Significance
Chaperonins GroEL and GroES facilitate the folding of diverse substrate proteins driven by ATP hydrolysis. GroEL subunits cycle through a series of allosteric states in a concerted manner, enabling work to be performed on substrate proteins. Removing two salt bridges that ordinarily break during the allosteric transitions of the WT permitted the structure of GroEL in the relaxed R state to be solved. Whereas the equatorial and intermediate domains display almost perfect sevenfold symmetry, the apical domains display remarkable asymmetry. Freed of intersubunit contacts, each subunit adopts a different conformation, suggesting a flexibility that permits interaction with diverse substrate proteins.
Keywords: protein machine, allostery, chaperonin
Abstract
The chaperonin proteins GroEL and GroES are cellular nanomachines driven by the hydrolysis of ATP that facilitate the folding of structurally diverse substrate proteins. In response to ligand binding, the subunits of a ring cycle in a concerted manner through a series of allosteric states (T, R, and R″), enabling work to be performed on the substrate protein. Removing two salt bridges that ordinarily break during the allosteric transitions of the WT permitted the structure of GroEL-ADP in the R state to be solved to 2.7 Å resolution. Whereas the equatorial domain displays almost perfect sevenfold symmetry, the apical domains, to which substrate proteins bind, and to a lesser extent, the intermediate domains display a remarkable asymmetry. Freed of intersubunit contacts, the apical domain of each subunit adopts a different conformation, suggesting a flexibility that permits interaction with diverse substrate proteins. This result contrasts with a previous cryo-EM study of a related allosteric ATP-bound state at lower resolution. After artificially imposing sevenfold symmetry it was concluded that a GroEL ring in the R-ATP state existed in six homogeneous but slightly different states. By imposing sevenfold symmetry on each of the subunits of the crystal structure of GroEL-ADP, we showed that the synthetic rings of (X-ray) GroEL-ADP and (cryo-EM) GroEL-ATP are structurally closely related. A deterministic model, the click stop mechanism, that implied temporal transitions between these states was proposed. Here, however, these conformational states are shown to exist as a structurally heterogeneous ensemble within a single ring.
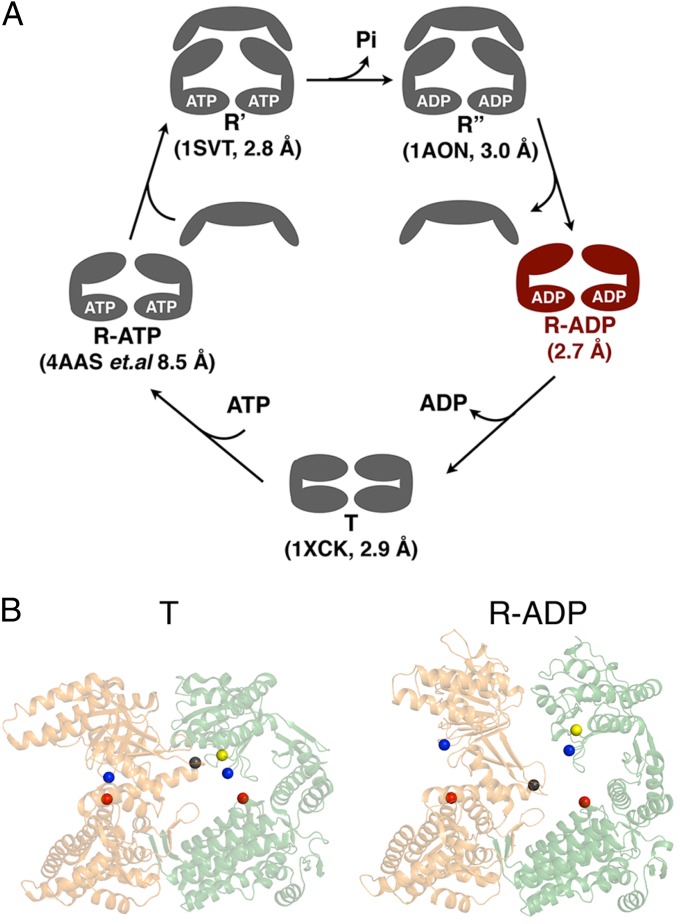
To assist protein folding, the GroEL/GroES chaperonin machine cycles through a series of conformational states in response to ligand binding (1–3). Two states, taut (T) and relaxed (R), are populated in the absence of GroES. Another two states, R′ and R″, that exist before and after ATP hydrolysis, respectively, are populated on GroES binding to the R state (1–3) (Fig. 1A). Crystal structures of T [Protein Data Bank (PDB) ID code 1OEL] (4) (PDB ID code 1XCK) (5), R′ (PDB ID code 1SVT) (6), and R″ (PDB ID code 1AON) (7) states are available.
Fig. 1.
(A) The conformational states visited during the chaperonin cycle (for simplicity, only one GroEL ring is shown). Three crystal structures (PDB ID codes 1SVT, 1AON, and 1XCK) have been solved by crystallography to a resolution of 3.0 Å or better. Cryo-EM structures (4AAS and five others) of the R-ATP state around 8.5 Å resolution also exist. The crystal structure of the R-ADP state is reported here. (B) The positions of the interdomain, intrasubunit D83-K327 (red and blue spheres) and interdomain, intersubunit R197-E386 (black and yellow spheres) salt bridges before and after the T to R allosteric transition.
As described by the theory of nested cooperativity, the conformational transitions within each heptameric ring are positively cooperative, whereas the transitions between the rings are negatively cooperative (8, 9). Multiple salt bridges, some within a subunit and others between subunits, stabilize these various conformational states. In such a dynamic system, these salt bridges must continually be broken and reformed at different points during the chaperonin cycle. Two interdomain salt bridges, an intrasubunit one (D83-K327) and an intersubunit one (R197-E386), stabilize the T state and are ordinarily broken during the T to R transition that follows the binding of ATP to the WT (Fig. 1B). Individually, the R197A mutation destabilizes the T state (10, 11), whereas replacing the D83-K327 salt bridge with a disulfide bond locks the ensemble in the T state (12, 13). Here, two mutations (D83A and R197A) are introduced to remove these salt bridges (D83-K327 and R197-E386), and therefore, the T state is destabilized. Steady state and presteady state analyses of GroELD83A/R197A reveal that it populates a T-like state in the absence of nucleotide but more readily transitions to the R state on nucleotide binding than the WT GroEL. We also report the crystal structure at 2.7 Å resolution of this GroELD83A/R197A containing bound ADP in the R conformational state. The crystal structure of the R state is distinct from the cryo-EM structures of GroEL-ATP (PDB ID code 4AAS, et al.) (14). It is similar, in some respects, to the cryo-EM (4AAS, etc.) structures of GroEL-ATP (14). However, the structure reported here differs by being strikingly asymmetric in the apical domain.
Results and Discussion
Removal of Two Salt Bridges Destabilizes the T Allosteric State.
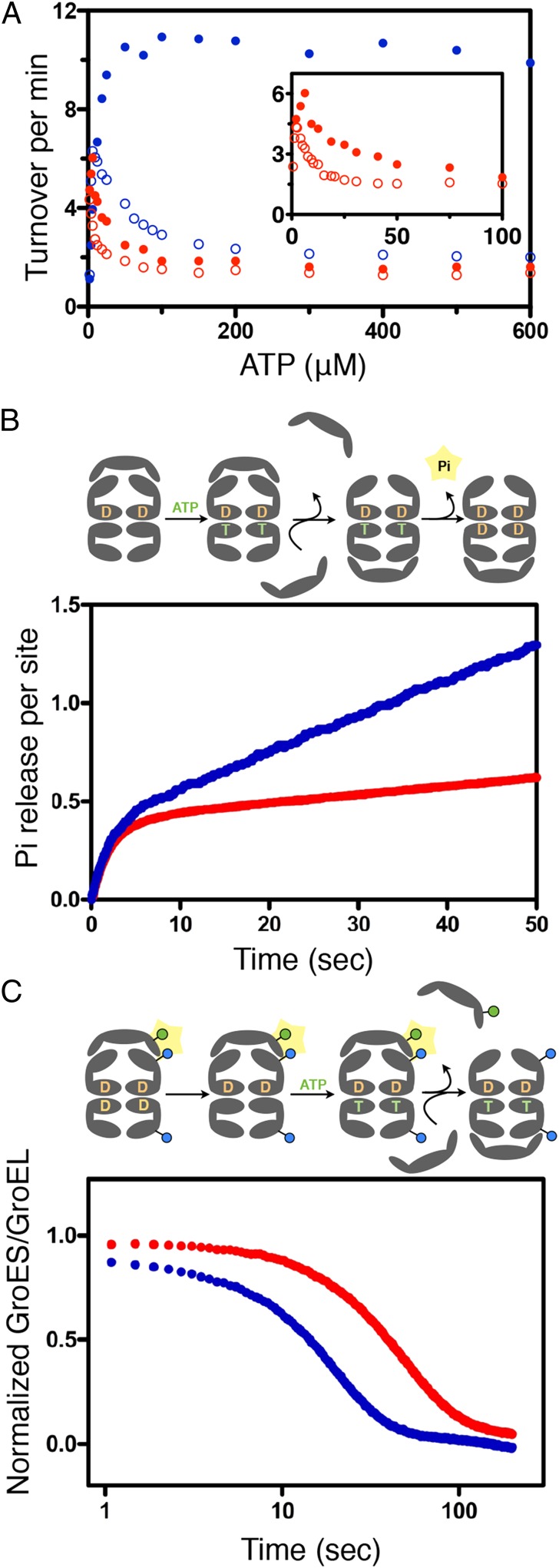
The steady state response of WT GroEL to ATP is characterized by two concerted, allosteric transitions from T7T7 to T7R7 to R7R7 (8, 9, 15). The first transition displays positive cooperativity between the subunits of a ring and results in an increase in the steady state rate of ATP hydrolysis. Because of negative cooperativity between the rings, the second transition occurs at higher ATP concentrations. In the R7R7 state, the rate-determining step becomes the dissociation of the product ADP (15); as a consequence, this second transition is associated with a decrease in the steady state rate of ATP hydrolysis (Fig. 2A, blue). The presence of substrate protein (SP) stabilizes the T state, such that the T7T7 to T7R7 to R7R7 transitions are suppressed, the steady state rate of ATP hydrolysis becomes increasingly noncooperative and the overall rate is enhanced 8- to 10-fold (9, 15).
Fig. 2.
Functional properties of GroELWT and the GroELD83A/R197A. (A) Steady state hydrolysis of ATP as a function of [ATP] by GroELWT (blue) or GroELD83A/R197A (red) measured as previously described in the absence (open dots) or presence (filled dots) of denatured α-lactalbumin (3.6 μM). These measurements were conducted at 37 °C with 100 mM K+ and 10 mM Mg2+. (B) Presteady state hydrolysis of ATP by GroELWT (blue) or GroELD83A/R197A (red) monitored by fluorescence of phosphate binding protein labeled with N-[2-(1-maleimidyl)ethyl]-7-(diethylamino)coumarin-3-carboxamide. The accepting state complex [cisGroEL-ADP-GroES][transGroEL] was mixed with ATP and labeled PBP to initiate the reaction. The rate constant for the burst phase was 0.49 s−1 (GroELWT) and 0.45 s−1 (GroELD83A/R197A). These measurements were conducted at 37 °C with 200 mM K+ and 10 mM Mg2+. (C) Dissociation of GroES from the cis ring of GroELE315C-IAEDANS (blue) or GroELD83A/R197A/E315C-IAEDANS (red) monitored by decrease of FRET signal. The resting state complex [cisGroELIAEDANS-ADP-GroESF5M][transGroELIAEDANS-ADP] was mixed with ATP and the excess amount of unlabeled GroES to initiate the reaction. The mean residence of GroESF5M dissociating from the wild type GroELIAEDANS resting state was ∼12 s and the D83A/R197A GroELIAEDANS resting state was ∼37 s. These measurements were conducted at 37 °C, 200 mM K+, and 10 mM Mg2+.
The removal of the D83-K327 and R197-E386 salt bridges destabilizes the T state, such that the T7T7 to T7R7 to R7R7 transitions occur at lower ATP concentrations than the WT (Fig. 2A, red). At [ATP] < 0.1 mM, the addition of SP (denatured α-lactalbumin) brought about a small (approximately twofold) stimulation in the steady state rate of ATP hydrolysis (Fig. 2A). However, unlike the WT, this small stimulation of the mutant ATPase became increasingly difficult to detect at [ATP] > 0.1 mM. Consequently, the ability of GroELD83A/R197A to populate the T state is limited but not completely abolished.
The Ability of GroELWT and GroELD83A/R197A to Hydrolyze ATP and to Bind and Release GroES Are Not Fundamentally Different.
i) Although the steady state rate of ATP hydrolysis by GroELD83A/R197A was somewhat reduced relative to the WT, it should be borne in mind that the steady state rate is a complex summation of several phenomena. Presteady state measurements of Pi (16) production using the fluorescent phosphate binding protein (PBP) (Fig. 2B) revealed that the ability of GroELD83A/R197A to hydrolyze ATP was not fundamentally different from the WT. In both cases, a burst phase occurred with an amplitude of ∼0.5 Pi/subunit, and similar kinetics (kWT = 0.49 ± 0.03 s−1; kD83A-R197A = 0.45 ± 0.02 s−1) followed by the linear steady state. The decreased steady state ATPase activity of GroELD83A/R197A is caused by a posthydrolysis event, namely the release of ADP.
ii) We and others (17–19) have shown that the ATP-induced dissociation of GroES from the cis ring of the asymmetric resting state complex [cisGroEL-ADP-GroES][transGroEL-ADP] is hindered by transADP. Decreasing the [K+] or adding SP reduces the affinity of the trans ring for ADP and promotes its reversion to the T state, a necessary condition for additional catalytic cycling. Because the T state of GroELD83A/R197A has been destabilized by removal of the salt bridges, it was to be expected that the ATP-induced dissociation of GroES from the cis ring would be inhibited. As shown in Fig. 2C, the mean residence time of the cis GroES was threefold longer in the case of GroELD83A/R197A. Note, however, that the amplitude of this transition was the same for WT and GroELD83A/R197A, indicating that the ability to form the resting state complex with GroES was common to both.
Structure of the GroELD83A/R197A-ADP in the Relaxed Allosteric State.
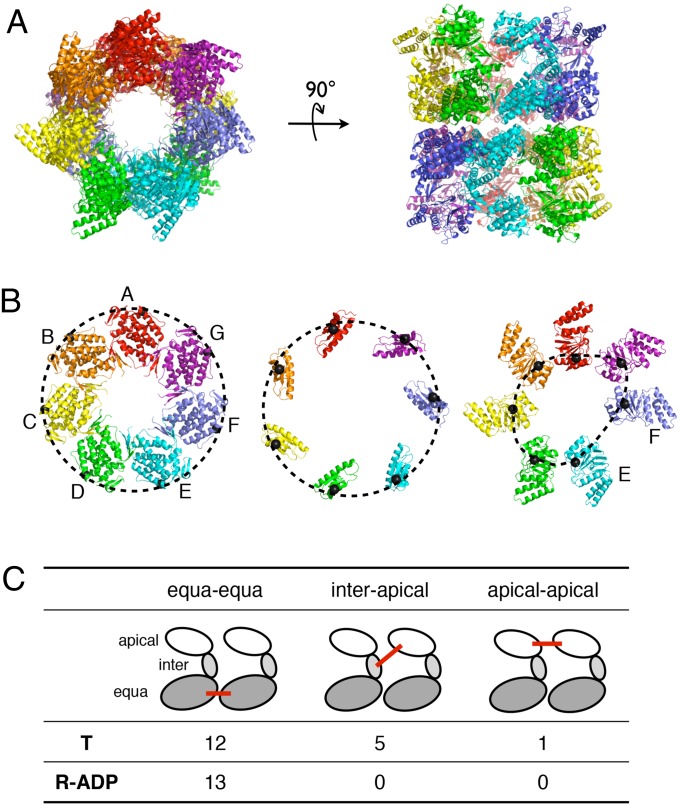
As expected, the overall basic architecture of GroEL is preserved. The 14 subunits are arranged into two heptameric rings and stacked back to back (Fig. 3A). Each subunit consists of three domains, equatorial, intermediate, and apical, that were previously described for other high-resolution structures of GroEL (4–7). Each nucleotide binding pocket, located in the equatorial domain, contains one ADP molecule (Fig. S1). Details of the structure refinement are listed in Tables S1 and S2.
Fig. 3.
Overall structure of the R-ADP and its asymmetry. (A) Overall structure of the R-ADP state viewed from (Left) the top and (Right) side. Seven subunits in one ring are colored in red, orange, yellow, green, cyan, blue, and purple. (B) Arrangement of (Left) equatorial, (Center) intermediate, and (Right) apical domains in the ring. E130 (in equatorial domain), V190 (in intermediate domain), and E310 (in apical domain) are connected (Cα values are shown as black spheres) to highlight the degree of asymmetry in different domains. (C) Numbers of intersubunit contacts between two neighboring subunits in the asymmetric R-ADP and symmetric T states. These contacts include salt bridges (<5 Å) and hydrogen bonds (<3.5 Å). Based on their locations, the contacts are categorized into three groups: between equatorial domains, between apical domains, and between intermediate apical domains.
Unexpected Asymmetry in the Apical Domains of the R-ADP is Revealed.
In contrast to previously solved GroEL structures in other conformational states, the R-ADP shows striking asymmetry. Although the equatorial domains and most of the intermediate domains are almost perfectly symmetrical, the apical domains break the sevenfold symmetry (Fig. 3B). A huge gap (∼1 nm) (Fig. 3B, Right) exists between the apical domains of the chains E and F.
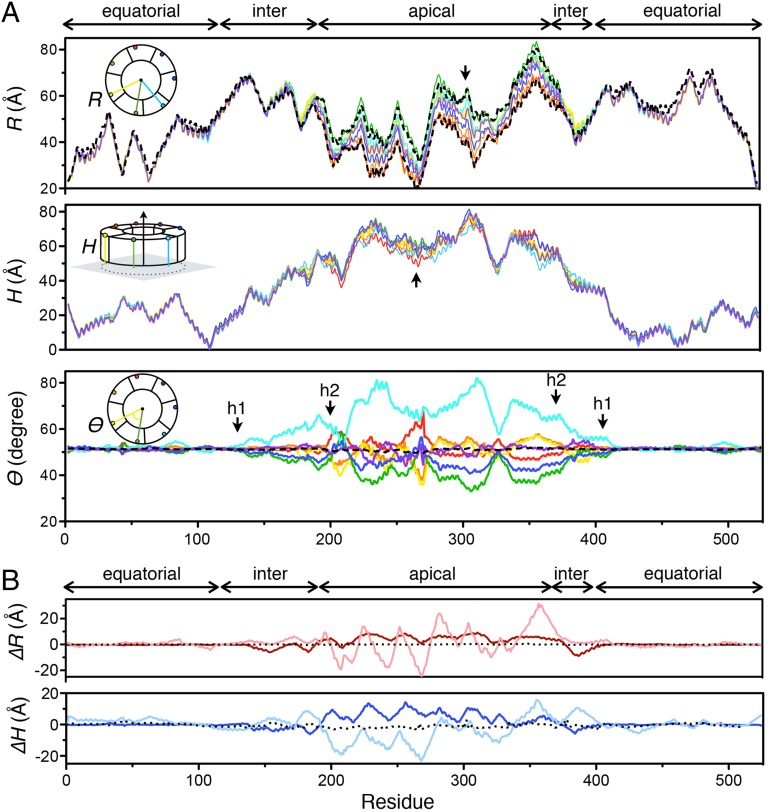
To quantify such asymmetry, we developed a cylindrical coordinate system to analyze the conformation of each GroEL subunit (Fig. 4A, Insets). R is the distance between the Cα of a given residue and the sevenfold symmetry axis (defined by the symmetrical equatorial domains). H is the height of the Cα of a given residue over the twofold plane of symmetry (between two rings). θ is the angle between two vectors; one connects the Cα of residue i to the axis of sevenfold symmetry, and one connects the Cα of residue i in the neighboring subunit to the axis of sevenfold symmetry. R, H, and θ together fully depict the unique conformation of each subunit, including possible asymmetry in three different directions: radial (R), vertical (H), and rotational (θ). Therefore, we call the R, H, and θ plots signature plots. If the seven subunits show perfect sevenfold symmetry for a given residue, R, H, and θ are expected to be the same for all seven subunits within a ring, and the signature plots of R, H, and θ of different subunits should overlap (θ plots overlap at 51.43°).
Fig. 4.
Quantitative analysis of the R-ADP using the cylindrical coordinate system. (A) Asymmetry between the subunits in the R-ADP is dissected (Insets in Top and Bottom) and quantified using (Top) R (distance from the Cα of a residue to the sevenfold axis), (Middle) H (distance from the Cα of a residue to the twofold plane), and (Bottom) θ (angle between two vectors: one from the Cα of a residue to the sevenfold axis and one from the Cα of same numbered residue to the sevenfold axis in the neighboring subunit). The colors of traces (from red to purple) reflect the arrangement of subunits and are consistent with the colors in Fig. 3B. Two black arrows in R and H indicate the largest degree of asymmetry in radial (R) or vertical (H) direction. Four black arrows in θ marked with h1 (P137 and G410) and h2 (G192 and G375) are hinge residues connecting the equatorial and intermediate domains and the intermediate and apical domains, respectively. (Top) For comparison, two R-ATP states determined by cryo-EM Rd2 (lower black dashed line; PDB ID code 4AB2) and Rdopen (upper black dashed line; PDB ID code 4AB3) are analyzed and plotted with the asymmetry of R in the R-ADP state. (Bottom) The T state with sevenfold symmetry (PDB ID code 1XCK) is also analyzed and plotted with the asymmetry of θ in the R-ADP state (black dashed line). (B) Conformation differences between the R-ADP and T or R″ states are analyzed in similar ways. Conformational difference in each residue is quantified by (Upper) ΔR and (Lower) ΔH. ΔR = <R> − <R′>, where R is the distance to the sevenfold axle in the R-ADP state, and R′ is the corresponding distance in the T (red; PDB ID code 1XCK) or R″ (pink; PDB ID code 1AON) state; <> denotes average over seven subunits. Similarly, ΔH = <H> − <H′>, where H is the height in the R-ADP state, and H′ is the corresponding height in a known state (T in blue and R″ in cyan). ΔR and ΔH between two T state structures (black dashed lines; PDB ID codes 1XCK and 2NWC) serve as the negative controls, showing that two crystal structures of the same conformational state of GroEL only have small differences.
Our quantitative analysis confirms the asymmetry, showing apparent heterogeneity in R, H, and θ (Fig. 4A). This heterogeneity or asymmetry is confined to the intermediate and apical domains. For a given residue, R can differ by up to 20 Å, and H can differ by up to 10 Å (indicated by arrows in Fig. 4A). For a given pair of residues, θ deviates from 51.43° (as in perfect sevenfold symmetry) between +30° (between chains E and F) and −20° (between chains D and E).
The signature plots show three features. First, the departure from symmetry that is evident in the intermediate and apical domains begins and ends at the two hinge points, hinges 1 (P137 and G410) and 2 (G192 and G375), which adjoin the intermediate and equatorial and the apical and intermediate domains, respectively (black arrows in Fig. 4A, θ plot), suggesting that the asymmetry arises by small differences in the rigid body motion that accompanies the T to R transition. Consistent with this idea, the plots corresponding to different subunits are parallel to one another (Fig. 4A, R plot), indicating that the secondary structure is conserved among subunits. Second, no two signature plots overlap at the apical domain (Fig. 4A), meaning that none of the two subunits adopts identical conformation. Third, for a given residue, the difference in R is smallest between neighboring subunits (Fig. 4A, R plot), indicating that the conformation of a given subunit influences the conformation of the neighboring subunits.
Crystal Structure of R-ADP Closely Resembles the cryo-EM Structure of R-ATP.
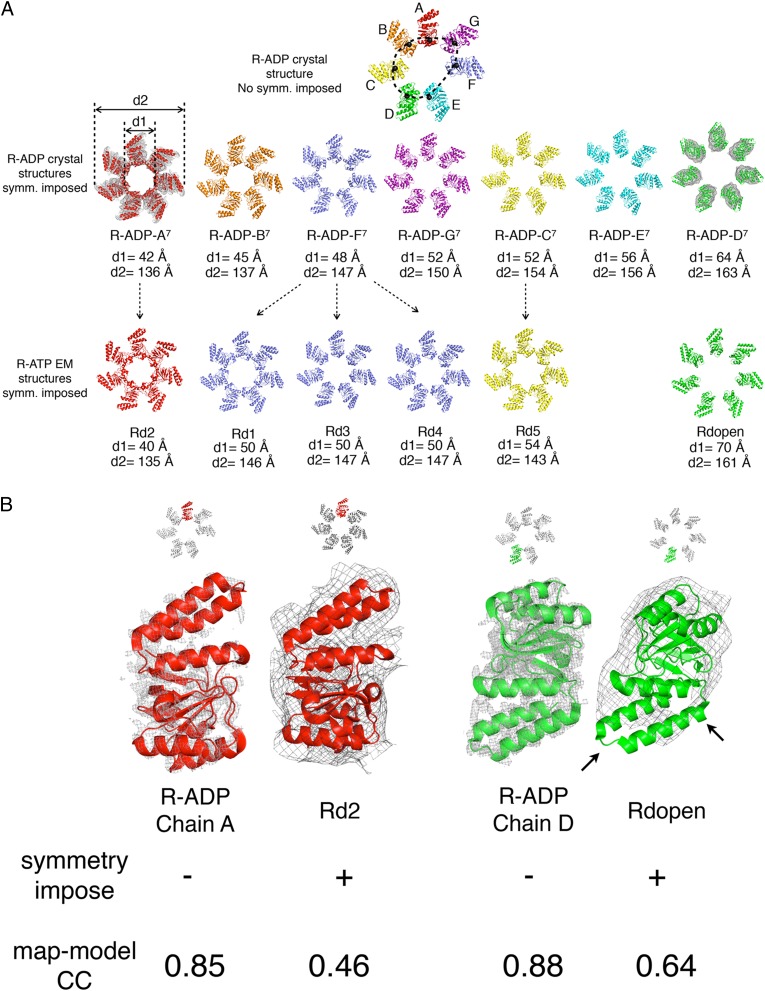
The flexibility of nucleotide-bound GroEL has recently been observed in the R-ATP by cryo-EM (14). After imposing sevenfold symmetry, it was concluded that six different symmetrical conformations of R-ATP existed in solution. To compare the variation of the R-ATP with R-ADP, we plot the residue-dependent R of the two most distinct conformations of the R-ATP state (two dotted lines in Fig. 4A, R plot). For >95% of the residues, the seven R signature plots of the asymmetric conformations in the R-ADP state (colored solid lines in Fig. 4A, R plot) are within the boundaries set by the Rd2 (lower dotted line in Fig. 4A, R plot) and the Rd-open (upper dotted line in Fig. 4A, R plot) conformations of the R-ATP state (14). Therefore, the flexibility of the R-ADP in the crystal structure is consistent with the variation of the R-ATP conformations observed previously in solution by cryo-EM. However, because of the imposition of sevenfold symmetry on each of the cryo-EM structures at 8.5 Å, the asymmetry evident in the crystal structure at 2.7 Å is lost.
To underscore the similarity of the R-ADP and R-ATP structures and their common asymmetry, we imposed sevenfold symmetry on each of the subunits A-G to create synthetic R-ADP-A7 to R-ADP-G7 rings (Fig. 5A), giving us the opportunity to compare these crystal-based synthetic ensembles with the synthetic cryo-EM–based ensembles. Despite the differences in resolution (2.7 vs. 8–9 Å) and correlation coefficient (0.89 vs. 0.69) for the X-ray and cryo-EM structures, respectively, it is obvious that the synthetic X-ray rings resemble the synthetic cryo-EM rings very closely, irrespective of the nature of the ligand (ADP or ATP) in the nucleotide binding site. The synthetic R-ADP rings can readily be ranked from the most closed structure (smallest value for d1) (Fig. 5A) to the most open structure (largest value for d1). Thus, the synthetic R-ADP-A7 corresponds to the cryo-EM–based structure Rd2. This comparison becomes especially evident when the two models for the apical domain are superimposed on their respective density maps (Fig. 5B, Left). Similarly, the synthetic R-ADP-D7 corresponds to the most open cryo-EM–based Rdopen (Fig. 5 A and B, Right). We conclude that, without imposition of sevenfold symmetry on the cryo-EM R-ATP structure, the asymmetry observed in the crystal structure of R-ADP would be found in solution in the cryo-EM structure of R-ATP.
Fig. 5.
Comparison between symmetry-imposed R-ADP to R-ATP (PDB ID codes: 4AAU, 4AB2, and 4AB3) structures determined by cryo-EM. (A) Each subunit in asymmetrical R-ADP (Top) is isolated and (Middle) symmetrized to generate synthetic R-ADP with sevenfold symmetry (upper line). These seven R-ADP structures are arranged from the least open one (R-ADP-A7) to the most open one (R-ADP-D7) according to their diameters (d1 and d2). Seven symmetrized R-ADP structures show good agreement with the cryo-EM densities of the R-ATP (Middle Left and Right: R-ADP-A7 vs. Rd2 and R-ADP-D7 vs. Rdopen) and they also closely resemble (Bottom) in diameters. The colors for symmetrized R-ADP are consistent with (Top) the asymmetrical R-ADP and reflect the arrangement of subunits in a ring. R-ATP structures are colored to the symmetrized R-ADP with the smallest RMSD. (B) Detailed view of model-density fitting in asymmetrical R-ADP and symmetrical R-ATP. At the cost of imposing symmetry, cryo-EM R-ATP models show rather poor correlated coefficients with maps and parts of the secondary structures showing no electron density (black arrows). Electron density maps are contoured at 1.0 σ.
T to R Transition Is Accompanied by the Loss of Intersubunit Stabilizing Contacts as the Origin of Asymmetry.
The flexibility of intermediate and apical domains in the R-ADP is correlated with the loss of intersubunit stabilizing contacts. In the T state, the intermediate and apical domains are stabilized by 42 intersubunit hydrogen bonds or salt bridges per ring (6 at each interface) (4, 5). However, in the crystal structure of R-ADP, these intersubunit contacts involving intermediate and apical domains are absent (Fig. 3C). As a result, the intermediate and apical domains in R-ADP can rotate relatively freely with respect to the equatorial domains, leading to the loss of sevenfold symmetry. Thus, the cause of flexibility and asymmetry in R-ADP (loss of intersubunit contacts) differs from the asymmetry observed in another case (20). In fact, the asymmetric arrangement that we observed in this crystal structure is only one possible conformation from an ensemble of flexible conformations that the R-ADP may access. An alternate source of asymmetry concerns the intrasubunit salt bridge between D155 and R395, both in the intermediate domain. The mutation D155A leads to an ATP-induced breakage of symmetry (21). However, because this particular salt bridge is maintained in R-ADP, it cannot be the source of the asymmetry observed here. Compared with the T or R″ state, the dramatically increased B factors for intermediate and apical domains in the R-ADP support this argument (Table S2). A computational study tracking the trajectories of the individual subunits of GroEL during the T to R transition reported this dynamic flexibility in the apical domain (22).
R-ADP is Distinct from Other Crystal Structures and cryo-EM Models of GroEL.
We quantified the averaged difference between the R-ADP state and previously solved crystal structures in the T (5) and R″ (7) states. We used the same cylindrical coordinate system as described above. For each residue, we first calculate the average R and H over seven subunits within a ring. ΔR and ΔH are the differences in R and H, respectively, between the R-ADP state and a previously solved state. Although the difference between T and R-ADP states is larger than 10 Å for some residues (red and blue in Fig. 4B), the difference between the R″ and R-ADP states exceeds 20 Å (pink and cyan in Fig. 4B). These differences are confined to the intermediate and apical domains.
The difference between the R-ADP and the crystal structures of T or R″ (solid lines in Fig. 4B) is much larger than the difference between two structural models of the same state (black dotted lines in Fig. 4B). Thus, the R-ADP state is distinct from previously known states. Furthermore, the difference is not solely caused by the flexibility of ADP-bound GroEL, because the difference does not vanish on averaging over seven subunits within a GroEL ring. The R-ADP is also significantly different from the crystal structure of the GroEL-ATPγS14 complex previously considered to be in the R state (23) (Figs. S1 and S2).
We also compared the R-ADP state GroEL with GroEL structures determined by cryo-EM at lower resolution. The R-ADP is similar to a series of cryo-EM R-ATP structures (14). For seven asymmetrical subunits in the R-ADP GroEL, chain A has the conformation closest to R-ATP state Rd2, whereas chain D is most similar to R-ATP state Rd-open (Fig. 4A, red, cyan, and two dashed black traces and Fig. 5A).
The conformational differences between the R-ADP and other structures are mainly caused by rigid body rotation of the intermediate and apical domains around hinge 1 (P137 and G410) and hinge 2 (G192 and G375) (7). For example, compared with the crystal structure of the T or R″ states, although the averaged conformational differences between the intermediate and apical domains can be greater than 20 Å (Fig. 4B), the secondary structure within these domains remains largely unchanged. The rmsd values of isolated intermediate domains are only 0.69 ± 0.28 (between the T and R-ADP) and 0.99 ± 0.33 Å (between the R″ and R-ADP). The corresponding deviations of isolated apical domains are 0.52 ± 0.12 and 1.11 ± 0.06 Å.
Rigid Body Rotation of the Intermediate Domain Controls Nucleotide Release: (i) the R″ to R-ADP Transition.
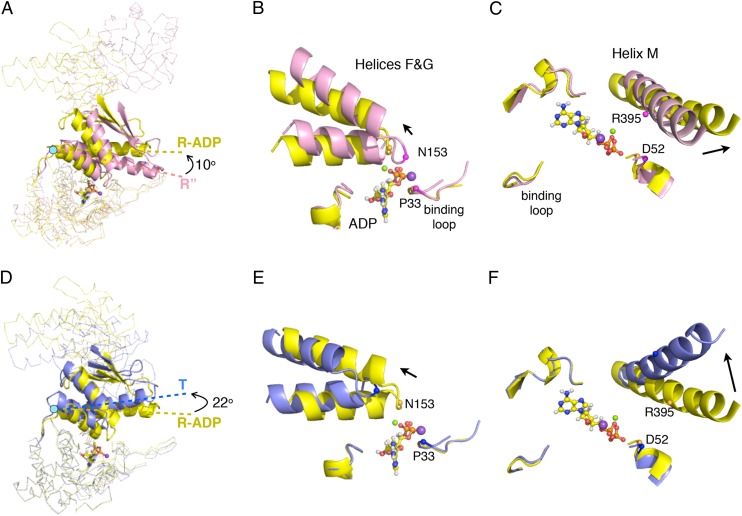
A structural comparison of the R″ and R-ADP states indicates that the rotation of the intermediate domain controls access to the nucleotide binding pocket. In progressing from the R″ to the R-ADP state, the intermediate domains undergo a 10° rotation away from the twofold symmetry plane with respect to hinge 1 (Fig. 6A). Consequently, the nucleotide binding pocket, which contains an intermediate domain lid (helices F, G, and M) and equatorial domain nucleotide binding loops, changes from a full- to a half-closed state. This movement can be measured from the change in distance between the lid helices and the nucleotide binding loops. The average distance from helices F/G (N153) to the loops (P33) increases from 5.5 ± 0.3 to 7.1 ± 0.4 Å (Fig. 6B), whereas the average distance from helix M (R395) to the loops (D52) increases from 7.6 ±0.1 to 10.1 ± 1.1 Å (Fig. 6C).
Fig. 6.
Conformational changes in the intermediate domains between the T (PDB ID code 1XCK), R-ADP, and R″ (PDB ID code 1AON) states. (A) A subunit from the R″ (pink) aligned with a subunit from the R-ADP (yellow) by superposition of the two equatorial domains. The conformation difference between the intermediate domains (in cartoon) in two structures is the result of domain rotation around hinge 1 (cyan dot). The axes of helix M are shown as dashed lines. Black arrows indicate directions of hinged rotations of intermediate domains. (B and C) Detailed views of helices F and G and helix M in the superimposed structures in A showing the distance from these helices to the nucleotide binding loops changes as the result of hinged rotation. Two pairs of residues (P33&N153 and D52&R395) are shown to highlight this distance change (Cαs as spheres). ADP is shown in ball-and-stick format, and Mg2+ and K+ are shown as green and purple spheres. Black arrows indicate the relative position change of these helices from R″ to R-ADP state. D–F are same as A–C, except showing the rotation of the intermediate domain and the position of helices F and G and helix M as GroEL switches from the R-ADP (yellow) to the T (blue) state.
The structural transition of the nucleotide binding site from fully closed in the R″ state to the half-closed R-ADP state is accompanied by an increase in the rate of ADP release. ADP, sequestered in the cis ring, in the R″ state does not exchange with free ligand in solution at all (24). However, ADP dissociates for the half-closed R-ADP state at a rate of ∼0.1 s−1 compared with the presteady state rate of ATP hydrolysis of ∼0.6 s−1 (Fig. 2B) (14, 16). In other words, ADP release from the half-closed R-ADP state, although faster than release from the R″ state, is still slower than ATP hydrolysis and thus, remains the rate-determining step in the chaperonin cycle in the absence of SP. By introducing the mutations that stabilized the half-closed R state, we expect ADP release to be even slower. We measured a threefold increase in the mean residence time of GroES dissociation that is induced on the addition of ATP to the resting state complex of GroELD83A/R197A and WT (Fig. 2C).
Rigid Body Rotation of Intermediate Domain Controls Nucleotide Release: (ii) the R-ADP to T Transition.
During the R-ADP to T transition, the intermediate domain rotates away from the cavity center and farther away from the twofold plane of symmetry by a total of 22° (Fig. 6D). The movement brings about a change in the nucleotide binding site from the half-closed to the open state. The average distance between the lid helices F/G (N153) and nucleotide binding loop (P33) increases from 7.1 ± 0.4 to 12.2 ± 0.2 Å (Fig. 6E). Similarly the average separation between lid helix M (R395) and nucleotide binding loop (D52) expanded from 10.1 ± 1.1 to 14.2 ± 0.1 Å (Fig. 6F). The opening of the nucleotide binding pocket associated with the reformation of the T state permits rapid ADP/ATP exchange that is necessary for continued cycling of the chaperonin machine.
Although the average distance between lid and loop is related to the rate of ADP release, the variance of distance in the R-ADP state explains SP-accelerated ADP release. The variance of the distance is significantly larger in the R-ADP than the other two states as the result of asymmetry (Fig. 5A). For example, the distance from helix M lid to loop varies from 8.5 to 11.9 Å in the R-ADP, approaching the average lid-to-loop distance in the open T state (14.2 ± 0.1 Å) on one hand and the full-closed R″ state (7.6 ± 0.1 Å) on the other hand. As described above, this variance is the result of the flexibility of the R-ADP, and it very likely exists in solution as well. Thus, it is likely that the nucleotide binding pockets in the R-ADP switch stochastically between the open and full-closed conformations in solution because of its structural flexibility.
We suggest that SP accelerates ADP/ATP exchange by selectively binding to GroEL with open nucleotide binding pockets. SP would consequently increase the population and lifetime of the open conformation, permitting rapid nucleotide exchange. This mechanism is similar to the out-of-equilibrium conformational cycling that others have proposed (25).
Rigid Body Rotation of Apical Domain Enables Selective Binding to GroES or Substrate Protein.
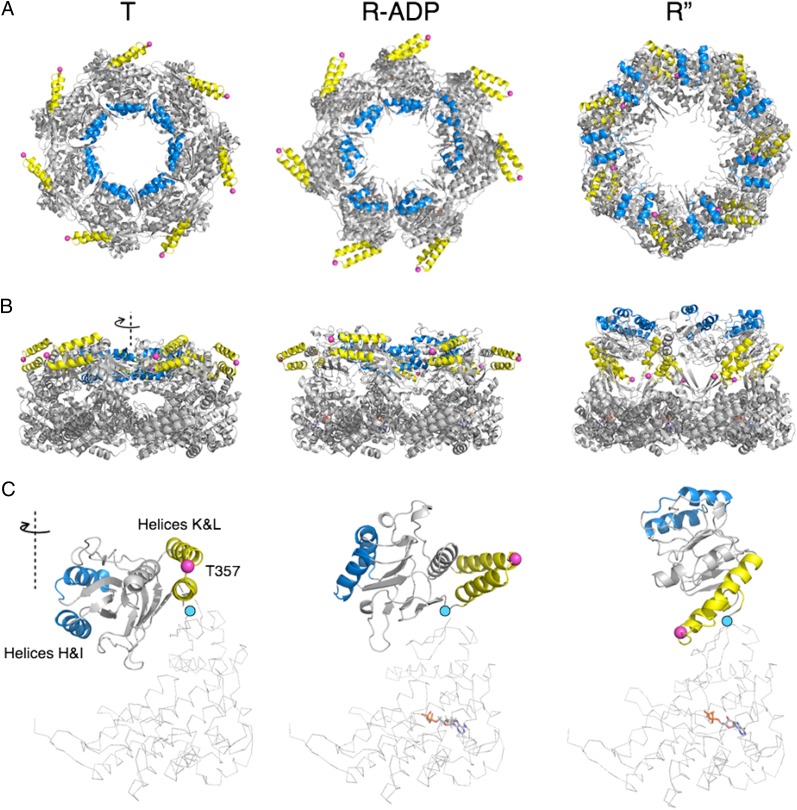
The apical domains rotate with respect to hinge 2 when GroEL cycles through the R″ to R-ADP to T states (cyan dots in Fig. 7C). The apical domains bind both SP and GroES that share the same binding site: the groove between helices H and I (7, 26). Hinged rigid body rotation changes the position of helices H and I and the binding grooves between two helices. Thus, GroEL binds to SP or GroES selectively at different stages of its cycle: the R″ state binds GroES, and the T binds SP, whereas the R-ADP state has a conformation intermediate.
Fig. 7.
Conformational changes in apical domains between the T (PDB ID code 1XCK), R-ADP, and R″ (PDB ID code 1AON) states. (A) Top and (B) sides views of a single GroEL ring in (Left) T, (Center) R-ADP, and (Right) R″ states. Positions of helices H and I (blue), helices K and L (yellow), and T357 (Cαs as pink spheres) are highlighted to trace apical domain rotations. (C) Detailed views of one subunit from each state. The conformation differences between the intermediate domains (in cartoon) are the result of domain rotation around hinge 2 (cyan dots). ADP is shown in sticks. Note in particular that the major motion of helices K and L, a flip that brings T357 from its position on the external surface of GroEL to one on the internal surface, occurs during the R to R″ transition.
(i) R″ to R-ADP Transition.
In progressing from the R state to the R-ADP state, the apical domains in the R″ state swing about 80° clockwise and move to the twofold plane (Fig. 7, Center and Right). As the apical domains move and the overall shape of GroEL changes, the position of the SP/GroES binding helices H and I changes dramatically. In R″ state, helices H and I present themselves at the upper surface of the GroEL ring, with the binding grooves between two helices fully exposed on the GroEL surface and well-separated from each other (Fig. 7 A, B, and C, Right). In R-ADP state, however, these two helices tilt toward the center of the cavity, partially exposing the binding grooves between helices H and I to the surface (Fig. 7A, Center). At the same time, in the R-ADP state, these helices move closer to each other and line the collar of the GroEL cavity (Fig. 7 A and B, Center).
The structure of the R-ADP suggests that it has lower affinity for GroES compared with the R″ state. The binding grooves in R″ favor GroES binding structurally by adopting a separated and exposed conformation, which adapt to the seven well-separated mobile loops of GroES dome (7). However, in the R-ADP state, the binding grooves rotate toward the center of the ring and become partially buried, and therefore, GroES binds less readily to the R-ADP state than to the R″ state. Reduced affinity for GroES would prevent it binding to R-ADP state GroEL before SP capture.
(ii) R-ADP to T Transition.
During the R-ADP to T transition, the apical domains swing about 30° counterclockwise and move closer to the twofold plate (Fig. 7, Left and Center). The partially exposed binding groove between helices H and I in the R-ADP state now reorient, such that they point toward the center of cavity in the T state (Fig. 7C, Left and Center). The circumference of the binding collar that contains seven pairs of helices H and I decreases (Fig. 7A, Left and Center), and as a consequence, the SP binding sites on adjacent subunits move 7 Å closer on average. Such conformational difference between the R-ADP and T states suggests that the T state binds SP tighter than the R-ADP state. Because SP needs to interact with multiple binding sites to bind GroEL with nanomolar affinity (27), the T state with smaller SP binding collar and shorter distance between adjacent binding sites is likely to have higher affinity to SP than the R-ADP state.
Biological Implications of Asymmetry in Oligomeric Chaperonin Rings.
In Escherichia coli, GroEL interacts with one-half of the soluble, misfolded proteins, which are diverse in sequence and structure (28). To capture so many heterogeneous proteins, the binding sites on GroEL have to be adaptive. Intersubunit apical domain contacts stabilize the T state. When these contacts are broken on the addition of ATP/ADP, the apical domains become free to express the intrinsic flexibility that is characteristic of the R state. The flexibility of apical domains would allow the R-ADP to adjust its binding collar according to a specific misfolded protein. Although it enables high promiscuity, the flexibility of the apical domain may lead to weaker affinity for SP. Nevertheless, SP must bind before GroEL can release bound ADP and proceed to the T state.
Hitherto, conventional wisdom dictated that the oligomeric class I chaperonin rings were fundamentally symmetrical and furthermore, that this symmetry was maintained during the various allosteric transitions of these nanomachines. However, asymmetry in the related class II chaperonins has been known for some time (29). Here, we present an example of how this symmetry is broken in the class I chaperonin, GroEL, merely by removing two salt bridges that are normally broken during the T to R transition. Intriguingly, other examples have very recently emerged in both classes I and II chaperonins (30, 31), raising the possibility that transient departures from structural symmetry may be an important and intrinsic part of the mechanism(s).
Comments on the Click Stop Mechanism.
In a recent cryo-EM study (14) of GroEL-ATP, sevenfold symmetry was artificially imposed, leading to the conclusion that the GroEL ring in the R state existed in six homogeneous but slightly different states. Although lacking supporting kinetic evidence, a deterministic model was proposed that implied temporal transitions between these states (14). Changes in several intersubunit salt bridges (K80-E386, E255-K207, E255-K245, E257-K245, and E257-K242) were suggested to form the basis of this click stop mechanism (table S2b in ref. 14). Skeptical that salt bridges could be reliably identified on the basis of a structure at 8–9 Å resolution, we reexamined this question using the 2.7 Å crystal structure of R-ADP (Table S3). Using established criteria (32, 33) for interatomic distances in protein salt bridges (<4 Å for a strong salt bridge and <6 Å for an N-O bridge), we find that only 4 of 35 ion pairs mentioned above are close enough to be considered as salt bridges. Most of the remaining pairs are tens of angstroms apart. The click stop mechanism, therefore, rests on the assumption of sevenfold symmetry, which is not supported by our data.
Materials and Methods
Construction of GroELD83A/R197A.
The mutants D83A/R197A, E315C, and D83A/R197A/E315C were prepared as described in SI Materials and Methods.
Steady State ATP Hydrolysis.
The steady state hydrolysis of ATP was measured at 37 °C as previously described (15).
Presteady State ATP Hydrolysis.
The steady state hydrolysis of ATP was measured using the inorganic PBP (16) at 37 °C as previously described (15).
Labeling GroELE315C and GroES98C.
Details are provided in SI Materials and Methods.
Stopped Flow Measurements of FRET.
All FRET measurements were carried out on an Applied Photophysics SX18 MV-R stopped flow apparatus. The instrument was configured with a 20-L flow cell with a path length of 2 mm and a 530-nm emission cutoff filter. The monochrometer slits were set to a width of 9 mm. The apparatus was thermostated at 30 °C. The excitation wavelength used for all measurements was 336 nm. Details are provided in SI Materials and Methods.
Structure Determination and Analysis of the R-ADP.
Details of the structure determination and analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Dorothy Beckett, Dr. Marius Clore, Dr. Michael Levitt, Dr. Dave Thirumalai, and Dr. Zhechun Zhang for constructive criticism. This work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which is supported by National Center for Research Resources Grant 5P41RR015301-10 and National Institute of General Medical Sciences Grant 8 P41 GM103403-10 from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by the Argonne National Laboratory, was supported by US Department of Energy Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4KI8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311996110/-/DCSupplemental.
References
- 1.Lin Z, Rye HS. GroEL-mediated protein folding: Making the impossible, possible. Crit Rev Biochem Mol Biol. 2006;41(4):211–239. doi: 10.1080/10409230600760382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horovitz A, Fridmann Y, Kafri G, Yifrach O. Review: Allostery in chaperonins. J Struct Biol. 2001;135(2):104–114. doi: 10.1006/jsbi.2001.4377. [DOI] [PubMed] [Google Scholar]
- 3.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30(2001):245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 4.Braig K, Adams PD, Brünger AT. Conformational variability in the refined structure of the chaperonin GroEL at 2.8 A resolution. Nat Struct Biol. 1995;2(12):1083–1094. doi: 10.1038/nsb1295-1083. [DOI] [PubMed] [Google Scholar]
- 5.Bartolucci C, Lamba D, Grazulis S, Manakova E, Heumann H. Crystal structure of wild-type chaperonin GroEL. J Mol Biol. 2005;354(4):940–951. doi: 10.1016/j.jmb.2005.09.096. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhry C, Horwich AL, Brunger AT, Adams PD. Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states. J Mol Biol. 2004;342(1):229–245. doi: 10.1016/j.jmb.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388(6644):741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 8.Yifrach O, Horovitz A. Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry. 1995;34(16):5303–5308. doi: 10.1021/bi00016a001. [DOI] [PubMed] [Google Scholar]
- 9.Yifrach O, Horovitz A. Allosteric control by ATP of non-folded protein binding to GroEL. J Mol Biol. 1996;255(3):356–361. doi: 10.1006/jmbi.1996.0028. [DOI] [PubMed] [Google Scholar]
- 10.Yifrach O, Horovitz A. Two lines of allosteric communication in the oligomeric chaperonin GroEL are revealed by the single mutation Arg 196-->Ala. J Mol Biol. 1994;243(3):397–401. doi: 10.1006/jmbi.1994.1667. [DOI] [PubMed] [Google Scholar]
- 11.White HE, et al. Structural basis of allosteric changes in the GroEL mutant Arg197—>Ala. Nat Struct Biol. 1997;4(9):690–694. doi: 10.1038/nsb0997-690. [DOI] [PubMed] [Google Scholar]
- 12.Murai N, Makino Y, Yoshida M. GroEL locked in a closed conformation by an interdomain cross-link can bind ATP and polypeptide but cannot process further reaction steps. J Biol Chem. 1996;271(45):28229–28234. doi: 10.1074/jbc.271.45.28229. [DOI] [PubMed] [Google Scholar]
- 13. Grason JP (2004) Allostery in GroEL: Its role in the refolding of protein substrate. PhD thesis (Univ of Maryland, College Park, MD)
- 14.Clare DK, et al. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell. 2012;149(1):113–123. doi: 10.1016/j.cell.2012.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grason JP, Gresham JS, Widjaja L, Wehri SC, Lorimer GH. Setting the chaperonin timer: The effects of K+ and substrate protein on ATP hydrolysis. Proc Natl Acad Sci USA. 2008;105(45):17334–17338. doi: 10.1073/pnas.0807429105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brune M, et al. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37(29):10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 17.Grason JP, Gresham JS, Lorimer GH. Setting the chaperonin timer: A two-stroke, two-speed, protein machine. Proc Natl Acad Sci USA. 2008;105(45):17339–17344. doi: 10.1073/pnas.0807418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sameshima T, et al. Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J Biol Chem. 2008;283(35):23765–23773. doi: 10.1074/jbc.M802541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridmann Y, Kafri G, Danziger O, Horovitz A. Dissociation of the GroEL-GroES asymmetric complex is accelerated by increased cooperativity in ATP binding to the GroEL ring distal to GroES. Biochemistry. 2002;41(18):5938–5944. doi: 10.1021/bi020117v. [DOI] [PubMed] [Google Scholar]
- 20.Shimamura T, et al. Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity. Structure. 2004;12(8):1471–1480. doi: 10.1016/j.str.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Danziger O, Rivenzon-Segal D, Wolf SG, Horovitz A. Conversion of the allosteric transition of GroEL from concerted to sequential by the single mutation Asp-155 -> Ala. Proc Natl Acad Sci USA. 2003;100(24):13797–13802. doi: 10.1073/pnas.2333925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyeon C, Lorimer GH, Thirumalai D. Dynamics of allosteric transitions in GroEL. Proc Natl Acad Sci USA. 2006;103(50):18939–18944. doi: 10.1073/pnas.0608759103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Boisvert DC. Structural basis for GroEL-assisted protein folding from the crystal structure of (GroEL-KMgATP)14 at 2.0A resolution. J Mol Biol. 2003;327(4):843–855. doi: 10.1016/s0022-2836(03)00184-0. [DOI] [PubMed] [Google Scholar]
- 24.Todd MJ, Viitanen PV, Lorimer GH. Dynamics of the chaperonin ATPase cycle: Implications for facilitated protein folding. Science. 1994;265(5172):659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 25.Frank GA, et al. Out-of-equilibrium conformational cycling of GroEL under saturating ATP concentrations. Proc Natl Acad Sci USA. 2010;107(14):6270–6274. doi: 10.1073/pnas.0910246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Sigler PB. The crystal structure of a GroEL/peptide complex: Plasticity as a basis for substrate diversity. Cell. 1999;99(7):757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- 27.Farr GW, et al. Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell. 2000;100(5):561–573. doi: 10.1016/s0092-8674(00)80692-3. [DOI] [PubMed] [Google Scholar]
- 28.Viitanen PV, Gatenby AA, Lorimer GH. Purified chaperonin 60 (groEL) interacts with the nonnative states of a multitude of Escherichia coli proteins. Protein Sci. 1992;1(3):363–369. doi: 10.1002/pro.5560010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transitions of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat Struct Mol Biol. 2005;12(3):233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- 30.Chen DH, et al. Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell. 2013;153(6):1354–1365. doi: 10.1016/j.cell.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalisman N, Schröder GF, Levitt M. The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure. 2013;21(4):540–549. doi: 10.1016/j.str.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlow DJ, Thornton JM. Ion-pairs in proteins. J Mol Biol. 1983;168(4):867–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Nussinov R. Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys J. 2002;83(3):1595–1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.