Abstract
Vascular endothelial growth factors (VEGFs) and their receptors (VEGFRs) are key drivers of blood and lymph vessel formation in development, but also in several pathological processes. VEGF-C signaling through VEGFR-3 promotes lymphangiogenesis, which is a clinically relevant target for treating lymphatic insufficiency and for blocking tumor angiogenesis and metastasis. The extracellular domain of VEGFRs consists of seven Ig homology domains; domains 1–3 (D1-3) are responsible for ligand binding, and the membrane-proximal domains 4–7 (D4-7) are involved in structural rearrangements essential for receptor dimerization and activation. Here we analyzed the crystal structures of VEGF-C in complex with VEGFR-3 domains D1-2 and of the VEGFR-3 D4-5 homodimer. The structures revealed a conserved ligand-binding interface in D2 and a unique mechanism for VEGFR dimerization and activation, with homotypic interactions in D5. Mutation of the conserved residues mediating the D5 interaction (Thr446 and Lys516) and the D7 interaction (Arg737) compromised VEGF-C induced VEGFR-3 activation. A thermodynamic analysis of VEGFR-3 deletion mutants showed that D3, D4-5, and D6-7 all contribute to ligand binding. A structural model of the VEGF-C/VEGFR-3 D1-7 complex derived from small-angle X-ray scattering data is consistent with the homotypic interactions in D5 and D7. Taken together, our data show that ligand-dependent homotypic interactions in D5 and D7 are essential for VEGFR activation, opening promising possibilities for the design of VEGFR-specific drugs.
Keywords: signal transduction, receptor tyrosine kinase
VEGFs stimulate angiogenesis and lymphangiogenesis via VEGF receptors (VEGFRs) in endothelial cells. VEGF-A signaling is mediated predominantly through activation of VEGFR-2, resulting in sprouting of blood vessels from preexisting vasculature (1). In contrast, VEGFR-1 seems to have an inhibitory role by sequestering VEGF-A and thereby preventing its interaction with VEGFR-2 (2). On the other hand, VEGFR-3 plays an indispensable role in lymphangiogenesis (3). VEGFRs are involved in various pathological conditions, including solid tumor growth, tumor metastasis, and vascular retinopathies (4, 5).
VEGF-C and VEGF-D compose a VEGFR-3–specific subfamily of VEGFs. They are produced with large N- and C-terminal propeptides and gain activity toward VEGFR-3 and VEGFR-2 on proteolytic processing (reviewed in ref. 5). VEGFR-3 maturation involves proteolytic cleavage of the extracellular domain (ECD) in D5 (6–8). Both VEGF-C and VEGFR-3 also interact with the coreceptor neuropilin-2 (9). Loss of the Vegfc gene results in embryonic lethality owing to a lack of lymphatic vessel formation (10), whereas mutations that interfere with VEGFR-3 signaling have been associated with hereditary lymphedema, and mice deficient in Vegfr3 die in utero due to abnormal development of the blood vasculature (11, 12). VEGFR-3 and its heterodimers with VEGFR-2 are also important for sprouting angiogenesis and vascular network formation (13–15).
VEGFRs are type V receptor tyrosine kinases (RTKs) composing a family of three transmembrane receptors, with the ECDs consisting of seven Ig homology domains (D1–D7). For ligand binding, VEGFRs use predominantly D2, with D3 providing additional binding affinity (16–18). In addition, D1 is required for VEGFR-3 ligand binding, but the exact role of this domain remains elusive (19, 20). VEGFRs and the closely related type III RTKs are activated by ligand-induced dimerization of the extracellular domain, followed by tyrosine autophosphorylation of the intracellular kinase domain to generate downstream signaling (21). As a prototype for these families, crystal structures of the Mast/stem cell growth factor receptor KIT ECD revealed ligand-induced homotypic interactions in D4 (22). Electron microscopy (EM), small-angle X-ray scattering (SAXS), and functional assays have established that VEGFR-2 D4-7 is essential for receptor activation by facilitating dimerization through homotypic contacts formed between D4-5 and D7 (23–26). In line with these findings, the crystal structure of a VEGFR-2 D7 homodimer reveals a pair of salt bridges in the juxtaposed loops between strands E and F (E-F loop), which are dispensable for receptor dimerization but not for receptor activation (24). Similar mutations in the E-F loop in D4 of VEGFR-2 do not affect VEGF-A-induced receptor activation (24), suggesting unique interactions in D4-5.
Blockage of VEGFR function has been shown to inhibit tumor angiogenesis, lymphangiogenesis, and metastasis in several mouse models (4, 5). Identified inhibitors include antibodies that block ligand/receptor binding. We recently reported an antibody against VEGFR-3 D5 that did not block ligand binding but did inhibit VEGFR-3 homodimer and VEGFR-3/VEGFR-2 heterodimer formation and signal transduction (8). Similarly, an antibody and designed ankyrin repeat proteins (DARPins) targeted against D4-7 of VEGFR-2 were found to inhibit ligand-induced receptor activation, but not dimerization (26, 27), suggesting that such binders represent a new generation of highly specific inhibitors of VEGFR signaling.
In this study, we provide the structural basis of ligand binding to D1-2 of VEGFR-3 and define a unique role of D4-5 for VEGFR dimerization and activation. Using receptor mutants, we show that homotypic interactions in D5 and D7 are essential for VEGFR-3 activation. Based on our data, we suggest a general mechanism for VEGFR dimerization and activation by ligand-induced homotypic interactions in D5 and D7.
Results
Structure of the VEGF-C/VEGFR-3 D1-2 Complex.
We expressed mature VEGF-C (C137A) and VEGFR-3 D1-2 in insect cells and crystallized the complex in space group I23 (a, b, c = 166.7 Å). The complex structure was solved at 4.2-Å resolution by molecular replacement using multiple isomorphous replacement with anomalous scattering (MIRAS) phases, and was refined to a crystallographic R value of 33.4% and a free R value of 37.1% (Fig. 1 and Table S1). The complex with WT VEGF-C was crystallized in the same crystal form, and cross-phasing at 6-Å resolution indicated identical packing and complex formation (Fig. S1A).
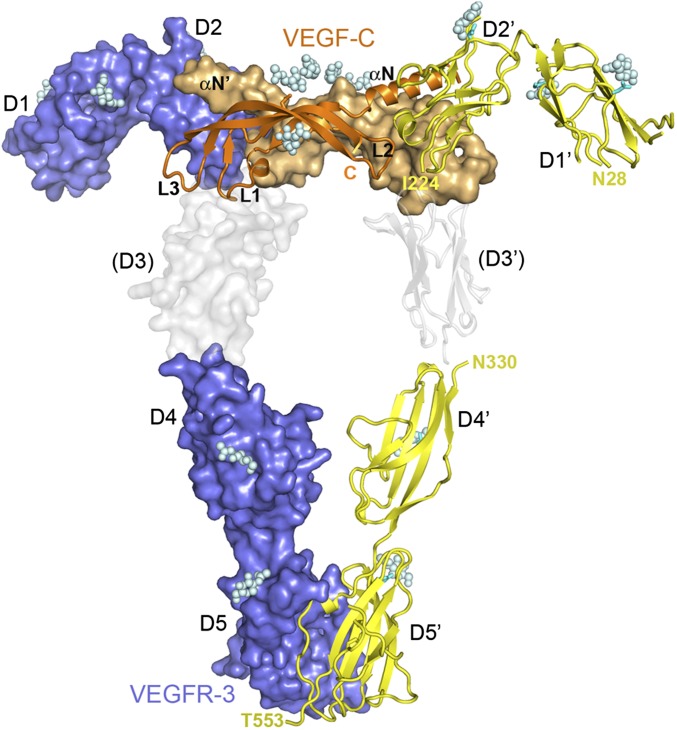
Fig. 1.
Crystal structures of the VEGF-C/VEGFR-3 D1-2 complex and the homodimer of VEGFR-3 D4-5. Shown are surface and cartoon representations with the two chains of VEGFR-3 in slate blue or yellow and the two chains of VEGF-C in orange or light orange. The VEGFR-3 complex, the D4-5 homodimer and our previous VEGF-C/VEGFR-2 D2-3 complex (PDB code 2X1X) were superimposed to the KIT/SCF complex (PDB code 2E9W). VEGFR-2 D3 is in gray. VEGFR Ig domains 1–5 (D1–D5), VEGF-C loops 1–3 (L1–L3), and the N-terminal helix (αN) are labeled. Glycan moieties are shown as spheres.
The overall architecture of the VEGFR-3 complex is very similar to the previously reported VEGFR-1 (16, 28, 29) and VEGFR-2 (18, 30) structures except, that in the VEGFR-3 complex structure, D1 is resolved as well. VEGF-C binding is limited to D2, with D1 protruding away from VEGF-C. Residues 28–134 of D1 are largely disordered, and we could build only the core of the apparent I-type Ig domain using KIT D1 (PDB ID code 2E9W) as a model. Our structure confirmed the presence of a disulfide bridge between Cys51 and Cys111 and N-glycosylation of Asn33 and Asn104. Residues 135–224 of D2 represent a smaller I-type Ig domain with a disulfide bridge between Cys158 and Cys206 and N-glycosylation of Asn166. Confronted with the low resolution, we could not define interactions at the binding interface. The structure clearly shows packing of the VEGF-C N-terminal helix against strand E and the C/C′ hairpin loop of D2. To confirm that these interactions are functional, we generated a double mutant of VEGF-C, D123A/Q130A, for VEGFR-3 binding studies (Fig. S1B). Our structure shows that loop 2 (L2) interacts with the loop connecting strands E and F of D2 and L3 with strands A′ and G.
The VEGF-C/VEGFR-3 D1-2 and VEGF-C/VEGFR-2 D2-3 complex structures can be aligned with an rmsd of 2.1 Å for 336 Cαatoms (Fig. S2 A–C). The N-terminal helix of VEGF-C in the VEGFR-3 complex is bent ∼9° toward D2, but L1–L3 adopt approximately the same conformations as in the VEGFR-2 complex.
Structure of VEGFR-3 D4-5.
We expressed VEGFR-3 D4-5 in insect cells and crystallized the purified protein in space group P3121 (a, b = 133.3 Å; c = 48.9 Å). The structure of D4-5 was determined at 2.5-Å resolution using single isomorphous replacement with anomalous scattering (SIRAS) phases, and was refined to a crystallographic R value of 21.0% and free R value of 25.3% (Fig. 1, Table S1, and Fig. S3 A and B).
D4, consisting of residues 330–419, is a small I-type Ig domain. Structural comparison shows that it is very similar to KIT D4; both domains can be aligned with an rmsd of 1.6 Å for 82 residues. However, unique to VEGFR-3 D4, strand D is missing, and strand C is connected directly to a short strand E on the opposite side of the molecule. D5, consisting of residues 420–553, is a larger I-type Ig domain characterized by a single 310 helical turn between strands A and A′, long strands D and E, and a long C-D loop of approximately 35 residues. Unlike D4, D5 has low homology to the corresponding D5 of KIT, with an rmsd of 2.6 Å for 74 residues. The overall temperature factor (B-factor) for D4 is far larger for D4 than for D5 (69.0Å2 vs. 55.1Å2; Fig. S3C). D4 has no disulfide bridges, whereas D5 has a buried disulfide bridge between the two sheets and an exposed disulfide bridge between Cys466 and Cys486 in the C-D loop, in which proteolytic cleavage occurs between Arg472 and Ser473 (7). The crystallized VEGFR-3 D4-5 appears to be only partially cleaved (Fig. S3D). Residues 470–483 in the C-D loop were disordered and thus were omitted from the final model. VEGFR-3 D4-5 has an overall extended structure connected by the linker peptide, a salt bridge between Glu344 and Lys539, and a hydrogen bond between Glu391 and Tyr448 (Fig. S3E).
VEGFR-3 D4-5 Homodimer and Homotypic Interactions in D5.
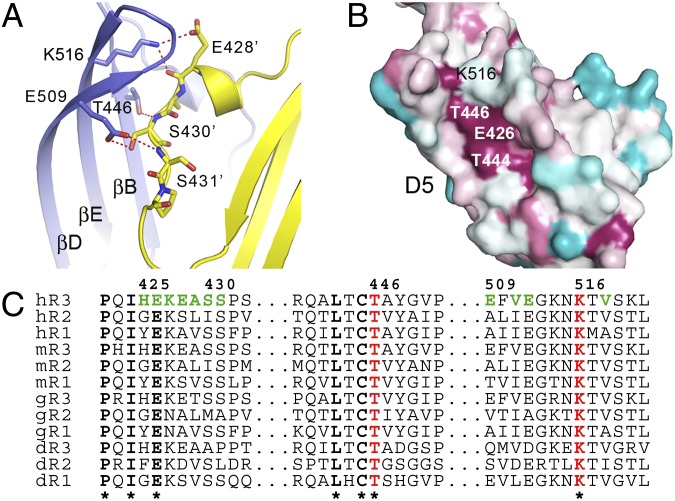
The D4-5 structure revealed homotypic interactions in D5, covering a solvent-accessible area of ∼570 Å2 per chain, but not in D4, where in contrast, Arg378 and Lys387 in the adjacent D4 face each other, which may lead to domain repulsion (Fig. S3F). The dimeric D4-5 molecules have a V-shaped arrangement at an angle of ∼45° and C and N termini at distances of approximately 14 Å and 65 Å, respectively (Fig. S3 F and G). The homotypic contacts in D5 between the sheets comprised of strands A′, B, D, and E are mediated by the helical protrusion between strands A and A′ occupying the cavity between the curved D-E loop and strands A and A′ in the other chain (Fig. 2A). The interactions are centered on the fully conserved Thr446 and Lys516, which create hydrogen bonds with the backbone atoms of Ser430 and Glu428 in strand A′ of the other chain (Fig. 2 B and C). His425, Glu428, and Glu509 contribute additional ionic interactions. Val518 and Ser430, and Thr446 and Ala429 form van der Waals contacts.
Fig. 2.
VEGFR-3 D5 homotypic interactions are centered to the conserved residues Thr446 and Lys516. (A) Homotypic interactions in D5. The two chains are shown in cartoon form, color-coded as in Fig. 1. Thr446, Glu509, Lys516, and their counterparts in strands A and A’ are shown as sticks. Hydrogen bonds are shown as red dashed lines. (B) D5, with conservation pattern in cyan through dark red for variable to conserved amino acids. Evolutionary rates of human, mouse, chicken, and zebrafish VEGFR sequences were were plotted using the ConSurf Web server. Thr444, Thr446, Glu426, and Lys516 compose a highly conserved patch on the D5 surface. (C) Representative amino acid sequence alignment from B. Residues involved in homotypic interactions are colored.
The VEGFR-3 D4-5 dimer structure agrees with the architecture of our previously reported ligand/VEGFR-2 D2-3 complex structures (PDB ID codes 2X1X, 3V6B, and 3V2A). Superposition of the VEGF-C/VEGFR-2 D2-3 and the VEGF-C/VEGFR-3 D1-2 structures on the KIT/SCF complex (PDB ID code 2E9W) creates a model of a VEGF/VEGFR D1-5 complex (Fig. 1 and Fig. S2C). In this model, the N termini of VEGFR-3 D4 (Asn330) are located within a few Ångstroms of the C termini of VEGFR-2 D3 (Glu326, the VEGFR-3 Glu329 counterpart).
Homotypic Interactions in D5 and D7 Are Essential for Ligand-Induced VEGFR-3 Activation.
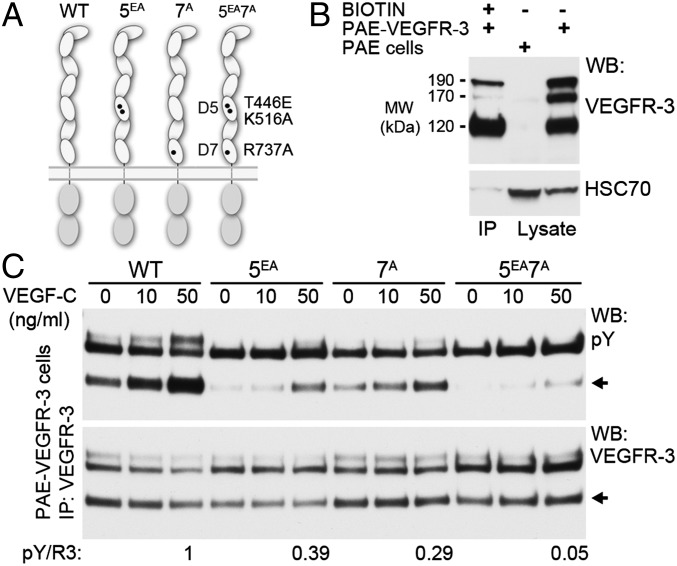
To determine the functional significance of the D5 and D7 interfaces in VEGF-C–induced VEGFR-3 activation, we generated a D5 double mutant, T446E/K516A (5EA), a D7 single mutant, R737A (7A) (24), and a D5/D7 triple mutant, T446E/K516A/R726 (5EA7A), of VEGFR-3 (Fig. 3A). To do so, we expressed WT VEGFR-3 and the mutant constructs in HEK293 cells and analyzed VEGF-C–induced VEGFR-3 activation using receptor immunoprecipitation, followed by anti-phosphotyrosine Western blot analysis (Fig. S4A). To quantify tyrosine autophosphorylation of VEGFR-3, we compared the level of phosphorylation of the band with an apparent Mr of 120 kDa on Western blots, corrected for VEGFR-3 expression level. This band represents the fully mature form of the receptor displayed on the cell surface (Fig. 3B). Our results indicate that the combination of D5 and D7 mutations (5EA7A) impairs VEGF-C–induced VEGFR-3 activation.
Fig. 3.
Homotypic interactions in D5 and D7 are mutually important for ligand-induced VEGFR-3 activation. (A) Schematic presentation of the WT VEGFR-3, 5EA, 7A, and 5EA7A mutants of VEGFR-3. (B) Biotinylation of cell surface-expressed VEGFR-3 isoforms. Biotinylated PAE-VEGFR-3 cell lysates were immunoprecipitated with streptavidin beads and blotted for VEGFR-3 and HSC70. Total lysates of PAE and PAE-VEGFR-3 cells were used as controls. (C) PAE cells stably expressing the VEGFR-3 constructs were stimulated with VEGF-C, and the cell lysates were immunoprecipitated with anti–VEGFR-3 and analyzed for VEGFR-3 autophosphorylation (pY) and expression (R3) by Western blot analysis. The pY/R3 ratio was quantified based on the ∼120-kDa band representing the fully processed VEGFR-3 (6, 7).
We performed a similar analysis in transfected porcine aortic endothelial (PAE) cells stably expressing the constructs (Fig. 3C and Fig. S4B). In these cells, both the D5 double mutant and D7 single mutant demonstrated reduced VEGFR-3 activity; however, effective VEGFR-3 blocking was obtained only with the triple mutant 5EA7A, suggesting the mutual importance of D5 and D7 interactions for VEGFR-3 activation.
Membrane-Proximal Domains of VEGFR-3 Provide Increased VEGF-C Affinity.
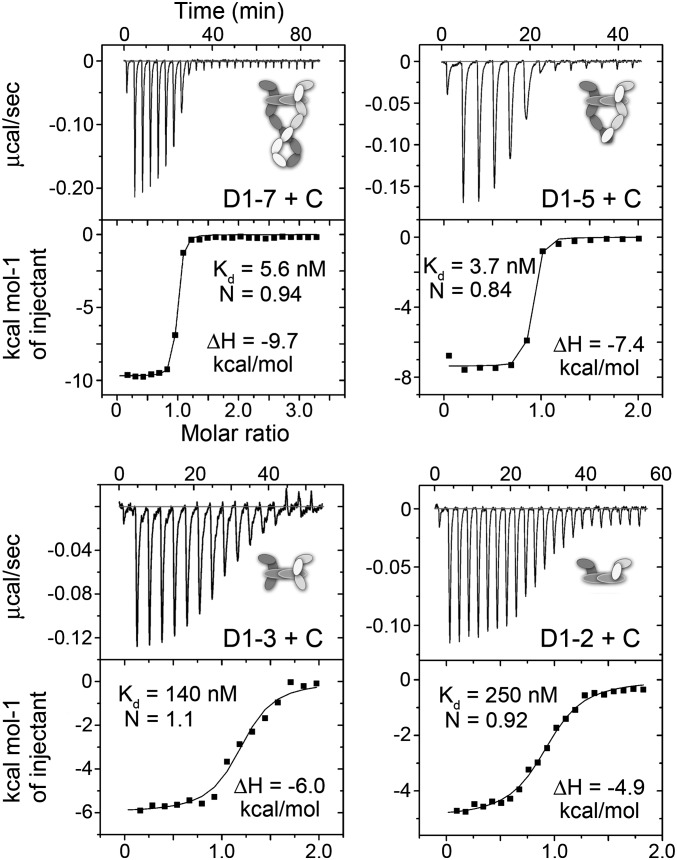
To explore the effect of the membrane-proximal domains of VEGFR-3 on VEGF-C binding, we generated C-terminal truncations of the soluble, monomeric VEGFR-3 ECD and measured VEGF-C binding by isothermal titration calorimetry (ITC). Our data show that VEGF-C binding to VEGFR-3 was enthalpically and entropically favorable, and that the presence of D3 and membrane-proximal D4-7 increased VEGF-C affinity (Fig. 4). In addition, deletion of D6-7, D4-5, and D3 gave rise to a gradual increase in binding enthalpy (ΔH), suggesting that these domains form additional interactions with the ligand in the case of D3, or ligand-induced dimerization in the case of D4-7. In addition, the D5 mutant demonstrated decreased affinity and a larger ΔH (Fig. S4 C and D).
Fig. 4.
Thermodynamic analysis of VEGF-C binding to VEGFR-3. Calorimetric titrations of VEGF-C to the monomeric VEGFR-3 ECD (D1-7) and its domain deletion mutants D1-2, D1-3 and D1-5 are shown. The enthalpy change (ΔH), dissociation constant (Kd), and stoichiometry (N) of the ITC assays are indicated.
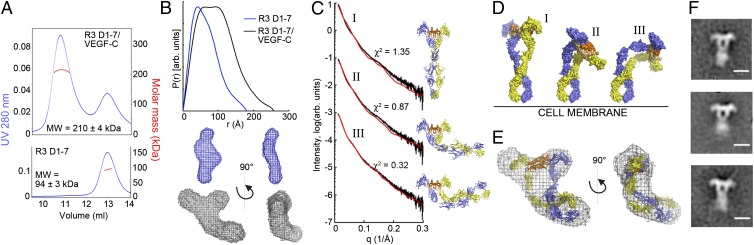
SAXS and Single-Particle EM Analysis of the VEGF-C/VEGFR-3 ECD Complex.
To confirm our structural models, we analyzed VEGFR-3 D1-7 and its complex with VEGF-C using multiangle laser light scattering (MALS) and SAXS (Fig. 5 A and B and Fig. S5 A and B). Our data show that the VEGFR-3 ECD is monomeric in solution and that dimerization is ligand-dependent. The SAXS-derived ab initio model of the complex revealed a bent, T-shaped molecular envelope, suggesting that the protrusions at the top of the molecules represent the D1 domains in the ligand-binding region and the main body represents the dimerized membrane-proximal domains.
Fig. 5.
Characterization of the VEGF-C/VEGFR-3 D1-7 complex in solution and in EM. (A) MALS analysis of the VEGF-C/VEGFR-3 D1-7 complex and VEGFR-3 D1-7. (B) The SAXS-derived distance distribution functions and averaged ab initio shape reconstructions of VEGFR-3 D1-7 and the VEGF-C/VEGFR-3 D1-7 complex. (C) Rigid-body models before and after refinement against the SAXS data were aligned using VEGF-C and are shown in cartoon form. The calculated (red) and experimental scattering curves (black) are compared. (I) The symmetrical model before refinement. (II) A representative model of the refinement with limited movement: D123C dimer–linker–D45 dimer–linker–D67 dimer. (III) Representative model of the refinement with increased movement: D12C dimer–linker–D3-D3–linker–D4-D4–linker–D5 dimer–linker–D67 dimer (SI Methods). (D) Rigid-body models from C aligned using the membrane-proximal D6-7 in vertical orientation and shown in a surface representation. (E) Overlay of the ab initio model of the complex from B and the final rigid-body model from C. (F) Representative class averages of the negative stain EM analysis of the VEGF-C/VEGFR-3 ECD complex. (Scale bar: 10 nm.)
Using the available VEGFR-2 and VEGFR-3 crystal structures and a homology model for D6, we prepared a model of the dimeric, glycosylated VEGF-C/VEGFR-3 ECD complex by applying rigid-body refinement of the SAXS data. The models with the best fit revealed strong bending in domains D3-5 that would bring VEGF-C closer to the plasma membrane (Fig. 5 C–E and Fig. S5C).
We also visualized the complex by negative-staining EM. The data agree with our structural models and clearly identify D1 protruding away from the complex (Fig. 5F and Fig. S6 A and B). Comparison of selected 2D projections of the D1-5 complex model with the EM structures also revealed a strong similarity with ligand-bound D2-5, although the membrane-proximal part was only partially resolved.
Discussion
Here we present structural data describing ligand binding to VEGFR-3 and propose a model for receptor activation. The structural data comprise crystal structures of a VEGF-C/VEGFR-3 D1-2 complex and the VEGFR-3 D4-5 homodimer, as well as an analysis of the complex by SAXS and EM. Our findings, complemented by a thermodynamic analysis of VEGF-C binding to VEGFR-3 and a cellular analysis of receptor constructs specifically mutated in D5 and D7, clearly show that the VEGFR-3 interactions observed in the crystal structures are functionally relevant for ligand-mediated dimerization and activation.
The VEGF-C/VEGFR-3 D1-2 complex structure provides a reliable model of the overall architecture of the ligand–receptor complex. Comparisons with the VEGFR-1 and VEGFR-2 complex structures show that all three VEGFRs share a conserved interface in D2 for ligand binding. VEGF-D has an extended N-terminal helix responsible for high-affinity binding and activation of VEGFR-3 (20), and we show here that mutation of the N-terminal Asp123 and Gln130 of VEGF-C decreases receptor binding, supporting the idea that the helix is an important determinant of VEGFR-3 ligand specificity (Figs. S1B and S4C). Additional structural elements of VEGF-C, including loops L1, L2, and L3, are involved in VEGFR-3 binding as well. L1 likely interacts with D3 (18, 30).
Our data also provide insight into D1. Despite its importance in VEGFR-3 ligand binding (19, 20), D1 protrudes away from VEGF-C and has no connections with it. The bent conformation of the D1-2 module is very similar to that observed in KIT, platelet-derived growth factor receptor beta, and macrophage colony-stimulating factor 1 receptor (Fig. S2C) (22, 31, 32). So far, KIT seems to be the only type III/V RTK that directly uses D1 for ligand binding (22). D1 may have a positive impact on D2 structural stability, as demonstrated by the poor resolution of the first strand of D2 (βA) in the multiple VEGFR-2 D2-3 structures, which lack D1 (18, 30). On the other hand, VEGFR-3 D1 has large disordered regions in the ligand complex, suggesting that it may interact with other binding partners in vivo, such as neuropilin-2 or the large N- and C-terminal propeptides of VEGF-C and VEGF-D.
VEGFR-3 maturation involves proteolytic cleavage in D5 (6, 7). This cleavage also occurred in the insect cell-expressed constructs, although the crystallized D4-5 was mainly uncleaved (Figs. S3D and S5A). The structure of the VEGFR-3 D4-5 homodimer suggests that proteolytic cleavage did not introduce significant changes in the D5 conformation, because it occurred in the C-D loop, which is disordered in the Cys466/Cys486 disulfide bridge. This loop is not involved in VEGFR-3 dimerization, leaving the function of this processing step elusive. Nonetheless, our structure suggests that the disulfide bridge restricts the flexibility of this loop, thereby increasing the stability of D5.
Calorimetric data for VEGF-C binding to our VEGFR-3 ECD mutant constructs indicate direct interaction of D3 with the ligand. In contrast, the membrane-proximal domains D4-5 and D6-7 modulate ligand binding indirectly. The contribution of D4-5 suggests that the homotypic interactions in D5 are energetically favorable. D6-7 does not increase VEGF-C affinity further; however, the decrease in ΔH indicates additional homotypic receptor contacts. This explains why mutations in D5 and D7 showed an additive inhibitory effect on VEGF-C–stimulated receptor phosphorylation in both HEK293 and PAE cells. The mutant and the ITC data agree with the observed interactions in D5 and with the putative salt bridges between Arg737 and Asp742 (human VEGFR-3 numbering) in VEGFR-3 D7. Taken together, the data indicate that the observed homotypic interactions in the membrane-proximal domains of VEGFRs are functionally highly relevant, as has been shown previously for VEGFR-2 (24, 26) and here for D5 and D7 of VEGFR-3.
VEGFRs are structurally highly similar and also signal as heterodimers (14, 33). Similar to the conserved D7 interface, the key residues involved in D5 dimerization (Thr446 and Lys516) are fully conserved, suggesting that similar D5 interactions exist in other VEGFRs. D4-directed homotypic interactions have been suggested previously based on the ligand-induced crossover in the middle of the EM structure of the VEGF-A/VEGFR-2 ECD complex (23). Considering the low resolution of the 2D projections of the EM data, these data are compatible with the involvement of D5 in receptor dimerization, which we report here for VEGFR-3. A previous binding study indicated that both D4 and D5 increase the affinity of VEGFR-2 for VEGF-A. Compared with D1-3, D1-5 showed 86-fold greater affinity and D1-4 showed 16-fold greater affinity (34). Similarly, both D4 and D4-5 of VEGFR-1 have been shown to enhance ligand-induced receptor dimerization in cross-linking experiments (35). Thus, the results of the VEGFR-1– and VEGFR-2–binding studies support a role for D5 in VEGFR dimerization, although additional interactions in D4 cannot be ruled out.
D4 may have an additional functional role, as suggested by previous studies showing that a swap of D4 with another domain, but not mutation of the D4 E-F loops facing each other, impaired ligand-induced VEGFR-2 activation (24, 26). D4 may facilitate proper orientation of the ligand-binding D1-3 in the active complex. D4 in VEGFRs and in type III RTKs lacks the intradomain disulfides (24), which have been shown to improve Ig domain stability (36). Compared with D5, the lack of disulfides in VEGFR-3 D4 is reflected in the higher overall B-factor level (Fig. S3C), suggesting increased domain flexibility in D4. This may be essential for the bending around D3-5 observed in the SAXS data. D3-4 and D4-5 hinge-like motions were also observed in the ligand-induced activation of KIT (22).
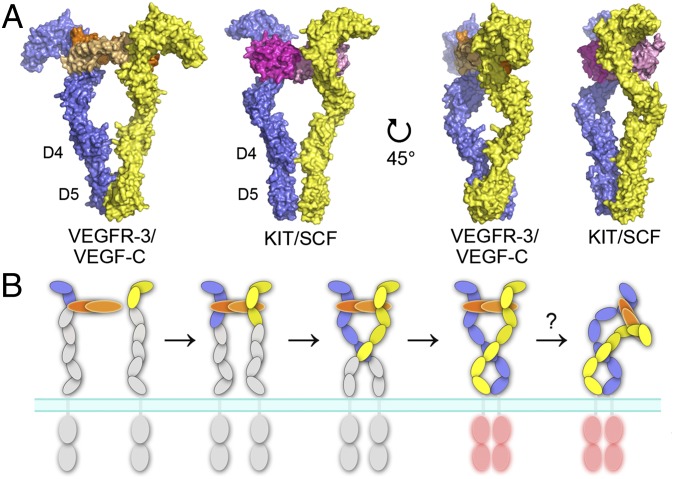
Based on high-resolution structural data for KIT, a model for the mechanism of ligand-induced activation of type III/V RTKs has been proposed (22). With this model, together with the available structures for VEGFR-2 and VEGFR-3, it is now possible to construct a similar model for VEGFR activation. Ligand-induced KIT dimerization promotes reorientation of D4-5, enabling lateral D4–D4 interactions essential for receptor activation (22). This seems to be a common mechanism in type III RTK activation (37–40). Previous studies found similarities in the ligand-binding modes for type III and IV RTKs and almost identical homotypic interactions in VEGFR-2 D7 and KIT D4 (18, 24, 32). The VEGFR-3 structures shown here indicate additional similarities to type III RTKs; however, our results also demonstrate some unique properties of these receptors that affect the mechanism of activation.
Despite the lack of homotypic interactions in VEGFR-3 D4, the arrangement of D4 in KIT and VEGFR-3 dimers is similar, with the E-F loops facing each other (Fig. S3). KIT D4-5 shows an almost parallel orientation in the ligand-bound receptor dimer, whereas VEGFR-3 D4-5 is twisted, resulting in separation of D4 and formation of unique D5 interactions (Fig. 6A). Because VEGFR activation also requires homotypic interactions in D7, the two additional Ig domains of VEGFRs give rise to a more complex, two-step mechanism for ligand-induced dimerization and receptor activation. This may be functionally relevant by allowing tight control of receptor activation.
Fig. 6.
The mechanism of ligand-induced VEGFR dimerization and activation. (A) Comparison of the ligand-induced dimerization of D1-5 of type III/V RTKs. The KIT/SCF complex (22) and the model of the VEGFR-3/VEGF-C complex are shown as surface representations in two orientations. SCF dimer is colored in magenta and in light magenta, and the two chains of KIT, VEGF-C, and VEGFR-3 are color-coded as in Fig. 1. (B) A proposed model of the ligand-induced dimerization and activation of VEGFRs. D1-2 represents the major ligand-binding unit. Ligand-induced D2-3 reconfiguration facilitates homotypic interactions in D5 and D7 that together are important for VEGFR activation. SAXS data indicates bending of the VEGF-C/VEGFR-3 complex around D3-5.
The subunit interactions in the D7 homodimer of VEGFR-2 cover a solvent-accessible area of only 480 Å2 per chain, and D7 has been shown to be monomeric in solution, indicating that the interactions are of low affinity (24). The dimerization interface in D5 is also relatively small (∼570 Å2 per chain), and the interface is dominated by ionic interactions. Like VEGFR-2 ECD, VEGFR-3 ECD is monomeric in solution, clearly showing that D5 and D7 are not capable of dimerizing the receptor in the absence of ligand (Fig. 5) (25). The restricted mobility of the receptors in the transmembrane domain may further aid the formation of such contacts in active receptor dimers.
A recent EM study revealed that the VEGFR-2 ECD is flexible and exists in multiple conformations in ligand-induced dimerization (23). Similarly, comparison of the available ligand/VEGFR-2 D2-3 complex structures found large variations in the D2-3 twist angles, indicating a D2-3 hinge-like rigid body motion (18, 30). Taken together with our data, these findings suggest a general mechanism for VEGFR activation in which ligand-induced D2-3 reorientation facilitates homotypic interactions in D5 and D7, resulting in specific positioning of the transmembrane and intracellular kinase domains in active receptor dimers (Fig. 6B). Interestingly, our SAXS data revealed strong bending of the receptor ECD complex in solution, which would bring VEGF-C closer to the plasma membrane when bound to the cell surface-expressed VEGFR-3. This bending, along with the distorted symmetry of receptor dimers, might be relevant for coreceptor binding.
VEGFRs also demonstrate differences in ligand-induced activation. D2 is the major ligand-binding domain, and the presence of D3 results in increased affinity in VEGF-A binding to VEGFR-2 and in placental growth factor binding to VEGFR-1 (17, 28). Consistent with the D2-3 reorientation, VEGFR-3 D3 also increases VEGF-C binding, whereas D4-5 and D6-7 further increase VEGFR-3 ligand-binding affinity. In contrast, the homotypic interactions in VEGFR-2 D4-7 are energetically unfavorable (30), suggesting that D3’s contribution to ligand binding is essential for VEGFR-2 dimerization and activation. VEGFR-1 D4-7 has only a small positive effect on ligand binding (16, 28).
VEGFR signaling has emerged as a key target for inhibiting tumor growth and metastasis by blocking tumor vascularization. Current therapeutics inhibiting VEGFR signaling include tyrosine kinase inhibitors, ligand traps, and antibodies blocking ligand binding to VEGFRs (4, 5). However, as is evident from previous studies, VEGFR and type III RTK activation requires additional specific homotypic interactions between the membrane-proximal domains (22–26, 38). Antibodies blocking these homotypic interactions are promising tools for therapeutic modulation of VEGFR activity (8, 26, 27). The experiments reported herein introduce a mechanism for VEGFR dimerization and activation via D5 and explain the efficacy of the VEGFR-3 D5-targeted antibodies (8). Our data suggest that the ligand-induced homotypic interactions in D5 and D7 are essential for VEGFR activation, providing strategies for the design of specific VEGFR inhibitors for use in combination with current anti-angiogenic inhibitors. Such an approach with two HER2 antibodies has proven successful in the treatment of patients with HER2-positive metastatic breast cancer (41).
Methods
Protein Expression and Purification.
Human VEGF-C, residues 103–215; C137A mutant of VEGF-C; human VEGFR-3 Ig D1-2, D1-3, D1-5, and D1-7; and the mutant 5EA in D1-5 were expressed in Sf21 insect cells using the baculovirus system and purified as described in SI Methods.
Binding Assays.
Calorimetric titrations of VEGF-C (C137A) to the monomeric VEGFR-3 deletion mutants were performed with a MicroCal VP-ITC calorimeter, as described in SI Methods. The data were processed using Origin 7.0 (MicroCal).
Crystallization and Structure Determination.
The VEGF-C/VEGFR3-D12 complex crystals were grown over a reservoir solution of 1.0–1.5 M ammonium sulfate (pH 8.5–9.5). Heavy-atom derivatives were prepared by soaking the crystals with methylmercury acetate, potassium tetrachloroplatinate, or hexatantalum tetradecabromide. VEGFR-3 D4-5 crystals were grown over a reservoir solution of 20–25% PEG 400 (pH 7.5–8.5), and a heavy-atom derivative was obtained with methylmercury acetate.
Complete datasets to 4.2-Å and 2.5-Å resolution were collected from the VEGF-C/VEGFR-3 D1-2 complex and the VEGFR-3 D4-5 crystals, respectively, at beamline X06SA at the Swiss Light Source (SLS). Anomalous data on the heavy-atom derivatives were collected at SLS beamlines X06SA and X06DA (Table S1). The sites were identified, and the phases were refined using autoSHARP (42). The structures were refined using PHENIX (43). Crystallographic details are provided in SI Methods.
SAXS Data Collection and Analysis.
The SAXS data were collected at SLS beamline cSAXS, and the data were processed using the ATSAS program package (44). The ab initio shape reconstructions were computed using DAMMIF, and rigid-body modeling was done using SASREF. Details of SAXS data collection and analysis are provided in SI Methods.
Negative-Staining EM and Image Analysis.
For the VEGF-C/VEGFR-3 D1-7 complex, data were acquired using a Philips CM10 transmission electron microscope equipped with an LaB6 filament. For projection analysis, 2,916 particles of the complex were selected from 487 images and classified into 30 classes. The EM analysis is described in detail in SI Methods.
Supplementary Material
Acknowledgments
We thank Tapio Tainola and Seppo Kaijalainen for excellent technical assistance, Dr. John H. Missimer for SAXS data evalution, and the staff at European Synchrotron Radiation Facility beamline ID29 and staff at Swiss Light Source beamlines X06SA, X06DA, and cSAXS for assistance and access to their facilities. This work was supported by grants from the Academy of Finland, the Sigrid Juselius Foundation, the Louis-Jeantet Foundation, the European Research Council (ERC-2010-AdG-268804-TX-FACTORS), Swiss National Science Foundation (Grant 31003A-130463), Oncosuisse (Grant OC2 01200-08-2007), NOVARTIS Stiftung für medizinisch-biologische Forschung (Grant 10C61), and the Swiss Initiative for Systems Biology (SystemsX.ch; grant CINA).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4BSJ and 4BSK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301415110/-/DCSupplemental.
References
- 1.Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Biochem J. 2011;437(2):169–183. doi: 10.1042/BJ20110301. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N. Vascular endothelial growth factor: Basic science and clinical progress. Endocr Rev. 2004;25(4):581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 3.Tammela T, Alitalo K. Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 2010;140(4):460–476. doi: 10.1016/j.cell.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438(7070):967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 5.Alitalo K. The lymphatic vasculature in disease. Nat Med. 2011;17(11):1371–1380. doi: 10.1038/nm.2545. [DOI] [PubMed] [Google Scholar]
- 6.Pajusola K, et al. Signalling properties of FLT4, a proteolytically processed receptor tyrosine kinase related to two VEGF receptors. Oncogene. 1994;9(12):3545–3555. [PubMed] [Google Scholar]
- 7.Lee J, et al. Vascular endothelial growth factor-related protein: A ligand and specific activator of the tyrosine kinase receptor Flt4. Proc Natl Acad Sci USA. 1996;93(5):1988–1992. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tvorogov D, et al. Effective suppression of vascular network formation by combination of antibodies blocking VEGFR ligand binding and receptor dimerization. Cancer Cell. 2010;18(6):630–640. doi: 10.1016/j.ccr.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Kärpänen T, et al. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J. 2006;20(9):1462–1472. doi: 10.1096/fj.05-5646com. [DOI] [PubMed] [Google Scholar]
- 10.Kärkkäinen MJ, et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat Immunol. 2004;5(1):74–80. doi: 10.1038/ni1013. [DOI] [PubMed] [Google Scholar]
- 11.Kärkkäinen MJ, et al. Missense mutations interfere with VEGFR-3 signalling in primary lymphoedema. Nat Genet. 2000;25(2):153–159. doi: 10.1038/75997. [DOI] [PubMed] [Google Scholar]
- 12.Dumont DJ, et al. Cardiovascular failure in mouse embryos deficient in VEGF receptor-3. Science. 1998;282(5390):946–949. doi: 10.1126/science.282.5390.946. [DOI] [PubMed] [Google Scholar]
- 13.Tammela T, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454(7204):656–660. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson I, et al. VEGF receptor 2/-3 heterodimers detected in situ by proximity ligation on angiogenic sprouts. EMBO J. 2010;29(8):1377–1388. doi: 10.1038/emboj.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tammela T, et al. VEGFR-3 controls tip to stalk conversion at vessel fusion sites by reinforcing Notch signalling. Nat Cell Biol. 2011;13(10):1202–1213. doi: 10.1038/ncb2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiesmann C, et al. Crystal structure at 1.7-Å resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91(5):695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 17.Fuh G, Li B, Crowley C, Cunningham B, Wells JA. Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor. J Biol Chem. 1998;273(18):11197–11204. doi: 10.1074/jbc.273.18.11197. [DOI] [PubMed] [Google Scholar]
- 18.Leppänen VM, et al. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci USA. 2010;107(6):2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeltsch M, et al. Vascular endothelial growth factor (VEGF)/VEGF-C mosaic molecules reveal specificity determinants and feature novel receptor binding patterns. J Biol Chem. 2006;281(17):12187–12195. doi: 10.1074/jbc.M511593200. [DOI] [PubMed] [Google Scholar]
- 20.Leppänen VM, et al. Structural determinants of vascular endothelial growth factor-D receptor binding and specificity. Blood. 2011;117(5):1507–1515. doi: 10.1182/blood-2010-08-301549. [DOI] [PubMed] [Google Scholar]
- 21.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuzawa S, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130(2):323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 23.Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007;14(3):249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc Natl Acad Sci USA. 2010;107(5):1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisko K, et al. Structural analysis of vascular endothelial growth factor receptor-2/ligand complexes by small-angle X-ray solution scattering. FASEB J. 2011;25(9):2980–2986. doi: 10.1096/fj.11-185397. [DOI] [PubMed] [Google Scholar]
- 26.Hyde CA, et al. Targeting extracellular domains D4 and D7 of vascular endothelial growth factor receptor 2 reveals allosteric receptor regulatory sites. Mol Cell Biol. 2012;32(19):3802–3813. doi: 10.1128/MCB.06787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kendrew J, et al. An antibody targeted to VEGFR-2 Ig domains 4-7 inhibits VEGFR-2 activation and VEGFR-2-dependent angiogenesis without affecting ligand binding. Mol Cancer Ther. 2011;10(5):770–783. doi: 10.1158/1535-7163.MCT-10-0876. [DOI] [PubMed] [Google Scholar]
- 28.Christinger HW, Fuh G, de Vos AM, Wiesmann C. The crystal structure of placental growth factor in complex with domain 2 of vascular endothelial growth factor receptor-1. J Biol Chem. 2004;279(11):10382–10388. doi: 10.1074/jbc.M313237200. [DOI] [PubMed] [Google Scholar]
- 29.Iyer S, Darley PI, Acharya KR. Structural insights into the binding of vascular endothelial growth factor-B by VEGFR-1(D2): Recognition and specificity. J Biol Chem. 2010;285(31):23779–23789. doi: 10.1074/jbc.M110.130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brozzo MS, et al. Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization. Blood. 2012;119(7):1781–1788. doi: 10.1182/blood-2011-11-390922. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony-stimulating factor bound to FMS: Diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci USA. 2008;105(47):18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shim AH, et al. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci USA. 2010;107(25):11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cudmore MJ, et al. The role of heterodimerization between VEGFR-1 and VEGFR-2 in the regulation of endothelial cell homeostasis. Nat Commun. 2012;3:972. doi: 10.1038/ncomms1977. [DOI] [PubMed] [Google Scholar]
- 34.Shinkai A, et al. Mapping of the sites involved in ligand association and dissociation at the extracellular domain of the kinase insert domain-containing receptor for vascular endothelial growth factor. J Biol Chem. 1998;273(47):31283–31288. doi: 10.1074/jbc.273.47.31283. [DOI] [PubMed] [Google Scholar]
- 35.Barleon B, et al. Mapping of the sites for ligand binding and receptor dimerization at the extracellular domain of the vascular endothelial growth factor receptor FLT-1. J Biol Chem. 1997;272(16):10382–10388. doi: 10.1074/jbc.272.16.10382. [DOI] [PubMed] [Google Scholar]
- 36.Carl P, Kwok CH, Manderson G, Speicher DW, Discher DE. Forced unfolding modulated by disulfide bonds in the Ig domains of a cell adhesion molecule. Proc Natl Acad Sci USA. 2001;98(4):1565–1570. doi: 10.1073/pnas.031409698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verstraete K, et al. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118(1):60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci USA. 2008;105(22):7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elegheert J, et al. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure. 2011;19(12):1762–1772. doi: 10.1016/j.str.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, et al. Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure. 2012;20(4):676–687. doi: 10.1016/j.str.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Swain SM, et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): Overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2013;14(6):461–471. doi: 10.1016/S1470-2045(13)70130-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr Opin Struct Biol. 2007;17(5):562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.