Abstract
Most current diagnostic tests for tuberculosis do not reveal the species or strain of pathogen causing pulmonary infection, which can lead to inappropriate treatment regimens and the spread of disease. Here, we report an assay for mycobacterial strain assignment based on genetically conserved mycobacterial sulfatases. We developed a sulfatase-activated probe, 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one)–sulfate, that detects enzyme activity in native protein gels, allowing the rapid detection of sulfatases in mycobacterial lysates. This assay revealed that mycobacterial strains have distinct sulfatase fingerprints that can be used to judge both the species and lineage. Our results demonstrate the potential of enzyme-activated probes for rapid pathogen discrimination for infectious diseases.
Keywords: Mycobacterium tuberculosis, chemical biology, fluorescence, enzyme assay, hydrolase
Tuberculosis (TB) is caused by members of the Mycobacterium tuberculosis complex (MTBC), which include Mycobacterium bovis, Mycobacterium africanum, Mycobacterium microti, and the hundreds of related strains of Mycobacterium tuberculosis. Pulmonary disease can also be caused by other mycobacterial species (e.g., Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium septicum, Mycobacterium kansasii) (1, 2). There is an emerging appreciation of the phenotypic variation across mycobacterial species and strains (3, 4), including the many significant differences in virulence and immune responses to different members of the MTBC (4, 5). Identification of the infecting species is critical because therapeutics have variable efficacy against the various pathogenic mycobacteria. For example, most strains of M. bovis are resistant to pyrazinamide (6), and pulmonary disease caused by the M. avium complex requires different treatment than TB caused by members of the MTBC (2). New assays for TB should be sought that enable various species and strains to be distinguished unambiguously based on easily detected variations in phenotype.
Enzyme probes have been developed to study human pathogens (7, 8); however, to date, they have not been explored as a means for mycobacterial strain assignment. Of particular use are probes that are enzyme-activatable and fluorogenic, going from a dim to bright fluorescent state on substrate turnover. Sensitivity is augmented because one enzyme can modify many probes, dramatically increasing the fluorescence signal that can be measured in complex samples. Furthermore, enzyme-activated probes can be used in a variety of assay platforms, including molecular imaging and high-throughput screens (9–11).
In our research, we have targeted the mycobacterial sulfotransferases and sulfatases, which control the sulfation states of biomolecules (12–14) (Fig. 1). Sulfated compounds have established roles in metabolism and virulence in mycobacteria (13–15). Sulfatases are conserved across mycobacterial genomes (13), although their substrates, structures, and functions are largely unknown. Since the 1950s, there have been efforts to use sulfatase activity to detect mycobacteria (16). One assay uses a 2-wk incubation with live mycobacteria in media containing the colorimetric sulfatase substrate phenolphthalein disulfate (17). Species and strain identification using this and other sulfatase probes [e.g., p-nitrophenyl sulfate (p-NPS) (18) and 4-methylumbelliferyl sulfate (4-MUS) (19); structures are shown in SI Appendix, Fig. S1] have been largely ineffective due to slow reaction kinetics, a high detection limit with these substrates, and the lack of a simple platform to resolve strain-specific enzymes.
Fig. 1.
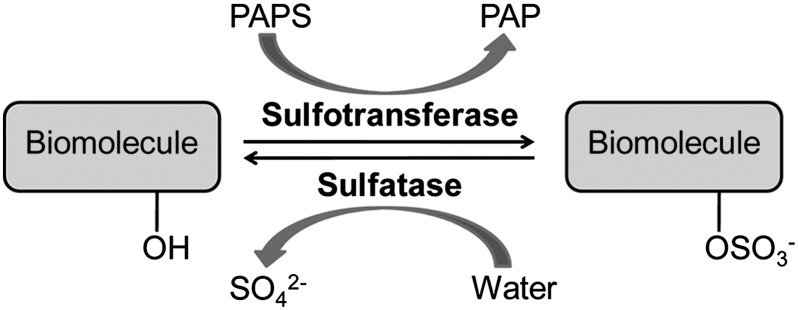
Sulfation is controlled by two classes of enzymes, sulfotransferases and sulfatases. Sulfotransferases catalyze sulfuryl group transfer to biomolecules using an activated donor called 3′-phosphoadenosine-5′-phosphosulfate (PAPS). Sulfate ester, or N-sulfate, hydrolysis is catalyzed by sulfatases. Sulfated biomolecules include steroids, lipids, carbohydrates, and proteins. PAP, 3′-phosphoadenosine-5′-phosphate.
In this paper, we describe a method for discriminating mycobacterial species and strains using the sulfatase-activated fluorophore 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO)–sulfate as an enzyme probe (Fig. 2). We separated mycobacterial lysates on native polyacrylamide gels and found, using DDAO fluorescence, unique sulfatase fingerprints that were dependent on the mycobacterial species and lineage. This in-gel assay was used to distinguish among different pathogenic species and M. tuberculosis strains rapidly, including a selection of M. tuberculosis strains from different geographical lineages.
Fig. 2.
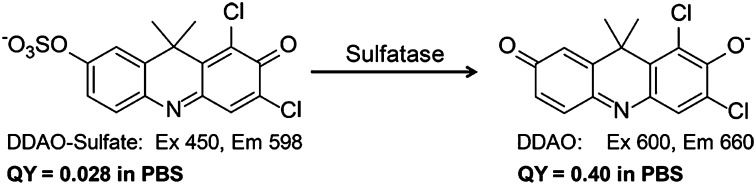
Sulfatase catalyzed hydrolysis of DDAO-sulfate to form DDAO. Hydrolysis of DDAO-sulfate results in red-shifted excitation (600 nm) and emission (660 nm) maxima. DDAO has a 14-fold increase in quantum yield (QY) compared with DDAO-sulfate.
Results and Discussion
Genetic Conservation of Mycobacterial Sulfatases.
Bioinformatics analysis of mycobacterial genomes suggests the presence of sulfatases from all three known classes. Types I and III were the most attractive enzyme targets to us because they catalyze the hydrolysis of sulfate esters (20–22) (Table 1) rather than the iron-dependent, oxidative process characteristic of the type II sulfatases (23, 24).
Table 1.
Genomic conservation of type I and III sulfatases
| Type 1 sulfatases |
Type III sulfatases |
|||||||
| AtsA | AtsB | AtsD | AtsF | AtsG | ZnSulf-A | ZnSulf-B | ZnSulf-C | |
| M. tuberculosis H37Rv* | Rv0711 | Rv3299 | Rv0663 | Rv3077 | Rv0296c | Rv3796 | Rv2407 | Rv3762c |
| M. tuberculosis CDC1551 | + | + | + | + | + | + | + | + |
| M. smegmatis | + | − | − | + | + | − | + | + |
| M. marinum | + | + | + | + | + | + | + | + |
| M. canetti | + | + | + | + | + | − | + | + |
| M. kansasii | + | + | + | + | + | − | + | + |
| M. avium | + | − | − | − | − | − | + | + |
| M. intracellulare | + | − | − | − | + | − | + | + |
| M. africanum | − | + | + | + | + | − | + | + |
| M. bovis (BCG) | + | + | + | + | + | + | + | + |
+, sulfatase gene is encoded in the genome; −, sulfatase gene is absent in the genome.
Rv numbers are provided for each sulfatase encoded in M. tuberculosis H37Rv.
Type I sulfatases have an unusual amino acid modification in their active site that confers catalytic activity. The formylglycine-generating enzyme (25–28) converts a cysteine residue within the consensus sequence -C-X-P-X-R- to a formylglycine residue. In water, the formylglycine aldehyde hydrates to form a geminal diol, which serves as a nucleophile during hydrolysis of the sulfate ester (20) (SI Appendix, Fig. S2). There are five putative type I sulfatases encoded in the M. tuberculosis H37Rv genome: Arylsulfatase (Ats)A (Rv0711), AtsB (Rv3299), AtsD (Rv0663), AtsF (Rv3077), and AtsG (Rv0296c).
Type III sulfatases are Zn2+-dependent metallo-β-lactamases. Enzymes of this type encode a conserved metal binding motif (21, 22), and there are three predicted in the genome of M. tuberculosis (Rv2407, Rv3796, and Rv3762c) (26). Rv3796 [zinc-dependent sulfatase-A (ZnSulf-A)] has a Zn2+-binding motif, but it was annotated previously as a type I sulfatase (13). Computational prediction of the protein structure using Phyre (protein homology/analogy recognition engine) (29) analysis supports the reclassification of ZnSulf-A as a member of the metallohydrolase superfamily.
Development of a Sulfatase-Activated Fluorophore.
The majority of enzyme-activated probes target proteases (30) and other hydrolases (31). In the latter category, examples abound for glycosidases, esterases, and phosphatases. Less explored are probes that target sulfatases (32). The hydrolysis of a sulfate ester can be used to modulate fluorescence, but the only scaffolds developed for fluorescent detection of this reaction are UV light-excited coumarins (e.g., 4-MUS) (33–35). As a commercially available fluorogenic probe for sulfatases, 4-MUS has been used extensively to assay sulfatase activity in vitro, but the high pKa of the hydrolysis product 4-methylumbelliferone (4-MU) and the high Km value of 4-MUS with most sulfatases limits its sensitivity and versatility.
We evaluated existing hydrolase-activated fluorophore scaffolds and selected a far-red fluorophore, DDAO (Fig. 2). DDAO’s excitation (600 nm) and emission (660 nm) maxima are near optimal for monitoring enzyme activity in biological samples, which can have high autofluorescence and light scattering at wavelengths below 600 nm. DDAO glycosides were reported as chromogenic glycosidase substrates for in vitro enzyme assays by Corey et al. (36). DDAO-β-d-galactoside has been used as a fluorogenic probe for monitoring β-galactosidase activity in vivo (37–39), and DDAO-phosphate has been used to monitor phosphatase activity in vitro (40).
To produce an analogous sulfatase substrate, DDAO-sulfate, the parent compound was sulfated using sulfur trioxide trimethylamine. In aqueous buffer, DDAO-sulfate (excitation = 450 nm, emission = 598 nm) had a large Stokes shift (150 nm) and the quantum yield (ΦDDAO-sulfate = 0.028) is decreased 14-fold compared with DDAO (ΦDDAO = 0.40). It is worth noting that this compound is chromogenic and converts from yellow to blue on sulfate ester hydrolysis. Although colorimetric assays are inherently less sensitive than fluorescent assays, monitoring hydrolysis by color change might be preferred for sample analysis in low-resource settings. [The concentration limit for a colorimetric assay can be calculated using the Beer–Lambert law. Assuming the lowest accurate absorption reading is 0.01, then the detection limit for DDAO is 244 nM.].
We validated DDAO-sulfate as an enzyme-activated probe by determining the kinetic parameters for this compound with commercially available sulfatases (SI Appendix, Table S1). Sulfatase activity from Aerobacter aerogenes was evaluated in buffer [50 mM Tris, 100 mM NaCl, 2.5 mM MgCl2, 250 μM CaCl2 (pH = 7.5)] at 37 °C, and the formation of DDAO was detected in real time. Hydrolysis of DDAO-sulfate in the absence of enzyme was negligible over the course of the experiment. The Km value for DDAO-sulfate was determined to be 164 ± 14 μM. For comparison, A. aerogenes sulfatase was evaluated using p-NPS and 4-MUS. The Km value for p-NPS was 1,800 ± 120 μM, and that for 4-MUS was 700 ± 160 μM (32).
The pKa of DDAO is 5.0 (41), and the formation of DDAO can be monitored continuously at any pH above this value. This is an important feature for a sulfatase probe because many sulfatases have an acidic pH optimum (12). In contrast, the pKa of 4-MU is 7.8 (42), and acidic enzyme reactions must be quenched by basification for detection of fluorescence. The high pKa of 4-MU prevents its use in continuous assays of enzymes at acidic pH. Sulfatases from three other sources were examined at their optimal pH, pH 5.0, and the Km values with DDAO-sulfate were determined (abalone entrails = 29 ± 3 μM, Helix pomatia = 13 ± 3 μM, Patella vulgata = 54 ± 0.00008 μM). By contrast, the reported Km values for P. vulgata with p-NPS and 4-MUS are 26,000 ± 2,000 μM and 2,500 ± 500 μM, respectively (34). Thus, DDAO-sulfate appears to be generally recognized as a substrate by a wide variety of sulfatases, which is an essential feature for screening purposes.
In Vitro Sulfatase Activity in Mycobacterial Lysates.
We used DDAO-sulfate to detect active sulfatases in lysates from 10 mycobacterial species. All mycobacteria were grown to midlogarithmic phase in vitro before lysis by mechanical disruption, as described in SI Appendix. Lysates were then incubated at 37 °C with 25 μM DDAO-sulfate. We examined lysates from M. tuberculosis (H37Rv, Erdman, and CDC1551), M. bovis [bacillus Calmette–Guérin (BCG)], M. kansasii, M. avium, M. intracellulare, M. africanum, Mycobacterium smegmatis, Mycobacterium marinum, Mycobacterium nonchromogenicum, and M. septicum. All species hydrolyzed DDAO-sulfate to a detectable amount of DDAO within 10 min (SI Appendix, Fig. S3). Most of these species were classified previously as lacking sulfatase activity (16). This activity was likely missed in earlier studies due to the use of substrates lacking the requisite sensitivity.
Sulfatase activity was confirmed in a parallel assay using 4-MUS. The signal-to-background ratio was lower, even though we used a higher probe concentration (1 mM 4-MUS). After a 30-min reaction, this assay confirmed sulfatase activity in every species tested (SI Appendix, Fig. S3). Although these two fluorescent assays demonstrated that sulfatase activity is detectable in mycobacterial lysates, they cannot be used to distinguish between MTBC species and M. tuberculosis strains accurately. The 96-well plate assay is also not suited to detecting activity from individual sulfatases (e.g., AtsB vs. AtsD).
An In-Gel Assay for Sulfatase Activity in Mycobacterial Lysates.
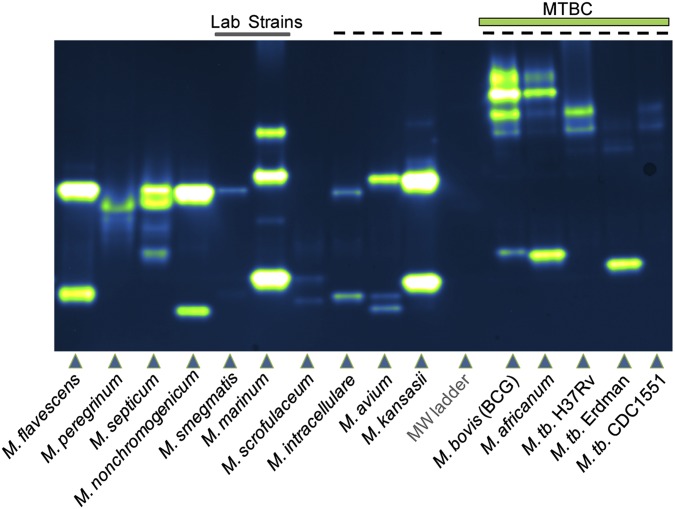
We developed an in-gel assay for profiling the activities of individual sulfatases that is analogous to zymography, a technique used for detecting protease activity following sample separation by polyarylamide gel electrophoresis (43, 44). Gels containing resolved mycobacterial lysates were soaked in 10–25 μM DDAO-sulfate for less than 15 min before imaging using a fluorescence scanner (excitation = 633 nm, emission = 670 bp 30). As shown in Fig. 3, every mycobacterial species analyzed possessed active sulfatases, as indicated by fluorescent bands, and discrete sulfatase “fingerprints” were identified that correlated with each species.
Fig. 3.
Sulfatase activity profiling of mycobacterial species. Lysates were resolved by native gel electrophoresis before exposure to 25 μM DDAO-sulfate in buffer [50 mM Tris (pH 7.5 at 37 °C); 100 mM NaCl; 250 μM MnCl2, MgCl2, and CaCl2]. Fluorescence imaging (excitation = 633, emission = 670 bp 30) revealed bands of DDAO fluorescence that are associated with sulfatase activity. Two laboratory strains, M. smegmatis and M. marinum, are indicated by a line at the top of the image. Species that can cause human pulmonary disease are indicated by a dashed line at the top of the gel. Members of the MTBC, including M. bovis, M. africanum, and M. tuberculosis, are labeled, with three strains of M. tuberculosis occupying the last three lanes. MW, molecular weight.
Some species (e.g., M. smegmatis, M. marinum) produced distinct patterns, whereas other species showed similar but not identical banding patterns (e.g., Mycobacterium flavescens, M. nonchromogenicum). We observed that the M. tuberculosis strains have unique banding patterns compared with other mycobacterial species, including M. avium, M. bovis (BCG), M. kansasii, M. intracellulare, and M. africanum. This is notable because each of these species causes pulmonary disease (45, 46). Among the M. tuberculosis strains, Erdman stood out because it possessed a bright band at a lower apparent molecular weight in addition to the higher molecular weight bands it shared with H37Rv and CDC1551.
We observed that M. avium and M. intracellulare produced different sulfatase patterns in the in-gel assay, and, as suggested by Falkinham (18), sulfatase activity might be used to distinguish between members of the M. avium complex. Falkinham demonstrated that sulfatase bands could be detected in resolved lysates of M. avium, M. intracellulare, and Mycobacterium scrofulaceum (18). In that work, sulfate ester hydrolysis was detected in a gel following an overnight incubation with 6-bromo-2-naphthyl sulfate, a chromogenic substrate. DDAO-sulfate enables more sulfatase bands to be observed in M. avium (three bands vs. one band) and M. scrofulaceum (two bands vs. one band). Both in-gel methods produced two bands in lysates from M. intracellulare. However, in the current work, sulfatase bands were observed after minutes of exposure to DDAO-sulfate, compared with the hours of exposure required for 6-bromo-2-naphthyl sulfate.
To explore the sensitivity of the DDAO-sulfate assay, we evaluated how much total protein lysate is required to detect sulfatase bands. The detection limit varied by species, and sulfatase bands could be observed with as little as 20 ng of total protein for M. africanum, M. marinum, M. kansasii, and M. flavescens. For M. nonchromogenicum and M. bovis (BCG), 40 ng of lysates was required for detection of sulfatases. A larger amount (200 ng) was loaded to detect M. tuberculosis (H37Rv and CDC1551). This requirement might reflect lower levels of sulfatase activity in these species or the longer processing time required to obtain sterile lysates from M. tuberculosis compared with other species (e.g., M. marinum).
There are 15 human sulfatases, most of which reside intracellularly (12), that we thought might produce fluorescent bands in our in-gel assay and thereby obscure bacterial strain assignments. To test their ability to hydrolyze DDAO-sulfate, we prepared and analyzed mammalian cell lysates from a variety of cell lines (e.g., CHO, Jurkat, RAW macrophages, COS-7, Rat-1 fibroblasts) (SI Appendix, Fig. S4). Furthermore, we evaluated M. tuberculosis-infected macrophages. We did not observe any fluorescent signal resulting from the activation of DDAO-sulfate by mammalian sulfatases. Most human sulfatases are lysosomal and, accordingly, have an acidic pH optimum; we examined sulfatase activity at pH 7.5, which may account for their lack of detection. Additionally, sulfatase activity may be lost or diminished during sample preparation for membrane-associated sulfatases, such as steroid sulfatase (47). The lack of detectable human sulfatase activity in the in-gel assay suggests that our assay will be useful for analyzing sulfatase activities in clinical samples, which could contain a mixture of human and mycobacterial cells.
Comparison of DDAO-Sulfate with p-NPS, 4-MUS, and 5-Bromo-4-Chloro-3-Indolyl Sulfate.
We used the in-gel assay to compare DDAO-sulfate with p-NPS, 4-MUS, and 5-bromo-4-chloro-3-indolyl sulfate (X-Sulf). We separated protein lysates on four identical native protein gels before a 15-min incubation with each substrate (SI Appendix, Fig. S5). DDAO-sulfate (25 μM) was able to reveal >45 fluorescent bands. X-Sulf (10 mM) rapidly produced six blue bands, whereas p-NPS (10 mM) revealed only four pale yellow bands. 4-MUS (1 mM) produced 20 fluorescent bands. DDAO-sulfate revealed more bands with 40-fold to 400-fold less probe. From this direct comparison, we conclude that DDAO-sulfate is rapidly activated by the greatest number of mycobacterial sulfatases and that it is the superior sulfatase substrate for mycobacterial species discrimination.
Identification of Active Sulfatases.
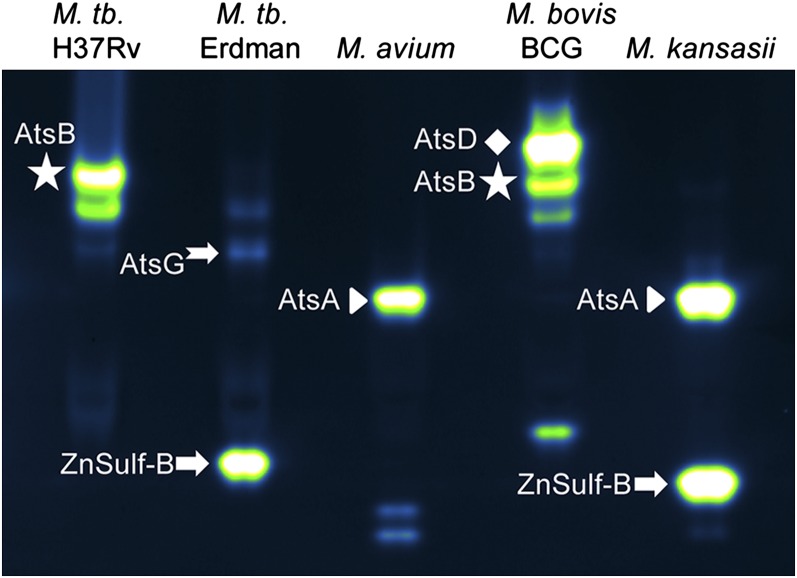
We used mass spectrometry (MS)-based proteomics methods to identify a subset of the sulfatases that hydrolyze DDAO-sulfate. We examined lysates from M. tuberculosis, M. bovis (BCG), M. kansasii, and M. avium. Crude lysate was partially enriched for sulfatases using an anion exchange column, resolved by native gel electrophoresis, and exposed to 10 μM DDAO-sulfate. Fluorescent sulfatase bands were excised and subjected to in-gel tryptic digestion and protein identification by MS. Sulfatase identities are indicated next to the fluorescent bands in the gel shown in Fig. 4, and identified peptides for each sulfatase are provided in SI Appendix, Table S4.
Fig. 4.
Assignment of sulfatase bands based on MS. Protein lysates were resolved and imaged. Fluorescent bands were excised and subjected to MS analysis. Sulfatase identities are indicated next to the fluorescent bands. Identified peptides for each sulfatase are provided in SI Appendix, Table S4.
In M. tuberculosis H37Rv, we identified AtsB (Rv3299) as the brightest fluorescent band. As described in SI Appendix, heterologous AtsB expression and transposon mutants confirmed that AtsB produces a set of fluorescent bands in M. tuberculosis lysates (SI Appendix, Fig. S6). We were able to isolate an M. tuberculosis Erdman band in the vicinity of the AtsB bands; this band had peptide matches with AtsG (Rv0296c). AtsB bands were also observed in lysates from M. africanum and M. bovis (BCG), and they were confirmed using MS analysis of the latter strain.
Notably, the M. tuberculosis Erdman strain stands out from two other laboratory strains, H37Rv and CDC1551, by the presence of a distinct sulfatase band at a lower apparent molecular weight. We used MS to determine that this activity is from the type III sulfatase ZnSulf-B (Rv2407). Although MS can be used to confirm which sulfatases are present in each species, we must emphasize that the banding patterns alone are sufficient to distinguish mycobacterial species rapidly.
AtsA (Rv0711) and AtsD (Rv0663) were also identified in mycobacterial lysates, as indicated in Fig. 4. ZnSulf-A (Rv3796) can hydrolyze DDAO-sulfate, but this enzyme was not observed in mycobacterial lysates.
Evaluation of Sulfatase Inhibitors.
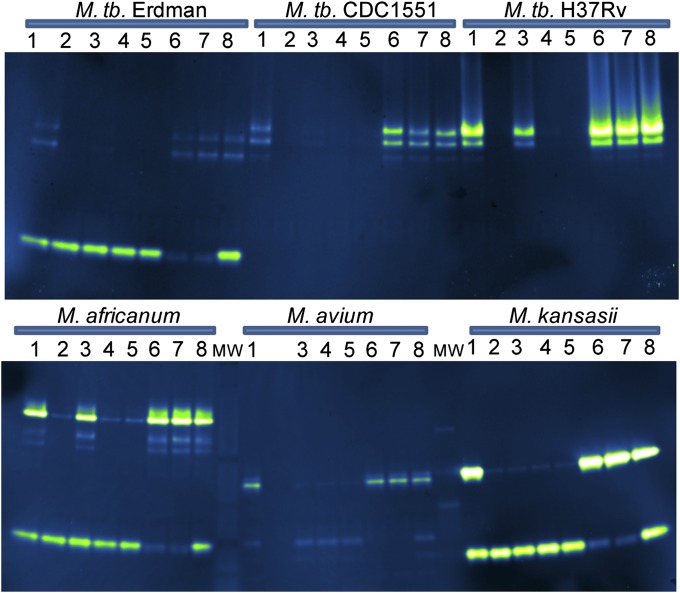
We sought further confirmation that the fluorescent bands observed were produced by active sulfatases using sulfatase inhibitors. First, we evaluated a set of arylsulfamates, which are known type I sulfatase inhibitors (48–51). Inhibition is irreversible and active site-directed, probably through covalent modification of the formylglycine residue (51). Four different arylsulfamates were examined for their ability to inhibit type I sulfatases in mycobacterial lysates. Lysates were incubated with 500 μM arylsulfamate for 1 h at 37 °C and then analyzed using the in-gel assay with DDAO-sulfate to detect remaining sulfatase activity (Fig. 5). Arylsulfamates vary in reactivity, with the highest reactivity assigned to compounds with lower pKa values for the parent phenol (51). Coumarin sulfamate (Stx64), a potent inhibitor of human steroid sulfatase (49), was the most reactive compound we examined, with a pKa of 7.8 for the parent phenol. In M. tuberculosis, incubation with Stx64 resulted in complete disappearance of the fluorescent bands associated with AtsB. Type I sulfatase bands in M. avium, M. africanum, and M. kansasii also disappeared. Inhibition of type I sulfatases was near complete following incubation with cyanosulfamate (pKa = 7.97) or chlorosulfamate (pKa = 9.41). Partial inhibition was observed for lysates incubated with phenylsulfamate, whose parent phenol has a pKa of 10 (51). The in vitro growth of M. tuberculosis was not inhibited by treatment with up to 1 mM arylsulfamate.
Fig. 5.
Evaluation of sulfatase inhibitors. Mycobacterial lysates (10 μg) were incubated for 1 h with phosphatase inhibitor mixture (lane 1), arylsulfamate (lanes 2–5), metal chelator (lanes 6–7), or water (lane 8). Lysates were resolved by electrophoresis before incubation with 10 μM DDAO-sulfate in 100 mM Tris⋅Cl (pH 7.5 at 37 °C) with 100 mM NaCl. Lane 1, phosphatase inhibitor mixture; lane 2, coumarin sulfamate (Stx64); lane 3, phenylsulfamate; lane 4, cyanosulfamate; lane 5, chlorosulfamate; lane 6, tetrakis-(2-pyridylmethyl)ethylenediamine; lane 7, 1,10-phenanthroline monohydrate; lane 8, water.
Arylsulfamates do not inhibit type III sulfatases, but metal chelators could affect substrate hydrolysis by sequestering the required Zn2+ ion. Accordingly, the fluorescence assigned to ZnSulf-B activity in M. tuberculosis Erdman and M. kansasii was diminished following treatment with metal chelators, including tetrakis-(2-pyridylmethyl)ethylenediamine (2 mM) and 1,10-phenanthroline monohydrate (4 mM) (Fig. 5). The fluorescence observed for the lowest apparent molecular weight sulfatase band in the lysates of both M. africanum and M. avium was diminished following treatment with either of these zinc chelators, indicating that these two species produce active type III sulfatases. DDAO-sulfate hydrolysis was unaffected by incubation with phosphatase inhibitors.
Lineage-Specific Sulfatase Patterns.
Clinical isolates and laboratory strains of M. tuberculosis have been assigned to six lineages based on comparative genomics. The lineages, which are named for particular geographic locations and human populations (52), are Indo-Oceanic, East Asian, East African-Indian, Euro-American, West African 1, and West African 2 (3). The two West African lineages include strains previously designated as M. africanum, which have an attenuated phenotype and are genetically related to M. bovis (53). The Beijing strains, some of which have been linked to higher virulence and drug resistance (54), are included in the East Asian lineage. H37Rv, CDC1551, and Erdman are assigned to the Euro-American lineage.
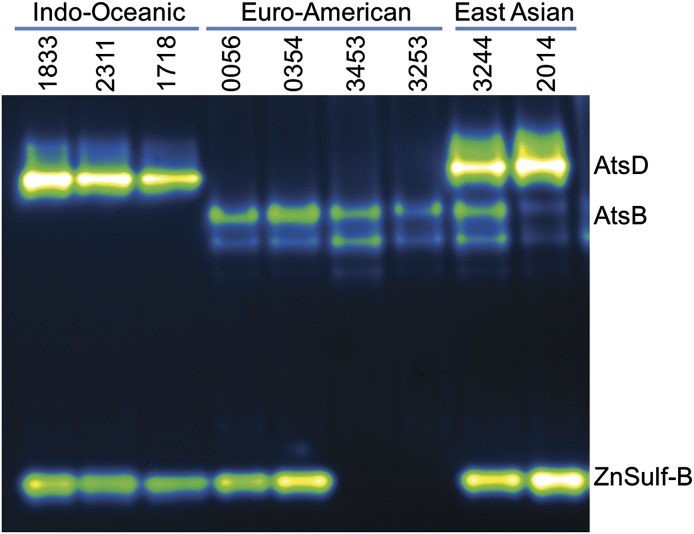
Using the in-gel assay, we first examined nine different strains from three lineages: Indo-Oceanic, Euro-American, and East Asian. These strains were derived from clinical samples collected at the University of California, San Francisco. For each of these strains, we observed lineage-specific banding patterns (Fig. 6). Resolved lysates from the Indo-Oceanic lineage lacked bands associated with AtsB but clearly displayed bands associated with AtsD and ZnSulf-B. The Euro-American lineage had AtsB activity and lacked AtsD activity. The ZnSulf-B activity varied by Euro-American strain, which is consistent with what we have observed for other members of this lineage (H37Rv, Erdman, and CDC1551). We examined strains from the East Asian lineage and observed activity in each strain for three different sulfatases: AtsB, AtsD, and ZnSulf-B.
Fig. 6.
M. tuberculosis strain-specific variation in sulfatase activity. Protein lysates were isolated from three distinct M. tuberculosis lineages: Indo-Oceanic, Euro-American, and East Asian. Lysates were resolved and imaged to reveal sulfatase activity. The three Indo-Oceanic samples, nos. 1833, 2311, and 1718, have bands that correspond to AtsD (Rv0633) and type III sulfatase (ZnSulf-B; Rv2407) activity. The Euro-American samples, nos. 0056, 0354, 3453, and 3253, all lack the highest AtsD band but have bands that match the pattern observed for AtsB (Rv3299). Two of the Euro-American samples, nos. 0056 and 0354, have the lowest band, corresponding to ZnSulf-B (Rv2407) activity. The East Asian strains, nos. 3244 and 2014, have bands corresponding to AtsD, AtsB, and ZnSulf-B activity.
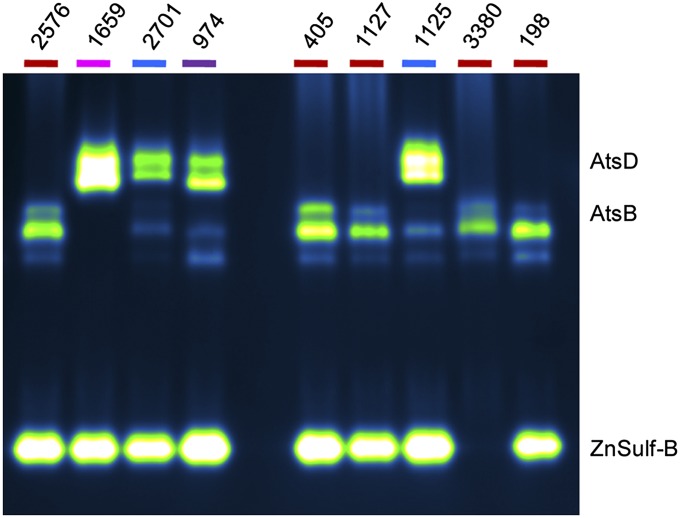
South Africa has a high TB burden, with patients infected by M. tuberculosis strains from several different lineages. The University of Stellenbosch, near Cape Town, has a collection of clinical isolates with a high degree of strain diversity (55, 56). Lysates derived from a selection of strains were evaluated using the in-gel assay (Fig. 7), and we found that the lineage-specific patterns are similar to those observed with the Indo-Oceanic, Euro-American, and East Asian strains shown in Fig. 6. For example, the M. tuberculosis strains from the East Asian lineage (nos. 1125 and 2701) displayed sulfatase activity at locations corresponding to AtsB, AtsD, and ZnSulf-B. Strain no. 198 has a drug-resistant phenotype (57) but still retained sulfatase bands consistent with other members of the Euro-American lineage (e.g., strain nos. 2576, 405, 1127, and 3380). These trends were observed in a subsequent set of analyzed strains (SI Appendix, Fig. S7). Our findings indicate that both the mycobacterial species and M. tuberculosis lineage can be assigned based on the sulfatase fingerprints revealed by our assay. For clinical settings, the ability of DDAO-sulfate to reveal lineage-specific patterns of sulfatase activity could be very powerful because, for human pulmonary disease, rapid species and strain assignment are essential for effective treatment.
Fig. 7.
M. tuberculosis strains from South Africa have varying patterns of sulfatase activity. The name of each lineage is color-coded: Euro-American (red; nos. 2576, 405, 1127, 3380, and 198), Indo-Oceanic (pink; no. 1659), East Asian (blue; nos. 2701 and 1125), and India and East Africa (purple; no. 974). Additional strains were evaluated and are shown in SI Appendix, Fig. S7.
Conclusion
We have described an activity-based sulfatase probe, DDAO-sulfate, that can be used in the context of an in-gel assay to discriminate mycobacterial species and strains rapidly. The assay is fast (5–15 min); requires only micromolar probe concentrations; and detects a variety of type I and III sulfatases, including AtsA, AtsB, AtsD, AtsG, and ZnSulf-B. Mycobacterial species produced fluorescent bands that varied in intensity and position on the gel, and the pattern of these bands might be used to identify a particular strain or species. As further evidence that mycobacterial sulfatases are a good target for clinical assays, M. avium, M. intracellulare, and M. kansasii (45, 46) could be distinguished from M. tuberculosis based on patterns of sulfatase activity.
Although genome sequencing provides information on the sulfatase genes present in various species, the activity of each sulfatase is more informative. For example, both M. tuberculosis strains H37Rv and CDC1551 encode nine sulfatases, yet only AtsB activity was observed in the in-gel assay. In contrast, the M. tuberculosis Erdman fingerprint revealed AtsB and ZnSulf-B activity. Similarly, the genomic encoding of AtsD is not predictive of activity in lysates, as observed using the in-gel assay to examine sulfatases in various strains of M. tuberculosis.
Strain variation among members of the MTBC is an area of active exploration (3, 4, 52). In addition to evaluating laboratory strains of M. tuberculosis (H37Rv, CDC1551, and Erdman), we examined a number of lysates derived from strains of M. tuberculosis collected from patients. Sulfatase patterns varied significantly among different lineages of M. tuberculosis and produced distinct fluorescent patterns for three lineages: Indo-Oceanic, East Asian, and Euro-American. Future work will focus on evaluating an expanded set of clinical isolates for sulfatase activities and in-gel activity patterns. Further efforts will focus on developing DDAO-sulfate assays that can be used by clinicians as diagnostic tools for TB.
Materials and Methods
Preparation of Mycobacterial Lysates.
Mycobacterial cultures were grown to log phase (OD600 = 0.5–1.5) before harvesting by centrifugation at 4 °C. The supernatant was discarded, and the pellet was frozen (−20 °C). Pellets were put in 50 mM Tris⋅Cl (pH 7 at 4 °C), 100 mM NaCl, 0.5 mM CaCl2, and 0.5 mM MgCl2, plus protease inhibitors. Cells were lysed by mechanical disruption on a bead-beating instrument. Crude lysates was clarified by centrifugation (10 min, 16,000 × g, 4 °C). Mycobacterial cell pellets from BioSafety level 3 pathogens (e.g., M. tuberculosis) were sterilized by filtration twice through a 0.2-μm filter.
In-Gel Activity Assay.
Mycobacterial lysates (1–20 μg) were resolved by native gel electrophoresis (4–15% Tris⋅Cl Criterion gel; Bio-Rad) in 1× Tris⋅glycine buffer. Gels were run at 180 V for 55 min on top of ice before soaking in 50 mM Tris (pH 7.5 at 37 °C); 100 mM NaCl; and 250 μM MnCl2, MgCl2, and CaCl2, with 10–25 μM DDAO-sulfate. Fluorescence of DDAO was detected on a fluorescence scanner (Typhoon 9410 Variable Mode Imager, GE Healthcare; excitation = 633 nm, emission = 670 bp 30, 100-μm resolution). Images were analyzed and false-colored (Green-Fire-Blue) using ImageJ (58).
Further information on synthetic methods, mycobacterial growth, lysate preparation, and sulfatase assays is available in SI Appendix. Ethics approval to collect strains of M. tuberculosis was obtained by Stellenbosch University from the Health Research Ethics Committee (N10/04/126).
Supplementary Material
Acknowledgments
We thank Jason Rush, Shana Topp, Kim Sogi, Kanna Palaniappan, Sarah Gilmore, Sloan Siegrist, Ed Ballister, and Prof. Sarah Stanley [University of California (UC), Berkeley] for valuable discussions. We thank Prof. Midori Kato-Maeda and Jordan Rose (UC, San Francisco) for beneficial discussions and for providing M. tuberculosis strains. Douglas Fox and Ellie Smith made critical suggestions on the manuscript. Funding was generously provided by National Institutes of Health (NIH) Postdoctoral Fellowship F32CA136109 (to K.E.B.), a Center for Emerging and Neglected Diseases fellowship (to B.M.S.), and NIH Grant AI51622 (to C.R.B.). Disease diagnostic training (K.E.B.) was provided by PATH and the University of Washington, Seattle. MS sample preparation and analysis were provided by Dr. Lori Kohlstaedt and the Vincent J. Coates Proteomics/Mass Spectrometry Laboratory (UC, Berkeley).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222041110/-/DCSupplemental.
References
- 1.Katoch VM. Infections due to non-tuberculous mycobacteria (NTM) Indian J Med Res. 2004;120(4):290–304. [PubMed] [Google Scholar]
- 2.McGrath EE, Anderson PB. The therapeutic approach to non-tuberculous mycobacterial infection of the lung. Pulm Pharmacol Ther. 2010;23(5):389–396. doi: 10.1016/j.pupt.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 2010;7(1):e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7(5):328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 5.Portevin D, Gagneux S, Comas I, Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7(3):e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konno K, Feldmann FM, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95(3):461–469. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 7.Raynaud C, Etienne G, Peyron P, Lanéelle M-A, Daffé M. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology. 1998;144(Pt 2):577–587. doi: 10.1099/00221287-144-2-577. [DOI] [PubMed] [Google Scholar]
- 8.Puri AW, Bogyo M. Using small molecules to dissect mechanisms of microbial pathogenesis. ACS Chem Biol. 2009;4(8):603–616. doi: 10.1021/cb9001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem Biol. 2008;3(3):142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Razgulin A, Ma N, Rao J. Strategies for in vivo imaging of enzyme activity: An overview and recent advances. Chem Soc Rev. 2011;40(7):4186–4216. doi: 10.1039/c1cs15035a. [DOI] [PubMed] [Google Scholar]
- 11. Weissleder R, Ross BD, Rehemtulla A, Gambhir SS (2010) Molecular Imaging: Principles and Practice (People’s Medical Publishing House, Shelton, CT.
- 12.Hanson SR, Best MD, Wong CH. Sulfatases: Structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed. 2004;43(43):5736–5763. doi: 10.1002/anie.200300632. [DOI] [PubMed] [Google Scholar]
- 13.Mougous JD, Green RE, Williams SJ, Brenner SE, Bertozzi CR. Sulfotransferases and sulfatases in mycobacteria. Chem Biol. 2002;9(7):767–776. doi: 10.1016/s1074-5521(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 14.Schelle MW, Bertozzi CR. Sulfate metabolism in mycobacteria. ChemBioChem. 2006;7(10):1516–1524. doi: 10.1002/cbic.200600224. [DOI] [PubMed] [Google Scholar]
- 15.Bhave DP, Muse WB, 3rd, Carroll KS. Drug targets in mycobacterial sulfur metabolism. Infect Disord Drug Targets. 2007;7(2):140–158. doi: 10.2174/187152607781001772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lévy-Frébault VV, Portaels F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42(2):315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 17.Whitehead JEM, Wildy P, Engbaek HC. Arylsulphatase activity of mycobacteria. J Pathol Bacteriol. 1953;65(2):451–460. doi: 10.1002/path.1700650217. [DOI] [PubMed] [Google Scholar]
- 18.Falkinham JO., 3rd Arylsulfatase activity of Mycobacterium avium, M. intracellulare, and M. scrofulaceum. Int J Syst Bacteriol. 1990;40(1):66–70. doi: 10.1099/00207713-40-1-66. [DOI] [PubMed] [Google Scholar]
- 19.Grange JM, Clark K. Use of umbelliferone derivatives in the study of enzyme activities of mycobacteria. J Clin Pathol. 1977;30(2):151–153. doi: 10.1136/jcp.30.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boltes I, et al. 1.3 A structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure. 2001;9(6):483–491. doi: 10.1016/s0969-2126(01)00609-8. [DOI] [PubMed] [Google Scholar]
- 21.Hagelueken G, et al. The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc Natl Acad Sci USA. 2006;103(20):7631–7636. doi: 10.1073/pnas.0510501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bebrone C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 2007;74(12):1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Kahnert A, Kertesz MA. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J Biol Chem. 2000;275(41):31661–31667. doi: 10.1074/jbc.M005820200. [DOI] [PubMed] [Google Scholar]
- 24.Müller I, et al. Crystal structure of the alkylsulfatase AtsK: Insights into the catalytic mechanism of the Fe(II) α-ketoglutarate-dependent dioxygenase superfamily. Biochemistry. 2004;43(11):3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- 25.Dierks T, et al. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(α)-formylglycine generating enzyme. Cell. 2003;113(4):435–444. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 26.Carlson BL, et al. Function and structure of a prokaryotic formylglycine-generating enzyme. J Biol Chem. 2008;283(29):20117–20125. doi: 10.1074/jbc.M800217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosma MP, et al. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113(4):445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 28.Rush JS, Bertozzi CR. New aldehyde tag sequences identified by screening formylglycine generating enzymes in vitro and in vivo. J Am Chem Soc. 2008;130(37):12240–12241. doi: 10.1021/ja804530w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.Tung CH. Fluorescent peptide probes for in vivo diagnostic imaging. Biopolymers. 2004;76(5):391–403. doi: 10.1002/bip.20139. [DOI] [PubMed] [Google Scholar]
- 31. Grimm JB, Heckman LM, Lavis LD (2013) The chemistry of small-molecule fluorogenic probes. Progress in Molecular Biology and Translational Science (Academic, Oxford, UK), Vol 113, pp 1–34. [DOI] [PubMed]
- 32.Rush JS, Beatty KE, Bertozzi CR. Bioluminescent probes of sulfatase activity. ChemBioChem. 2010;11(15):2096–2099. doi: 10.1002/cbic.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed V, Ispahany M, Ruttgaizer S, Guillemette G, Taylor SD. A fluorogenic substrate for the continuous assaying of aryl sulfatases. Anal Biochem. 2005;340(1):80–88. doi: 10.1016/j.ab.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Sherman WR, Stanfield EF. Measurement of the arylsulphatase of Patella vulgata with 4-methylumbelliferone sulphate. Biochem J. 1967;102(3):905–909. doi: 10.1042/bj1020905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bilban M, Billich A, Auer M, Nussbaumer P. New fluorogenic substrate for the first continuous steroid sulfatase assay. Bioorg Med Chem Lett. 2000;10(9):967–969. doi: 10.1016/s0960-894x(00)00144-x. [DOI] [PubMed] [Google Scholar]
- 36.Corey PF, Trimmer RW, Biddlecom WG. A new chromogenic beta-galactosidase substrate—7-beta-D-galactopyranosyloxy-9,9-dimethyl-9h-acridin-2-one. Angew Chem Int Edit USA. 1991;30(12):1646–1648. [Google Scholar]
- 37.Tung CH, et al. In vivo imaging of beta-galactosidase activity using far red fluorescent switch. Cancer Res. 2004;64(5):1579–1583. doi: 10.1158/0008-5472.can-03-3226. [DOI] [PubMed] [Google Scholar]
- 38.Josserand V, Texier-Nogues I, Huber P, Favrot MC, Coll JL. Non-invasive in vivo optical imaging of the lacZ and luc gene expression in mice. Gene Ther. 2007;14(22):1587–1593. doi: 10.1038/sj.gt.3303028. [DOI] [PubMed] [Google Scholar]
- 39.Zinselmeyer BH, Beggbie N, Uchegbu IF, Schätzlein AG. Quantification of β-galactosidase activity after non-viral transfection in vivo. J Control Release. 2003;91(1-2):201–208. doi: 10.1016/s0168-3659(03)00234-7. [DOI] [PubMed] [Google Scholar]
- 40.Leira F, Vieites JM, Vieytes MR, Botana LM. Characterization of 9H-(1,3-dichlor-9, 9-dimethylacridin-2-ona-7-yl)-phosphate (DDAO) as substrate of PP-2A in a fluorimetric microplate assay for diarrhetic shellfish toxins (DSP) Toxicon. 2000;38(12):1833–1844. doi: 10.1016/s0041-0101(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 41.Warther D, et al. Live-cell one- and two-photon uncaging of a far-red emitting acridinone fluorophore. J Am Chem Soc. 2010;132(8):2585–2590. doi: 10.1021/ja9074562. [DOI] [PubMed] [Google Scholar]
- 42.Sun WC, Gee KR, Haugland RP. Synthesis of novel fluorinated coumarins: Excellent UV-light excitable fluorescent dyes. Bioorg Med Chem Lett. 1998;8(22):3107–3110. doi: 10.1016/s0960-894x(98)00578-2. [DOI] [PubMed] [Google Scholar]
- 43.Snoek-van Beurden PAM, Von den Hoff JW. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques. 2005;38(1):73–83. doi: 10.2144/05381RV01. [DOI] [PubMed] [Google Scholar]
- 44.Lantz MS, Ciborowski P. Zymographic techniques for detection and characterization of microbial proteases. Methods Enzymol. 1994;235:563–594. doi: 10.1016/0076-6879(94)35171-6. [DOI] [PubMed] [Google Scholar]
- 45.Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10(2):103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 46.Warren RM, et al. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S Afr Med J. 1996;86(1):45–49. [PubMed] [Google Scholar]
- 47.Reed MJ, Purohit A, Woo LWL, Newman SP, Potter BVL. Steroid sulfatase: Molecular biology, regulation, and inhibition. Endocr Rev. 2005;26(2):171–202. doi: 10.1210/er.2004-0003. [DOI] [PubMed] [Google Scholar]
- 48.Bojarová P, Williams SJ. Aryl sulfamates are broad spectrum inactivators of sulfatases: Effects on sulfatases from various sources. Bioorg Med Chem Lett. 2009;19(2):477–480. doi: 10.1016/j.bmcl.2008.11.059. [DOI] [PubMed] [Google Scholar]
- 49.Woo LL, Purohit A, Malini B, Reed MJ, Potter BVL. Potent active site-directed inhibition of steroid sulphatase by tricyclic coumarin-based sulphamates. Chem Biol. 2000;7(10):773–791. doi: 10.1016/s1074-5521(00)00023-5. [DOI] [PubMed] [Google Scholar]
- 50.Schelwies M, et al. Glucosamine-6-sulfamate analogues of heparan sulfate as inhibitors of endosulfatases. ChemBioChem. 2010;11(17):2393–2397. doi: 10.1002/cbic.201000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bojarová P, et al. Direct evidence for ArO-S bond cleavage upon inactivation of Pseudomonas aeruginosa arylsulfatase by aryl sulfamates. ChemBioChem. 2008;9(4):613–623. doi: 10.1002/cbic.200700579. [DOI] [PubMed] [Google Scholar]
- 52.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(8):2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bentley SD, et al. (2012) The genome of Mycobacterium africanum West African 2 reveals a lineage-specific locus and genome erosion common to the M. tuberculosis complex. PLoS Neglect Trop D 6(2):e1552. [DOI] [PMC free article] [PubMed]
- 54.Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10(2):103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 55.Warren RM, et al. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S Afr Med J. 1996;86(1):45–49. [PubMed] [Google Scholar]
- 56.Richardson M, et al. Historic and recent events contribute to the disease dynamics of Beijing-like Mycobacterium tuberculosis isolates in a high incidence region. Int J Tuberc Lung Dis. 2002;6(11):1001–1011. [PubMed] [Google Scholar]
- 57.Victor TC, et al. Spread of an emerging Mycobacterium tuberculosis drug-resistant strain in the western Cape of South Africa. Int J Tuberc Lung Dis. 2007;11(2):195–201. [PubMed] [Google Scholar]
- 58. Rasband WS (1997–2007) ImageJ (US National Institutes of Health, Bethesda). Available at http://imagej.nih.gov/ij/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.