Abstract
The thorniest problem in comparative neurobiology is the identification of the particular brain region of birds and reptiles that corresponds to the mammalian neocortex [Butler AB, Reiner A, Karten HJ (2011) Ann N Y Acad Sci 1225:14–27; Wang Y, Brzozowska-Prechtl A, Karten HJ (2010) Proc Natl Acad Sci USA 107(28):12676–12681]. We explored which genes are actively transcribed in the regions of controversial ancestry in a representative bird (chicken) and mammal (mouse) at adult stages. We conducted four analyses comparing the expression patterns of their 5,130 most highly expressed one-to-one orthologous genes that considered global patterns of expression specificity, strong gene markers, and coexpression networks. Our study demonstrates transcriptomic divergence, plausible convergence, and, in two exceptional cases, conservation between specialized avian and mammalian telencephalic regions. This large-scale study potentially resolves the complex relationship between developmental homology and functional characteristics on the molecular level and settles long-standing evolutionary debates.
Keywords: cerebral cortex, Wulst, equivalent circuit hypothesis, brain evolution, dorsal ventricular ridge
Despite recent advances in our knowledge of comparative aspects of cortical neurogenesis, migration, clonal relationships, and gene expression patterns, there is no consensus on how these processes evolved together to determine the adult brain structures across diverse amniotes (1–5). Anatomical, hodological, embryological, and gene expression data (based on few select genes) provide conflicting answers on brain homology across vertebrates. Studies of embryonic neurogenesis and cell migration have informed homology of developmental territories (6–8), but the striking similarities in lamination, connectivity, and physiological properties observed between adult forms derived from noncorresponding pallial regions remain unexplained (3–5, 9). Comparative transcriptomics is a powerful approach to interrogating regional correspondence without resorting to limited lists of selected genetic markers. Recent methodological advances in profiling mammalian cerebral cortical layer transcriptomes (10–12) could objectively test the validity of proposed, yet controversial, relationships between regions of adult mammalian and avian brains. These comparisons are motivated by our understanding of the evolution of mammals from a reptilian subclass, represented by the synapsid condition (with a cranial opening in the cheek region of the skull), and of birds from another reptilian subclass, represented by the diapsid condition (two postorbital skull openings).
Results
We extended our previous transcriptomic analysis of cortical layers in the adult mouse (13) to additional structures, 16 in total, and compared these with seven regions of the adult chicken brain (Fig. 1A and SI Appendix, Figs. S1 and S2). All dissected regions in both species (except the striatum, which is subpallial) develop primarily from one of four morphogenetically delineated sectors of the pallium (the dorsal portion of the telencephalon): medial, dorsal, lateral, and ventral. Striatum and hippocampus (a medial pallium derivative) were dissected from both species as control regions of undisputed homology and conserved function. The additional chicken regions were hyperpallium, a dorsal pallial derivative; mesopallium, a lateral pallial derivative; dorsolateral corticoid area, a dorsal or lateral pallial derivative; and nidopallium and arcopallium, both ventral pallial derivatives. Mouse samples were dissected from layers of dorsal and lateral cortex, both dorsal pallial derivatives; and claustrum-endopiriform complex and pallial amygdala, both primarily ventral pallial derivatives (although pallial amygdala has a component from lateral pallium). We validated the chicken dissections with in situ hybridization of resulting marker genes (SI Appendix, Fig. S8 and Table S4) and the mouse dissections using hybridizations from the Allen Mouse Brain Atlas (SI Appendix, Figs. S3–S7). We deep sequenced cDNA libraries from pooled polyadenylated RNA from 8 mice (postnatal 56 d) and 18 chickens (12 wk) and compared the expression patterns in each species of the most highly expressed 5,130 one-to-one orthologs (SI Appendix, Tables S1–S3) following extensive quality checks (SI Appendix, Figs. S23–S44 and Tables S9–S12).
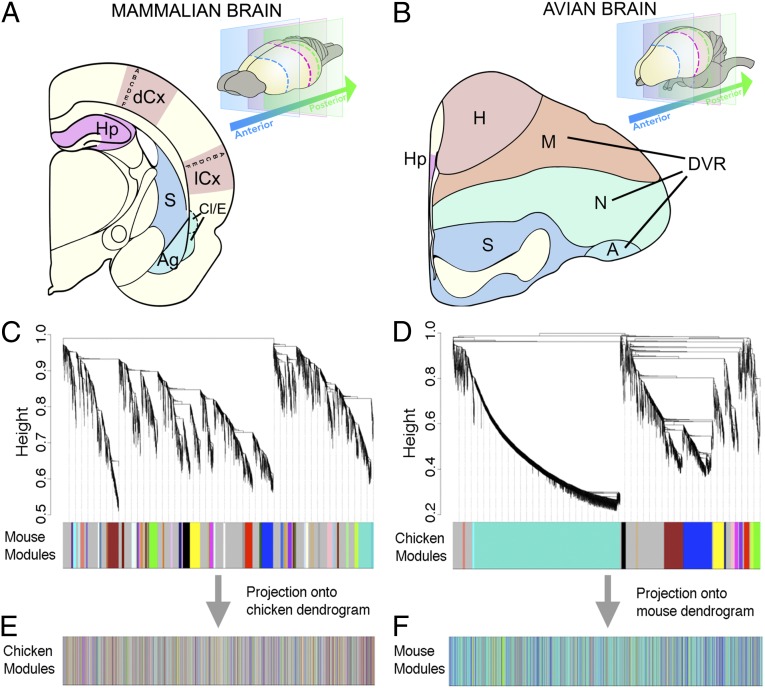
Fig. 1.
Studied regions and little conservation of gene coexpression between avian and mammalian brains. Brain regions involved in the evolutionary debate of neocortex homology studied in mouse (A) and chicken (B) were dissected from serial sections in the anteroposterior axis; the claustrum (Cl, dashed lines) is located in more anterior sections of mammalian brains, and the arcopallium (A) is located in more posterior sections of avian brains. The avian dorsolateral corticoid area is shown in SI Appendix, Fig. S8. (C) Topological dissimilarity dendrogram and coexpressed mouse modules. Gene pairs that branch from one another lower on this dissimilarity dendrogram have more similar expression patterns across the dissected samples than other pairs. A color band corresponds to a group of genes sharing a similar expression pattern across samples; thus, larger bands reflect more genes that share a pattern. (D) Topological dissimilarity dendrogram and coexpressed chicken modules. (E) Relative positions in the chicken dendrogram (represented in B) of one-to-one orthologs of genes in the mouse clustering (represented in A). (F) Relative positions in the mouse dendrogram (represented in A) of one-to-one orthologs of genes in the chicken clustering (represented in B). A to F in mouse, cortical layers; Ag, basolateral amygdala; dCx, dorsal cerebral cortex; E, endopiriform complex; H, hyperpallium; Hp, hippocampus; lCx, lateral cerebral cortex; M, mesopallium; N, nidopallium; S, striatum.
We used four approaches to quantify expression relationships, yielding consistent and complementary results. First, we considered if there was a particular alignment of expression specificities of all genes in one region in one species with any region in the other species (the “specificity comparison”). Second, we objectively defined the top marker genes in each region of one species (each in turn) and assessed if they showed a tendency to be expressed more likely than expected by chance in any particular region of the other species (the “marker comparison”). Third, we looked for cross-species overlap of gene coexpression modules using Weighted Gene Coexpression Network Analysis (10–12) (the “network comparison”; Fig. 1 B–E and SI Appendix, Figs. S10, S11, S45–S50 and Tables S7–S9, S13, S14, and S20). Fourth, we examined the cross-species preservation of modules (the “module preservation comparison”) (SI Appendix, Tables S5, S6, and S15). In these comparisons, the average rank of gene expression level was moderately conserved between mouse and chicken (r = 0.49, P < 10−200; Fig. S9). This correlation is slightly lower than previously reported for brain transcriptomes between mouse and human (12, 14) (r = 0.60, P < 10−400), species with fourfold less divergence time. In the network comparison, we found some robust cross-species markers for oligodendrocytes, striatum, and hippocampus (SI Appendix, Figs. S16 and S17). The marker comparison returned the expected transcriptional conservation in striatum and hippocampus and thus validated our dissection methods (SI Appendix, Figs. S52 and S53). Likewise, the module preservation comparison revealed a preservation of coexpressed striatal genes (SI Appendix, Tables S5 and S6 and Fig. S51).
All four sets of analyses, however, failed to identify any degree of similarity between nonhippocampal pallial regions that reflected homology. In fact, no significant similarities were identified from the specificity comparison, which we know from simulations to have the power to detect a transcriptome-wide similarity of a mere ∼1% greater than random pairings (SI Appendix, Materials and Methods). This suggests that the highly significant similarities identified using the marker and network comparisons are driven by subsets of genes. An apparent marker gene in one species is more likely to be an apparent marker gene in the other species: the maximum specificity for the chicken ortholog of the median marker gene in mouse was at the 79th percentile among all genes in chicken and at the 95th percentile of marker genes in mouse (the median chicken marker was at the 99th percentile in chicken and its ortholog at the 85th percentile in mouse). When considered jointly, there is little discernible pattern in how these genes are expressed, suggesting that a strong argument for one-to-one homologies between birds and mammals should not be based on a few adult marker genes. Nonetheless, comparative studies based on the expression of a few select marker genes have been used to support specific, and sometimes contradictory, one-to-one homologies between nonhippocampal pallial regions of adult birds and mammals that are apparently developmentally unrelated (8, 15). As a recent example, deep cortical layers (largely layer V) were proposed to be homologous to the avian parahippocampal area (8) on the basis of two genes that were used separately to support an alternative homology with avian arcopallium (15). Our results explain why this approach leads to contradictory conclusions.
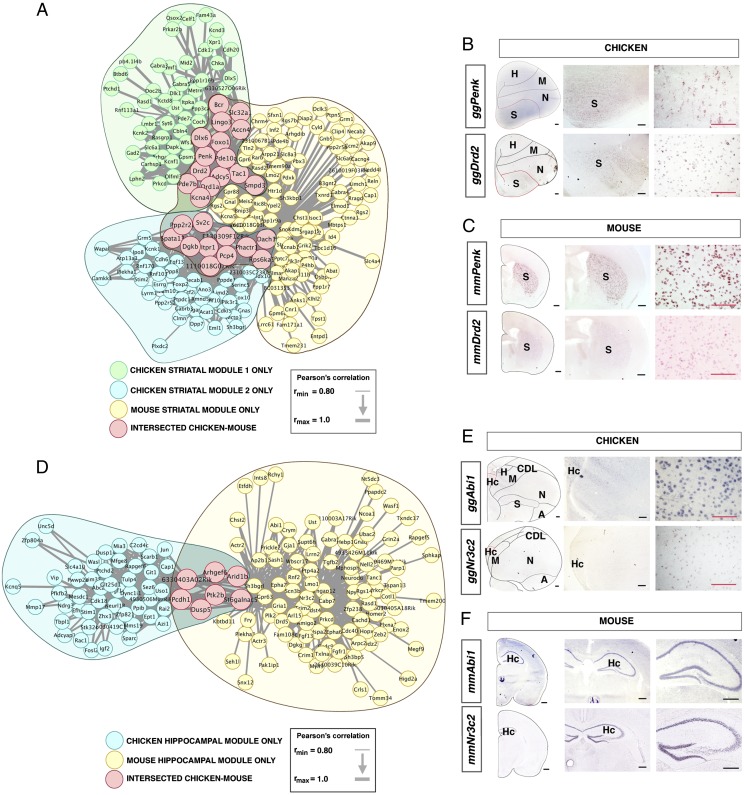
We compared the gene coexpression modules (Fig. 1) constructed in mouse (49 modules) and chick (15 modules) with quality controls (SI Appendix, Fig. S54 and Tables S16–S19). Eleven modules (six from chicken and five from mouse) significantly overlapped between the two species, even after applying a conservative Bonferroni correction (SI Appendix, Figs. S18 and S19). As expected for a region of uncontroversial homology (9) (which may be considered to be a positive control), significantly similar gene sets are expressed in the striatum of both species. A module of mouse striatal genes overlapped significantly with two chicken modules of striatal genes having slightly different expression patterns (Fig. 2 A–C). This result was confirmed using a more conservative analysis in which the lowest quality samples were removed (SI Appendix, Materials and Methods and Results). Functional studies further confirm that these genes with conserved striatal expression are significantly associated with striatal molecular and cellular neurobiology, obesity and exercise, drug sensitization, and pain modulation and have been associated with Parkinson disease models (SI Appendix, Results). The second positive control dissection, mouse and chicken hippocampus, also yielded the expected overlap of modules (Fig. 2 D–F). Despite their different anatomical appearance, lesions in avian hippocampus cause the same constellation of impairments as lesions in the mammalian hippocampus, supporting conserved function in these clades (16). We find that these functional similarities are also reflected in these regions’ transcriptomes. There has been little previous work on these conserved hippocampus marker genes’ hippocampal functions or the behavioral effects following their disruption.
Fig. 2.
Conserved striatal and hippocampal gene coexpression modules in chicken and mouse. (A) Expression correlations between genes in two striatal chicken gene coexpression modules and one striatal gene coexpression module in mouse with significant overlap between the two species (chicken striatal module 1 overlapping with Bonferroni-corrected P = 8.7 × 10−21, chicken striatal module 2 overlapping with Bonferroni-corrected P = 1.4 × 10−4; both hypergeometric tests). (B) RNA in situ hybridization (ISH) of conserved markers of chicken striatum. (C) RNA-ISH of conserved markers of mouse striatum. (D) Expression correlations between genes in significantly overlapping chicken and mouse hippocampal gene coexpression modules (Bonferroni-corrected P = 3.6 × 10−3, hypergeometric test). (E) RNA-ISH of conserved markers of chicken hippocampus. (F) RNA-ISH of conserved markers of mouse hippocampus. CDL, corticoid dorsolateral area. (Scale bars: black, 500 μm; red, 200 μm.)
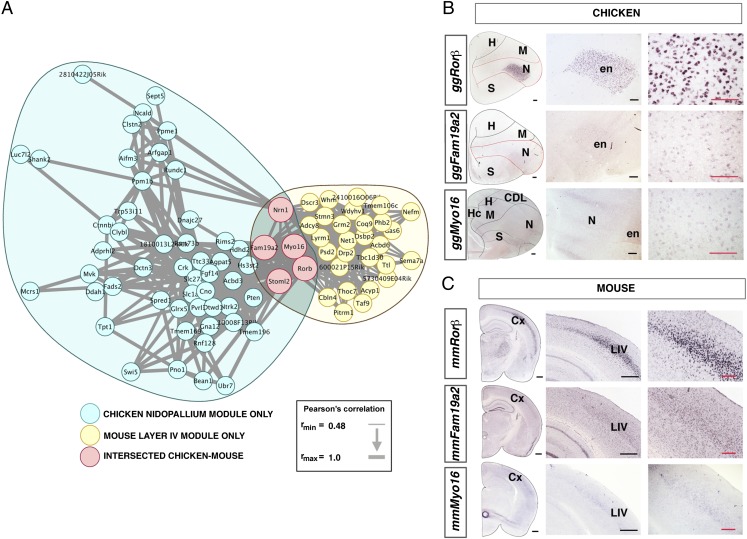
Owing to the ongoing debate on mammalian neocortical origin (1–3, 6, 15), one interesting finding is the statistically significant overlap between genes marking chick nidopallium and mouse layer IV neocortex found in both the network comparison and the marker comparison (Fig. 3 A–C). Indeed, some of these genes have already been used to support homology between nidopallium and layer IV (4, 9, 15, 17–19). However, we do not consider this a strong argument for homology (evolution by vertical descent) of these two regions for three reasons. First, just five genes explained this particular significant overlap in the network comparison (0.5 expected, reflecting a 10-fold enrichment; Fig. 3A). Gene coexpression modules can sometimes indicate a larger trend. However, the significant overlap of two modules need not necessarily imply that genes other than the hub genes (i.e., those genes whose expression is most correlated with the other genes in the module) are under similar regulatory control. Furthermore, upon repeating the network analysis removing samples with low RNA integrity number (RIN) (hyperpallium and arcopallium) to assess robustness, we did not recapitulate this result although we did again find strong striatal overlap (SI Appendix, Results, Figs. S12–S15, and Tables S2–S19 and Dataset S1). Second, three of the top hub genes for which the Allen Mouse Brain Atlas had in situ hybridization images are expressed in a variety of very different brain regions, including complex and divergent expression patterns outside the dissected areas in structures involved in processing or analyzing streams of sensory input: family with sequence similarity 19, member A2 (Fam19a2) in olfactory-related areas, dynactin 3 (Dctn3) in olfactory and somatosensory areas, and RAR-related orphan receptor β (Rorβ) in olfactory and visual regions (SI Appendix, Figs. S20 and S21). There is also some functional evidence consistent with a sensory-processing Rorβ protein overexpression being sufficient to induce clustering of layer IV neurons and the attraction of thalamic innervation (20). Fam19a2 is hypothesized to have a role in axonal guidance (21) and it is known that both chick nidopallium and mouse layer IV are thalamic recipients. In addition, the chick nidopallium derives from the [empty spiracles homeobox 1 (Emx1)-negative] ventral pallium (22), whereas mouse neocortex is generally thought to derive from the (Emx1-positive) dorsal or dorsal-and-lateral pallium (1, 6, 22). Finally, if these genes indicated homologous regions from an as-yet-undiscovered definitive tangential migration of excitatory neurons into layer IV of neocortex, one might also expect these genes to be found in other regions containing ventral pallial derivatives, namely pallial amygdala, claustrum, and endopiriform nuclei. This, however, was not observed in our analysis. A transient population of developing brain homeobox 1 (Dbx1) excitatory neurons migrates from the ventral pallium through the cortical subventricular zone, stimulating proliferative activity at embryonic stages. Subsequently, these Dbx1-positive neurons invade all cortical layers from the early postnatal period, but then largely disappear by adulthood (6, 23). We thus favor the alternative explanation that this result is due to convergence of expression levels for genes that underlie common functional repertoires between nidopallium and neocortical layer IV.
Fig. 3.
Convergent gene coexpression modules marking chicken nidopallium and mouse layer IV. (A) Expression correlations between genes in significantly overlapping gene coexpression modules expressed in chicken nidopallium and mouse layer IV (Bonferroni-corrected P = 4.6 × 10−3, hypergeometric test). (B) RNA-ISH of convergent nidopallium markers. (C) RNA-ISH of convergent layer IV markers. (Scale bars: black, 500 μm; gray, 200 μm.) Cx, neocortex; en, entopallium; LIV, neocortical layer IV. (Scale bars: black, 500 μm; red, 200 μm.)
Discussion
Embryonic gene expression and supposed cellular migrations have supported the hypothesis that the neocortex derives from dorsal pallium, whereas the avian dorsal ventricular ridge (DVR, Fig. 1A) derives from the lateroventral pallium (6, 9). However, studies focusing on connectivity (17, 18), lamination (4, 9, 18), and the expression of some genes (8, 9, 15) note striking similarities between the ventral-pallial–derivative nidopallium and the neocortex, which were originally postulated to be homologous (9, 15, 17, 18).
We reveal species-specific and cross-species regional markers for pallial and striatal domains; for further exploitation, we provide a Web resource for adult forebrain expression in mouse and chicken at http://genserv.anat.ox.ac.uk/brainevo. This dataset not only will enable insights into telencephalic gene expression evolution, but also can facilitate future functional investigations. For example, the cross-species marker lists for striatum and hippocampus can readily be expanded using the advanced search feature on the website.
Using more than 5,000 genes, we find that adult gene expression does not cleanly and consistently fit any model based exclusively on developmental cell lineage or cellular characteristics. Unexpectedly, regions that have common developmental origins, but show functional divergence, exhibited no greater transcriptomic similarity than developmentally unrelated but functionally comparable regions. In contrast, striatum or hippocampus, regions with undisputed homology and similar functions in both species, each exhibited conserved groups of coexpressed genes and characteristic region-specific markers. Functionally analogous and hodologically similar ventral and dorsal pallial derivatives, such as nidopallium and neocortical layer IV (3, 4, 6), showed weak but significant convergence in gene expression despite their very different developmental trajectories and agrees with previous hodological and functional characteristics. Our results suggest that the pallium has undergone major transcriptomic reorganization, with traces of both molecular homoplasy and homology (24) (SI Appendix, Fig. S22). These results do not imply that there is no homology among other pallial sectors, only that homology is not a dominant factor in their adult gene expression patterns. Homology might have a greater impact on pallial gene expression if we had studied a greater number of smaller regions or even individual cells. If the function imparted by these recently derived pallial structures is governed largely by physiology and connectivity, intrapallial spatial constraints on adult gene expression may have been relaxed. Considering these results, subsequent investigations into the evolution of the neocortex should complement studies of homology based on cell lineage with multiple levels of information in various taxa toward a holistic understanding of how its molecular programs were repurposed (1, 2, 24), resulting in such cognitive convergence (5).
Materials and Methods
Brain Tissue.
Regions were dissected from brain sections derived from postnatal day 56 C57BL/6J male mice and 12-wk-old mixed-sex chickens.
RNA-seq.
Total RNA was extracted from dissected tissues pooled by region using a QIAGEN RNeasy Lipid Tissue Mini kit according to the manufacturer’s protocol. Per the standard Illumina RNA-seq library preparation protocol, this was poly(A)-selected, reverse transcribed to cDNA, and sequenced on the Illumina GAIIx.
Anatomical Validations.
We confirmed dissection accuracy by reviewing apparent regional marker genes in the Allen Mouse Brain Atlas and performing in situ hybridizations in chicken.
Expression Analyses.
We implemented Weighted Gene Coexpression Network Analyses, assessed cross-species marker gene overlaps, and constructed an estimator of transcriptome-wide specificity as described in the SI Appendix, Material and Methods.
A detailed SI Appendix, Material and Methods, accompanies this paper.
Supplementary Material
Acknowledgments
We thank E. D. Green and R. A. Chodroff for their roles in project conceptualization, T. M. Sirey and L. Bluy for RNA extractions, H. Ozel Abaan for the chicken poly(A) selection, the National Institutes of Health (NIH) Intramural Sequencing Center for library preparation and sequencing, J. L. Becker and M. W. Lesko for maintaining computational infrastructure, L. Goodstadt for ruffus, L. Puelles and C. Nellåker for helpful discussions, H. Kaune and A. F. P. Cheung for help in chicken brain collection, and M. Neubrandt for help in brain sectioning. This work was supported by the Intramural Program of the National Human Genome Research Institute, the Medical Research Council, Biotechnology and Biological Sciences Research Council, a Human Frontiers Science Program postdoctoral fellowship, Parkinson’s UK, the European Research Council, the Marshall Scholarship, New College, Oxford, the NIH-Oxford-Cambridge Scholars Program, and the Becas Chile Postdoctoral Fellowship.
Footnotes
Conflict of interest statement: E.H.M. is an employee of Illumina, Inc.
This article is a PNAS Direct Submission.
Data deposition: Species-specific and cross-species regional markers for pallial and striatal domains and for further exploitation of adult forebrain expression in mouse and chicken have been deposited at Mouse-Chicken Transcriptome Comparisons, http://genserv.anat.ox.ac.uk/brainevo. All sequence data have been deposited in the NCBI SRA under BioProject accession no. PRJNA210655.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307444110/-/DCSupplemental.
References
- 1.Molnár Z, Butler AB. Neuronal changes during forebrain evolution in amniotes: An evolutionary developmental perspective. Prog Brain Res. 2002;136:21–38. doi: 10.1016/s0079-6123(02)36005-9. [DOI] [PubMed] [Google Scholar]
- 2. Aboitiz F, Morales D, Montiel J (2003) The evolutionary origin of the mammalian isocortex: Towards an integrated developmental and functional approach. Behav Brain Sci 26:(5)535–552; discussion 552–585. [DOI] [PubMed]
- 3.Butler AB, Reiner A, Karten HJ. Evolution of the amniote pallium and the origins of mammalian neocortex. Ann N Y Acad Sci. 2011;1225:14–27. doi: 10.1111/j.1749-6632.2011.06006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Brzozowska-Prechtl A, Karten HJ. Laminar and columnar auditory cortex in avian brain. Proc Natl Acad Sci USA. 2010;107(28):12676–12681. doi: 10.1073/pnas.1006645107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Güntürkün O. The convergent evolution of neural substrates for cognition. Psychol Res. 2012;76(2):212–219. doi: 10.1007/s00426-011-0377-9. [DOI] [PubMed] [Google Scholar]
- 6.Puelles L. Pallio-pallial tangential migrations and growth signaling: New scenario for cortical evolution? Brain Behav Evol. 2011;78(1):108–127. doi: 10.1159/000327905. [DOI] [PubMed] [Google Scholar]
- 7.Nomura T, Takahashi M, Hara Y, Osumi N. Patterns of neurogenesis and amplitude of Reelin expression are essential for making a mammalian-type cortex. PLoS ONE. 2008;3(1):e1454. doi: 10.1371/journal.pone.0001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki IK, Kawasaki T, Gojobori T, Hirata T. The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev Cell. 2012;22(4):863–870. doi: 10.1016/j.devcel.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Jarvis ED, et al. Avian Brain Nomenclature Consortium Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci. 2005;6(2):151–159. doi: 10.1038/nrn1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103(46):17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA. 2010;107(28):12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belgard TG, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard A, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73(6):1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dugas-Ford J, Rowell JJ, Ragsdale CW. Cell-type homologies and the origins of the neocortex. Proc Natl Acad Sci USA. 2012;109(42):16974–16979. doi: 10.1073/pnas.1204773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo M, Broadbent N. Is the avian hippocampus a functional homologue of the mammalian hippocampus? Neurosci Biobehav Rev. 2000;24(4):465–484. doi: 10.1016/s0149-7634(00)00016-6. [DOI] [PubMed] [Google Scholar]
- 17.Karten HJ. The organization of the avian telencephalon and some speculations on the phylogeny of the amniote telencephalon. Ann N Y Acad Sci. 1969;167:164–179. [Google Scholar]
- 18.Karten HJ. Evolutionary developmental biology meets the brain: The origins of mammalian cortex. Proc Natl Acad Sci USA. 1997;94(7):2800–2804. doi: 10.1073/pnas.94.7.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karten HJ. Neocortical evolution: Neuronal circuits arise independently of lamination. Curr Biol. 2013;23(1):R12–R15. doi: 10.1016/j.cub.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Jabaudon D, Shnider SJ, Tischfield DJ, Galazo MJ, Macklis JD. RORβ induces barrel-like neuronal clusters in the developing neocortex. Cereb Cortex. 2012;22(5):996–1006. doi: 10.1093/cercor/bhr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tom Tang Y, et al. TAFA: A novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics. 2004;83(4):727–734. doi: 10.1016/j.ygeno.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez AS, Pieau C, Repérant J, Boncinelli E, Wassef M. Expression of the Emx-1 and Dlx-1 homeobox genes define three molecularly Distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: implications for the evolution of telencephalic subdivisions in amniotes. Development. 1998;125(11):2099–2111. doi: 10.1242/dev.125.11.2099. [DOI] [PubMed] [Google Scholar]
- 23.Teissier A, et al. A novel transient glutamatergic population migrating from the pallial-subpallial boundary contributes to neocortical development. J Neurosci. 2010;30(31):10563–10574. doi: 10.1523/JNEUROSCI.0776-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northcutt RG. Evolution of the vertebrate central nervous system: Patterns and processes. Am Zool. 1984;24(3):701–716. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.