuzumab is attributable to the ability of the antibody to disrupt active MET signaling as early as 6 h postdosing.
To model intracranial GBM tumor growth with systemic onartuzumab treatment, orthotopic U-87 MG tumor xenografts were established via direct inoculation of U-87 MG cells into the brains of nude mice. In this context, the antibody must localize to brain tumors, circumventing the blood–brain barrier (BBB) if intact. GBM growth can disrupt BBB integrity (36), as is known to occur with orthotopic U-87 MG tumors (37), thereby allowing entry of some antibody into tumors. Single doses of onartuzumab (7.5, 30, 60, and 120 mg/kg i.p.) were given and the effect on tumor volume [measured by micro-magnetic resonance imaging (µMRI) at pretreatment (P), 1 and 2 wk posttreatment], and survival was examined. Onartuzumab delayed U-87 MG tumor growth by about 1 wk at a dose of 7.5 mg/kg (Fig. 3E), resulting in a 76% improvement in median survival (from 21 to 37 d; Fig. 3F). At doses ≥30 mg/kg, tumor volumes were static or reduced in size (Fig. 3E), with a 2.5-fold increase in median survival (30 mg/kg, 52 d; 60 mg/kg, 53 d; 120 mg/kg, 51 d; Fig. 3F).
Because murine HGF does not activate huMET (38, 39), onartuzumab was evaluated in paracrine-driven human xenograft models grown in hHGFtg-C3H-SCID mice (40). The NCI-H596 (METexon14del) NSCLC cell line harbors a deletion in exon 14 of the MET gene. This removes the Cbl-binding site within MET that is responsible for MET ubiquitination and turnover following HGF activation, thereby converting MET into a transforming protein (41, 42). Growth of NCI-H596 xenograft tumors was enhanced in hHGFtg-SCID mice relative to littermate C3H-SCID control mice, and onartuzumab treatment (15–90 mg/kg, single i.p. dose) resulted in significant tumor growth inhibition (Fig. 3G). In excised NCI-H596 tumor samples from hHGFTg-C3H-SCID mice treated with onartuzumab (30 mg/kg, i.p.), there was a 72% decrease in p-MET levels, with no apparent decrease in total MET levels at 72 h (Fig. 3H). Collectively, these data indicate that doses of 15–30 mg/kg onartuzumab are sufficient to inhibit HGF/MET-dependent tumor growth in autocrine (s.c. and intracranial) and paracrine-driven tumor models, enabling therapeutic inhibition of HGF/MET-dependent signaling with PK characteristics close to those of other therapeutic antibodies.
Mechanism of MET Inhibition by Onartuzumab.
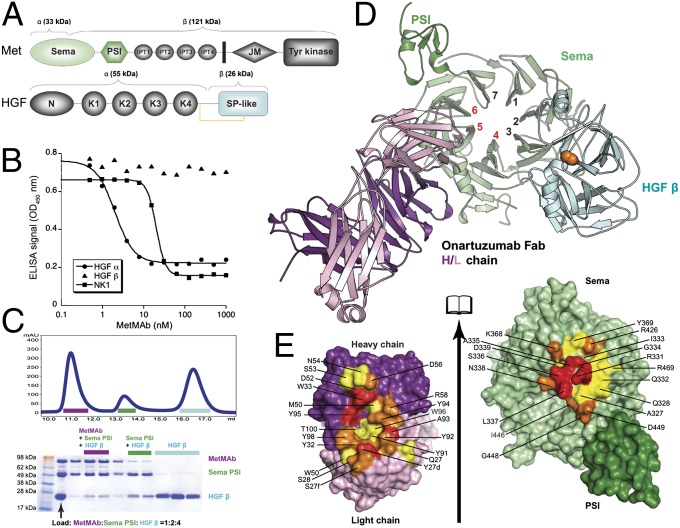
The 5D5 Fab was shown previously to interact with the MET Sema domain (15); however, to understand how onartuzumab disrupts the interaction between MET and the HGF α/β-heterodimer (Fig. 4A), competitive-binding assays were performed. No competition was observed between onartuzumab and HGF-β, whereas HGF-α binding was blocked by onartuzumab, with an IC50 of 2 nM (Fig. 4B). To further localize binding, we tested a shorter fragment of the α-chain composed of the N-terminal and first kringle domain (NK1) (44), which was also blocked by onartuzumab, with an IC50 of 20 nM (Fig. 4B). Cocrystallization of onartuzumab with MET was pursued to determine the structural and functional basis of MET antagonism by onartuzumab. Despite using multiple strategies, the MET Sema-PSI was recalcitrant to cocrystallize with onartuzumab or its Fab or scFv fragments. Because onartuzumab and the HGF β-chain can bind MET Sema-PSI simultaneously (Fig. 4C), we sought to crystallize a ternary complex using the Fab format, which resulted in a 2.8-Å resolution structure (Fig. 4D and Table S1). The structure of MET Sema-PSI/HGF-β within the ternary complex is essentially identical to that in the binary complex described previously (1.1-Å Cα r.m.s.d. for 721 structurally aligned residues) (45). The onartuzumab (hu5D5.v2) Fab binds MET at a distinct site using all three heavy-chain CDRs and the first and third complementary determining regions of the light-chain (CDR-L1 and CDR-L3) to bind to blades 4, 5, and 6 of the β-propeller Sema domain (Fig. 4 D and E). The large (1870-Å2) interface is dominated by electrostatic and hydrogen-bonding interactions, with limited hydrophobic character (Table S1 and Fig. S5).
Fig. 4.
Structural basis for antagonism of MET signaling by onartuzumab. (A) Schematic of MET and HGF domain architecture. Functional domains are indicated: Sema domain; PSI domain; Ig-like fold shared by plexins and transcriptional factors (IPT) domain; juxtamembrane (JM) domain; N-terminal (N) domain; kringle (K) domain; serine protease-like (SP-like) domain. The disulfide between the HGF α- and β-chains is shown as a yellow line. Colors used for individual domains in A match those used in D. (B) Competition-binding assays with MET ECD-Fc (encoding MET residues 25–929, which encode the MET Sema domain, the PSI domain, and all four IPT domains) immobilized on plates indicates that onartuzumab blocks HGF-α and NK1, but not HGF-β, binding to MET. (C) Analytical size-exclusion chromatography analysis of onartuzumab, MET Sema-PSI, and HGF-β mixed at 1:2:4 ratio. The three peaks in the chromatogram were visualized by SDS/PAGE and correspond to the onartuzumab/MET Sema-PSI/HGF-β ternary complex, onartuzumab Fab/HGF-β binary complex, and excess HGF-β. (D) Cartoon representation of the 2.8-Å crystal structure of the onartuzumab Fab/MET Sema-PSI/HGF-β ternary complex (PDB ID code 4K3J) showing the Fab heavy chains (deep purple), Fab light chains (pink), MET Sema (light green) and PSI (deep green) domains, and HGF-β (cyan). The Sema β-propeller is viewed from the “bottom” face and the blades are numbered. (E) Open-book view of the interface between onartuzumab Fab (Left) and human MET Sema-PSI (Right). Residues in the interface were colored according to their percentage of reduction in accessible surface area upon complex formation (15–44%, yellow; 45–74%, orange; >75%, red). Interface analysis was performed using the protein interfaces, surfaces and assemblies (PISA) server (43).
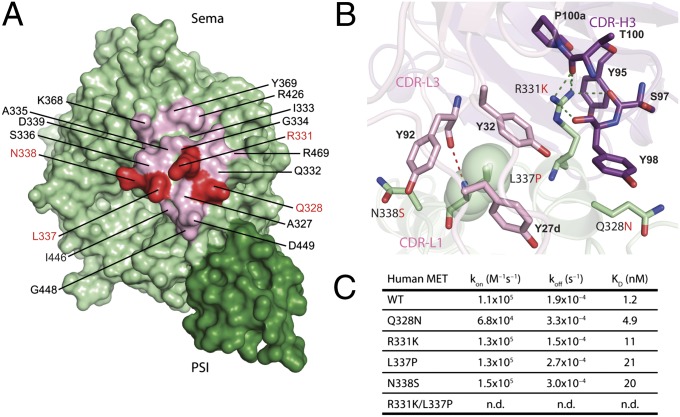
Despite human and murine MET (muMET) having over 87% sequence identity in their Sema-PSI domains, the 5D5 antibody does not recognize muMET, presumably because of four amino acid differences within the onartuzumab epitope (Fig. 5A). Single substitution of these residues to their murine counterparts (Q328N, R331K, L337P, and N338S) resulted in 4- to 17-fold reduced antibody binding, whereas the R331K/L337P double mutant resulted in abrogation of binding (Fig. 5 B and C). Because these four residues make side chain-specific interactions with CDR-H3, CDR-L1, and CDR-L3 of the onartuzumab Fab, the functional binding data are in agreement with the onartuzumab-binding site revealed in our crystal structure.
Fig. 5.
Key differences between human and murine MET within the onartuzumab epitope. (A) Surface representation of MET Sema-PSI highlighting four nonconserved amino acid changes (red) between human and murine MET within the onartuzumab epitope (light pink). (B) Close-up of the intermolecular-interactions around these four key amino acids in onartuzumab/MET Sema-PSI complex. Green and red dotted lines indicate hydrogen bonds between MET and onartuzumab heavy chain and light chain. (C) Summary of the biotinylated onartuzumab binding experiments to wild-type (WT) human MET Sema-PSI, four single mutants, and one double mutant. The rate of association (kon), dissociation (koff), and binding affinity (KD) of the kinetic experiments are shown. n.d. indicates that binding was not detected.
Discussion
Protein engineering of onartuzumab has led to an antibody with unique properties. We have shown that a potent anti-MET agonistic antibody can be engineered into a “one-armed” monovalent antibody that prevents antibody-induced dimerization and results in potent antagonism of HGF/MET signaling (21). Although our primary focus was developing an antibody as a pure antagonist of ligand binding to MET, its therapeutic impact required several critical engineered properties.
Firstly, the use of the knob and hole mutations resulted in efficient Fc heterodimerization and high levels of the desired antibody, facilitating purification for clinical use. Secondly, the Fc of onartuzumab provided native-like FcRn binding, providing serum clearance rates close to human IgGs in mouse (Fig. S4A and Fig. 3A). Although attempts at generating monovalent antibodies with slow clearance rates have been reported (46, 47), most have a nonnative structure and increased risk of immunogenicity. The knob and hole mutations are buried in the CH3-CH3 interface and completely hidden from the surface (Fig. S6). Indeed, no anti-onartuzumab antibodies toward the knob and hole regions of onartuzumab in cynomolgus monkeys or humans have been identified (32–34, 48). Finally, onartuzumab is expressed in E. coli, thus aglycosylated and devoid of Fc effector functions (27), such as ADCC or CDC (Fig. S4 B and C). The lack of onartuzumab binding to either Fcγ receptors or C1q minimizes any immune-mediated dimerization and MET activation, which is key to ensuring that onartuzumab cannot be cross-linked, which could result in MET oligomerization. The absence of ADCC and CDC activity might also be important for safety as MET is a broadly expressed target. Although the lack of effector function was an engineered feature for onartuzumab, this may represent a limitation of E. coli-produced antibodies for targets in which effector function is needed for biological activity unless known Fc variants are incorporated (49). Expression of monovalent antibodies in mammalian cells could potentially address this limitation, enabling the therapeutic targeting with pure antagonists with the addition of effector function. Taken together, these attributes enable effective inhibition of HGF/MET signaling in autocrine- and paracrine-activated tumor models, leading to significant inhibition of tumor growth (Fig. 3 B–H).
The ternary structure for the onartuzumab Fab binding to the MET Sema-PSI domain in the presence of the HGF β-chain reveals several important findings. We now understand the molecular basis for high-affinity binding of onartuzumab to huMET and lack of binding to muMET. Both HGF-β and onartuzumab bind to the same side of the MET β-propeller domain (Fig. 4D and Fig. S7) and have no overlapping binding sites, consistent with our competitive-binding and size-exclusion chromatography results (Fig. 4 B and C) (45). In contrast, comparison of the MET-binding sites for onartuzumab and Internalin (Inl)B, an invasive protein from Listeria monocytogenes, shows an extensive overlap (50) (Fig. S7). Given that InlB, which is structurally distinct from onartuzumab, partially competes with HGF for binding to MET (50), this suggests that HGF-α, onartuzumab, and InlB bind sites on MET that at least partially overlap. Interestingly, there is a relatively highly conserved epitope on the Sema-PSI domain close to the onartuzumab epitope, which could play a role in HGF α-chain binding (Fig. S8). Thus, although the exact binding site for the α-chain of HGF on MET remains elusive, our data reveal that inhibition of high-affinity HGF α-chain and NK1 domain binding to MET underscores the molecular mechanism of action for onartuzumab as an antagonist of MET signaling.
Although our data and those published elsewhere (51) show MET as a target sensitive to antibody-dependent dimerization, the use of the technology described here has potential for broad applicability in drug development and diagnostics or where control of antibody valency is desirable. Bivalent antibodies frequently induce receptor oligomerization and initiation of signaling. For example, Fab fragments derived from agonist antibodies against β2-adrenoreceptor act as potent antagonists of the pathway, highlighting the potential for monovalent antibodies as therapeutic or research tools (52). Similarly, antibodies against the tropomyosin-related kinase (Trk)B act as ligand mimetics, replicating the effects of its ligand, brain-derived neurotrophic factor (BDNF), and highlighting the therapeutic potential for treating TrkB-related neurodegenerative diseases (53). In contrast, there could be therapeutic potential of monovalent antagonistic antibodies derived from these agonist mAbs in targeting BDNF/TrkB signaling thought to influence growth and survival of several cancers (54, 55). One possibility with the knob and hole heterodimerization technology is the potential to generate bispecific antibodies, either against the same target with two distinct epitopes or against different targets (4). We explored this possibility using MET and epidermal growth factor receptor as cotargets and found the system to be exceptionally modular, enabling successful inhibition of disparate targets in a single-antibody format (56).
In summary, we show that onartuzumab represents the founding member of a class of monovalent therapeutic antibody with native-like antibody architecture that, unlike bivalent antibodies, acts as a potent antagonist. We describe how these antibodies are engineered to enable large-scale manufacturing. Finally, we describe the mechanism by which onartuzumab engages MET, thus revealing the putative binding site for NK1 and HGF α-chain, and can rationalize the necessity for monovalency, which enables it to function as a potent antagonist of the MET pathway.
Materials and Methods
Animal Study Ethics.
All animal studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health (NIH) (NIH Publication 8523, revised 1985). The Institutional Animal Care and Use Committee (IACUC) at Genentech reviewed and approved all animal protocols.
Cell Lines.
Cell lines were obtained from the American Tissue Type Collection (ATCC), the German Collection of Microorganisms and Cell Cultures (DSMZ), the Japanese Collection of Research Bioresources, or the Riken Bioresource Center Cell Bank. The human cell lines used included GBM U-87 MG (ATCC) and NCI-H596 (ATCC). Full details of all of the human cell lines used are given in SI Materials and Methods. The mouse cell line Ba/F3-huMET was stably transfected at Genentech and grown as described previously (22). All cell lines were maintained according to cell bank’s recommendations or in normal growth medium [RPMI; 2 mM glutamine; 10% (vol/vol) FCS] at 37 °C and 5% (vol/vol) CO2.
Generation of Anti-MET Antibodies.
BALB/c mice were injected in each rear footpad with soluble MET-IgG (7). Four days after the last immunization, lymph node cells were harvested and fused with P3/X63 Ag8U1 myeloma cells (57) using 35% polyethylene glycol. Hybridoma cell lines that produced an antibody specifically against MET were identified by a capture ELISA and screened by flow cytometry using Ba/F3 cells stably transfected with and expressing MET (22). Selected hybridomas were tested for their ability to inhibit binding of biotinylated HGF to MET-IgG. Hybridomas were cloned twice by limiting dilution and characterized for their antagonistic and agonistic activity.
Humanization of 5D5.
A CDR graft of 5D5, displayed monovalently on phage, was created by grafting 5D5 (5D5) positions 24–34 (L1), 50–56 (L2), and 89–97 (L3) in VL and positions 26–35 (H1), 49–65 (H2), and 95–102 (H3) in VH into a human consensus κI and a VH3 framework used for the humanization of trastuzumab (5D5 graft; Fig. 2) (58). This was performed by Kunkel mutagenesis using a separate oligonucleotide for each hypervariable region. Correct clones were assessed by DNA sequencing. To restore binding in the 5D5 graft, a CDR repair approach was taken (59). Sequence diversity was introduced into each CDR using a soft randomization strategy that maintained a bias toward the CDR graft sequence using Kunkel mutagenesis and a poisoned oligonucleotide synthesis strategy (60). Library sizes ranged from 1 to 2 × 109 independent clones. Random clones from the initial libraries were sequenced to assess library quality. Phage libraries were sorted using a solution-sorting method with biotinylated MET-IgG, and selected clones were screened for improved binding using a phage ELISA (61).
Affinity Maturation of Humanized 5D5.v1.
Six phage display libraries, each targeting a single CDR, were generated in the background of humanized 5D5.v1 (R94S) for affinity maturation. A solution sorting method was used to increase the stringency of the affinity-based phage selection process (61). This allowed control of the biotinylated target concentration, a reduction in the phage capture time to lower backgrounds, and the addition of unbiotinylated MET to eliminate clones with faster off rates. Selected clones were screened by phage ELISA and expressed as Fab (61).
Affinity Determination.
Affinity determinations were performed by surface plasmon resonance using a BIAcoreTM-2000 (GE Healthcare). MET-IgG was immobilized [∼1,000 response units (RU)] on a CM5 chip and the binding of Fab (1.5–3,000 nM) in PBS containing 0.1% Tween-20 was assessed. After each injection the chip was regenerated using 100 mM HCl. Binding response was corrected by subtracting the RU from a blank flow cell. A 1:1 Languir model of simultaneous fitting of kon and koff was used for kinetic analysis.
Production and Scale-Up of the Monovalent 5D5 Antibodies.
The plasmid used for the production of the monovalent 5D5 one-armed anti-MET antibody (huOA5D5) was based on the separate cistron vectors described for the expression of full-length antibodies in E. coli (27). Three bacterial alkaline phosphatase gene (phoA)-promoter cistrons were included on the same plasmid for the transcription of the 5D5 antibody light and heavy chain and IgG1 Fc fragment. All three chains were targeted to the periplasmic space with the heat-stable enterotoxin II signal sequence, and each was translationally controlled at the initiation stage using a signal sequence variant with a relative strength of one (27). To promote heterodimerization in the CH3 domains, the 5D5 heavy chain was modified to encode the T366S:L368A:Y407V mutations, and the Fc fragment was modified to contain the T366W mutation (24).
Small-Scale Expression and Purification of One-Armed 5D5.
Derivatives of E. coli strain W3110 were used for the expression of the one-armed 5D5 antibody and onartuzumab (27). Cells were lysed using a microfluidizer (Microfluidics), and 0.1% (vol/vol) PEI was added to the lysate and stirred for 1 h at 4 °C. After centrifugation at 15,000 × g, the supernatant was combined with a protein A-affinity resin and stirred overnight at 4 °C, and the resin was poured into a column. The column was washed with 10 mM Tris⋅HCl, 1 mM EDTA buffer (pH 7.5), followed by 0.5 M NaCl in the same buffer, and eluted with a gradient from pH 6.0–2.0 in 50 mM sodium citrate, 0.1 M NaCl. Eluted fractions were adjusted to a final concentration of 2 M urea (pH 5.4) and analyzed by SDS/PAGE. Those containing one-armed 5D5 were pooled and subjected to cation exchange chromatography using SP Sepharose (GE Healthcare) equilibrated with 25 mM Mes [(2-N-morpholino)ethanesulfonic acid], 2 M urea (pH 5.4). The column was eluted with a gradient of 0–1 M NaCl in 25 mM Mes (pH 5.4). Following SDS/PAGE, pooled fractions were adjusted with 0.4 M sodium sulfate (pH 6) and loaded onto a Hi Propyl (Mallinckrodt Baker) column equilibrated with 0.4 M sodium sulfate, 25 mM Mes (pH 6.0). The column was eluted with a gradient of 0.4 to 0 M sodium sulfate in 25 mM Mes (pH 6.0). Fractions containing the antibody were pooled, concentrated using CentriPrep 10 (Millipore), and subjected to size-exclusion chromatography using a Superdex 200 column (Amersham Biosciences) equilibrated with 10 mM sodium succinate, 0.15 M NaCl (pH 5.0).
Electrochemiluminescence Assay for Onartuzumab Blocking of HGF/MET Minding.
Purified huMET-IgG protein (PUR9250; Genentech) was incubated with 20-fold molar excess NHS-X-biotin (biotinylamidocaproic acid-N-hydroxysuccinimide ester) in 0.1 M NaHCO3 (pH 8.5) using biotin-X-NHS (Research Organics). Purified human 2-chain HGF produced at Genentech was labeled with BV-TAG via NHS-ester chemistry (BioVeris). The huMET-IgG-biotin (500 ng/mL), HGF-Ru Tag (250 ng/mL), and titrations of onartuzumab (or chOA5D5) antibody ranging from 0.1 to 200 nM were incubated in a volume of 100 µL of assay diluent: PBS plus 0.5% BSA/0.5% Tween-20/0.033% Proclin. The mixtures were incubated in sealed polypropylene round-bottom, 96-well plates (Corning) for 2–4 h at room temperature with shaking. Streptavidin magnetic beads (Dynabeads; BioVeris) were added. After a 45-min incubation with vigorous shaking, the plates were read using a BioVeris M-Series instrument.
MET KIRA Assay.
A549 cells were maintained in growth medium (Ham’s F-12/DMEM 50:50; Gibco) containing 10% FBS (Sigma). Cells from confluent cultures were detached using Accutase (ICN) and seeded in 96-well plates (50,000 cells per well). After overnight incubation at 37 °C, growth media were removed, and cells were serum-starved for 30–60 min in medium containing 0.1% FBS. To determine the activity of onartuzumab (or derivatives), antibodies were serially diluted beginning at a concentration of 200 nM in medium plus 0.1% FBS and added to the assay plates. After a 30-min incubation at 37 °C, 1 nM HGF was added to all assay wells, and plates were incubated for an additional 10 min. The media were removed, and a cell lysis buffer was added [Cell Signaling Technologies; catalog no. 9803; supplemented with a protease inhibitor mixture (Calbiochem; catalog no. 539131)]. Lysates were analyzed for phosphorylated MET via an ECL (Amersham Biosciences) assay using a BioVeris M-Series instrument (BioVeris,). An anti-phosphotyrosine mAb (clone 4G10; Millipore) was labeled with BV-TAG via NHS-ester chemistry (BioVeris). A mAb (produced at Genentech; clone 1928) against the MET extracellular domain was biotinylated using biotin-X-NHS (Research Organics). The BV-TAG–labeled 4G10 and biotinylated anti-MET mAb were diluted in assay buffer (PBS/0.5% Tween-10/0.5% BSA) and added to the cell lysates. After a 1.5- to 2-h incubation at room temperature with vigorous shaking, streptavidin magnetic beads were added. Following a 45-min incubation, the plates were read on the BioVeris instrument. EC50 values were determined by nonlinear regression analysis with a four-parameter model (KaleidaGraph; Synergy Software).
Cell-Proliferation Assays.
Cell-proliferation studies were performed using [3H]thymidine-incorporation assays, CellTiter-Glo, or Alamar Blue assays (Promega). Cell lines were plated between 2–5 × 105 cells per milliliter in a volume of 100 µL in normal growth media without FCS, with or without HGF (1–500 ng/mL) in 96-well plates. Proliferation assays were run for 72 h before reading out. Following initial screening of cell lines for HGF-induced proliferation to a dose range of 1–500 ng/mL, inhibition of cell proliferation was determined using conditions that resulted in the maximum-fold induction of HGF-driven cell proliferation, typically at 50 ng/mL of HGF and 1 µg/mL of heparin with the indicated concentrations of antibody.
Cell-Migration Assays.
Cell-migration assays were performed using transwell migration assays with BD FluoroBlok 8-µm pore, 96-well inserts (catalog no. 351164; BD Biosciences). Briefly, 1.25 × 104 cells were added to the apical side of prewetted transwell chambers in normal growth media without FCS and with 0.1% BSA and HGF (10–50 ng/mL). Plates were incubated at 37 °C and 5% CO2 overnight, and then media were carefully removed. Cells were fixed to the bottom of the chamber using 70% ethanol for 10 min and stained with SYTOX Green Nucleic Acid Stain (Invitrogen-Molecular Probes). Plates were imaged using the ImageXpress Micro High Content Screening System (Molecular Devices). Alternatively, cells were assessed in wound healing scratch assays using the IncuCyte (Essen Bioscience). Briefly, 4 × 105 cells were plated on the 96-well ImageLock plates (Essen BioScience; catalog no. 4379) and incubated in the complete media (RPMI with 10% FBS and 2 mM l-glutamine) for 18 h to allow a confluent monolayer to form. Wounds were made the following day using the 96-pin WoundMaker (Essen BioScience), washed twice with RPMI media (with no supplements), and incubated with RPMI containing 0.1% BSA, 2 mM l-glutamine, 5 µg/mL heparin, and 50 ng/mL HGF with or without increasing concentrations of onartuzumab. Cell migration was monitored in real time by IncuCyte, and confluence was measured by the IncuCyte software. Graphs and IC50 values were generated using Prism 5 (GraphPad Software), based upon the calculated relative wound density.
Immunoprecipitation and Immunoblotting.
Cells were washed once with cold PBS and lysed in 1× Cell Extraction Buffer (Biosource). For frozen tumor samples, tumors were pulverized on dry ice using a small Bessman tissue pulverizer (Spectrum Laboratories) and prepared as described by the manufacturer.
Antibodies to cleaved MET, pAkt, Akt, pERK1/2, ERK1/2, P70-S6K, and p-Tyr were obtained from Cell Signaling. Anti-MET (C28) beads used for immunoprecipitation were from Santa Cruz Biotechnology. The β-actin and GAPDH antibodies were obtained from Sigma. The anti-Fab and anti-Fc Abs were from Jackson ImmunoResearch. Specific antigen–antibody interaction was detected with a HRP-conjugated secondary antibody IgG using ECL detection reagents (Amersham Biosciences). The p-MET and total MET band intensities were quantified using the LI-COR Odyssey immunoblotting system. Immunoprecipitations for MET and p-MET were performed as described previously (17).
PK Study in Mice.
Athymic nude (nu/nu) mice received a single i.v. dose of 5D5 Fab (n = 16) or chOA5D5 (n = 40) at 5 mg/kg. Serum samples were collected at various time points up to 24 h for 5D5 Fab and up to 14 d for chOA5D5, with four mice per time point, and stored at −70 °C until assayed by ELISA. Noncompartmental analysis was used to analyze PK data for both 5D5 Fab and chOA5D5 (WinNonlin Version 5.2.1; Pharsight). A naïve pooled approach was used to provide one estimate for each molecule.
OA5D5 and Onartuzumab ELISA.
Microtiter 384-well Maxisorp plates (Nunc) were coated overnight at 4 °C with 2 μg/mL His8-conjugated human Met-ECD in coat buffer (0.05 M bicarbonate buffer; pH 9.6) and blocked with block buffer (0.5% BSA in PBS). Plates were washed six times with 300 µL of wash buffer (PBS, 0.05% Tween-20; Sigma Aldrich) between each subsequent step. Dilutions of OA5D5 or onartuzumab standard, controls, and test samples were prepared in high-salt assay diluent (0.5% BSA; 10 ppm Proclin; Supelco), 0.05% Tween-20, 0.2% bovine γ-globulin (BioCell), 0.25% CHAPS (Sigma Aldrich), 5 mM EDTA, and 0.35 M sodium chloride in PBS (pH 7.4), and 25 µL was incubated on coated and blocked plates and then detected with 0.05 μg/mL peroxidase-labeled Fc specific F(ab′)2 fragments of goat anti-human IgG, followed by 3,3′,5,5′-tetramethylbenzidine (TMB). Plates were stopped with phosphoric acid and read as for the HGF assay described above. The assay range is 40 ng/mL to 160 pg/mL, and the limit of quantitation is 16 ng/mL for cyno serum samples, with a minimum dilution of 1:100. Up to 10 μg/mL of human Met ECD and up to 1 mg/mL of human HGF did not interfere with detection of onartuzumab in the assay.
Subcutaneous Tumor Models.
A total of 5 × 106 U-87 MG cells were mixed in Hank’s balanced salt solution and matrigel (growth factor reduced; catalog no. 356231; BD Biosciences) inoculated s.c. in the rear right flank of nude (nu/nu) mice. The NCI-H596 tumor model was performed as described previously (40, 54). Briefly, 2 × 106 NCI-H596 tumor cells were inoculated s.c. in the shaved right rear flank or hHGFTg-C3H-SCID or littermate C3H-SCID mice. Tumor volumes were determined using digital calipers (Fred V. Fowler Company) using the formula (length × width × width)/2. Tumor growth inhibition (%TGI) was calculated as the percentage of the area under the fitted curve (AUC) for the respective dose group per day in relation to the vehicle, such that %TGI = 100 × [1 − (AUCtreatment/day)/(AUCvehicle/day)]. Curve fitting was applied to log2-transformed individual tumor volume data using a linear mixed-effects model using the R package non-linear mixed effects Version 3.1-97 in R Version 2.12.0.
U-87 MG Intracranial Tumor Model.
A total of 2.5 × 105 U-87 MG cells were injected via stereotaxic surgery into the right striatum of athymic nude (nu/nu) mice in a volume of 5 µL. Animals were anesthetized with 2% isoflurane and placed into the stereotactic apparatus. The head was cleaned with povidone and alcohol. A midline incision was made exposing the underlying frontal and parietal bones that were scraped clean of periosteal membrane. A 1-mm burr hole was drilled through the parietal bone overlying the right striatum at stereotaxic coordinates: (i) apical, +0.2–0.15 mm from the bregma; (ii) mediolateral, 2 mm; and (iii) dorsoventral, 3.2–3.5 mm flat skull. Using a 10-µL Hamilton syringe fitted with a 28-gauge stainless steel cannula, 5 µL of cells was injected to a depth of 3.2–3.5 mm into the striatum at a rate of 0.5 µL per 30 s, and with the cannula left in place for 3–5 min before slowly retracting it. The wound was cleaned with povidone-alcohol, and the incision closed with tissue glue, sutures, or wound clips. Buprenorphine (0.05–0.1 mg/kg s.c.) and topical lidocaine were given at the end of the surgery, and animals were allowed to recover over a water-circulating heating blanket and returned to the holding area once ambulatory. Mice were randomized into groups of 10 to obtain comparable group mean tumor volumes and distribution, as determined by µMRI. Body weight and condition was monitored twice per week during study, and animals were euthanized if body weight loss was >20%. Tumor volumes were monitored by T2 µMRI on a Varian 9.4T MRI system with a 30-mm quadrature volume coil.
Protein Purification.
The human MET receptor Sema-PSI fragment (residues 25–567) containing the N-terminal gp67 secretion signal, and C-terminal His6 tag was cloned into the pFastBac-1 vector (Invitrogen) and used to generate high-titer recombinant baculovirus using the Bac-to-Bac system according to the manufacturer’s protocol. MET Sema-PSI single mutants Q328N, R331K, L337P, and N338S and double mutant R331K/L337P were introduced by QuikChange mutagenesis (Agilent Technologies) and used to generate recombinant baculovirus as described above. Human NK1 was expressed in E. coli and purified (62). Human HGF β-chain (residues 495–728; C604S) and α-chain (residues 32–494) were expressed as described previously (45, 63). Briefly, viral stocks were prepared from Sf9 insect cells transfected with recombinant pAcGP67A vectors encoding each protein fused with C-terminal His6 tag. High-titer recombinant virus stocks were produced after three rounds of amplification and used directly for expression in Tni PRO insect cells growing in ESF 921 media (Expression Systems LLC). Supernatants were harvested 48 h posttransfection; treated with 1 mM NiCl2, 5 mM CaCl2, and 50 mM Tris⋅HCl (pH 8.0); and clarified through a 0.2-μm filter. Recombinant MET Sema-PSI, HGF-α, and HGF-β were purified sequentially by Ni-NTA (Qiagen) and size-exclusion (HiLoad 16/60 Superdex 200; GE Healthcare) chromatography. Fractions containing eluted proteins were analyzed by SDS/PAGE and pooled. Protein concentration was determined by the absorbance at 280 nm (A280) (64).
HGF and Onartuzumab Competition-Binding ELISA.
Microtiter plates (Nunc) were coated overnight at 4 °C with 2 µg/mL rabbit anti-human IgG Fc-specific antibody (Jackson ImmunoResearch Laboratory) in 50 mM sodium carbonate buffer (pH 9.6). After blocking with 1% BSA in HBS buffer [50 mM Hepes (pH 7.2), 150 mM NaCl, 5 mM CaCl2, and 0.1% Tween-20], 1 μg/mL MET-IgG fusion protein (7) was added, and plates were incubated for 1 h with gentle shaking at room temperature. After washing with HBS buffer, HGF-α or HGF-β proteins preincubated with various concentrations of onartuzumab were added for 1 h. Bound HGF proteins were detected using anti–His-HRP (Qiagen), followed by addition of TMB/H2O2 substrate (KPL). The reaction was stopped with 1 M H3PO4, and the A450 was measured on a Molecular Devices SpectraMax Plus384 microplate reader. The effective concentration of onartuzumab to give IC50 was determined by a four-parameter fit using Kaleidagraph (Synergy Software).
Analytical Size-Exclusion Chromatography.
MET Sema-PSI (158 μM), HGF-β (440 μM), and onartuzumab (71 μM), each in a volume of 50 µL, were mixed and incubated for 10 min before loading onto a Superdex 200 10/300 GL (GE Healthcare) equilibrated in 20 mM Hepes (pH 7.2) and 150 mM NaCl. The column was run at 0.5 mL/min and followed by the A280. Fractions of 0.25 mL were collected and analyzed by SDS/PAGE.
Crystallization and Data Collection.
MET Sema-PSI, HGF-β, and onartuzumab (5D5.v2) Fab fragment were mixed at 1:1:1 ratio and subjected to a final size-exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare). Diffracting crystals of the ternary complex were obtained directly from Protein Complex Suite B09 [0.1 M sodium cacodylate pH 6, 15% (wt/vol) PEG 4000; Qiagen] at 19 °C and, after minor optimization, led to crystals diffracting to ∼4 Å in-house. Crystals used for data collection grew at 19 °C by equilibrating equal volumes of protein (20 mg/mL) and reservoir solution [0.1 M sodium cacodylate pH 6.2, 20% (wt/vol) PEG 4000] by the sitting-drop vapor diffusion method. Before data collection, crystals were cryoprotected in reservoir solution containing 30% glycerol and flash-frozen in liquid nitrogen. Data were collected from a single crystal at the Advanced Light Source beamline 5.0.2, set at a wavelength of 1.0 Å, and processed with HKL2000 (65).
Structure Determination and Refinement.
The structure of the onartuzumab Fab/MET Sema-PSI/HGF-β ternary complex published here [Protein Data Bank (PDB) ID code 4K3J] was solved at 2.8-Å resolution by molecular replacement with PHASER (66) using the previously solved MET Sema-PSI/HGF-β complex (PDB ID code 1SHY) (45) and anti-VEGF G6 Fab (PDB ID code 2FJF) (67) as starting models. Onartuzumab CDR loops of the Fab fragment were manually rebuilt with the crystallographic object-oriented toolkit (68) using simulated annealing composite omit map implemented in Phenix (69). Subsequent rounds of model building and refinement with Refmac program (70) were carried out until convergence. The asymmetric unit contains one entire ternary complex consisting of one MET Sema-PSI fragment, one HGF β-chain, and one onartuzumab Fab region. The structure is nearly isomorphous with that of the MET Sema-PSI/HGF-β binary complex, although accommodation of the onartuzumab Fab fragment resulted in slightly different cell parameters. Despite medium resolution of the structure, the final electron density is of sufficient quality, and key features such as the onartuzumab CDR loops and epitopes on MET are unambiguous and allowed detailed interrogation of this interface. The final model includes residues 495–721 of HGF-β; residues 39–301, 311–397, 415–497, and 499–564 of MET; and residues 1–215 and 1–211 of onartuzumab Fab fragment H chain and L chain, respectively; 1 N-acetyl-d-glucosamine covalently attached to Met Asn45 and 183 water molecules. The Rfactor/Rfree is 21.1%/25.3%, with excellent geometry as assessed with MolProbity (Table S1) (70); residues in favored, allowed, and outlier regions are 96.03%, 3.79%, and 0.18%, respectively.
Binding Measurements of MET Sema-PSI Mutants to Onartuzumab.
The onartuzumab Fab (5D5.v2 Fab) was biotinylated in a 1:1 molar ratio using EZ-Link Sulfo-NHS-LC-Biotin kit (Thermo Fisher). Biotinylated onartuzumab Fab at 25 μg/mL in kinetic buffer (Fortebio) was captured by streptavidin biosensors on an OctetRED384 (Fortebio) during a 5-min incubation, which was followed by a 5-min wash in kinetic buffer. The association and dissociation binding kinetics for eight different concentrations (starting at 1,000 nM; twofold serial dilution) of wild-type and each mutant MET Sema-PSI protein were measured for 10 min and evaluated with OctetRED Evaluation Software 7 using a 1:1 binding model to derive kon, koff, and KD values.
Supplementary Material
Acknowledgments
We thank all members of the onartuzumab development teams who have contributed to the development of this antibody. We acknowledge Ling Chang, Kelly Dodge, Sean Kelly, Teresa Davancaze, Chae Reed, Mark Ultsch, Charlie Eigenbrot, and Paul Carter for their contributions to the development of onartuzumab and this manuscript. We thank Charlotte Kennerley and all of the staff at Gardiner-Caldwell Communications for editorial support in generation of this manuscript. We thank the staff at the Advanced Light Source for their assistance with sample handling and data collection; ALS beamline 5.0.2 and the Berkeley Center for Structural Biology are supported by the National Institute of General Medical Sciences and the Department of Energy. All studies were funded by Genentech, Inc.; editorial support was funded by Genentech, Inc.
Footnotes
Conflict of interest statement: All authors except Y.-W.Z., Y.S., D.W.K., and G.F.V.W. are or were employees of Genentech, Inc., at the time when they performed the work presented in this manuscript.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4K3J).
8Deceased August 26, 2005.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302725110/-/DCSupplemental.
References
- 1.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9(10):767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 3.Niemann HH. Structural insights into Met receptor activation. Eur J Cell Biol. 2011;90(11):972–981. doi: 10.1016/j.ejcb.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9(7):617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 5.Donate LE, et al. Molecular evolution and domain structure of plasminogen-related growth factors (HGF/SF and HGF1/MSP) Protein Sci. 1994;3(12):2378–2394. doi: 10.1002/pro.5560031222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann G, et al. A functional domain in the heavy chain of scatter factor/hepatocyte growth factor binds the c-Met receptor and induces cell dissociation but not mitogenesis. Proc Natl Acad Sci USA. 1992;89(23):11574–11578. doi: 10.1073/pnas.89.23.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lokker NA, et al. Structure-function analysis of hepatocyte growth factor: Identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J. 1992;11(7):2503–2510. doi: 10.1002/j.1460-2075.1992.tb05315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4(12):915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 9.Birchmeier C, Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol. 1998;8(10):404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 10.Neuss S, Becher E, Wöltje M, Tietze L, Jahnen-Dechent W. Functional expression of HGF and HGF receptor/c-met in adult human mesenchymal stem cells suggests a role in cell mobilization, tissue repair, and wound healing. Stem Cells. 2004;22(3):405–414. doi: 10.1634/stemcells.22-3-405. [DOI] [PubMed] [Google Scholar]
- 11.Huh CG, et al. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci USA. 2004;101(13):4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borowiak M, et al. Met provides essential signals for liver regeneration. Proc Natl Acad Sci USA. 2004;101(29):10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chmielowiec J, et al. c-Met is essential for wound healing in the skin. J Cell Biol. 2007;177(1):151–162. doi: 10.1083/jcb.200701086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: Rationale and progress. Nat Rev Cancer. 2012;12(2):89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 15.Kong-Beltran M, Stamos J, Wickramasinghe D. The Sema domain of Met is necessary for receptor dimerization and activation. Cancer Cell. 2004;6(1):75–84. doi: 10.1016/j.ccr.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Martens T, et al. A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo. Clin Cancer Res. 2006;12(20 Pt 1):6144–6152. doi: 10.1158/1078-0432.CCR-05-1418. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, et al. MetMAb, the one-armed 5D5 anti-c-Met antibody, inhibits orthotopic pancreatic tumor growth and improves survival. Cancer Res. 2008;68(11):4360–4368. doi: 10.1158/0008-5472.CAN-07-5960. [DOI] [PubMed] [Google Scholar]
- 18.Moss R, et al. 2010. Complete results from phase 1 dose escalation study of MetMAb, a monovalent antagonist antibody to the receptor Met, dosed as a single agent and in combination with bevacizumab in patients with advanced solid malignancies. Ann Oncol 21(Suppl 8):viii165.
- 19.Spigel DR, et al. Final efficacy results from OAM4558g, a randomized phase II study evaluating MetMAb or placebo in combination with erlotinib in advanced NSCLC. J Clin Oncol. 2011;29(Suppl 15):477s. [Google Scholar]
- 20.Petrelli A, et al. Ab-induced ectodomain shedding mediates hepatocyte growth factor receptor down-regulation and hampers biological activity. Proc Natl Acad Sci USA. 2006;103(13):5090–5095. doi: 10.1073/pnas.0508156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prat M, Crepaldi T, Pennacchietti S, Bussolino F, Comoglio PM. Agonistic monoclonal antibodies against the Met receptor dissect the biological responses to HGF. J Cell Sci. 1998;111(Pt 2):237–247. doi: 10.1242/jcs.111.2.237. [DOI] [PubMed] [Google Scholar]
- 22.Schwall RH, et al. Heparin induces dimerization and confers proliferative activity onto the hepatocyte growth factor antagonists NK1 and NK2. J Cell Biol. 1996;133(3):709–718. doi: 10.1083/jcb.133.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam SH, Sassoli PM, Jordan RE, Nakada MT. Abciximab (ReoPro, chimeric 7E3 Fab) demonstrates equivalent affinity and functional blockade of glycoprotein IIb/IIIa and αvβ3 integrins. Circulation. 1998;98(11):1085–1091. doi: 10.1161/01.cir.98.11.1085. [DOI] [PubMed] [Google Scholar]
- 24.Merchant AM, et al. An efficient route to human bispecific IgG. Nat Biotechnol. 1998;16(7):677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 25.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of Immunological Interest. Bethesda, MD: Public Health Service, National Institute of Health; 1991. [Google Scholar]
- 26.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: Contact analysis and binding site topography. J Mol Biol. 1996;262(5):732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 27.Simmons LC, et al. Expression of full-length immunoglobulins in Escherichia coli: Rapid and efficient production of aglycosylated antibodies. J Immunol Methods. 2002;263(1-2):133–147. doi: 10.1016/s0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- 28.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93(23):13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goebl NA, et al. Neonatal Fc receptor mediates internalization of Fc in transfected human endothelial cells. Mol Biol Cell. 2008;19(12):5490–5505. doi: 10.1091/mbc.E07-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junghans RP, Anderson CL. The protection receptor for IgG catabolism is the β2-microglobulin-containing neonatal intestinal transport receptor. Proc Natl Acad Sci USA. 1996;93(11):5512–5516. doi: 10.1073/pnas.93.11.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radaev S, Sun PD. Recognition of IgG by Fcgamma receptor. The role of Fc glycosylation and the binding of peptide inhibitors. J Biol Chem. 2001;276(19):16478–16483. doi: 10.1074/jbc.M100351200. [DOI] [PubMed] [Google Scholar]
- 32.Bender BC, et al. Translational pharmacokinetic (PK), pharmacodynamic (PD) modeling and simulation analysis of MetMAb. EJS Suppl. 2008;6(12):170. [Google Scholar]
- 33.Xiang H, et al. Supporting MetMAb entry into the clinic with nonclinical pharmacokinetic (PK) and pharmacodynamic (PD) information. EJC Suppl. 2008;6(12):167. [Google Scholar]
- 34.Bai S, et al. Population pharmacokinetic analysis from phase I and phase II studies of the humanized monovalent antibody, MetMAb, in patients with advanced solid tumors. J Clin Oncol. 2011;29(Suppl 15):182s. doi: 10.1002/jcph.148. [DOI] [PubMed] [Google Scholar]
- 35.Salgia R, et al. A phase I, open-label, dose-escalation study of the safety and pharmacology of MetMAb, a monovalent antagonist antibody to the receptor c-Met, administered IV in patients with locally advanced or metastatic solid tumors. EJC Suppl. 2008;6(12):129. [Google Scholar]
- 36.Schneider SW, et al. Glioblastoma cells release factors that disrupt blood-brain barrier features. Acta Neuropathol. 2004;107(3):272–276. doi: 10.1007/s00401-003-0810-2. [DOI] [PubMed] [Google Scholar]
- 37.de Vries NA, Beijnen JH, van Tellingen O. High-grade glioma mouse models and their applicability for preclinical testing. Cancer Treat Rev. 2009;35(8):714–723. doi: 10.1016/j.ctrv.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Rong S, et al. Tumorigenicity of the met proto-oncogene and the gene for hepatocyte growth factor. Mol Cell Biol. 1992;12(11):5152–5158. doi: 10.1128/mcb.12.11.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhargava M, et al. Scatter factor and hepatocyte growth factor: Activities, properties, and mechanism. Cell Growth Differ. 1992;3(1):11–20. [PubMed] [Google Scholar]
- 40.Zhang YW, et al. Enhanced growth of human met-expressing xenografts in a new strain of immunocompromised mice transgenic for human hepatocyte growth factor/scatter factor. Oncogene. 2005;24(1):101–106. doi: 10.1038/sj.onc.1208181. [DOI] [PubMed] [Google Scholar]
- 41.Peschard P, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8(5):995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 42.Kong-Beltran M, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66(1):283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 43.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Lokker NA, Godowski PJ. Generation and characterization of a competitive antagonist of human hepatocyte growth factor, HGF/NK1. J Biol Chem. 1993;268(23):17145–17150. [PubMed] [Google Scholar]
- 45.Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. Crystal structure of the HGF β-chain in complex with the Sema domain of the Met receptor. EMBO J. 2004;23(12):2325–2335. doi: 10.1038/sj.emboj.7600243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunasekaran K, et al. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: Applications to bispecific molecules and monovalent IgG. J Biol Chem. 2010;285(25):19637–19646. doi: 10.1074/jbc.M110.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muda M, et al. Therapeutic assessment of SEED: A new engineered antibody platform designed to generate mono- and bispecific antibodies. Protein Eng Des Sel. 2011;24(5):447–454. doi: 10.1093/protein/gzq123. [DOI] [PubMed] [Google Scholar]
- 48.Salgia R, et al. Complete results from a phase Ia dose-escalation and dose-expansion study of single-agent MetMAb, a monovalent antagonist antibody to the receptor Met, administered intravenously in patients with locally advanced or metastatic solid tumors. Cancer Res. 2010;70(8 Suppl):2774. [Google Scholar]
- 49.Jung ST, et al. Aglycosylated IgG variants expressed in bacteria that selectively bind FcgammaRI potentiate tumor cell killing by monocyte-dendritic cells. Proc Natl Acad Sci USA. 2010;107(2):604–609. doi: 10.1073/pnas.0908590107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niemann HH, et al. Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell. 2007;130(2):235–246. doi: 10.1016/j.cell.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 51.Pacchiana G, et al. Monovalency unleashes the full therapeutic potential of the DN-30 anti-Met antibody. J Biol Chem. 2010;285(46):36149–36157. doi: 10.1074/jbc.M110.134031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mijares A, Lebesgue D, Wallukat G, Hoebeke J. From agonist to antagonist: Fab fragments of an agonist-like monoclonal anti-β2-adrenoceptor antibody behave as antagonists. Mol Pharmacol. 2000;58(2):373–379. doi: 10.1124/mol.58.2.373. [DOI] [PubMed] [Google Scholar]
- 53.Qian MD, et al. Novel agonist monoclonal antibodies activate TrkB receptors and demonstrate potent neurotrophic activities. J Neurosci. 2006;26(37):9394–9403. doi: 10.1523/JNEUROSCI.1118-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y, Zhang S, Wang X, Yang Z, Ou G. Overexpression of TrkB promotes the progression of colon cancer. APMIS. 2010;118(3):188–195. doi: 10.1111/j.1600-0463.2009.02577.x. [DOI] [PubMed] [Google Scholar]
- 55.Geiger TR, Peeper DS. Critical role for TrkB kinase function in anoikis suppression, tumorigenesis, and metastasis. Cancer Res. 2007;67(13):6221–6229. doi: 10.1158/0008-5472.CAN-07-0121. [DOI] [PubMed] [Google Scholar]
- 56.Spiess C, et al. Bispecific antibodies with natural architecture produced by co-culture of bacterial expressing two distinct half-antibodies. Nat Biotechnol. 2013 doi: 10.1038/nbt.2621. in press. [DOI] [PubMed] [Google Scholar]
- 57.Margulies DH, Kuehl WM, Scharff MD. Somatic cell hybridization of mouse myeloma cells. Cell. 1976;8(3):405–415. doi: 10.1016/0092-8674(76)90153-7. [DOI] [PubMed] [Google Scholar]
- 58.Carter P, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89(10):4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dennis MS. Current Trends in Monoclonal Antibody Development and Manufacturing. 2010. Humanization by CDR repair in pharmaceutical aspects of monoclonal antibodies. Shire SJ, Gombotz W, Bechtold-Peters K, Andya J, eds (Association for Pharmaceutical Scientists and Springer, New York), pp 9–28. [Google Scholar]
- 60.Gallop MA, Barrett RW, Dower WJ, Fodor SP, Gordon EM. Applications of combinatorial technologies to drug discovery. 1. Background and peptide combinatorial libraries. J Med Chem. 1994;37(9):1233–1251. doi: 10.1021/jm00035a001. [DOI] [PubMed] [Google Scholar]
- 61.Lee CV, et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340(5):1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 62.Ultsch M, Lokker NA, Godowski PJ, de Vos AM. Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution. Structure. 1998;6(11):1383–1393. doi: 10.1016/s0969-2126(98)00138-5. [DOI] [PubMed] [Google Scholar]
- 63.Kirchhofer D, et al. Structural and functional basis of the serine protease-like hepatocyte growth factor β-chain in Met binding and signaling. J Biol Chem. 2004;279(38):39915–39924. doi: 10.1074/jbc.M404795200. [DOI] [PubMed] [Google Scholar]
- 64.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4(11):2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 66.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuh G, et al. Structure-function studies of two synthetic anti-vascular endothelial growth factor Fabs and comparison with the Avastin Fab. J Biol Chem. 2006;281(10):6625–6631. doi: 10.1074/jbc.M507783200. [DOI] [PubMed] [Google Scholar]
- 68.Emsley P, Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132.
- 69.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.