Abstract
PII proteins are one of the most widespread families of signal transduction proteins in nature, being ubiquitous throughout bacteria, archaea, and plants. In all these organisms, PII proteins coordinate many facets of nitrogen metabolism by interacting with and regulating the activities of enzymes, transcription factors, and membrane transport proteins. The primary mode of signal perception by PII proteins derives from their ability to bind the effector molecules 2-oxoglutarate (2-OG) and ATP or ADP. The role of 2-OG as an indicator of cellular nitrogen status is well understood, but the function of ATP/ADP binding has remained unresolved. We have now shown that the Escherichia coli PII protein, GlnK, has an ATPase activity that is inhibited by 2-OG. Hence, when a drop in the cellular 2-OG pool signals nitrogen sufficiency, 2-OG depletion of GlnK causes bound ATP to be hydrolyzed to ADP, leading to a conformational change in the protein. We propose that the role of ATP/ADP binding in E. coli GlnK is to effect a 2-OG-dependent molecular switch that drives a conformational change in the T loops of the PII protein. We have further shown that two other PII proteins, Azospirillum brasilense GlnZ and Arabidopsis thaliana PII, have a similar ATPase activity, and we therefore suggest that this switch mechanism is likely to be a general property of most members of the PII protein family.
Keywords: nitrogen regulation, metabolic signalling, ATP hydrolysis
The PII proteins are some of the most widely distributed signal transduction proteins in nature. They are ubiquitous in bacteria, archaea, and plants, where they are involved in the regulation of many aspects of nitrogen metabolism. They function by protein–protein interaction, whereby they control the activities of enzymes, transcription factors, and membrane transport proteins (1–5).
PII proteins are homotrimers composed of 12- to 13-kDa subunits with a highly conserved structure. The body of the protein is a compact cylinder from which three long loops (the T loops) protrude. These loops have considerable structural flexibility, and they constitute the interaction interface for many of the PII target proteins (5). Signal perception by PII proteins can occur at two levels. The primary mode of signal perception appears to be almost universal and involves the binding of the effector molecules 2-oxoglutarate (2-OG) and ATP/ADP within the lateral intersubunit clefts. A secondary mode of signal perception, which is less conserved, involves covalent modification of a residue within the T loop.
In the primary mode of signal perception, the binding of 2-OG and ATP is strongly synergistic (6). This synergy is explicable from the structures of the Azospirillum brasilense GlnK ortholog, GlnZ, the Synechococcus elongatus PII protein, and Archaeoglobus fulgidus GlnK3, each with bound 2-OG and MgATP (7–9). In all cases, 2-OG binds close to MgATP within the lateral cleft. The Mg2+ ion is coordinated by the 2-oxo moiety of 2-OG, together with the three phosphate oxygens of ATP and the side chain of the highly conserved residue Gln39 (A. brasilense residue numbering) at the base of the T loop. The 5-carboxy group of 2-OG forms a salt bridge with another highly conserved residue, Lys58. One of the best-characterized PII interactions is its binding to the integral membrane ammonia channel protein AmtB, thereby controlling the flux of ammonia through AmtB and into the cell. The binding mode of ADP to PII proteins was revealed from the structure of Escherichia coli GlnK bound to AmtB, in which ADP occupies the same nucleotide-binding site as ATP but Mg2+ and 2-OG are absent (10). The absence of 2-OG allows Gln39 to reorientate and form a bond to Lys58 with a concomitant conformational change in the T loop.
As the intracellular 2-OG pool is directly related to the cellular nitrogen-status, 2-OG is a logical effector molecule for PII, but the role of ATP or ADP as PII effector molecules has remained unclear. It was long considered that ATP could not play a regulatory role because its intracellular concentration is typically 1–5 mM, whereas the affinity of PII proteins for the nucleotide is in the micromole range (Kd ∼50 μM) (11, 12). However, the subsequent recognition that ADP is also a physiological effector (10) led to a reevaluation of the role of nucleotides, and a number of studies concluded that PII proteins might also act as sensors of cellular energy status, as reflected by fluctuations in the ATP/ADP ratio (13–18).
A full understanding of the mode of action of PII effector molecules requires a well-defined model system that can be studied both in vivo and in vitro and for which structural information is also available. The interaction of the E. coli PII protein, GlnK, with its cognate target, the ammonia channel AmtB, offers just such a model (10, 19, 20). Furthermore, phylogenetic analysis suggests that the regulation by GlnK of ammonia influx into the cell through AmtB is likely to represent the ancestral role of PII proteins (1).
Studies of the E. coli GlnK-AmtB system have shown that when cells are nitrogen-limited, GlnK is cytoplasmically located and uridylylated within the T loops, the cellular ATP and 2-OG pools are high, and GlnK is expected to contain a single molecule of 2-OG, Mg2+, and ATP in each of the intersubunit clefts (7, 20, 21). When nitrogen-limited cells are subject to an extracellular ammonium shock, the 2-OG pool drops rapidly from 1.4 to 0.3 mM. GlnK is rapidly deuridylylated and binds in a 1:1 stoichiometry to the cytoplasmic face of the AmtB trimer (19, 20). The conformation of the GlnK T loops changes to adopt an extended form, thereby allowing them to protrude into the cytoplasmic ends of the AmtB conduction channels and block further ammonium uptake (10). When isolated directly from cells, the GlnK–AmtB complex contains a single molecule of ADP, rather than ATP, per GlnK subunit and no 2-OG or Mg2+ (10). These in vivo changes can be replicated in vitro, confirming that complex formation is promoted by ADP and is inhibited by the presence of 2-OG, Mg2+, and ATP (20). Although these studies confirmed 2-OG as a key effector molecule, they did not fully rationalize the role of ATP and ADP.
We have now carried out a series of studies that lead us to propose that E. coli GlnK has an inherent ATP hydrolysis activity that is inhibited by 2-OG. This activity is conserved in other PII proteins, and we therefore suggest that the primary role of nucleotide binding is to facilitate a 2-OG-dependent conformational switch and that this is likely to be a characteristic of most PII proteins.
Results
GlnK Is an ATPase that Is Inhibited by 2-OG.
In our previous in vitro studies investigating the roles of ATP and ADP in regulating complex formation between GlnK and AmtB, we used slightly different experimental protocols in which GlnK was only sometimes preincubated with the nucleotide (19, 20). We subsequently recognized a correlation between the observed effects of ATP and the inclusion of a preincubation step. This led us to devise a new series of studies on the effects of both ATP and ADP on the association and dissociation of the GlnK–AmtB complex in vitro.
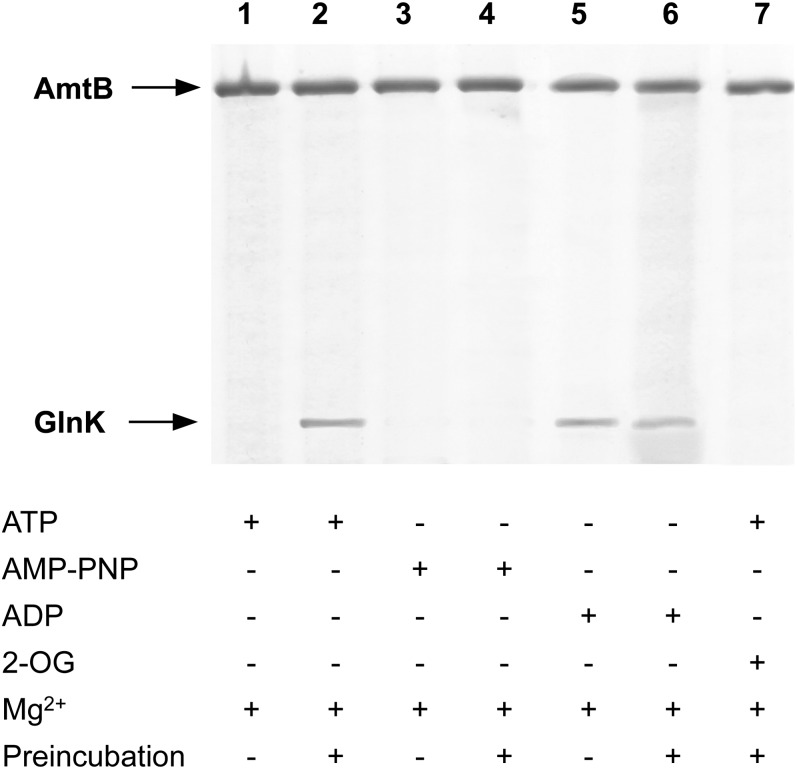
In our in vitro assay, we used wild-type E. coli GlnK that had been purified to >99% purity, using chromatography on heparin followed by gel filtration (Materials and Methods). When this GlnK was preincubated in the presence of the nucleotide, either ATP or ADP promoted complex formation with AmtB (Fig. 1, lanes 2 and 6), but without preincubation, only ADP was effective (Fig. 1, lanes 1 and 5). ATP did not promote complex formation when preincubated with the proteins in the presence of Mg2+ and 2-OG (Fig. 1, lane 7) (20). We therefore hypothesized that preincubation of GlnK with ATP allowed ADP to be formed, but only in the absence of Mg2+ and 2-OG. This would explain the observations by Durand and Merrick (19), who used a preincubation step and found that ATP promoted complex formation. Furthermore, consistent with this hypothesis, the nonhydrolysable ATP analog, gamma-imino-ATP (AMP-PNP), did not promote complex formation with or without preincubation (Fig. 1, lanes 3 and 4).
Fig. 1.
Nucleotide-dependence of GlnK–AmtB complex formation. Complex formation between His6-AmtB and GlnK in the presence of various effectors was assessed by elution from a His-Select Spin column. The effectors present for each lane are indicated in the table below the figure. Preincubation indicates that GlnK was preincubated with the effectors for 15 min at 30 °C before loading onto the column.
The purity and stability of our ATP preparation was assessed by HPLC, from which we determined that the ATP solution was >99% pure and that no significant hydrolysis occurred during the preincubation period of 15 min at 30 °C (Fig. S1). We then considered two hypotheses for the origin of the ATP hydrolysis. First, our GlnK preparation could contain a contaminating ATPase, and such contamination had previously been reported to be present in preparations of the E. coli PII protein GlnB (12). Alternatively, GlnK is itself an ATPase, but it only exhibits this activity in the absence of 2-OG.
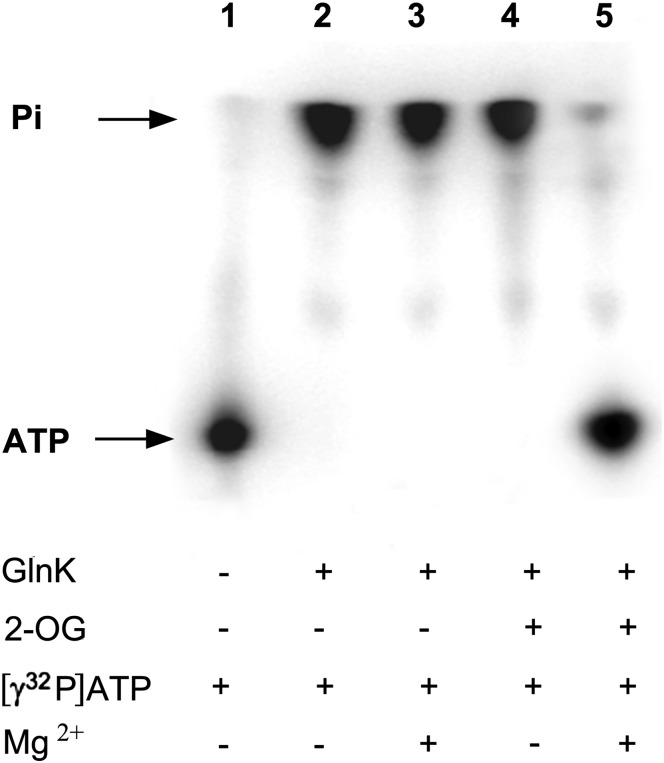
We therefore subjected wild-type GlnK to a conventional ATPase assay (Materials and Methods), with [γ32P]-ATP as substrate, and we analyzed the release of [32Pi] by TLC. This protocol assumes a relatively high turnover rate, and consequently it requires that only a small fraction of the substrate be radiolabeled [32P]-ATP. Using such a protocol, we failed to detect any significant hydrolysis by GlnK (Fig. S2). However, we then considered potential analogies with the signal-transducing Gα proteins that switch between a GTP- and a GDP-bound form and that typically exhibit low rates of GTP hydrolysis and GDP dissociation (22). Using this analogy, we reasoned that if GlnK had a low turnover rate of ATP hydrolysis, and if the subsequent rate of dissociation of ADP was also low, such that after hydrolysis ADP effectively remains bound to the protein, we might not detect hydrolysis of the [32P]-ATP with a conventional assay. We therefore repeated the assay, omitting the unlabeled ATP and using only radiolabeled ATP (present either as [γ32P]-ATP or [α32P]-ATP). As a consequence, we had a much higher specific radioactivity in the assay, and with the concentration of substrate (3 μM) significantly lower than that of the protein (100 μM), all the added ATP could potentially be hydrolyzed. Under these conditions, the GlnK protein preparation showed a clear ATPase activity with hydrolysis of ATP to ADP and free phosphate (Fig. 2 and Fig. S3). This activity was independent of the presence of Mg2+ (Fig. 2 and Fig. S3). Our studies on in vitro formation of the GlnK–AmtB complex had also indicated that if the ATPase activity was a property of GlnK, then ADP-bound GlnK would only be formed in 2-OG-limited conditions and ATP hydrolysis should be inhibited in the presence of 2-OG. This proved to be the case, and furthermore, 2-OG was only inhibitory in the presence of Mg2+ (Fig. 2 and Fig. S3). These data strongly support the proposal that the ATP hydrolysis activity is a property of GlnK.
Fig. 2.
ATPase activity of E. coli GlnK. Analysis of ATP hydrolysis using [γ32P]-ATP as described in Materials and Methods. Components of the assay are listed below the figure. Release of free Pi is shown in lanes 2–4.
GlnK Residues that Influence ATPase Activity.
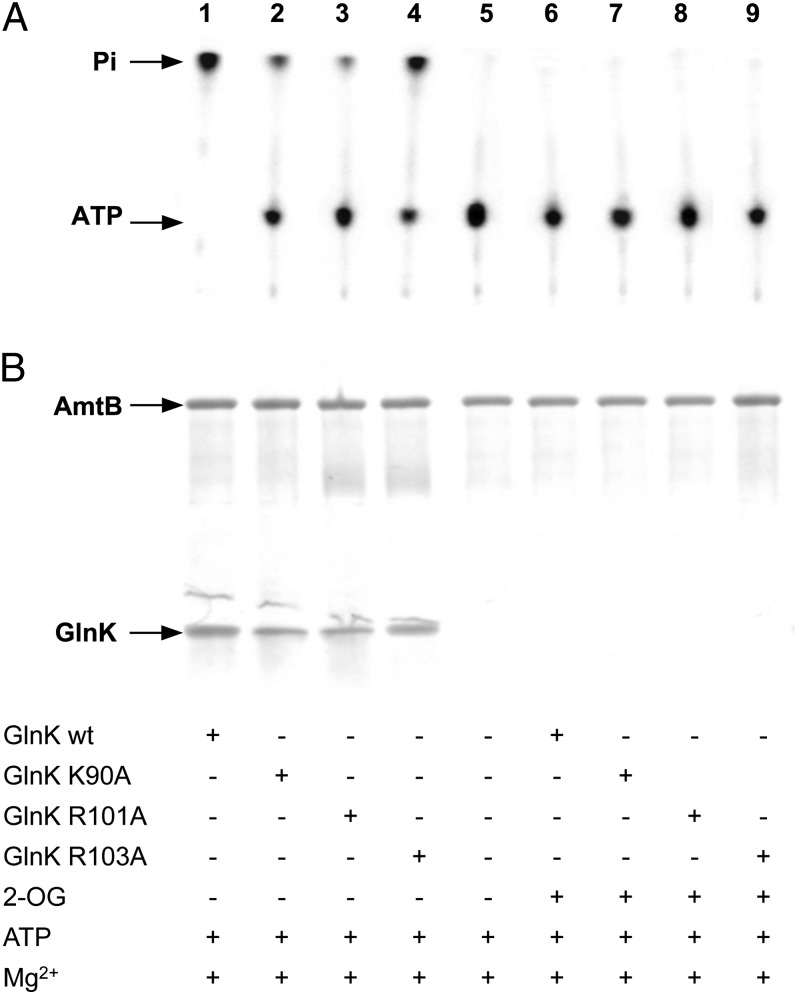
We reasoned that changes in one or more key residues in GlnK might be expected to influence its proposed ATPase activity. PII proteins contain a consensus sequence (T83GKIGDGKIF92) that shares significant homology with the Walker A motif (xGxxGxGKTxx) found in many nucleotide-binding proteins (1, 23), as well as two conserved arginine residues (R101 and R103) that coordinate the phosphate groups and stabilize the negative charges of the nucleotide triphosphate (23). Furthermore, from observation of the ADP-binding site in the GlnK–AmtB complex, Gruswitz et al. had previously suggested that K90, R101, and R103, together with a highly coordinated buried water molecule, might facilitate ATP hydrolysis by GlnK (24). We therefore examined the properties of K90A, R101A, and R103A variants of GlnK with respect to their ATPase activities and their abilities to form a GlnK–AmtB complex. In addition, we examined variants Q39A, Q39E, and K58A because 2-OG binding to GlnK inhibited ATPase activity, and both Q39 and K58 are involved in 2-OG binding (7, 25).
The K90A, R101A, and R103A variants of GlnK were all impaired in their ATPase activity compared with wild-type GlnK protein, but similar to the wild-type, all of them showed complete inhibition in the presence of 2-OG and Mg2+ (Fig. 3A). We hypothesized that the reduced activity of these variants could reflect an effect of the altered residue on the protein’s ability to bind ATP, rather than its catalytic activity. We therefore measured ATP binding of each of these variants by isothermal calorimetry and found all three to be significantly impaired in ATP-binding compared with wild-type GlnK. The Kds for ATP were wild-type, 5 µM; K90A, 168 µM; R101A, 260 µM; and R103A, 228 µM. All three variants were also assessed for their ability to form a complex with AmtB in vitro, and their phenotypes in this assay mirrored their ATPase activities. All three variants formed complexes to some degree, with R101A being the least effective and R103A the most effective (Fig. 3B).
Fig. 3.
ATPase activity of E. coli GlnK variants K90A, R101A, and R103A. (A) Analysis of ATP hydrolysis by TLC. (B) Complex formation between His6-AmtB and GlnK in vitro, assessed by elution from a His-Select Spin column. Components of the assay are listed below the figure.
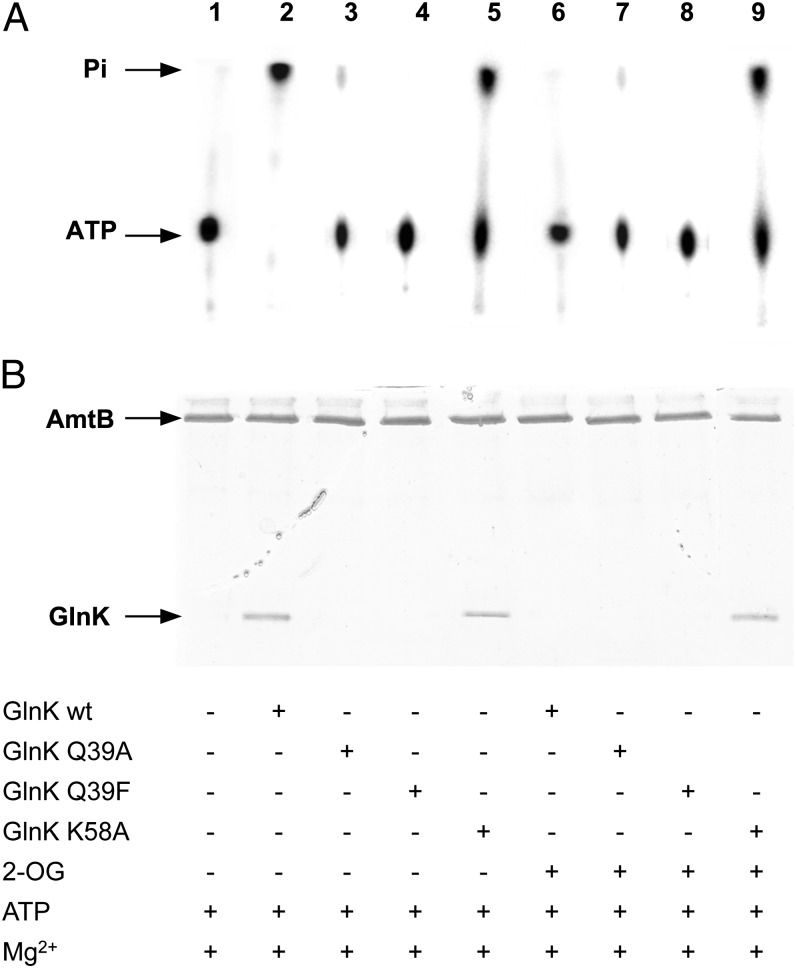
GlnK K58A had a markedly reduced ATPase activity compared with wild-type, but this activity was not inhibited by the presence of 2-OG (Fig. 4A). In contrast, GlnK Q39A and Q39E showed little or no ATPase activity (Fig. 4A). Both structural data (7) and previous studies of the effector-binding properties of a Q39E variant of E. coli GlnB (25) indicate that alterations to Q39 will impair 2-OG binding, but not ATP binding. Hence, these data suggest that Q39 could play a key role in the mechanism of ATP hydrolysis. Consistent with the observed ATPase activities, GlnK Q39A and Q39E failed to form complexes with AmtB, irrespective of whether 2-OG was present, whereas GlnK K58A still formed complexes, albeit less effectively than wild-type GlnK (Fig. 4B).
Fig. 4.
ATPase activity of E. coli GlnK variants Q39A, Q39E, and K58A. (A) Analysis of ATP hydrolysis by TLC. (B) Complex formation between His6-AmtB and GlnK in vitro, assessed by elution from a His-Select Spin column. Components of the assay are listed below the figure.
The pH Dependence of ATP Hydrolysis.
A number of crystal structures for PII proteins with bound ATP have been reported (4), but they have notably used crystallization conditions below pH 7.0. We therefore examined the pH dependence of ATP hydrolysis by GlnK and determined that the activity was significantly impaired at pH 6.0 and completely inhibited at pH 4.0 (Fig. S4). Hence, we suggest that the ability to “trap” the ATP-bound form of PII proteins in the absence of 2-OG is dependent on the crystallization being carried out at low pH where ATP hydrolysis is inhibited.
ATP Hydrolysis by Other PII Proteins.
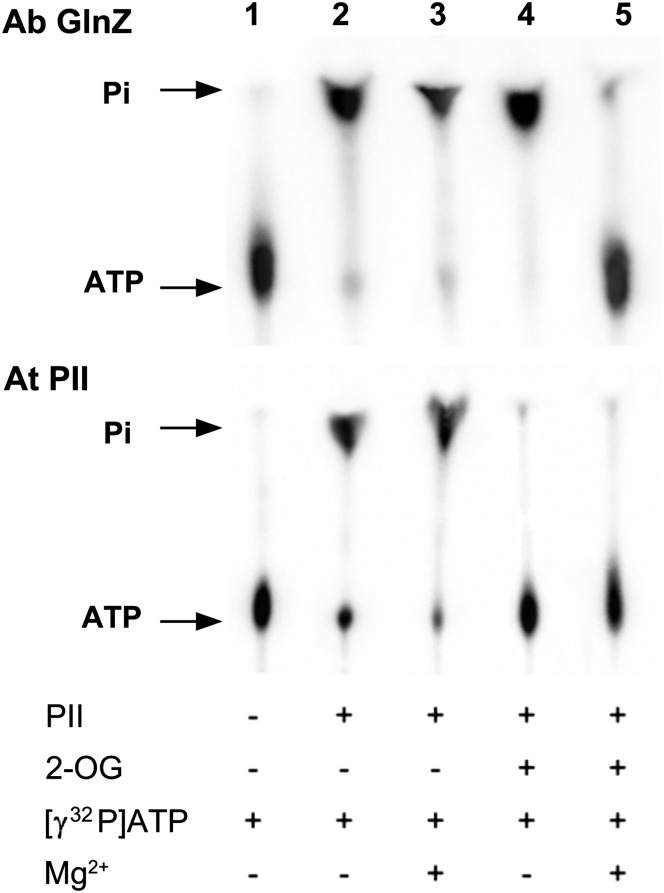
The ability to bind 2-OG and ATP/ADP is an almost universal characteristic of PII proteins, and consequently we considered the possibility that other PII proteins would also exhibit an ATPase activity. We tested the ATPase activity of A. brasilense GlnZ (a GlnK ortholog) and of the PII protein from the model plant, Arabidopsis thaliana, using the same assay we applied to E. coli GlnK. Both these PII proteins also had ATPase activity. The activity of A. brasilense GlnZ had an identical profile to that observed with E. coli GlnK, whereas A. thaliana PII showed inhibition of the ATPase activity by 2-OG in both the presence and absence of Mg2+ (Fig. 5).
Fig. 5.
ATPase activities of A. brasilense GlnZ and A. thaliana PII. Analysis of ATP hydrolysis by TLC. Components of the assay are listed below the figure. ATPase activity is indicated by release of free Pi from [γ32P]-ATP.
Discussion
In this study, we have shown that the E. coli GlnK protein has an ATPase activity that is only revealed in the absence of 2-OG. This observation is a previously unrecognized facet of PII biology and resolves a long-standing question as to the likely role of ATP binding to PII proteins. In nitrogen-limited cells, PII proteins are expected to have 2-OG, Mg2+, and ATP bound in each of the three ligand-binding pockets, and the T loops of the protein will be relatively unstructured, as seen in the A. brasilense GlnZ structure (7, 20). In this condition, ATP hydrolysis is inhibited. Improvement in the cellular nitrogen-status results in a marked drop in the 2-OG pool (20), and as a consequence, we expect that 2-OG occupancy of PII will fall and the bound ATP will be concomitantly hydrolyzed to ADP. Crystal structures of E. coli GlnK, A. brasilense GlnZ, and S. elongatus PII with either Mg-ATP and 2-OG bound or with ADP bound offer snapshots of the likely structures before and after ATP hydrolysis. From these structures, we can deduce that the concomitant loss of 2-OG and hydrolysis of Mg-ATP to ADP liberates residue Gln39 from its coordination of Mg-ATP and allows it to undergo a major conformational change to occupy the position vacated by 2-OG and to establish a bond with conserved residue Lys58 (7). Because Gln39 is located at the base of the T loop, such a switch is predicted to result in a conformational change in the T loops, which then adopt an extended more rigid structure (7, 8, 10). The energy released on ATP hydrolysis could help drive this conformational change. The ability of ADP to act as a potent inhibitor of ATP binding in low 2-OG has previously been demonstrated in vitro (13, 20). As a consequence, once the PII protein is in the ADP-bound form, its replacement by Mg-ATP is not favored until the cellular 2-OG pool rises again. This 2-OG dependence of ATP binding, together with the hydrolysis of ATP to ADP in the absence of 2-OG, provides a robust 2-OG-dependent conformational switch.
The precise mechanism of ATP hydrolysis by GlnK will require further detailed study. However, the key feature in this study is that in all three PII proteins examined here, ATPase activity is inhibited by 2-OG. For both E. coli GlnK and A. brasilense GlnZ, that inhibition also requires Mg2+, which is predictable given the known role of Mg2+ in coordinating the binding of 2-OG (7, 8). However, under the conditions used, 2-OG inhibition was apparently not dependent on Mg2+ in A. thaliana PII. Residue Gln39 is very highly conserved in all PII proteins (4) and is required for ATP hydrolysis, suggesting it plays a key catalytic role.
It is interesting to consider the functional similarities between the role of ATP and ADP, as proposed here for the regulation of PII protein conformation, and that of GTP and GDP in regulating the conformational switch in the signal-transducing Gα proteins (26). These small GTPases transition between a MgGTP-bound and a GDP-bound conformation and typically exhibit slow rates of GTP hydrolysis and GDP dissociation (22). The mechanism of GTP hydrolysis by this family of proteins is believed to involve a conserved glutamine residue that contributes to catalysis by orientating a nucleophilic water molecule for attack on the γ-phosphate of GTP (27). Interestingly, the side chain of the catalytic Gln residue only exhibits catalytic geometry in the transition state (28).
Sequence comparisons among many PII proteins, together with in vivo and in vitro studies in a number of model systems, suggest that ATP/ADP binding is likely to be a universal feature of the PII family and that 2-OG binding is also highly conserved (7, 13, 20, 23, 29, 30). We have demonstrated that the ATPase activity of E. coli GlnK extends to A. brasilense GlnZ and A. thaliana PII, suggesting that this is indeed likely to be a characteristic of most PII proteins. However, it should be noted that Bacillus subtilis GlnK has been reported to bind 2-OG only weakly (31), and A. fulgidus GlnK2 apparently shows no 2-OG binding (32), suggesting that these proteins might have a different mode of action.
These studies rationalize a number of previous observations. Crystallization studies on PII proteins in the presence of ATP have been found to produce crystals containing ADP, rather than ATP. The crystal structure of the GlnK–AmtB complex described by Gruswitz et al. (24) was achieved by cocrystallization of separately purified GlnK and AmtB proteins at an equimolar concentration of 0.22 mM in the presence of 2 mM ATP. However, the resultant crystal complex contained ADP bound in all three GlnK nucleotide-binding sites. Gruswitz et al. (24) speculated that their observation may be explained by an innate ATPase activity of GlnK, and our data support that view. Likewise, our previous observation that complex formation in vitro is facilitated by ATP (19) can now be explained by GlnK-mediated ADP formation from ATP during the preincubation period in those experiments.
Crystal structures for PII proteins with bound ATP but no 2-OG have been reported in a number of cases including A. fulgidus GlnK2 and GlnK3 (9, 32), E. coli GlnK (23), Methanococcus jannaschii GlnK1 (33), Mycobacterium tuberculosis PII (34), S. elongatus PII (35), and Thermatoga thermophilus GlnK (36). On the basis of our observations, we would deduce that in these cases, either there was no significant ATPase activity during the crystallization or the particular PII protein does not exhibit such an activity. However, it is notable that with one exception (S. elongatus PII with bound ATP–PDB: 2XBP), all the structures listed earlier were derived in crystallization conditions with a pH below 7.0 (pH range, 3.5–6.0). Consistent with this, we have shown that the ATP hydrolysis activity of GlnK is indeed inhibited below pH 7.0, and we suggest that this offers an explanation for the ability of a number of groups to obtain PII-ATP structures. In contrast, the GlnK–AmtB structure derived by Gruswitz and colleagues, in which the proteins were cocrystallized with ATP but ADP was present in the resultant complex, was obtained with a crystallization buffer at pH 8.0, where we would expect GlnK to show ATP hydrolysis activity (24).
A number of authors also have proposed that in addition to sensing the cellular nitrogen-status, PII proteins may also act as sensors of cellular adenylate energy charge; that is, as monitors of the ATP:ADP ratio in the cell (13–18). The majority of those studies report either the differential binding of ATP and ADP to PII proteins in vitro or the in vitro effects of ATP and ADP on interactions of PII proteins with a number of their targets. However, the cellular adenylate energy charge is quite strongly buffered around a value of 0.9, and the cellular ATP concentration is typically six to 10 times greater than that of ADP (37, 38). For the published in vitro studies to translate into sensing energy charge in vivo, there need to be conditions under which the cellular ATP pool drops to a level at which ADP would compete efficiently for PII binding. Such fluctuations in adenylate pools are challenging to demonstrate experimentally, and attempts to manipulate these pools in Rhodospirillum rubrum so as to provoke an effect on the regulatory activity of PII were not successful (16). Thus, although the possibility remains that PII proteins might act as sensors of the cellular ATP:ADP ratio, this remains to be proven, and our studies suggest that the primary source of ADP as a PII ligand is likely to be as a result of PII-mediated ATP hydrolysis.
Materials and Methods
Protein Purification.
E. coli His6-AmtB was purified from strain GT1000 (pAD4), as described previously (19). Wild-type E. coli GlnK was purified from BL21(DE3)pLysS (pJT25), using a one-step purification heparin chromatography protocol described previously (20), which gave protein of >95% purity. This was followed by a second gel-filtration chromatography step to give protein of >99% purity. GlnK fractions from the heparin column were pooled and concentrated by centrifugal filtration (Amicon Ultra 10K, Millipore). This sample was then purified on a Superose 12 10/300 GL column (GE Healthcare), in 50 mM Tris at pH 7.5, 100 mM KCl at 0.5 mL/min.
Mutant alleles of E. coli glnK were made by site-directed mutagenesis of plasmid pJT25 (20), and the resultant plasmids were used to overexpress GlnK variant proteins Q39A, Q39E, K58A, K90A, R101A, and R103A in E. coli BL21(DE3)pLysS. However, the variant proteins failed to bind effectively to heparin, as observed for the wild-type, and consequently an alternative purification method was used. Cells were grown in 20 mL LB broth supplemented with 100 μg/mL carbenicillin for 8 h at 37 °C. This culture was inoculated into 1 L autoinduction medium (ForMedium Ltd), grown overnight at 37 °C, harvested at 6,000 Χ g, and resuspended in 50 mM Tris⋅HCl at pH 7.5, 100 mM KCl, and 20% (vol/vol) glycerol. Whole-cell extracts of the culture were heated to 80 °C for 4 min, cooled on ice for 10 min, and harvested by centrifugation at 28,000 × g. The supernatant was loaded onto a 30-mL HiLoad 16/10 Phenyl Sepharose HP column (GE Healthcare); equilibrated with 50 mM Tris at pH 7.6, 100 mM NaCl, 1 mM EDTA, and ammonium sulfate to 25% (wt/vol) saturation; and run at 1 mL/min. GlnK was eluted with a gradient to 0% ammonium sulfate over 40 mL at 2 mL/min. GlnK fractions were pooled and ammonium sulfate added to 60% (wt/vol) saturation with stirring at 4 °C to precipitate GlnK. The pellet was harvested by centrifugation at 7,000 × g for 20 min at 4 °C and then resuspended into a minimal volume of 50 mM Tris⋅HCl at pH 7.5, 100 mM KCl, and centrifuged at 4 °C to remove any remaining solids. A sample volume of 0.5 mL was loaded onto a Superose 12 10/300 GL column (GE Healthcare) equilibrated with 50 mM Tris at pH 7.5, 100 mM KCl, and run at 0.5 mL/min. Pure fractions of GlnK were identified on 12.5% (wt/vol) SDS-polyacrylamide gel and stored at −80 °C. Whereas heparin chromatography resulted in GlnK fractions that contained no contaminating ATPase activity, fractions coming from phenyl Sepharose chromatography had to be monitored by TLC for a contaminating ATPase activity that came off the gel-filtration column just ahead of GlnK. Only the later fractions were used in subsequent assays.
A. brasilense GlnZ was purified as described previously (7) and was the kind gift of Xiao-Dan Li (Paul Scherrer Institute, Villigen, Switzerland). A. thaliana PII was purified from strain BL21(DE3)pLysS pET29-PII-HT. Cells were grown in 1 L autoinduction medium (ForMedium Ltd) supplemented with 15 μg/mL kanamycin overnight at 28 °C, harvested at 6,000 × g, and resuspended in 50 mM Tris⋅HCl at pH 7.5, 150 mM NaCl, 10 mM imidazole, and 5% (vol/vol) glycerol. Whole-cell extracts of the culture were applied to a 5-mL His Trap (GE Healthcare) column equilibrated with 50 mM Tris⋅HCl at pH 7.5, 150 mM NaCl, and 10 mM imidazole. The column was washed with 50 mM Tris⋅HCl at pH 7.5, 1,000 mM NaCl, 35 mM imidazole, and 0.1% Tween 20 to remove contaminating proteins, and the A. thaliana PII was eluted with 50 mM Tris⋅HCl at pH 7.5, 150 mM NaCl, and 500 mM imidazole. Fractions of 1 mL were collected, and the flow rate used was 1 mL/min throughout. Fractions from the elution peak were pooled and dialyzed for 1 h against 50 mM Tris at pH 7.6, 100 mM NaCl, 1 mM EDTA, and then ammonium sulfate was added to 25% (wt/vol) saturation. Protein was applied to a 30-mL HiLoad 16/10 Phenyl Sepharose HP column (GE Healthcare) equilibrated with 50 mM Tris at pH 7.6, 100 mM NaCl, 1 mM EDTA, and ammonium sulfate to 25% (wt/vol) saturation and run at 1 mL/min. PII was eluted with a gradient to 0% ammonium sulfate over 40 mL at 2 mL/min. Pure fractions were analyzed on a 15% (wt/vol) SDS-polyacrylamide gel; pooled; buffer exchanged by filtration into 50 mM Tris at pH 7.5, 100 mM KCl; and then concentrated to 38 μM.
ATPase Assay by TLC.
Initial ATPase assays were performed in 50 mM Tris⋅HCI at pH 8, 100 mM NaCl, 10% (vol/vol) glycerol, and 0.05% (wt/vol) lauryl dimethylamine oxide. Assays were performed at 30 °C for 1 h in a final reaction volume of 10 μL. Reactions were initiated by the addition of ATP (0.8 μL of 100 mM ATP plus 1 μL [γ32P]-ATP; 3000 Ci mmol, Amersham Biosciences). The final ATP concentration in the assay was 4 mM, and the protein concentration was 150 μM. The reactions were terminated by adding one-tenth vol of 5% (wt/vol) SDS, 20 mM EDTA, followed by freezing in liquid nitrogen.
Subsequent ATPase assays contained only [γ32P]-ATP or [α32P]-ATP with no unlabeled ATP, and unless otherwise indicated, reactions were carried out in 50 mM Tris⋅HCl at pH 8.0, 100 mM KCl in a total volume of 10 μL. For experiments to analyze the pH sensitivity of the ATPase, reactions were carried out in 50 mM citrate buffer at pH 4.0 or pH 5.0, 50 mM Mes NaOH buffer at pH 6.0 or pH 7.0, and standard 50 mM Tris⋅HCl at pH 8.0. PII protein, and effector molecules where required (2-OG 1.5 mM, MgCl2 6 mM), were mixed, and the reaction was initiated by the addition of [γ32P] or [α32P]-ATP (Perkin-Elmer 3000 Ci/mmole) to a final concentration of 3 μM. The final protein concentration varied between 100 and 150 μM, according to the experiment. Reactions were incubated for 30 min at 30 °C.
In all cases, the chromatography membrane (Polygram CEL300 PEI/UV254 20 × 20cm, Macherey-Nagel) was prerun in water and dried before loading. One to 2 μL of the reaction was loaded onto the membrane and rapidly dried (∼5 s) with a hair dryer. The membrane was then run in 0.75 M KH2PO4 at pH 3.5 and dried, and the resultant signals were detected by a 2-min exposure to a BAS-MP 2040S image plate (Fujifilm) and analyzed using a Phosphor-Imager (FLA-7000 Image reader, Fujifilm).
In Vitro GlnK–AmtB Complex Formation.
In vitro GlnK–AmtB complex formation was carried out as described previously (20). The final concentrations of effectors used in these experiments were 5 mM ATP, 1.5 mM 2-OG, and 6 mM Mg2+.
ATP Analysis by HPLC.
Aqueous solutions of adenosine phosphate standards or samples were injected onto a Poros HQ50 strong anion exchange column for analysis by HPLC (Dionex Ultimate 3000). The column was first equilibrated with 5 column volumes of 5 mM ammonium bicarbonate buffer, and then elution profiles for each standard or sample were developed over a linear gradient of ammonium bicarbonate from 5 t o 250 mM at a flow rate of 8 mL/min. An online detector was used to monitor A265.
Isothermal Calorimetry.
Isothermal calorimetry was performed as described previously (20).
Supplementary Material
Acknowledgments
We thank Xiao-Dan Li for the gift of purified A. brasilense GlnZ, Michael Hodges for plasmid pET29-PII-HT, Juliana Ibana and Luciano Huergo for introducing us to the concept of the one-step purification protocol for GlnK, Martin Rejzek for help with HPLC, and Anne Durand for conducting the initial ATPase assays described in this article. This work was funded by Biotechnology and Biological Sciences Research Council Grant BB/E022308/1 (to M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 12863.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304386110/-/DCSupplemental.
References
- 1.Sant’Anna FH, et al. The PII superfamily revised: A novel group and evolutionary insights. J Mol Evol. 2009;68(4):322–336. doi: 10.1007/s00239-009-9209-6. [DOI] [PubMed] [Google Scholar]
- 2.Forchhammer K. PII signal transducers: Novel functional and structural insights. Trends Microbiol. 2008;16(2):65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Uhrig RG, Ng KK, Moorhead GB. PII in higher plants: A modern role for an ancient protein. Trends Plant Sci. 2009;14(9):505–511. doi: 10.1016/j.tplants.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Huergo LF, Chandra G, Merrick M. PII signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol Rev. 2013;37(2):251–283. doi: 10.1111/j.1574-6976.2012.00351.x. [DOI] [PubMed] [Google Scholar]
- 5.Radchenko M, Merrick M. The role of effector molecules in signal transduction by PII proteins. Biochem Soc Trans. 2011;39(1):189–194. doi: 10.1042/BST0390189. [DOI] [PubMed] [Google Scholar]
- 6.Ninfa AJ, Jiang P. PII signal transduction proteins: Sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8(2):168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Truan D, et al. A new PII protein structure identifies the 2-oxoglutarate binding site. J Mol Biol. 2010;400(3):531–539. doi: 10.1016/j.jmb.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Fokina O, Chellamuthu VR, Forchhammer K, Zeth K. Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc Natl Acad Sci USA. 2010;107(46):19760–19765. doi: 10.1073/pnas.1007653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier S, et al. Mechanism of disruption of the Amt-GlnK complex by PII-mediated sensing of 2-oxoglutarate. PLoS ONE. 2011;6(10):e26327. doi: 10.1371/journal.pone.0026327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conroy MJ, et al. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104(4):1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamberov ES, Atkinson MR, Ninfa AJ. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J Biol Chem. 1995;270(30):17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, Peliska JA, Ninfa AJ. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry. 1998;37(37):12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 13.Jiang P, Ninfa AJ. Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in vitro. Biochemistry. 2007;46(45):12979–12996. doi: 10.1021/bi701062t. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Pohlmann EL, Ludden PW, Roberts GP. Functional characterization of three GlnB homologs in the photosynthetic bacterium Rhodospirillum rubrum: Roles in sensing ammonium and energy status. J Bacteriol. 2001;183(21):6159–6168. doi: 10.1128/JB.183.21.6159-6168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeira PF, Jonsson A, Frank M, Wang H, Nordlund S. Interaction of the signal transduction protein GlnJ with the cellular targets AmtB1, GlnE and GlnD in Rhodospirillum rubrum: Dependence on manganese, 2-oxoglutarate and the ADP/ATP ratio. Microbiology. 2008;154(Pt 8):2336–2347. doi: 10.1099/mic.0.2008/017533-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Pohlmann EL, Roberts GP. Effect of perturbation of ATP level on the activity and regulation of nitrogenase in Rhodospirillum rubrum. J Bacteriol. 2009;191(17):5526–5537. doi: 10.1128/JB.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe DM, Zhang Y, Roberts GP. Specificity and regulation of interaction between the PII and AmtB1 proteins in Rhodospirillum rubrum. J Bacteriol. 2007;189(19):6861–6869. doi: 10.1128/JB.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang P, Ninfa AJ. Sensation and signaling of alpha-ketoglutarate and adenylylate energy charge by the Escherichia coli PII signal transduction protein require cooperation of the three ligand-binding sites within the PII trimer. Biochemistry. 2009;48(48):11522–11531. doi: 10.1021/bi9011594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand A, Merrick M. In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate. J Biol Chem. 2006;281(40):29558–29567. doi: 10.1074/jbc.M602477200. [DOI] [PubMed] [Google Scholar]
- 20.Radchenko MV, Thornton J, Merrick M. Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J Biol Chem. 2010;285(40):31037–31045. doi: 10.1074/jbc.M110.153908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coutts G, Thomas G, Blakey D, Merrick M. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 2002;21(4):536–545. doi: 10.1093/emboj/21.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: A conserved switch for diverse cell functions. Nature. 1990;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, et al. GlnK, a PII-homologue: Structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J Mol Biol. 1998;282(1):149–165. doi: 10.1006/jmbi.1998.1979. [DOI] [PubMed] [Google Scholar]
- 24.Gruswitz F, O’Connell J, 3rd, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc Natl Acad Sci USA. 2007;104(1):42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang P, et al. Structure/function analysis of the PII signal transduction protein of Escherichia coli: Genetic separation of interactions with protein receptors. J Bacteriol. 1997;179(13):4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilman AG. G proteins: Transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 27.Coleman DE, et al. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265(5177):1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 28.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389(6652):758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 29.Smith CS, Weljie AM, Moorhead GB. Molecular properties of the putative nitrogen sensor PII from Arabidopsis thaliana. Plant J. 2003;33(2):353–360. doi: 10.1046/j.1365-313x.2003.01634.x. [DOI] [PubMed] [Google Scholar]
- 30.Huergo LF, et al. In vitro interactions between the PII proteins and the nitrogenase regulatory enzymes dinitrogenase reductase ADP-ribosyltransferase (DraT) and dinitrogenase reductase-activating glycohydrolase (DraG) in Azospirillum brasilense. J Biol Chem. 2009;284(11):6674–6682. doi: 10.1074/jbc.M807378200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinrich A, et al. Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J Biol Chem. 2006;281(46):34909–34917. doi: 10.1074/jbc.M607582200. [DOI] [PubMed] [Google Scholar]
- 32.Helfmann S, Lü W, Litz C, Andrade SL. Cooperative binding of MgATP and MgADP in the trimeric PII protein GlnK2 from Archaeoglobus fulgidus. J Mol Biol. 2010;402(1):165–177. doi: 10.1016/j.jmb.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Yildiz O, Kalthoff C, Raunser S, Kühlbrandt W. Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake. EMBO J. 2007;26(2):589–599. doi: 10.1038/sj.emboj.7601492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shetty ND, Reddy MC, Palaninathan SK, Owen JL, Sacchettini JC. Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein. Protein Sci. 2010;19(8):1513–1524. doi: 10.1002/pro.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fokina O, Chellamuthu VR, Zeth K, Forchhammer K. A novel signal transduction protein PII variant from Synechococcus elongatus PCC 7942 indicates a two-step process for NAGK-PII complex formation. J Mol Biol. 2010;399(3):410–421. doi: 10.1016/j.jmb.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 36.Sakai H, et al. Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8. J Struct Biol. 2005;149(1):99–110. doi: 10.1016/j.jsb.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman AG, Fall L, Atkinson DE. Adenylate energy charge in Escherichia coli during growth and starvation. J Bacteriol. 1971;108(3):1072–1086. doi: 10.1128/jb.108.3.1072-1086.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.