Abstract
Bacterial survival requires the rapid propagation of signals through gene networks during stress, but how this is achieved is not well understood. This study systematically characterizes the signaling dynamics of a cascade of RNA–protein interactions in the CsrA system, which regulates stress responses and biofilm formation in Escherichia coli. Noncoding RNAs are at the center of the CsrA system; target mRNAs are bound by CsrA proteins that inhibit their translation, CsrA proteins are sequestered by CsrB noncoding RNAs, and the degradation of CsrB RNAs is increased by CsrD proteins. Here, we show using in vivo experiments and quantitative modeling that the CsrA system integrates three strategies to achieve rapid and robust signaling. These strategies include: (i) the sequestration of stable proteins by noncoding RNAs, which rapidly inactivates protein activity; (ii) the degradation of stable noncoding RNAs, which enables their rapid removal; and (iii) a negative-feedback loop created by CsrA repression of CsrD production, which reduces the time for the system to achieve steady state. We also demonstrate that sequestration in the CsrA system results in signaling that is robust to growth rates because it does not rely on the slow dilution of molecules via cell division; therefore, signaling can occur even during growth arrest induced by starvation or antibiotic treatment.
Keywords: gene circuit, reverse engineering, synthetic biology, systems biology
A quantitative understanding of signaling dynamics is critical to determining how bacteria adapt to sudden environmental changes, combating pathogenesis, and designing synthetic circuits with specific dynamic properties. Recent studies have characterized signaling by transcription factor proteins (1–4) and by small RNAs that bind to target mRNAs to modulate their translation and/or degradation (3–6). However, the signaling properties of noncoding RNAs that sequester proteins have not been defined and are of particular interest because of their theoretical potential for very rapid signaling (Results).
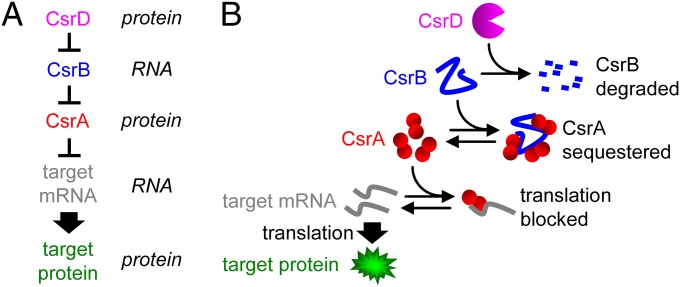
In this study, we chose the CsrA system in Escherichia coli as a model to investigate signaling by protein-sequestering noncoding RNAs (Fig. 1). CsrA regulation is important for carbon storage, motility, biofilm formation, and pathogenesis, and is evolutionarily conserved across distant groups of bacteria (7–9). The system consists of CsrA protein, CsrB and CsrC noncoding RNAs, and CsrD protein. CsrA binds as a dimer to the 5′-UTR of target mRNAs to silence their translation (7–9). CsrB and CsrC, which have binding sites for nine and three to four CsrA dimers, respectively (10), sequester CsrA to prevent it from silencing target mRNA translation (10, 11). CsrB and CsrC concentrations are in turn regulated by CsrD, which acts as a specificity factor to increase their degradation by RNase E (12).
Fig. 1.
CsrA system. (A) Simplified schematic of the synthetic CsrA system. (B) Mechanistic description of the synthetic CsrA system (see main text).
This study has three major parts. The first part describes the dynamics of signaling in a synthetic CsrA cascade without native control mechanisms in response to turning on and off transcription at each level in the cascade (target gene, csrA, csrB, and csrD). Our model and experiments show that sequestration of CsrA and degradation of CsrB enable rapid signaling by eliminating the need for multiple generations of cell division to dilute out these stable molecules. The second part describes how the sequestration of CsrA by CsrB enables signaling to occur in cells with growth arrest caused by starvation- and antibiotic-induced stress. The third part describes the systematic reintroduction of wild-type components with their native transcriptional and translational regulatory sequences into the CsrA cascade. These experiments show that negative-feedback control enables signaling in the native system to be even faster than in the synthetic system. Together, these findings highlight general strategies for rapid intracellular signaling that are important for reprogramming gene expression during stress.
Results
Modeling the Dynamics.
We constructed a simple and general mathematical model to qualitatively predict and interpret how turning on and off the production of CsrA, CsrB, and CsrD affect the dynamics of target gene expression (Fig. S1 and Table S1). Briefly, the model uses ordinary differential equations (details in SI Text) to describe the production, clearance, association, dissociation, and/or catalytic activity of the target mRNA, target protein, CsrA, CsrB, CsrD, and their complexes. For simplicity, all binding reactions are independent and the production of CsrA dimers and CsrD occur in a single step. CsrA primarily exists as a dimer in solution (13); therefore, dimerization is presumably rapid compared with the CsrA turnover rate (SI Text). Inclusion of separate transcription, translation, and dimerization steps is unnecessary because these reactions are assumed to be fast relative to the overall dynamics. The model is later extended to include feedback regulation.
Parameter values were obtained from our data or the literature (SI Text). In the synthetic CsrA systems (Figs. 2–4), the only parameters fitted to our dynamics data were the production rates. Each production rate was fitted in a circuit where it was the only free parameter, and once fitted, the parameter was constant across all simulations. In addition, we validated the model and parameter values by predicting the effect of steady-state CsrA, CsrB, and CsrD concentrations ([CsrA], [CsrB], [CsrD], respectively) on target protein (GlgC-GFP) expression and then confirmed the predictions qualitatively with in vivo measurements (Figs. 2D, 3D, and 4D). In the native CsrA system, parameter values for feedback and saturation of CsrD activity were also obtained from the dynamics data.
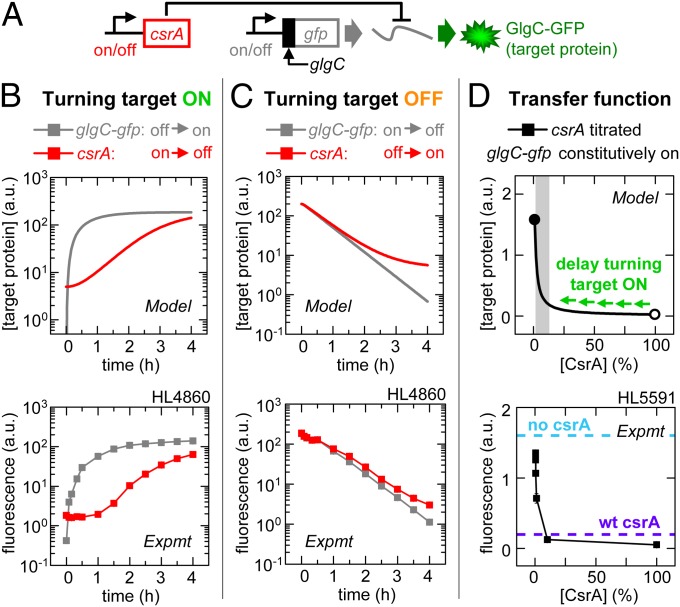
Fig. 2.
CsrA signaling: Stable signaling molecules can cause delays. Error bars are SEM of duplicate measurements. (A) Experimental schematic. (B) Target expression turned on directly by turning on glgC-gfp transcription (IPTG added) or indirectly by turning off csrA transcription (aTc removed). (C) Target expression turned off directly by turning off glgC-gfp transcription (IPTG removed) or indirectly by turning on csrA transcription (aTc added). (D) GlgC-GFP expression as a function of percentage maximum csrA transcription (calibrated using PLlacO-1:st7:gfp; Fig. S2Q). Target expression was also measured in strains without csrA (HL5594; cyan dashed line) or native (wt) csrA (HL5562 and HL5596; purple dashed line indicates both as the data overlay). The gray shading indicates the range over which the CsrA concentration has a significant effect on target expression (“regulatory range”). The open and closed circles are 100% and 0% of maximum [CsrA], respectively.
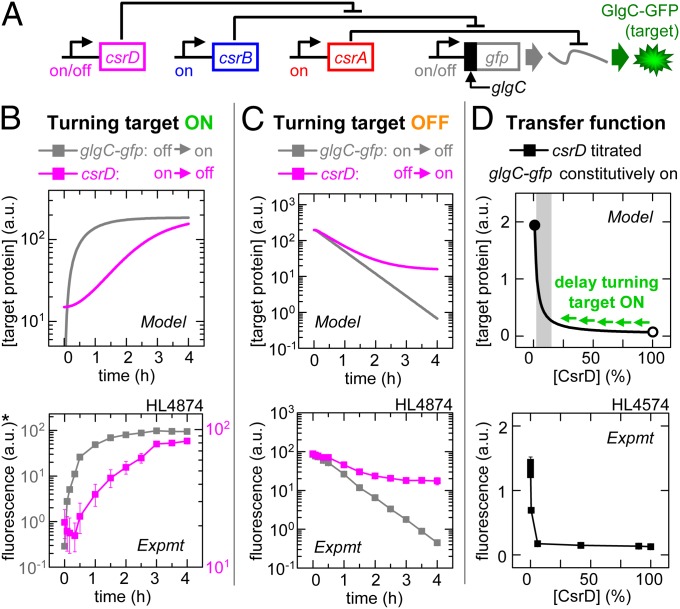
Fig. 4.
CsrD signaling: Degradation can prevent downstream delays. Error bars are SEM of duplicate measurements. (A) Experimental schematic. (B) Target expression turned on directly by turning on glgC-gfp transcription (IPTG added) or indirectly by turning off csrD transcription (aTc removed). *Left and right y axis correspond to turning on glgC-gfp (gray) and turning off csrD (magenta), respectively. (C) Target expression turned off directly by turning off glgC-gfp transcription (IPTG removed) or indirectly by turning on csrD transcription (aTc added). (D) GlgC-GFP expression as a function of percentage maximum csrD transcription (calibrated using PLlacO-1:st7:gfp; Fig. S2Q). The gray shading indicates the regulatory range for CsrD. The open and closed circles are 100% and 0% of the maximum [CsrD], respectively.
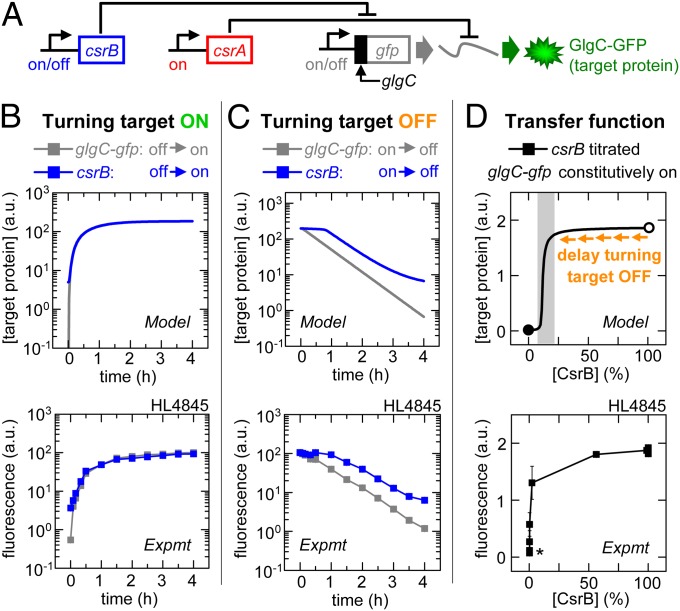
Fig. 3.
CsrB signaling: Sequestration can bypass downstream delays. Error bars are SEM of duplicate measurements. (A) Experimental schematic. (B) Target expression turned on directly by turning on glgC-gfp transcription (IPTG added) or indirectly by turning on csrB transcription (aTc added). (C) Target expression turned off directly by turning off glgC-gfp transcription (IPTG removed) or indirectly by turning off csrB transcription (aTc removed). (D) GlgC-GFP expression as a function of percentage maximum csrB transcription (calibrated using PLtetO-1:st7:gfp; Fig. S2R). The gray shading indicates the regulatory range for CsrB. The open and closed circles are 100% and 0% of maximum [CsrB], respectively. *Incomplete silencing of GlgC-GFP expression occurs if the total [CsrA] is less than the total [target mRNA] or if there is “leaky” CsrB expression.
Experimental System and Signaling Metrics.
To test the model’s predictions, signaling was initially measured in a completely synthetic CsrA system (SI Text). In this system, CsrA binding sites and flanking sequences from glgC (−61 to +8 nucleotides relative to the start codon) (14) were fused to gfp, thereby enabling GlgC-GFP expression (“target expression”) to be quantified by fluorescence [measured in arbitrary units (a.u.)]. The transcription of each component (glgC-gfp, csrA, csrB, and/or csrD) was controlled by an inducible promoter (PLlacO-1 or PLtetO-1) (15) or a constitutive promoter [variants of Pcon/O3 (16)]. CsrB and CsrC are believed to behave similarly; therefore, only the more potent (CsrB) was used. These synthetic circuits were constructed on plasmids and transformed into strains with chromosomal csrA, csrB, csrC, csrD, glgCAP, and/or pgaABCD deleted. Deletion of glgCAP was necessary for csrA knockouts to survive (17), and deletion of pgaABCD was required to prevent the overproduction of biofilm adhesins (18) and to enable efficient transformation.
Dynamics experiments were performed by turning transcription on or off for each component in the synthetic cascade and measuring GlgC-GFP expression at regular intervals. The rate of change in target expression reflects the convolved effects of target protein clearance, the difference between the initial and final steady states for the target protein, and the time required for the CsrA cascade upstream of the target protein to reach equilibrium (4). Because the target protein degradation rate is constant and we rescaled the initial and final steady states of target expression so the dynamic range is the same for all experiments, any observed differences in the dynamics are due to the delay in the CsrA cascade reaching equilibrium. We quantified the delay by measuring the time to reach 20%, 50%, and 80% of the maximum GFP level (τ20, τ50, and τ80) (Fig. S2 and Table S2). Because target expression was often slow to turn off or on (and therefore did not fall or rise to 50% of maximum expression in the experimental timeframe), τ80 or τ20, respectively, were used to quantify changes in target expression.
We compared time delays using stable GFP (with constant and predictable clearance by dilution) to destabilized GFP [with a tail-specific degradation tag (19) that decreases its half-life by up to 3.8 ± 0.5-fold]. We chose not to use the latter because it did not improve the resolution of the time delays, and it convolves saturation effects for active degradation with the delay in the CsrA system (SI Text).
CsrA Signaling: Stable Signaling Molecules Can Cause Delays.
In our first set of experiments, we turned on target expression directly by turning on glgC-gfp transcription (with csrA kept off) or indirectly by turning off csrA transcription (with glgC-gfp kept on) (Fig. 2 A and B). The model predicts that turning on transcription of glgC-gfp mRNA will increase target expression after a very short period, whereas a long delay will occur between turning off csrA transcription and a significant increase in target expression. The delay will occur for two major reasons: (i) clearance of a stable protein such as CsrA occurs slowly via dilution due to cell growth (20) (Fig. S1); and (ii) the transfer function reveals a high initial CsrA concentration at 100% transcription that must first be cleared before a significant increase in target expression can occur. That is, when moving from high to low CsrA concentration (open to solid circle), there is minimal effect on target expression until the CsrA level is quite low (shaded region) (Fig. 2D). We tested the predictions in our experimental system (Fig. 2B) and confirmed that target expression took longer to rise to 20% of maximum when csrA transcription was turned off compared with when glgC-gfp transcription was turned on (τ20 = 187 ± 1 and 39 ± 1 min, respectively; Fig. S2).
We next turned off target expression directly by turning off glgC-gfp transcription (with csrA kept off) or indirectly by turning on csrA transcription (with glgC-gfp kept on) (Fig. 2 A and C). The model predicts similar signaling delays for the two mechanisms (note: the curves eventually diverge because they have different steady states). Turning off glgC-gfp transcription causes target expression to fall almost immediately because preexisting target mRNAs are quickly degraded (Fig. S1). Turning on csrA transcription quickly decreases target expression despite the signal having to propagate through an extra regulatory step. This is because a small amount of CsrA is sufficient to silence most of the glgC-gfp mRNA [due to high affinity binding (14) and the stability of CsrA] as shown by the transfer function (Fig. 2D). We confirmed the predictions experimentally; turning off glgC-gfp transcription and turning on csrA transcription caused target expression to fall to 80% of maximum on similar timescales (τ80 = 9 ± 1 and 8 ± 3 min, respectively; Fig. S2).
We measured the level of target gene expression in a strain with native csrA instead of synthetic csrA (Fig. 2D, purple dashed line), and this indicated that physiological levels of CsrA are close to but not quite at a saturating concentration. That is, for target mRNAs at high concentrations, the native CsrA concentration minimizes delays in signaling without a significant trade-off in effectiveness (i.e., if CsrA levels were reduced too much, then the dynamic range of CsrA activity would be severely diminished). These findings may not only apply to highly transcribed individual target mRNAs, but also to target mRNAs that are transcribed concurrently with many others (i.e., when the total target mRNA pool is large due to a global transcriptional response to stress). However, in the case of target mRNAs that are at low concentrations, native CsrA concentrations are saturating (Fig. S3) and this is expected to cause long signaling delays when CsrA is removed solely by dilution (hence the importance of CsrB; see below).
In summary, our model and experiments demonstrate that stable proteins such as CsrA can introduce long delays when signaling depends on their removal; this is consistent with predictions (6, 21) and experiments in other systems (4, 22).
CsrB Signaling: Sequestration Can Bypass Downstream Delays.
In our second set of experiments, we turned on target expression directly by turning on glgC-gfp transcription (with csrA and csrB kept on) or indirectly by turning on csrB transcription (with glgC-gfp and csrA kept on) (Fig. 3 A and B). The latter decreases free CsrA levels to increase glgC-gfp mRNA translation. Our model predicts similar signaling times for turning on target expression via these two mechanisms. The transfer function shows that CsrB can quickly reach a level that is sufficient to sequester CsrA away from its target mRNAs (Fig. 3D) due to its rapid production, long half-life (12), and multiple CsrA binding sites (Fig. S1). We experimentally confirmed that turning on glgC-gfp transcription and turning on csrB transcription caused target expression to rise to 20% of maximum in comparable periods (τ20 = 27 ± 1 and 26 ± 1 min, respectively; Fig. S2).
We next turned off target expression either directly by turning off glgC-gfp transcription (with csrA and csrB kept on) or indirectly by turning off csrB transcription (with glgC-gfp and csrA kept on) (Fig. 3 A and C). Our model predicts a significant delay in signaling for the latter. Turning off csrB transcription decreases sequestration of CsrA and consequently decreases glgC-gfp mRNA translation. The delay will occur for the same basic reasons that CsrA activity is slow to turn off: (i) CsrB is slowly cleared from the cell in the absence of CsrD (12); and (ii) the CsrB concentration at 100% transcription corresponds to the saturating part of the transfer function (Fig. 3D, open circle), and therefore most of this excess CsrB must be cleared before it reaches a concentration that significantly decreases target expression (Fig. 3D, shaded region). Our in vivo experiments confirmed this prediction; turning off glgC-gfp transcription caused target expression to fall to 80% of maximum in less time than turning off csrB transcription (τ80 = 13 ± 1 and 67 ± 3 min, respectively; Fig. S2).
Our model and experiments demonstrate that CsrB can rapidly sequester and turn off the activity of the downstream CsrA, thereby bypassing delays due to slow CsrA clearance. However, clearance of CsrB is itself slow; therefore, it can delay signal propagation when turned off.
CsrD Signaling: Degradation Can Prevent Downstream Delays.
In our third set of experiments, we investigated signaling using CsrD, a specificity factor that decreases the CsrB half-life from >30 to <2 min at wild-type CsrD levels (12). We turned on target expression directly by turning on glgC-gfp transcription (with csrA and csrB kept on and csrD kept off) or indirectly by turning off csrD transcription (with glgC-gfp, csrA, and csrB kept on) (Fig. 4 A and B). The model predicts it will take longer to turn on target expression by turning off csrD transcription because (i) CsrD is cleared slowly by dilution, and (ii) the initial CsrD concentration corresponds to the saturating part of the transfer function (Fig. 4D, open circle); therefore, most of it must be cleared before there will be a significant increase in target expression (Fig. 4D, shaded region). Our experimental results agree with the model. Target expression took longer to increase to 20% of maximum when csrD transcription was turned off than when glgC-gfp transcription was turned on (τ20 = 77 ± 18 and 28 ± 1 min, respectively; Fig. S2).
We next turned off target expression directly by turning off glgC-gfp transcription (with csrA and csrB kept on and csrD kept off) or indirectly by turning on csrD transcription (with glgC-gfp, csrA, and csrB kept on) (Fig. 4 A and C). The model predicts that turning on csrD transcription will have an almost immediate effect on target expression. This is because a very small amount of CsrD is sufficient to dramatically increase CsrB degradation and consequently decrease CsrA sequestration and target mRNA translation (Fig. 4D). Our in vivo experiments confirmed this prediction. Turning on csrD transcription and turning off glgC-gfp transcription caused target expression to fall to 80% of maximum after a similar delay (τ80 = 17 ± 2 and 14 ± 1 min, respectively; Fig. S2).
Faster Signaling in Longer Cascades Under Some Conditions.
We have shown that turning on csrB transcription caused target expression to turn on faster than turning off csrA transcription (Figs. 2B and 3B). Similarly, turning on csrD transcription caused target expression to turn off faster than turning off csrB transcription (Figs. 3C and 4C). These results show, somewhat counterintuitively, that regulating an upstream molecule can alter target expression more rapidly than regulating a downstream molecule under some conditions (i.e., when the shorter cascade requires the slow clearance of a stable molecule and the longer cascade does not). Because these measurements were in different strains, we constructed circuits to directly compare signaling between CsrA and CsrB, or between CsrB and CsrD in the same strain (Fig. S2 G and H); these measurements confirmed our earlier findings. Therefore, the CsrA cascade differs from purely transcriptional cascades in that the number of regulatory connections in the cascade does not predict signaling delay (2, 21).
Robust Signaling During Stress.
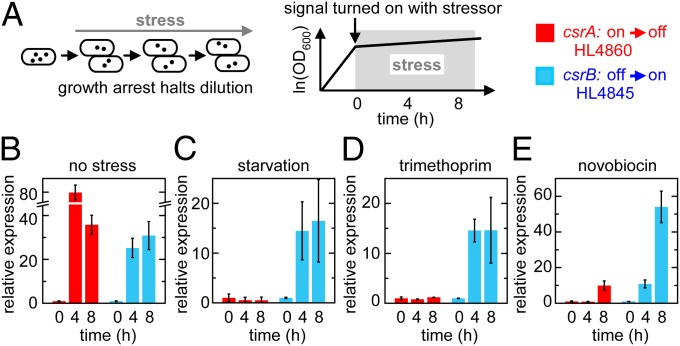
Rapid cell signaling is most crucial when cells are exposed to potentially lethal environmental stress. However, stress often causes growth arrest, which prevents the clearance of stable molecules by dilution causing signaling lockup. In theory, signaling by sequestration bypasses this problem. To test this proposal, we attempted to turn on target expression by turning off csrA transcription and turning on csrB transcription during starvation (M9 media without a carbon source) or in the presence of antibiotics for which the cells were not resistant.
We tested several classes of antibiotics including the following: sulfamethoxazole (100–500 µg/mL) and trimethoprim (1.5–15 µg/mL), which disrupt folate synthesis; novobiocin (200–3,000 µg/mL) and norfloxacin (12.5–1,250 ng/mL), which inhibit DNA gyrase; and polymyxin B (0.25–50 µg/mL), which destabilizes the outer membrane. Trimethoprim (5 µg/mL) and novobiocin (200 μg/mL) in LB media caused significant growth arrest without rapid lysis. Doubling times in M9, novobiocin, and trimethoprim were 700 ± 300, 400 ± 100, and 180 ± 30 min, respectively (mean ± SEM). In all three stress conditions, target expression could be turned on by inducing csrB transcription to sequester CsrA but not by turning off csrA transcription (Fig. 5).
Fig. 5.
Robust signaling during stress. Error bars are SEM of two or more measurements. (A) Experimental schematic. (B–E) Experiments performed using circuits shown in Figs. 2A and 3A without stress treatment (B), in M9 with no carbohydrate (C), in LB with 5 µg/mL trimethoprim (D), or in LB with 200 µg/mL novobiocin (E). In HL4860, csrA transcription was kept off (control) or turned off at t = 0. In HL4845, csrB transcription was kept on (control) or turned on at t = 0. Fluorescence levels were normalized to their respective control at each time point to correct for general effects of stress. The normalized values were rescaled to the initial measurement to determine the fold change in expression (“relative expression”).
These experiments with three independent sources of stress confirm the generality of our prediction that sequestration can be essential for signaling in pathways with stable signaling molecules during stress.
Feedback in the Native CsrA System.
We next probed the architecture of the native CsrA cascade using the synthetic CsrA system as a benchmark. Specifically, we sought to determine whether putative transcriptional and translational feedback loops described in the native system affect signaling dynamics under our experimental conditions (7, 23). Feedback is known to influence signaling dynamics (1) and CsrA has been reported to (i) positively and negatively regulate its own expression via mechanisms that have not been fully elucidated (23), (ii) inhibit its own activity by increasing transcription of CsrB and CsrC (10, 24), and (iii) inhibit its own activity by decreasing CsrD production, which increases CsrB (25).
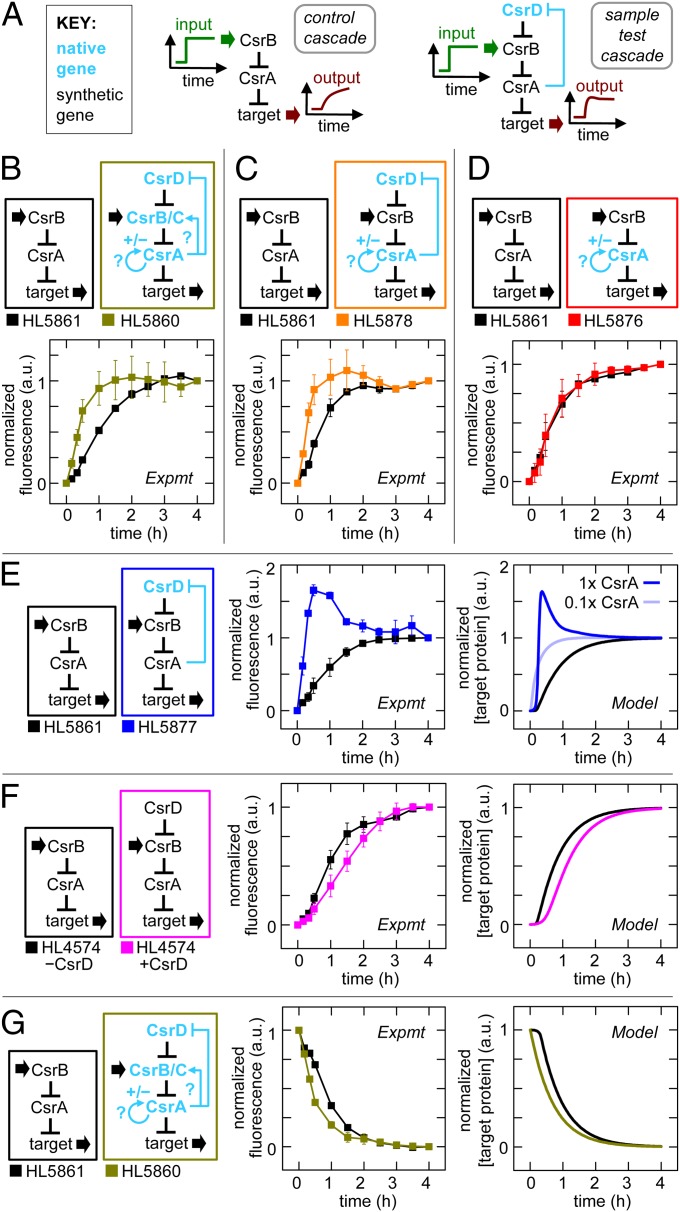
In these experiments, we systematically replaced components of the synthetic cascade with native genes in the chromosome that have intact regulatory sequences. We turned on and off synthetic csrB transcription (input) and measured the effect of the native gene(s) on the dynamics of target expression (output) (Fig. 6A). Initially, we started with all four native genes (csrA, csrB, csrC, csrD) present (Fig. 6B), and remarkably we found that turning on csrB transcription with the entire native CsrA cascade caused target expression to turn on more rapidly than the synthetic cascade (Fig. 6B). To isolate the regulatory interaction responsible for this “enhanced signaling” speed in the native CsrA cascade, we incrementally replaced or removed native genes (Fig. 6 C–F).
Fig. 6.
Feedback in the native CsrA system. Error bars are SEM of duplicate measurements. (A) Experimental schematic with synthetic (black) and native genes with reported feedback loops (blue). The “synthetic cascade,” which was a benchmark for comparison, was composed of synthetic csrA and csrB. Normalized fluorescence was determined by dividing each value by the fluorescence value in a control with csrB transcribed constitutively; the resulting ratio was rescaled so the start and end points were 0 and 1, respectively (Fig. S2). (B–F) Comparison of systems with synthetic and native genes where synthetic csrB was induced at t = 0. (B) Cascade with native csrA, csrB, csrC, and csrD, and synthetic csrB (gold) versus synthetic cascade (black). (C) Cascade with native csrA and csrD (orange) versus synthetic cascade (black). (D) Cascade with native csrA (red) versus synthetic cascade (black). (E) Cascade with native csrD (blue) versus synthetic cascade (black). Native csrD is modeled at low (light blue) and high (dark blue) CsrA levels. (F) Cascade with and without synthetic csrD expression (magenta and black, respectively). (G) Comparison of cascade with native csrA, csrB, csrC and csrD, and synthetic csrB (gold) versus synthetic cascade (black) where synthetic csrB was turned off at t = 0.
We started by removing native csrB and csrC and found their removal did not eliminate enhanced signaling (Fig. 6C). However, when we also removed native csrD, enhanced signaling no longer occurred (Fig. 6D). Therefore, native csrD was necessary for enhanced signaling. Enhanced signaling was restored when native csrD was reinstated with synthetic csrA (Fig. 6E). This latter strain also showed a transient overshoot of the steady-state target expression. Given that enhanced signaling and overshoot are known to be generated by negative feedback under certain conditions (1) and that native csrD was necessary for these behaviors, our findings are consistent with the reported negative-feedback regulation in which CsrA represses the production of CsrD. In further support of this, we demonstrated that enhanced signaling does not occur with the synthetic csrD gene, which lacks the flanking sequences (including the native csrD promoter and 5′-UTR sequences) that are necessary for CsrA repression of CsrD production (12, 25). Instead, we found that the induction of a small, constant amount of CsrD from synthetic csrD resulted in slower signaling than in the control cascade without CsrD induction (Fig. 6F). Additionally, we showed by quantitative RT-PCR that the CsrD mRNA concentration decreases with increased CsrA production (SI Text), which is consistent with negative feedback and previous reports (12, 25).
We used the model to determine whether negative feedback explains the enhanced signaling and overshoot observed. The model was modified to include repression of CsrD production by CsrA and the capacity for CsrD binding to be saturated by CsrB (SI Text). We found that an increase in csrB transcription is countered by negative feedback, which decreases CsrB levels after a delay (Fig. S1 J and K). The net effect is a pulse of CsrB resulting in a brief surge in target mRNA translation that causes target expression to reach its final steady state faster (Fig. 6E, Right). If the delay in the negative feedback is sufficiently large, which depends on the total CsrA concentration, then target expression can briefly overshoot the new steady state (Fig. S1J) (1). This explains the overshoot with synthetic csrA and why it does not occur with native csrA that produces approximately 1/10th the CsrA concentration (Figs. 2D and 6 B, C, and E). Negative feedback can also allow target expression to reach a new steady state sooner when csrB transcription is decreased. In this case, the response is faster because increased CsrD concentrations minimize the accumulation of saturating amounts of CsrB in the first place and increase the CsrB clearance rate (Fig. 6G). The model also explains why slower signaling is observed with synthetic csrD (Fig. 6F). Initially, the CsrB concentration increases slowly because it is cleared by CsrD; therefore, target expression increases more slowly than in the control cascade (Fig. S1L). However, as CsrB accumulates, CsrD becomes saturated, causing the overall half-life of CsrB and the CsrB concentration to increase at a faster rate, resulting in a rapid increase in target expression. That is, there is a slow initial increase in target expression followed by a fast increase with synthetic csrD, which is the opposite of native csrD with negative feedback.
In summary, characterization of the native CsrA system reveals that negative feedback (i.e., inhibition of CsrD production by CsrA) enhances the speed of turning on and off target protein expression.
Discussion
In this study, we reverse engineered and quantitatively modeled the CsrA regulatory network to obtain a detailed and coherent picture of its signaling dynamics. We found that it is capable of rapid signal propagation due to three general principles: (i) sequestration of stable signaling molecules, (ii) degradation of stable signaling molecules, and (iii) negative-feedback regulation. Sequestration and degradation of CsrA and CsrB, respectively, avoids long delays due to the slow removal of these stable signaling molecules by dilution through cell division. Negative-feedback regulation (due to CsrA inhibition of CsrD production) enables target expression to reach its new steady state faster in response to changes in csrB transcription (as explained above).
How might rapid signaling in the CsrA system aid adaptation to stress? Unfortunately, the pathways that sense environmental stresses and transmit this information to the CsrA system have not been fully elucidated. However, one regulator that has been identified to activate the CsrA cascade is the BarA/UvrY two-component system. Two component systems also control expression of CsrB and CsrC homologs in other bacteria (e.g., GacS/GacA in Pseudomonas aeruginosa, LetS/LetA in Legionella pneumophila, and VarS/VarA in Vibrio cholerae) (8). Extracellular weak acids are thought to activate BarA, which alters the phosphorylation state of UvrY; UvrY then interacts with the CsrA system by increasing csrB and csrC transcription (7–9). As with other two component systems, the BarA/UvrY response to a signal is believed to be extremely rapid; therefore, propagation of the signal through the CsrA cascade is likely to be rate limiting. As a consequence, increasing the speed of signaling through the CsrA system should directly shorten delays in adapting gene expression patterns and phenotypes to environmental perturbations. Our findings with stable and destabilized GFP show that shortening the time delay in the CsrA system benefits the dynamics of target proteins with short lifetimes as well as those with long lifetimes (Fig. S4).
There are many parallels between the CsrA system and the architecture of other bacterial stress response pathways. In the Salmonella ChiP and E. coli YbfM pathways, a small RNA silences target mRNA translation (ChiX and MicM, respectively) in a manner analogous to CsrA silencing of mRNAs (26, 27). These small RNAs are themselves sequestered by a sRNA or mRNA to turn off their activity (in a manner analogous to CsrB). In Bacillus subtillus, a CsrA homolog and AarB (another global regulator of biofilm formation) are also regulated by sequestration but by proteins (FliW and AbbA, respectively) instead of noncoding RNAs (28, 29). In E. coli, the regulation of the extracytoplasmic stress sigma factor (σE) that controls the transcription of genes in stress response pathways also has features in common with the CsrA system (30). σE is inactivated by sequestration to the membrane by the RseA protein (analogous to CsrA and CsrB, respectively) and reactivated when RseA is cleaved by DegS and YeaL (analogous to CsrD) (30). These examples highlight the general importance of sequestration and degradation in regulating the dynamics of stress response pathways.
The regulation of signaling molecules by sequestration and degradation not only increases signaling speed but it also makes signaling robust to growth rates. As we showed, this is important for preventing signaling lockup when stresses such as starvation and antibiotics lead to growth arrest.
In conclusion, this study shows that CsrB, which is at the center of the CsrA network, synergistically integrates multiple mechanisms to achieve rapid and robust signaling. These findings provide further evidence that noncoding RNAs, which include small RNAs, have evolved a prominent role in connecting genetic pathways due to their general ability to rapidly propagate signals needed for prompt adaptation to stress.
Methods
Bacterial Strains and Plasmids.
Plasmids, strains, and oligonucleotide sequences are in Tables S3 and S4. Plasmid maps are in Fig. S5. Construction details are in SI Text.
Gene Expression Measurement and Analysis.
Steady-state experiments (e.g., measuring transfer functions) were performed by inoculating 5–50 μL of overnight culture in 5 mL of LB media with 100 µg/mL ampicillin and/or 50 µg/mL kanamycin. Cultures were grown for 3.5 h with shaking at 37 °C and 200 rpm, and then 5 μL of culture was inoculated into 5 mL of fresh LB with antibiotics and isopropyl β-d-1-thiogalactopyranoside (IPTG) (0.01–1 mM), anhydrotetracycline (aTc) (0.01–1 µM), both or neither. Harvested cells were placed on ice and GFP expression was measured using a Coulter EPICS-XL flow cytometer (488-nm/15-mW argon ion laser) and analyzed as described (31). GFP distributions were unimodal for all measurements in this study. However, bimodality was observed at very high free CsrA concentrations that exceeded levels presented in this study and caused aberrant morphologies consistent with severe cellular stress.
Dynamics experiments were performed as above except as follows. There was a preinduction step (to achieve steady-state on or off transcription) and a postinduction step (to reverse or maintain the transcription states of the preinduction step). In the preinduction step, cells were grown with IPTG (0.1–1 mM), aTc (1 µM), both or neither for 3.5 h and then inoculated into 2.5 mL of fresh media (with the same antibiotic and inducer concentrations) to produce an OD600 = 0.01–0.05. The culture was grown for 30 min, diluted 1 in 2 in the same fresh media, and grown for an additional 30 min. Postinduction cultures were created by taking preinduction cells, centrifuging them (16,100 × g for 1 min), discarding the supernatant, and resuspending the pellets in LB with the same antibiotics and IPTG (0.1–1 mM), aTc (1 µM), both or neither. The cultures were grown for 4 h and diluted ∼1 in 2 every 30 min to maintain OD600. HL5877 was inoculated from glycerol stock instead of overnight cultures to prevent selection of a “locked-on” phenotype with constitutively high GlgC-GFP expression.
Growth arrest experiments were performed as the dynamics experiments except as follows. Preinduction cultures were 10 mL and grown for 4 h with IPTG (1 mM), aTc (1 µM), both or neither. Postinduction cultures were created by resuspending preinduction cells in M9 media with no carbohydrate or in LB with trimethoprim or novobiocin. IPTG, aTc, both or neither, were added to the media. Cells were harvested after 4 and 8 h of stress.
Mathematical Models.
Differential equations, parameter values, and initial conditions are discussed in Results, Table S1, and SI Text. Simulations were performed using MATLAB (Mathworks).
Supplementary Material
Acknowledgments
We thank Razika Hussein for helpful discussions. This work was supported by a National Science Foundation Graduate Research Fellowship (to D.N.A.) and University of California, Berkeley start-up funds (to H.N.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308476110/-/DCSupplemental.
References
- 1.Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323(5):785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 2.Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci USA. 2005;102(10):3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta P, Goyal S, Wingreen NS. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Syst Biol. 2008;4:221. doi: 10.1038/msb.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussein R, Lim HN. Direct comparison of small RNA and transcription factor signaling. Nucleic Acids Res. 2012;40(15):7269–7279. doi: 10.1093/nar/gks439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5(9):e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimoni Y, et al. Regulation of gene expression by small non-coding RNAs: A quantitative view. Mol Syst Biol. 2007;3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67(17):2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology. 2008;154(Pt 1):16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- 9.Romeo T, Vakulskas CA, Babitzke P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ Microbiol. 2013;15(2):313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weilbacher T, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48(3):657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu MY, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272(28):17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20(18):2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez P, et al. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol. 2005;187(10):3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44(6):1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 15.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25(6):1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmermans J, Van Melderen L. Conditional essentiality of the csrA gene in Escherichia coli. J Bacteriol. 2009;191(5):1722–1724. doi: 10.1128/JB.01573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56(6):1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 19.Andersen JB, et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64(6):2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 21.Rosenfeld N, Alon U. Response delays and the structure of transcription networks. J Mol Biol. 2003;329(4):645–654. doi: 10.1016/s0022-2836(03)00506-0. [DOI] [PubMed] [Google Scholar]
- 22.Grilly C, Stricker J, Pang WL, Bennett MR, Hasty J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol Syst Biol. 2007;3:127. doi: 10.1038/msb4100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yakhnin H, et al. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Eσ70 and Eσ(S), and indirect transcriptional activation by CsrA. Mol Microbiol. 2011;81(3):689–704. doi: 10.1111/j.1365-2958.2011.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: The RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183(20):6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonas K, et al. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70(1):236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73(5):790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 27.Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23(17):2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee S, et al. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol. 2011;82(2):447–461. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banse AV, Chastanet A, Rahn-Lee L, Hobbs EC, Losick R. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc Natl Acad Sci USA. 2008;105(40):15547–15552. doi: 10.1073/pnas.0805203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52(3):613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 31.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA. 2011;108(3):1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.