Abstract
The concentration of ribonucleoside triphosphates (rNTPs) in cells is far greater than the concentration of deoxyribonucleoside triphosphates (dNTPs), and this pool imbalance presents a challenge for DNA polymerases (Pols) to select their proper substrate. This report examines the effect of nucleotide pool imbalance on the rate and fidelity of the Escherichia coli replisome. We find that rNTPs decrease replication fork rate by competing with dNTPs at the active site of the C-family Pol III replicase at a step that does not require correct base-pairing. The effect of rNTPs on Pol rate generalizes to B-family eukaryotic replicases, Pols δ and ε. Imbalance of the dNTP pool also slows the replisome and thus is not specific to rNTPs. We observe a measurable frequency of rNMP incorporation that predicts one rNTP incorporated every 2.3 kb during chromosome replication. Given the frequency of rNMP incorporation, the repair of rNMPs is likely rapid. RNase HII nicks DNA at single rNMP residues to initiate replacement with dNMP. Considering that rNMPs will mark the new strand, RNase HII may direct strand-specificity for mismatch repair (MMR). How the newly synthesized strand is recognized for MMR is uncertain in eukaryotes and most bacteria, which lack a methyl-directed nicking system. Here we demonstrate that Bacillus subtilis incorporates rNMPs in vivo, that RNase HII plays a role in their removal, and the RNase HII gene deletion enhances mutagenesis, suggesting a possible role of incorporated rNMPs in MMR.
The structures of ribonucleoside triphosphates (rNTPs) and deoxyribonucleoside triphosphates (dNTPs) differ by a single atom, yet each must be distinguished by DNA and RNA polymerases (Pols). This is more challenging for DNA Pols because the intracellular concentration of rNTPs is 10–100-fold higher than that of dNTPs (1–3). DNA is more stabile than RNA, and rNMP residues in DNA could lead to spontaneous strand breaks. Hence, it is important to genomic integrity that DNA Pols exclude rNMPs from incorporation into the genome (1, 4).
Structural studies reveal that DNA Pols distinguish ribo and deoxyribo sugars via a “steric gate,” in which a bulky residue or main chain atom sterically occludes binding of the ribo 2′OH (5, 6). However, the single-atom difference between rNTPs and dNTPs imposes an upper limit to sugar recognition, and thus DNA Pols incorporate rNMPs at a low frequency. For example, studies in yeast demonstrate that rNMPs are incorporated in vivo, and studies in vitro demonstrate a frequency of rNMP incorporation predicting that 10,000 rNMPs or more may be incorporated each replication cycle (2, 7, 8). Given their abundant incorporation, there are probably multiple pathways to remove rNMPs, as their persistence is associated with genomic instability in yeast (7, 9).
The current study reconstitutes the Escherichia coli replisome and examines the cost of rNTP/dNTP nucleotide pool imbalance on the frequency of rNMP incorporation and the rate of fork progression. We find that a nucleotide pool imbalance slows the replisome two- to threefold by competing with dNTPs at the active site of the Pol III replicase at a step that does not require correct base-pairing. We determine the rNMP incorporation frequency and estimate that at intracellular concentrations of nucleotides, Pol III incorporates one rNMP every 2.3 kb, for a total of about 2,000 rNMPs per daughter chromosome. We also find that the replisome pauses 4–30-fold at a template rNMP, which could possibly promote genomic instability if rNMPs were not removed before the next round of replication.
Despite detrimental aspects of the rNTP/dNTP pool imbalance, there exists a potential benefit of rNMP incorporation: It has been proposed that replicative Pols may incorporate rNMPs for particular tasks, one of which may mark the newly replicated DNA strand for mismatch repair (MMR) (2). Some bacteria (e.g., E. coli) direct MMR using a DNA methylase and MutH endonuclease to recognize and nick the newly replicated strand (1, 10, 11). However, eukaryotes and most bacteria (e.g., Bacillus subtilis) do not use methyl direction for MMR. Because rNMP incorporation is specific to the newly synthesized strand, the nick generated during its repair could be recruited to direct MMR. We investigated this hypothesis in B. subtilis, a Gram-positive bacterium that lacks a methyl-directed MMR system, and report an increase in mutagenesis after deletion of two RNase H genes.
Results
Replisome Progression Is Slowed by rNTPs.
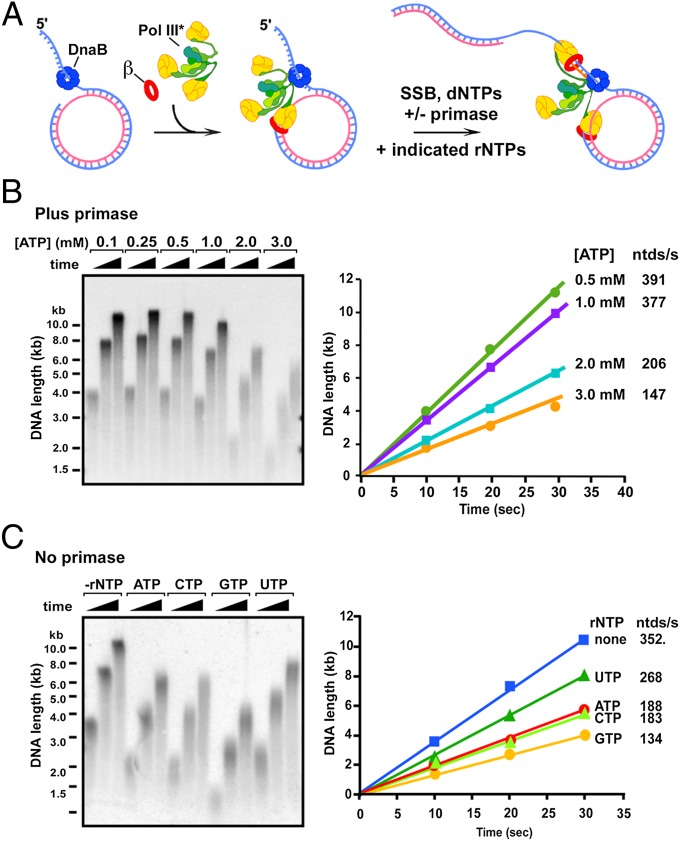
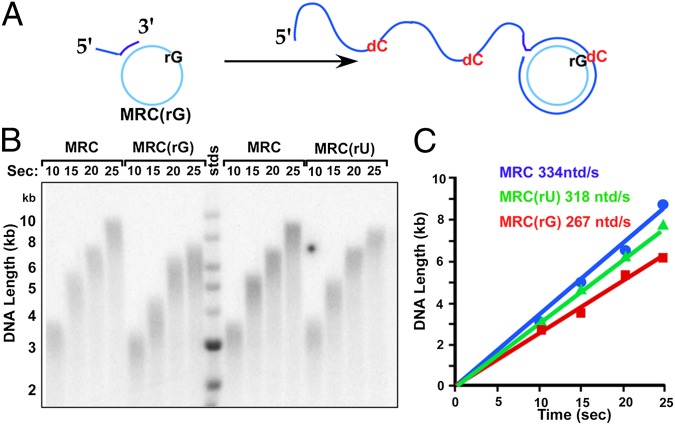
To study the effect of rNTPs on the rate of the E. coli replisome, we used a 100mer rolling-circle DNA that lacks dTMP on a single strand, allowing specific labeling of the leading strand. The replisome is assembled by adding DnaB helicase for 30s (Fig. 1A), and then Pol III* and the β clamp are added along with dCTP and dGTP for 1 min to form Pol III*-β on DNA. Pol III* contains three Pol III cores connected to the three τ subunits of the clamp loader [(Pol III core)3τ3δδ’χψ]; two Pol III cores are used on the lagging strand (12, 13). Replication is then initiated on adding SSB, primase, 32P-α-dTTP, dATP, and the presence or absence of rNTPs. Because of the rapid speed of the replisome [650 nucleotides (ntd)/s at 37 °C] (14), studies are performed at 25 °C to enable accurate rate measurements of replisome progression.
Fig. 1.
rNTPs decrease the rate of the replisome. (A) Scheme of the rolling circle assay. (B) ATP is titrated into replication reactions. (Left) Alkaline agarose gel. (Right) Quantitation. (C) Leading strand replisome reactions (i.e., no primase) are performed with no additional rNTP or with 3 mM of the indicated rNTP. (Left) Alkaline gel. (Right) Quantitation. Time points were 10, 20, and 30 s.
In the experiment of Fig. 1B, we examined the rate of the E. coli replisome at different concentrations of rATP. As rATP is increased to 3 mM, the rate of the replisome is diminished about 2.7-fold. A simple explanation is that rATP competes with dATP for the active site of the Pol, lowering its rate of forward progression. However, primase and DnaB helicase also use rNTP substrates and could conceivably be targets of rATP competition with other nucleotides. To determine whether primase underlies the rATP-mediated rate decrease, we omitted primase to eliminate lagging strand synthesis and analyzed the effect of rATP on progression of the leading strand replisome. The result shows that 3 mM rATP decreases the rate of the leading strand replisome to a similar extent as the coupled leading/lagging strand replisome (Fig. 1C, compare lanes 1–3 with 4–6). Hence, primase would not appear to underlie the rATP-mediated decrease in rate of fork progression. Each of the other three rNTPs also slow the leading strand replisome (Fig. 1 C and D). rUTP is the least inhibitory, possibly because of the molecular difference in its base in addition to the 2′OH. The DnaB helicase cannot be omitted from replisome reactions, but DnaB is unlikely to be affected by the rNTP/dNTP ratio, as it only uses rNTPs and is equally active with each rNTP (15). We will show later that rNTPs target Pol III*-β in experiments with no DnaB or primase.
As a further test of the theory that the decrease in replisome rate is a result of competition of rNTPs with dNTPs, we increased the concentration of dNTPs relative to rNTP, which should lower the severity of the competition and result in a faster rate. As predicted, an increase in dNTP concentration gives a partial relief of rNTP-mediated replication fork slowdown (Fig. S1). It is interesting to note that the rolling circle substrate has no dTMP in the leading strand template, yet rATP inhibits replisome rate (Fig. 1B). This observation suggests that rNTPs bind and compete for dNTPs at the Pol active site at a step that does not require correct base-pairing.
To examine whether rATP inhibition may be a result of mismatch incorporation, we used a mutant Pol III* lacking 3′-5′ proofreading activity. If mismatches are formed, the replisome should be further slowed by the mutant Pol III*, which cannot excise a mismatch. However, the result showed a similar rate of replisome advance as use of wt Pol III*, indicating that mismatched incorporation of rNMPs does not underlie the effect of rNTPs on replisome rate (Fig. S2). We also examined rADP, rAMP, and cAMP (Fig. S3). rADP inhibits the rate of fork progression about twofold more than rATP, whereas rAMP does not inhibit at all. cAMP was not effective either. Because replisome rate is affected by rADP more than by rATP, it is possible that the energy state of the cell influences the rate of replication. We presume the dADP inhibition is a result of competition for the Pol active site; this interesting observation will require further study. Because DNA Pols have only a single NTP binding site, one may predict that 3 mM dATP (with no rNTPs) will also compete with correct dNTPs and result in a rate decrease. Indeed, 3 mM dATP gave a similar rate effect as 3 mM rATP (Fig. S4). This report focuses on the effects of rNTPs, as rNTPs create the nucleotide pool imbalance in vivo.
Pol III Holoenzyme Is the Target of rNTP Slow Down at the Replication Fork.
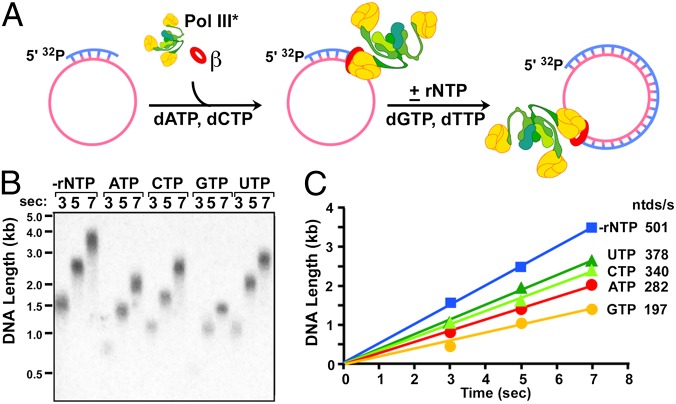
Although there have been many studies of rNTP incorporation by DNA Pols, we do not know of a study showing that a rNTP/dNTP pool imbalance slows the rate of DNA polymerization. To more firmly establish that rNTPs slow the replisome by targeting the Pol, we studied synthesis in the absence of DnaB and primase by assembling Pol III*-β onto a 5.4-kb circular ϕX174 ssDNA primed with a DNA 30mer (scheme in Fig. 2A). Synchronous synthesis is initiated on addition of α-32P-TTP, and timed aliquots are quenched and analyzed in an alkaline agarose gel. Reactions are performed either with no rNTPs or with 3 mM of each individual rNTP. The result, shown in Fig. 2B, shows that Pol III*-β is slowed by each rNTP. One might question whether the rate decrease is an artifact caused by rNMP incorporation and subsequent strand breakage during alkaline gel analysis. However, a control reaction using DNA containing rNMPs gives no detectable cleavage in alkaline gels without pretreatment with 300 mM alkali at elevated temperature (Fig. S5). These results identify Pol III*-β as the primary target of the rNTP-mediated decrease in replisome rate.
Fig. 2.
rNTPs decrease the rate of Pol III*-β on SSB coated ssDNA. (A) Scheme of the assay. ϕX174 ssDNA is coated with SSB and primed with a DNA 30mer. Pol III* and β are preincubated with 20 μM dCTP and dATP to form the Pol III*-β holoenzyme, and replication is initiated on adding dCTP and dTTP, along with 3 mM of the indicated rNTP. (B) Alkaline gel analysis. (C) Quantitation of the data.
rNTPs also Decrease the Rate of Eukaryotic B Family Replicases.
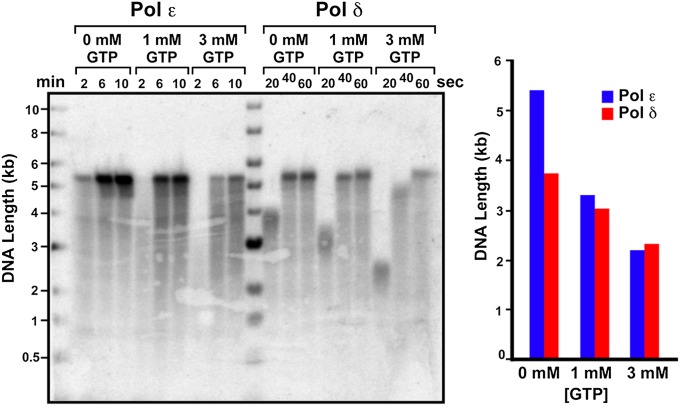
The eukaryotic genome is replicated by two different DNA Pols: Pol ε (leading strand) and Pol δ (lagging strand) (16, 17). Both Pols are stimulated by the proliferating cell nuclear antigen (PCNA) clamp and replication factor C (RFC) clamp loader. However, eukaryotic Pols δ and ε belong to the B family of DNA Pols, whereas bacterial Pol III (Pol C) belongs to the C family (18). In addition, replication forks in yeast proceed at about 30 ntd/s (19), which is much slower than the 650 ntd/s rate of the E. coli replisome (14). Perhaps the slower rate of eukaryotic Pols gives them more time to discriminate a rNTP over dNTP, and thus their rates of elongation may not be affected by the rNTP/dNTP pool imbalance.
To determine whether rNTPs decrease the rate of the Pol δ and Pol ε holoenzymes, we assembled yeast Pols δ and ε with PCNA and RFC on primed ϕX174 ssDNA in a preincubation step and then initiated synchronous extension in the presence of 0, 1, or 3 mM rGTP (Fig. 3). The result shows that both Pol ε and Pol δ holoenzymes are slowed twofold by 3 mM rGTP, indicating that the effect of rNTPs on the rate of the C-family Pol III*-β generalizes to the B-family eukaryotic Pol δ-PCNA and Pol ε-PCNA.
Fig. 3.
rNTPs decrease the rate of eukaryotic Pol δ and Pol ε holoenzymes. (Upper) The PCNA clamp is loaded onto singly primed ϕX174 ssDNA by the RFC clamp loader, and either Pol δ or Pol ε is added to assemble the Pol δ or Pol ε holoenzyme. Only two dNTPs are present to prevent elongation, and then synchronous synthesis is initiated on adding remaining dNTPs. (Left) Alkaline gel. (Right) Histogram of product lengths observed at 20 s.
rNTPs Are Incorporated by Pol III at a Low Frequency.
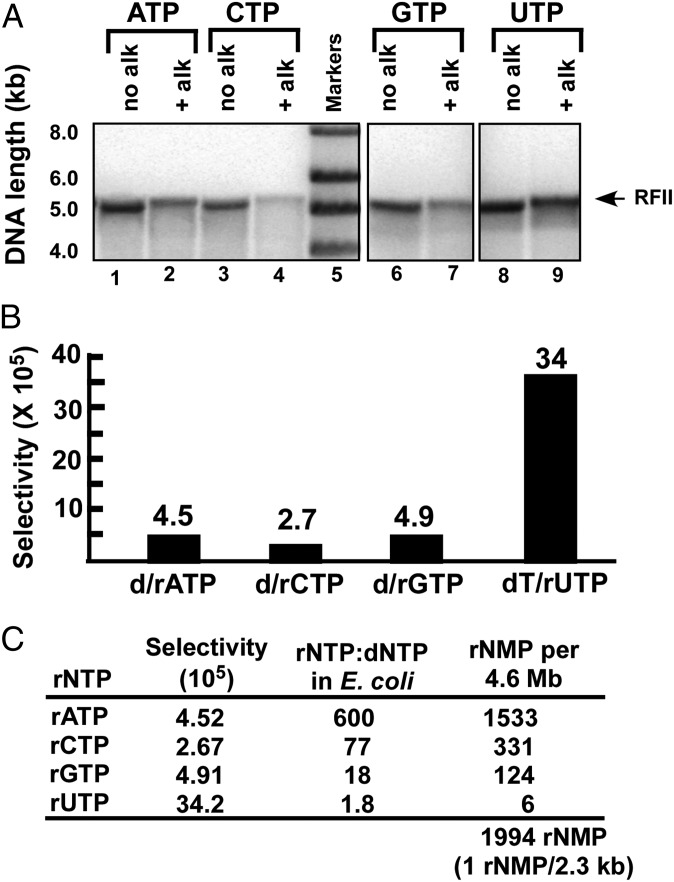
In addition to the effect of rNTPs on the rate of DNA Pols, rNMPs are incorporated at a low level. To measure the frequency of rNMP incorporation, we used an assay similar to a study of yeast Pols (8). A 5.4-kb SSB coated primed ϕX174 ssDNA was replicated by Pol III*-β in the presence or absence of 3 mM of rATP, rGTP, rCTP, or rUTP, followed by treatment with alkali under conditions for complete cleavage at each incorporated rNMP. Samples were then analyzed on an alkaline agarose gel (Fig. 4A). Quantitation of the full-length replicative form II (RFII) duplex ϕX174 DNA that remains after alkali treatment reveals the percentage of molecules with an incorporated rNTP. For example, if 50% of RFII products are cleaved by alkali, the frequency of misincorporation is 0.5 rNMP every 5.4 kb (or 1 rNTP per 10.8 kb). The observed percentage cleavage could be corrected for the theoretical percentage of two or more cleavages (e.g., for 50% cleavage, about 8% of total DNA would contain two or more rNMPs). The data are not corrected for this possibility.
Fig. 4.
rNMP misincorporation by Pol III holoenzyme. (A) Replication of 5.4 kb 5′ 32P-primed ϕX174 ssDNA using 20 μM NTPs, with or without 3 mM of the indicated rNTP, to produce full-length 5.4-kb duplex circles (RFII). Reaction products were incubated with or without alkali to cleave the 5.4-kb RFII at incorporated rNMPs, and then analyzed in an agarose gel. Cleavage of alkali-treated products relative to untreated products are rATP, 44.7%; rCTP, 75.5%; rGTP, 41.2%; and rUTP, 5.9%. (B) Bar plot of rNTP/dNTP selectivity numbers assuming 1:1 rNTP:dNTP ratio. (C) Predicted rNMP residues incorporated in a single daughter chromosome during E. coli replication, calculated from the measured selectivity numbers and the in vivo rNTP and dNTP concentrations, as described in Experimental Procedures.
Application of this assay to Pol III*-β is shown in Fig. 4A. Selectivity numbers for the incorporation of each rNMP, assuming equal concentration to its corresponding dNTP, can be calculated from the data (Fig. 4B). For example, 3 mM rATP results in 44.7% cleavage of RFII, and thus the Pol extends DNA about 12,049 bp for each rAMP incorporated (i.e., 1/0.447 = 2.24; 2.23 × 5,386 bp ϕX174 ssDNA = 12,064 bp). About a quarter of this length will be dAMP residues, and thus Pol III*-β inserts 3,012 dAMP before incorporating an rAMP. The ratio of 3 mM rATP and 20 μM dATP in the assay is 150. Therefore, the selectivity number for rAMP vs. dAMP is 150 × 3,102 = 4.65 × 105. Likewise, selectivity numbers of rC, rG, and rU can be calculated (Fig. 4B). Interestingly, selectivity of the C-family Pol III is 18–30-fold higher for dTMP/rUMP compared with other dNMP/rNMPs, unlike the eukaryotic B family Pols (2). Indeed, rUTP is also the least inhibitory rNTP on DNA synthesis rate (Figs. 1 and 2). Presumably, selectivity by C-family Pols is aided by the additional methyl group in the dTTP base compared with dUTP.
Incorporation of 2,000 rNMPs Predicted During Replication of the E. coli Genome.
The selectivity values can be used to calculate the amount of rNMPs incorporated during replication, using a known ratio of rNTP/dNTP in vivo. Fig. 4C shows the calculated rNMP incorporation during E. coli genome duplication, using intracellular nucleotide concentrations determined by mass spectrometry, reported to circumvent nucleotide instability using other methods (20). Using this method, the total rNTP:dNTP ratio in E. coli is within twofold of the total rNTP:dNTP ratio in yeast (2), as well as in other cells (3). rNMP incorporation into a 4.6-Mb E. coli daughter chromosome can be determined by multiplying each selectivity number by the measured intracellular rNTP:dNTP ratio. The calculation suggests that about 2,000 rNMPs are incorporated in each daughter chromosome at an average frequency of 1 rNMP every 2.3 kb.
The Replisome Pauses at a Template rNMP.
Considering the potential for thousands of rNMPs incorporated during replication, repair reactions that replace rNMPs with dNMPs may not keep pace with the rapid rate of E. coli replication. Thus, the replisome may sometimes encounter a rNMP in the template strand during the next round of replication. To assess the rate of a Pol III replisome as it moves past a template rNMP, we synthesized 100mer rolling circle substrates containing either a rGMP or rUMP in the leading strand template (Fig. 5A) and compared the rate of synthesis to the same substrate that lacks a rNMP residue. The replisome must traverse the template rNMP once every 100 bp. Therefore, the rate over a known distance enables calculation of the average delay in traversing the template rNMP. Time courses of replication show a detectable difference in rate, using templates containing a rNMP compared with a control with no rNMP residue. We approximate the delay in traversing a template rGMP to be 90 ms, and a template rUMP to be 14 ms, compared with the average rate of about 3 ms to traverse template dNMPs. Pausing is likely very dependent on sequence context, but these two singular results suggest that replication over template rNMPs probably causes replisome pausing in vivo.
Fig. 5.
The replisome pauses at template rNMP residues. (A) Illustration of rolling circle replication containing a template rGMP on the inner circle (leading strand template). (B) Agarose gel of time courses of replication on rolling circle templates containing no rNMP (lanes 1–4 and lanes 10–13), rGMP (lanes, 1–8), or rUMP (lanes 14–17). Lane 9 is a size standard. (C) Plots of the time courses in B.
RNase HII Does Not Prevent Mutations in E. coli.
Considering the frequency of rNMP incorporation by the chromosomal replicase, it seems possible that rNMP residues could signal the newly synthesized strand for MMR, as suggested (2), or cause mutagenesis when imbedded. A role for rNMP residues in MMR is unlikely for E. coli, which has a well-defined MMR system, with a MutH enzyme that nicks the newly synthesized DNA strand at hemimethylated GATC sites to direct repair to the new strand (10, 11). RNase HII nicks at single rNMP residues, unlike RNase HI. Hence, we mutated the single RNase HII gene (rnhB) of E. coli to determine whether it is involved in mutagenesis. The rnhB mutant grew as fast as WT (Fig. S6) and displayed no increase in spontaneous mutagenesis (Table 1). Mutagenesis was determined by resistance to rifampicin, which mainly scores for single base substitutions, and thus other types of mutations may not be detected. We also examined a double mutant, rnhB /mutH, but observed no difference in mutagenesis over a mutH strain. The results are not surprising, given the methyl-directed MMR system of E. coli, and also suggest that rNMP residues persisting in the E. coli genome are nonmutagenic.
Table 1.
Rifampicin resistance mutation rate in E. coli WT DH12S cells and its knock-out derivatives
| Strain | Mutation rate (×10−7) |
| DH12S | 0.33 ± 0.06 |
| DH12SΔrnhB | 0.37 ± 0.09 |
| DH12SΔmutH | 39.7 ± 5.3 |
| DH12SΔrnhBΔmutH | 36.2 ± 5.1 |
Margins of error represent the 95% confidence interval.
RNase HII Prevents Mutagenesis in B. subtilis.
Many bacteria and all eukaryotes lack MutH (21). Instead, their MutL contains an endonuclease activity not present in E. coli MutL (22–24). The proposed mechanism of MMR requires a strand-specific signal to direct incision by MutL to the newly synthesized strand (22, 25). Strand interruptions between Okazaki fragments may provide a signal for the lagging strand, but not the leading strand.
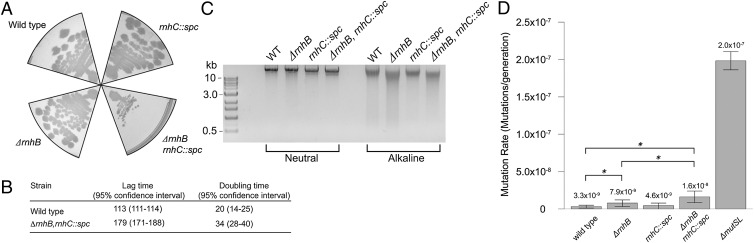
To investigate the possibility that rNMP incorporation may play a role in strand recognition for MMR in organisms lacking a methylation-directed system, we inactivated the RNase H-encoding genes of B. subtilis. B. subtilis encodes two genes that complement loss of RNase H activity in E. coli, rnhB, and rnhC (26). RnhB resembles RNase HII, and RnhC has been described as RNase HIII (27). The substrate specificities of B. subtilis RnhB and RnhC show differences in efficiency and cleavage site selection; however, current evidence suggests that B. subtilis RnhB is most similar to E. coli RNase HII, and B. subtilis RnhC appears functionally more similar to E. coli RNase HI (26). Because the roles of rnhB and rnhC are uncertain, we chose to characterize both in B. subtilis. We cleanly deleted rnhB and disrupted rnhC, and we also generated a strain lacking both rnhB and rnhC. On the basis of colony size (Fig. 6A) and growth rate, loss of rnhB or rnhC is without affect (Fig. 6A). The double mutant, however, grows poorly (Fig. 6 A and B and Fig. S7) and exhibits a ∼fivefold decrease in cell viability (26). To determine whether rnhB and rnhC are indeed involved in rNMP removal in vivo, we purified genomic DNA from B. subtilis cells for analysis by alkali cleavage. We find substantial DNA cleavage in cells deleted for rnhB, demonstrating that rNMPs are incorporated into genomic DNA. Loss of rnhC shows no difference in DNA fragmentation relative to WT, nor did loss of rnhC exacerbate the DNA fragmentation observed with ΔrnhB, suggesting that rnhB is primarily responsible for removal of single rNMPs in vivo.
Fig. 6.
B. subtilis incorporates rNMPs during replication in vivo. (A) Representative images of the indicated strains after plating and growth at 37 °C for 12 h. For strains containing the rnhC::spc allele, growth was in the presence of 100 µg/mL spectinomycin. (B) Table showing lag time and fastest doubling time of WT and ΔrnhB, rnhC::spc strains. (C) Alkaline analysis of genomic DNA from the indicated strains electrophoresed in a 1.0% agarose gel. (D) Mutation rate analysis as measured by formation of rifampin-resistant colonies. Error bars represent the 95% confidence interval, and asterisks indicate significance as measured with a one-tailed P value < 0.05.
To test whether rNMP incorporation may contribute to MMR in B. subtilis, we measured spontaneous mutation rates (28–30). We find that deletion of rnhB results in a 2.4-fold increase in mutation rate, whereas loss of rnhC appears nonmutagenic with a 1.3-fold increase relative to WT (P = 0.03 and 0.2, respectively). Measurement of spontaneous mutation rate for the double mutant showed a fivefold increase compared with WT (Fig. 6C), which is significant with respect to WT and the ΔrnhB strain (P = 0.0017 and P = 0.03, respectively). For comparison, we estimated the mutation rate of ΔmutSL cells to be 2.0 × 10−7 per generation, which is 60-fold higher than WT (P < 4.0 × 10−5) and 25-fold higher than the rnhB and rnhC double mutant. Thus, B. subtilis deficient in rnhB shows an increase in spontaneous mutagenesis, as well as mutagenesis increases in the double mutant. We attempted to test whether mutagenesis caused by loss of rnhB is in the MMR pathway by determining whether rnhB is epistatic to ΔmutSL. Despite repeated attempts, we have been unable to construct a strain with clean deletions in both rnhB and mutSL. With these results, we find that loss of rnhB or loss of rnhB and rnhC increases spontaneous mutagenesis, providing a link between impaired removal of rNMPs and an increase in spontaneous mutagenesis.
Discussion
The Cost of rNTP/dNTP Pool Imbalance on Replisome Rate.
The current report demonstrates that high intracellular levels of rNTPs compared with dNTPs decreases the rate of DNA replication and likely increases the rate of mutagenesis via rNMP incorporation. Competition of rNTPs for dNTPs at the active site of the Pol III replicase underlies the rNTP-mediated rate decrease of the replisome. We find that a decrease in rate by rNTP/dNTP competition generalizes to eukaryotic Pols δ and ε. On hindsight, it may not seem a surprise that the natural nucleotide pool imbalance would slow the Pol within the replisome. However, before this report, this topic has not been directly addressed in a moving system to our knowledge.
Competition of rNTPs with dNTP occurs even in the absence of correct base-pairing with the template strand, illustrated here using a synthetic rolling circle substrate composed of only three of the dNTPs (Fig. 1C). The result suggests that rNTP/dNTP competition may occur at a step that precedes base-pairing of the dNTP to the template base. rUTP is the least-efficient rNTP in slowing the replisome. This may be because rUTP is the only rNTP with a difference in the nucleotide base compared with the corresponding dTTP. B-family Pols (eukaryotic replicases) appear to lack this discrimination. Another point of interest is that ADP is twice as potent an inhibitor as ATP. Thus, replisome rate may be linked to the energy state of the cell.
The Cost of rNTP/dNTP Pool Imbalance on the Fidelity of the Replisome.
This report also examines the frequency with which the bacterial Pol III replicase (Pol III*-β) incorporates rNMP residues. The average incorporation frequency of Pol III*-β predicts one rNMP incorporation every 2.3 kb, assuming the intracellular concentrations of rNTPs and dNTPs determined in ref. 20. At this frequency, replication of the E. coli genome is predicted to incorporate about 2,000 rNMPs into each of the two 4.6-Mb daughter chromosomes. This can be compared with the 10,000 rNMPs estimated to be incorporated in the yeast 12.5-Mb genome (2), and the more than 1 million rNMP during replication of the mouse genome (31). rNTPs are also incorporated in Schizosaccharomyces pombe (32). Hence, the current study shows that rNMP incorporation is evolutionarily conserved from eukaryotes to bacteria.
The current report notes that bacterial replisome progression is delayed 4–30-fold by a template rNMP. Although most or all rNMPs are probably repaired before the next replication cycle, some rNMPs may remain and cause Pol pausing, which could possibly lead to genomic instability. Frequent lesions are often repaired by multiple means, as evidenced by the presence of multiple paths that repair 8-oxo-guanine, abasic sites, and RNA primers used to initiate Okazaki fragments (33–35). Considering the abundance of rNMPs incorporated during replication, one may expect various pathways exist to repair them, consistent with the presence of multiple RNase H enzymes in cells (36).
Are Incorporated rNMPs Used as a Signal for MMR?
Incorporation of rNMPs within genomic DNA may sometimes benefit the life of an organism. For example, eukaryotic Pol mu is highly proficient at rNTP incorporation and is suggested to use rNTPs during nonhomologous end joining of ds breaks at low dNTP concentration (37). Indeed, it appears that ligases involved in nonhomologous end joining are more proficient in using a 3′ terminal rNMP (38, 39). Moreover, in S. pombe, two rNMPs are incorporated in S phase and retained during the next round of replication, which enhances the mating type switch recombination event (39). It has been hypothesized by the Kunkel laboratory that the high frequency of rNMP incorporation by yeast replicases may be harnessed for DNA metabolic actions, including use as marks of the newly replicated strand of DNA (2). The current study demonstrates that rNMP residues are also a frequent event in bacteria, suggesting this activity is widely conserved. Hence, the use of incorporated rNMP residues to mark the newly synthesized DNA strand for MMR seems a logical possibility in organisms lacking DNA adenine methylase (Dam) and MutH. In fact, the rNMP would not need to remain long. Consider MMR in E. coli: Dam methylase is continually present, and the hemimethylated state of the GATC site sequence is thought to be transient, but of sufficient duration to be harnessed by MMR. Hence, rNMP residues would not necessarily need to remain through a second round of replication to direct MMR, a prerequisite for a mutagenic phenotype. We also show in E. coli that loss of rnhB does not increase mutagenesis, and therefore rNMP misincorporation could serve as a nonmutagenic signal for the nascent strand, much like hemimethylation does during MMR in E. coli.
In our initial exploration of this hypothesis, we performed genetic knockouts of RNase HII, an enzyme known to nick at single rNMPs within DNA (1). All eukaryotes, and most bacteria, lack methyl direction (i.e., they do not contain MutH or Dam methylase), instead containing a MutL with endonuclease activity. This report includes an examination of B. subtilis, a Gram-positive bacterium that contains a MutL with endonuclease activity and does not use methyl direction for MMR (24). Even though B. subtilis MutL contains an intrinsic endonuclease activity, MMR requires a signal to specify MutL action to the newly replicated strand (22, 25). Thus, it seemed possible that rNMP incorporation during replication may direct MMR. We demonstrate here that B. subtilis incorporates rNMP residues during cell growth, as demonstrated in yeast (7). If incorporated rNMPs are used to specify the newly synthesized DNA strand, mutation of an RNase HII may result in enhanced spontaneous mutation. B. subtilis contains two genes that can remove rNMP residues, and in fact mutation of both genes yields about a sixfold increase in mutagenesis compared with wt cells. Hence, RNase H enzymes may indeed be involved in MMR. If mutagenesis resulting from loss of rnhB and rnhC in B. subtilis is caused by partial loss of MMR, such a loss would account for 9.3% of MMR in B. subtilis. It should also be noted that the double mutant in B. subtilis lowers viability, suggesting that the sixfold increase in mutagenesis we measured may be an underestimate. Considering this observation and that frequent mutagenic events often have multiple pathways of repair (34), it is reasonable to expect that other enzymes, yet to be identified, also cleave at newly incorporated rNMPs, and the resulting nicks could be used for MMR. This perspective of rNMP repair, combined with the RNase HII data of this report, suggest that the canonical RNase HII enzymes contribute to ∼10% of MMR.
Experimental Procedures
Rolling Circle Replication.
Purification of the subunits and reconstitution of Pol III* (Pol III core3τ3δδ'χψ) was as described (40). Primase, SSB, and DnaB were purified as described (40). Reactions contained 7.69 nM 100mer rolling circle DNA and 219 nM DnaB that were preincubated for 30s at 37 °C in buffer A [20 mM Tris-HCl at pH 7.5, 5 mM DTT, 40 µg/mL BSA, 4% (vol/vol) glycerol] with 8 mM MgOAc2; 50 mM K-glutamate; 100 μM ATP; 60 μM each dCTP and dGTP; 240 μM each CTP, GTP, and UTP; and 148 nM β2. Then 7.32 nM Pol III* was added, followed by a 1-min incubation at 37 °C, after which the reaction was lowered to 25 °C for 1 min. Replication was initiated on adding 506.8 nM SSB4, 240 nM primase, 60 μM dATP, 20 μM 32P-dTTP, and amounts of ATP indicated in the figures. Leading strand reactions were performed similarly, except the 100mer DNA (10 nM) was 5′ end-labeled with 32P instead of using 32P-dTTP. The dNTPs were 20 μM, and rNTPs (as indicated) were 3 mM. Timed aliquots were removed and quenched on addition to an equal volume of 1% SDS and 40 mM EDTA. Quenched reactions were divided: one half was analyzed in a 0.5% native agarose gel, and the other half was analyzed in a 0.5% alkaline agarose gel, using a Typhoon phosphoimager.
Replication on Primed ssDNA.
Reactions contained 5.4 kb ϕX174 ssDNA (1.2 nM) primed with a single 5′ 32P-DNA 30mer, 433 nM SSB4, 4.88 nM Pol III*, 14.8 nM β2, 8 mM MgOAc2, and 20 μM each dATP and dCTP in buffer A. After a 30 s incubation at 37 °C, the temperature was lowered to 25 °C and replication was initiated with 20 μM dGTP and dTTP, along with the indicated rNTP. Analysis of yeast Pols ε and δ contained 1.5 nM 5′ 32P-primed ϕX174 ssDNA, 500 nM SSB4, 8 mM MgOAc2, 50 mM K-glutamate, 0.1 mM ATP, and 60 μM each dCTP and dGTP, 4.8 nM PCNA3, 4.0 nM RFC, and either 20 nM Pol ε or 11 nM Pol δ. Reactions were incubated at 30 °C for 5 min, and replication was initiated with 60 μM dATP and 20 μM dTTP, along with 1 mM, 3 mM, or no GTP. Reactions were quenched at the indicated times and analyzed as described for rolling circle reactions.
rNTP Incorporation by E. coli Pol III.
Reactions contained 1.2 nM 5′ 32P-primed ϕX174 ssDNA, 515 nM SSB4, 8 mM MgOAc2, 40 mM NaCl, 20 μM each dATP and dCTP, 9.8 nM Pol III*, and 125 nM β2 in buffer A. After 1 min at 37 °C, replication was initiated on adding 20 μM dGTP and 20 μM dTTP, along with 3 mM of the indicated rNTP, and continued for 10 min at 37 °C. Reactions were quenched with an equal volume of 1% SDS and 40 mM EDTA and then divided; one half was incubated in 0.3 M NaOH at 37 °C for 1 h, and the other half was untreated. Both halves were analyzed in a 0.5% alkaline agarose gel and percentage full-length (RFII) product remaining after alkali treatment was determined, using a Typhoon imager. Selectivity for insertion of rNTP vs. dNTP was calculated as described in the text. Predicted frequency of rNMP incorporation into the genome was calculated by multiplying the selectivity numbers by the in vivo ratio of dNTP/rNTP pairs reported in ref. 20. Because the concentration of dGTP was undetermined, we assumed 92 μM, as in ref. 41.
Analysis of rNMP Incorporation in B. subtilis DNA.
rNMPs were detected in B. subtilis genomic DNA as described, with slight modifications (7). Cultures were grown in LB at 37 °C to midlog growth (OD600 = 0.6), and genomic DNA was purified using standard procedures, with the addition of RNase A (50 μg/mL final concentration) and a 15-min incubation at 37 °C before cell lysis. For each strain, genomic DNA was aliquoted into two 30 μL reactions (2 μg each): one for treatment with 0.3 M KCl as a control and the other with 0.3 M KOH to hydrolyze DNA at positions containing an rNMP. Reactions were incubated at 55 °C for 30 min, followed by analysis in a 1% agarose gel.
Supplementary Material
Acknowledgments
The authors are grateful for support from the National Institutes of Health (NIH) (GM38831), MCB1050948 (to L.A.S.), and NIH Genetics Training Grant T32GM007544 (to J.W.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309506110/-/DCSupplemental.
References
- 1.Kornberg A, Baker TA. DNA Replication. 2nd Ed. New York: W. H. Freeman; 1992. [Google Scholar]
- 2.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107(11):4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 4.Clark AB, Kunkel TA. The importance of being DNA. Cell Cycle. 2010;9(22):4422–4424. doi: 10.4161/cc.9.22.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31(14):4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyce CM. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc Natl Acad Sci USA. 1997;94(5):1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6(10):774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47(6):980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332(6037):1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106(2):302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 11.Larrea AA, Lujan SA, Kunkel TA. 2010. SnapShot: DNA mismatch repair. Cell 141(4):730.e1. [DOI] [PubMed]
- 12.Georgescu RE, Kurth I, O’Donnell ME. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat Struct Mol Biol. 2012;19(1):113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lia G, Michel B, Allemand JF. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science. 2012;335(6066):328–331. doi: 10.1126/science.1210400. [DOI] [PubMed] [Google Scholar]
- 14.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci USA. 2005;102(11):3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261(10):4738–4748. [PubMed] [Google Scholar]
- 16.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317(5834):127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, et al. Insights into base selectivity from the 1.8 Å resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2011;50(4):581–590. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekedat MD, et al. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol. 2010;6:353. doi: 10.1038/msb.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435(3):171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126(2):297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 23.Kadyrov FA, et al. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J Biol Chem. 2007;282(51):37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillon MC, et al. Structure of the endonuclease domain of MutL: Unlicensed to cut. Mol Cell. 2010;39(1):145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pluciennik A, et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107(37):16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itaya M, et al. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J Bacteriol. 1999;181(7):2118–2123. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohtani N, et al. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: Classification of RNases H into three families. Biochemistry. 1999;38(2):605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 28.Bolz NJ, Lenhart JS, Weindorf SC, Simmons LA. Residues in the N-terminal domain of MutL required for mismatch repair in Bacillus subtilis. J Bacteriol. 2012;194(19):5361–5367. doi: 10.1128/JB.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupes NM, et al. Mutations in the Bacillus subtilis beta clamp that separate its roles in DNA replication from mismatch repair. J Bacteriol. 2010;192(13):3452–3463. doi: 10.1128/JB.01435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenhart JS, Sharma A, Hingorani MM, Simmons LA. DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Mol Microbiol. 2013;87(3):553–568. doi: 10.1111/mmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reijns MA, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149(5):1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7(12):e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 34. Friedberg EC, Walker GC, Siede W, et al. (2006) DNA Repair and Mutagenesis (ASM Press, Washington, DC), 2nd Ed.
- 35.Rossi ML, Purohit V, Brandt PD, Bambara RA. Lagging strand replication proteins in genome stability and DNA repair. Chem Rev. 2006;106(2):453–473. doi: 10.1021/cr040497l. [DOI] [PubMed] [Google Scholar]
- 36.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23(7):2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J Biol Chem. 2008;283(13):8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7(1):59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27(4):527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 41.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190(2):718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.