Abstract
Peptidylarginine deiminases (PADs) play a critical role in generating autoantigens in rheumatoid arthritis (RA), but the mechanisms underlying their dysregulation in this disease remain unknown. Although PADs require supraphysiologic concentrations of calcium for activity in vitro, the enzymes are clearly active in vivo (e.g. in RA synovial fluid) where calcium concentrations are much lower. In this study, we have discovered a novel subset of anti-PAD4 autoantibodies (identified by their cross-reactivity with PAD3) which strikingly increase the catalytic efficiency of PAD4 by decreasing the enzyme’s requirement for calcium into the physiologic range. Patients with these novel PAD3/PAD4 cross-reactive autoantibodies had higher baseline radiographic damage scores and a higher likelihood of radiographic progression compared to individuals negative for these antibodies. The ability of autoantibodies to activate an enzyme that itself generates citrullinated autoantigens identifies an important feed-forward loop which may drive the erosive outcome observed in RA patients with these autoantibodies. PAD3 autoantibodies may therefore identify RA patients who would benefit from early aggressive treatment or addition of PAD-inhibitor therapy.
Introduction
Peptidylarginine deiminases (PADs) have emerged as key participants in the pathogenesis of rheumatoid arthritis (RA), a common autoimmune disease characterized by chronic inflammation of the joints and synovial tissue, leading to pain, swelling, bone erosions, and disability [1]. PADs catalyze the post-translational deimination of peptidyl-arginine to citrulline [2], generating the hallmark targets of the autoantibody response in RA [3]. In addition, PAD2 and PAD4 are expressed by neutrophils and monocytes [4] and are present at high levels in RA synovial tissue in regions co-expressing citrullinated proteins [4,5].
PAD4 requires calcium for catalytic activity, and calcium activation of PAD4 displays positive cooperativity [6]. Although in vitro citrullination assays typically use 5-10 mM calcium to achieve maximal PAD4 activation [6-8], it is not possible that such high calcium concentrations are present during PAD4 activation in vivo. Indeed, extracellular free calcium concentrations are estimated to be 0.49-0.98 mM in synovial fluid and 1.1-1.3 mM in plasma [9], and the maximum intracellular calcium concentration achieved by primary human cells even after stimulation with various stimuli does not exceed 100 μM [9-11]. The discrepancy between the in vitro requirements and in vivo availability of calcium suggests that undiscovered factors may modulate PAD4 calcium sensitivity in vivo during homeostasis and RA pathology.
Several studies have demonstrated that in addition to its role in protein citrullination, PAD4 is also a frequent antigenic target in RA [12-14]. PAD4 autoantibodies are detectable prior to disease onset [15] and are associated with more erosive RA that persists despite treatment with TNFα inhibitors [13,15,16]. Although anti-PAD4 antibodies mark a subset of RA patients with severe disease, there is heterogeneity in disease severity observed amongst this group and a pathogenic role for these antibodies remains undefined. We recently showed that peripheral blood neutrophils express PAD3 protein, which is capable of citrullinating intracellular targets [17], and addressed whether PAD3 was also an autoantigen in RA.
These studies reveal that anti-PAD3 autoantibodies are present in 12-18% of RA patients and 0% of healthy controls. Anti-PAD3 antibodies are only detected in anti-PAD4 positive sera, and competition experiments demonstrated that these are PAD3/PAD4 cross-reactive autoantibodies. Anti-PAD3/PAD4 positive RA patients have the most erosive joint disease when compared to anti-PAD negative patients or patients with anti-PAD4 antibodies only. Using histone H3 as a macromolecular substrate for citrullination, we demonstrated that cross-reactive antibodies strikingly enhance PAD4 activity. Autoantibody-induced changes in the calcium sensitivity of PAD4 that mimic calcium-ion binding augment enzymatic activity at physiologic calcium concentrations, and may be an important driver of dysregulated protein citrullination in RA. Such properties have mechanistic and therapeutic implications.
Results
A subset of anti-PAD4 positive RA patients has antibodies recognizing PAD3
In order to screen for anti-PAD3 antibodies, sera from a convenience sample of RA patients were initially used to immunoprecipitate 35S-methionine labeled in vitro transcribed and translated (IVTT) PAD3 protein. This approach has been used previously to detect anti-PAD4 antibodies with high sensitivity and specificity compared to ELISA [13]. PAD3 autoantibodies were detected in 18% (8/44) of sera (Figure 1A and S1A). Anti-PAD3 was exclusively present in patients with PAD4 antibodies and was observed in 40% of anti-PAD4 positive sera (Fig. S1A). In order to determine the prevalence of anti-PAD3 in a large, well-defined group of established RA patients, PAD3 immunoprecipitation was performed on 194 sera from the ESCAPE RA cohort (a prospective observational cohort study of subclinical cardiovascular disease in RA, for which extensive clinical and serologic data was available) [13]. Again, anti-PAD3 was only detected in the serum of patients with anti-PAD4 antibodies (Fig. 1A and B, representative positives shown in lanes 3, 4, 10, 13 and 18). The overall prevalence of anti-PAD3 antibodies was 12% in this cohort, and these antibodies were present in 32% of anti-PAD4 positive individuals. Anti-PAD3 was not detected in sera from 36 healthy controls or 30 patients with psoriatic arthritis (Fig. 1A and S1B). Anti-PAD3 antibodies thus appear to be specific for RA and are uniformly associated with anti-PAD4 antibodies.
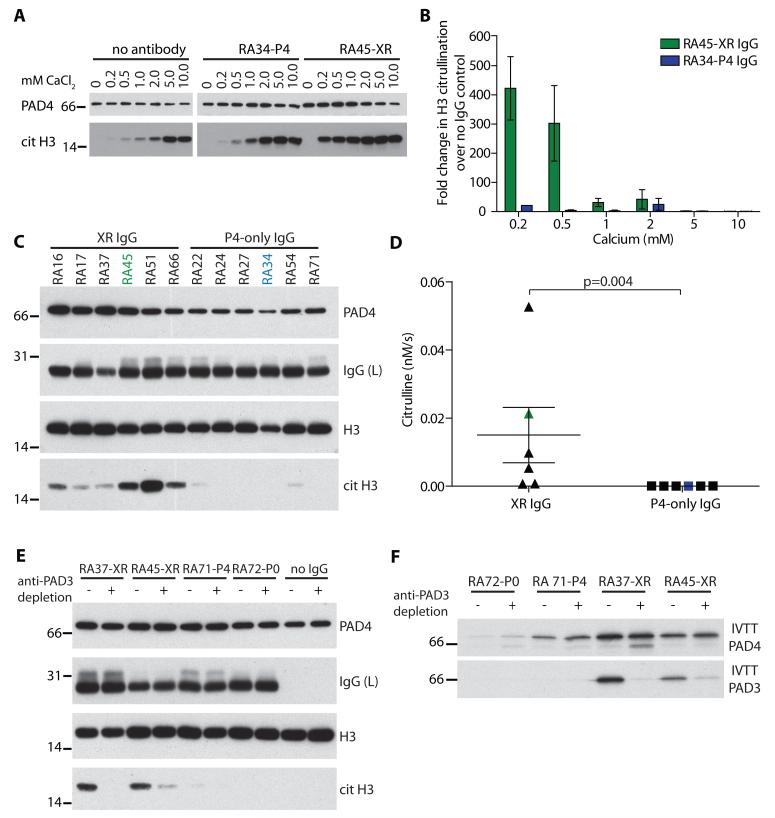
Figure. 1. PAD3/PAD4 cross-reactive autoantibodies are present in a subset of RA patients.
(A) Sera from the ESCAPE RA cohort and convenience samples of RA, psoriatic arthritis, and healthy controls were used to immunoprecipitate IVTT PAD3. “Normalized anti-PAD3 units” were calculated by densitometry and samples with a value ≥0.01 were considered positive. (B) Representative radiographs from 18 anti-PAD4 positive (upper panel) and 18 anti-PAD4 negative (lower panel) ESCAPE RA patient sera are shown. (C) Anti-PAD3/anti-PAD4 positive patient sera were blocked with rPAD3 or rPAD4 prior to performing IVTT PAD3 or IVTT PAD4 immunoprecipitations. (D) The “% IVTT PAD4 IP blocked by recombinant protein” was calculated by densitometry for 10 PAD3/PAD4 positive and 8 PAD4-only patients and the two groups were compared using a two-tailed t-test.
Anti-PAD3 antibodies cross-react with PAD4
In order to determine if anti-PAD3 and anti-PAD4 antibodies were recognizing the same or distinct antigens, competition experiments were performed. Recognition of radiolabeled IVTT PAD3 was abrogated by pre-incubation of anti-PAD3 positive sera with unlabeled recombinant PAD3 (rPAD3) or PAD4 (rPAD4), demonstrating that these antibodies are indeed cross-reactive (Fig. 1C upper panel and S1C). Interestingly, the antibody response to PAD4 is polyclonal and only a subset of PAD4 autoantibodies in the sera from these patients cross-react with PAD3. Thus, pre-incubation of cross-reactive sera with unlabeled rPAD3 reduced the immunoprecipitation of IVTT PAD4 by an average of 33.0±15.8% (Fig. 1C, lower panel, and D). This suggests that the anti-PAD3 cross-reactive antibody subset comprises 2.2-83% of the PAD4 autoantibody pool, depending on the individual serum tested. In contrast, in sera from anti-PAD4 positive individuals who lacked anti-PAD3 antibodies, pre-incubation with rPAD3 reduced the immunoprecipitation of IVTT PAD4 by only 4.9±3.8% (p=0.03)(Fig. 1D and S1D). Importantly, PAD3/PAD4 cross-reactive antibody-positive patients exhibited no or minimal recognition of the homologous protein PAD2 (Fig. S1A). These data demonstrate that a subset of RA patients have antibodies which bind to an epitope shared by PAD3 and PAD4; we will refer to these as PAD3/PAD4 cross-reactive antibodies.
Cross-reactive antibodies are associated with severity of erosive joint disease
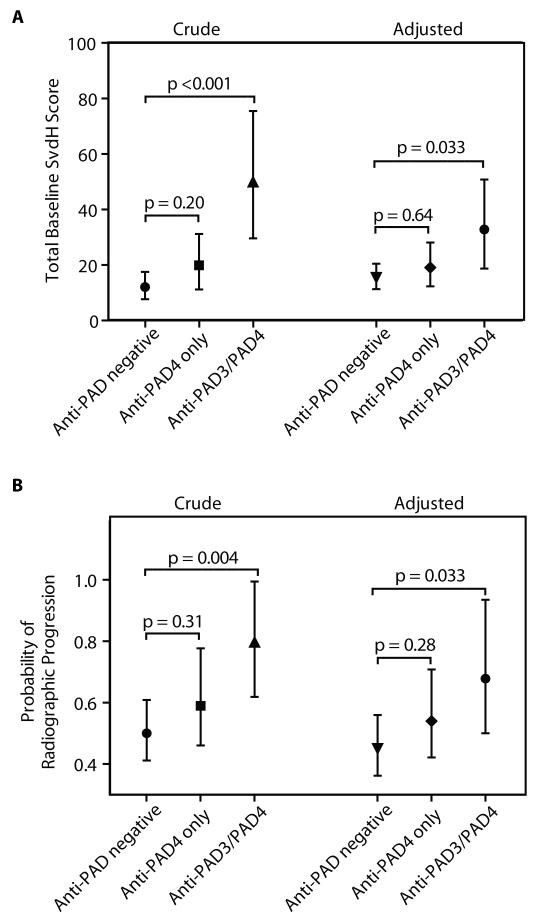
Several previous studies have demonstrated that anti-PAD4 antibodies are associated with increased disease severity in RA and anti-CCP antibodies [12-14]. Due to the cross-reactivity of anti-PAD3 with PAD4, we sought to determine if these associations were maintained in RA patients with PAD3/PAD4 cross-reactive antibodies and if novel associations existed. Patients from the ESCAPE RA cohort were divided into three groups based on PAD antibody status (Group 1: anti-PAD negative, Group 2: anti-PAD4 only, and Group 3: anti-PAD3/PAD4). Importantly, the previous association of anti-PAD4 and anti-CCP antibodies was maintained in the anti-PAD3 positive subset, with over 80% of individuals in both the anti-PAD4 only and antiPAD3/PAD4 groups being anti-CCP positive (Table 1). The total Sharp van der Heidje score (SvdH) (a cumulative measure of disease severity that scores the degree of radiographic joint space narrowing and bone erosion in multiple joints of the hands, wrists, and feet [18]) was available for all 194 patients at the baseline visit. For 150 individuals, this score was also available at follow-up, an average of 39 ± 4 months later. Patients with anti-PAD3 antibodies had a more than 5-fold higher baseline SvdH score than those with anti-PAD4 only (p=0.015) and a more than 8-fold higher score than PAD antibody negative patients (p<0.001) (Fig. 2A and Table 1). Additionally, anti-PAD3 positive patients were significantly more likely to have radiographic progression compared to PAD antibody negative individuals (80 vs. 50%, respectively; p=0.004) despite equivalent treatment with DMARDs and glucocorticoids (Fig. 2B and Table 1). The associations of anti-PAD3 antibodies with disease severity and mean probability of progression were maintained after adjusting for cohort-specific indicators of radiographic damage (age, sex, disease duration, anti-CCP positivity, presence of shared-epitope alleles, and CRP) (Fig. 2), indicating that the anti-PAD3 cross-reactive subset is an independent contributor to the previously defined association of anti-PAD4 autoantibodies with erosive disease.
Table 1.
Characteristics of ESCAPE RA patients by anti-PAD3 and anti-PAD4 status
| Anti-PAD negative (P0) n=122 |
anti-PAD4only (P4) n=49 |
Anti-PAD3/PAD4 (XR) n = 23 |
XR vs. P0 p |
P4 vs. P0 p |
XR vs. P4 p |
|
|---|---|---|---|---|---|---|
| Age, years | 59 ± 8 | 58 ± 9 | 62 ± 8 | 0.26 | 0.42 | 0.14 |
| Male gender, n (%) | 43 (35) | 27 (55) | 9 (39) | 0.72 | 0.017 | 0.21 |
| Caucasian, n (%) | 102(84) | 46 (94) | 18 (78) | 0.53 | 0.075 | 0.049 |
| Ever smoking, n (%) | 80 (66) | 23 (47) | 11 (50) | 0.16 | 0.024 | 0.81 |
| RA duration, years | 7 (4 – 15) | 13 (6 – 19) | 21 (11 – 28) | <0.001 | 0.017 | 0.021 |
| RF seropositivity, n (%) | 75 (61) | 36 (73) | 17 (74) | 0.26 | 0.14 | 0.97 |
| Anti-CCP seropositivity, n (%) | 77 (63) | 42 (86) | 19(83) | 0.070 | 0.004 | 0.73 |
| Any HLA-DRB1 SE Alleles | 78 (65) | 36 (73) | 21 (91) | 0.012 | 0.29 | 0.082 |
| DAS28, median (IQR) | 3.6 (2.9 – 4.3) | 3.4 (2.9 – 4.3) | 3.8 (3.3 – 4.5) | 0.43 | 0.42 | 0.25 |
| HAQ score (0 – 3) , median (IQR) | 0.63 (0.13 – 1.25) | 0.63 (0 – 1.25) | 1.13 (0.13 – 2.00) | 0.21 | 0.44 | 0.10 |
| CRP, median (IQR) | 2.09 (0.97 – 6.74) | 3.32 (1.42 – 7.95) | 2.64 (1.62 – 9.27) | 0.19 | 0.12 | 0.74 |
| SvdH Score, median (IQR) | 6 (0 – 26) | 9 (1 – 60) | 49 (12 – 98) | <0.001 | 0.20 | 0.015 |
| Any increase in SvdH, n (%)* | 49 (50) | 22 (59) | 12 (80) | 0.004 | 0.31 | 0.16 |
| Non-biologic DMARDs, n (%) | 102(84) | 44 (90) | 17 (74) | 0.23 | 0.35 | 0.081 |
| Biologic DMARDs, n (%) | 53 (44) | 23 (47) | 11 (48) | 0.72 | 0.71 | 0.94 |
| Glucocorticoids, n (%) | 44 (36) | 21 (43) | 9 (39) | 0.78 | 0.41 | 0.77 |
150 individuals had radiographs at follow-up (n=98 in group 1; n=37 in group 2; and n=15 in group 3)
IQR= interquartile range
SvdH = Sharp van der Heijde Score
DMARDs= disease modifying antirheumatic drugs
Figure. 2. Patients with cross-reactive antibodies have the most severe and progressive disease.
(A) 194 patients in the ESCAPE RA cohort were divided into three groups according to their autoantibody status (Anti-PAD negative, Anti-PAD4 only, or Anti-PAD3/PAD4). The mean total baseline SvdH score for each group was determined and revealed a significant association of PAD3/PAD4 cross-reactive antibodies with Sharp score (Crude). (B) Sharp scores were available on 150 patients at follow-up, with progression depicted as the proportion of patients with any increase in SvdH score from baseline (Crude). These associations were maintained after adjusting for age, gender, RA duration, shared epitope alleles, anti-CCP status, and log CRP levels (Adjusted).
In addition to the associations with disease severity and anti-CCP status, anti-PAD3 antibodies were associated with significantly longer RA duration compared to the anti-PAD4 only (p=0.021) and PAD antibody negative groups (p<0.001) (Table 1). The association of anti-PAD3 antibodies with RA duration was independent of age at disease onset and each additional year post RA diagnosis conferred an 8% higher odds of observing anti-PAD3 antibodies (p=0.009) (Table S1). As noted above, the association of anti-PAD3 autoantibodies with disease progression was maintained after adjusting for disease duration. Additionally, 91% of patients with anti-PAD3 antibodies had one or more HLA-DRB1 shared epitope alleles compared to only 73% of patients with anti-PAD4 only (p=0.082) and 65% without PAD antibodies (p=0.012). Interestingly, these associations were not observed in previous studies of anti-PAD4 antibodies [12-14], again reinforcing that anti-PAD3 is marking a distinct patient subset.
PAD3/PAD4 cross-reactive antibodies increase the sensitivity of PAD4 to Ca2+
The finding that patients with PAD3/PAD4 cross-reactive antibodies have the most severe disease suggested that these antibodies may play a pathogenic role in vivo. We hypothesized that these antibodies contribute to disease pathogenesis by modulating PAD4 enzymatic function. This was explored using the well-defined physiologic PAD4 substrate, histone H3, at a range of calcium concentrations.
In the absence of antibody, PAD4 activity was maximal at 5 mM calcium and declined abruptly below 2 mM, such that activity at physiologic calcium concentrations was markedly diminished (Figure 3A). A cross-reactive antibody (RA45) strikingly expanded the range of calcium concentrations able to support PAD4 catalytic activity, with robust histone citrullination observed over a physiologic range of calcium concentrations (0.2-1 mM calcium). In fact, this cross-reactive antibody increased the rate of histone H3 citrullination over 400-fold at 0.2 mM calcium, compared to 10.7-fold with a PAD4-only IgG (Figure 3B).
Figure. 3. PAD3/PAD4 cross-reactive antibodies increase the sensitivity of PAD4 to Ca2+.
PAD4 was pre-incubated with no antibody, anti-PAD4 only (RA34-P4), or PAD3/PAD4 cross-reactive (RA45-XR) IgG prior to use in citrullination assays. (A) Calcium titrations were performed using a sub-saturating concentration of histone H3 (700 nM) and H3 citrullination was determined by immunoblotting. Representative data from 5 independent experiments is shown. (B) The “Fold change in H3 citrullination over no IgG control” was determined by densitometry. (C) H3 was citrullinated at 0.2mM calcium in the presence of 6 cross-reactive IgGs (XR IgG) or 6 PAD4-only IgGs. Representative data from 4 experiments is shown. (D) Citrullination was quantified by densitometry and the mean citrullination of the two groups was compared using a two-tailed t-test.(E) Anti-PAD3 antibodies were depleted prior to use in H3 citrullination assays. H3 citrullination and equal PAD4 and IgG loading were determined by immunoblotting. Representative data from 2 experiments is shown. (F) Specificity of the anti-PAD3 depletion was confirmed by performing IVTT PAD4 or IVTT PAD3 immunoprecipitations.
The ability of cross-reactive antibodies to activate PAD4 at low calcium concentrations was further explored by citrullinating H3 in the presence of IgG from six cross-reactive antibody-positive and -negative individuals at 0.2 mM Ca2+ (Fig. 3C). IgG from cross-reactive patients increased the rate of H3 citrullination by an average of 500-fold compared to that observed with PAD4-only sera, from 0.03 ± 0.2 pM/s to 15.0 ± 10.0 pM/s (p=0.004) (Fig. 3D). The increase in PAD4 activity was specifically dependent upon the PAD3/PAD4 cross-reactive IgG subset because depletion of anti-PAD3 antibodies on solid-immobilized rPAD3 reduced H3 citrullination by 92-100% (Fig. 3E and F). Thus, PAD3/PAD4 cross-reactive antibodies increase the sensitivity of PAD4 to calcium and greatly augment histone citrullination at Ca2+ concentrations that are relevant physiologically.
Cross-reactive antibodies decrease the calcium-binding cooperativity of PAD4
To better understand the molecular interactions behind these rate enhancements, kinetic constants were determined for histone H3 and compared to those for the model small-molecule substrate benzoyl-arginine ethyl ester (BAEE). The kinetic constants for these two substrates represent two extremes: the histone has an exceptionally high binding affinity (Km = 0.256 μM) compared to BAEE (Km = 960 μM), but is turned over much more slowly (kcat = 0.006 s−1) than BAEE (kcat = 0.242 s−1). Unexpectedly, the cross-reactive antibodies were found to have almost no effect on Km and kcat for either substrate at saturating calcium concentrations (Figure 4 A and B and Table 2).
Figure. 4. Cross-reactive antibodies decrease the calcium-binding cooperativity of PAD4.
PAD4 was pre-incubated with no IgG, control IgG, anti-PAD4 only (P4) IgG, or PAD3/PAD4 cross-reactive (XR) IgG prior to use in citrullination assays. (A) Histone H3 titrations were performed at 5 mM calcium and citrullinated H3 was detected by immunoblotting. (B) BAEE titrations were performed at 2mM calcium and citrullination was calculated using an L-citrulline standard curve. The data were fit to the Michaelis-Menten equation. (C) Calcium titrations were performed and data were fit to the Hill equation. Representative data from two experiments P4-IgG (RA27 and RA34), XR-IgG (RA45 and RA16), and no IgG is compiled and shown.
Table 2.
Kinetic parameters for substrate dose responses at saturating calcium
| Substrate | no IgG | Control IgG | RA27-P4 IgG | RA45-XR IgG |
|---|---|---|---|---|
| Histone H3 | ||||
| Km (M) | 2.56 × 10−7 | NA | 3.73 × 10−7 | 3.44 × 10−7 |
| kcat (s−1) | 6.05 × 10−3 | NA | 2.36 × 10−3 | 8.88 × 10−3 |
| kcat/Km (M−1s−1) | 23600 | NA | 6330 | 25800 |
| BAEE | ||||
| Km (M) | 0.96 × 10−3 | 1.13 × 10−3 | 1.36 × 10−3 | 0.25 × 10−3 |
| kcat (s−1) | 0.242 | 0.271 | 0.415 | 0.113 |
| kcat/Km (M−1s−1) | 252 | 241 | 306 | 448 |
NA= not assayed
In the absence of antibody, these two substrates differed markedly in Ca2+ sensitivity: the calcium concentration required for half-maximal citrullination (K0.5) of BAEE is known to be 0.5 mM [6] but for histone H3 half-maximal PAD4 activity required 3.3 ± 0.8 mM Ca2+, well above physiologic concentrations (Figure 4C and Table 3). Cross-reactive antibodies had no effect on the calcium sensitivity for catalysis of BAEE (Figure S2A). However, the prototypic cross-reactive antibody (RA45) produced a significant decrease in the calcium concentration required for half-maximal histone H3 citrullination to 0.5 ± 0.1 mM (p=0.04), while an anti-PAD4 only antibody (RA34, Figure 4C and Table 3) or control antibody (Figure S2B and Table 3) did not (p=0.14). These striking increases in activity are a consequence of the large Hill coefficient and cooperative nature of calcium binding, which ordinarily causes the enzyme to become active over an unusually narrow range of calcium ion concentrations.
Table 3.
Kinetic parameters for histone H3 calcium titrations
| Histone H3 | no IgG* | Control IgG | RA34-P4 IgG* | RA45-XR IgG* |
|---|---|---|---|---|
| K05 Calcium (mM) | 3.3 ± 0.8 | 3.36 | 1.6 ± 0.2 | 0.5 ± 0.1ψ |
| Hill coefficient (n) | 4.3 ± 0.4 | 6.48 | 5.1 ± 0.1 | 2.0 ± 0.6ψ |
Data represents average of 2-4 independent experiments
p<0.05 compared to “no IgG” control using a two-tailed t-test
In the absence of antibodies, PAD4 displayed the expected response, with a Hill coefficient (nominally representing the minimum number of calcium ions involved) of 4.3 ± 0.4. The cross-reactive antibody, RA45, significantly decreased the Hill coefficient to 2.0 ± 0.6 (p=0.03) while PAD4-only (RA34) and control IgG did not, suggesting a decrease in calcium-binding cooperativity (Table 3 and Figure S2B). The smaller Hill coefficient for the cross-reactive antibody implies either a decrease in the number of calcium ions involved, or a decrease in their interaction factors, or both. Consequently, the principal effect of these cross-reactive antibodies appears to lie in restoring the calcium sensitivity (K0.5) of the histone reaction to that for BAEE (and other small molecule substrates [6]), not in changes to the fundamental catalytic structure.
Cross-reactive antibodies modify accessibility to protease cleavage at D388
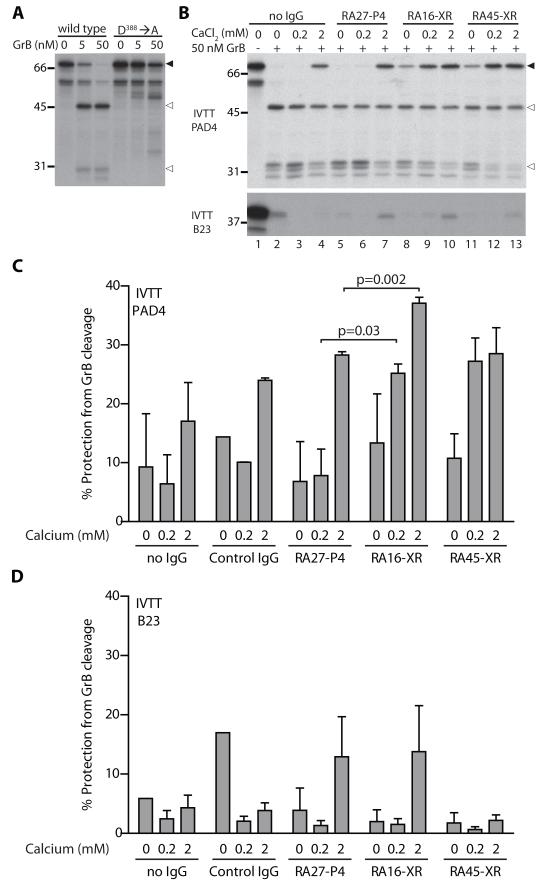
The observation that cross-reactive antibodies enhanced PAD4 activity by modulating calcium sensitivity suggested that they may act to stabilize regions of the protein ordinarily stabilized by calcium ion-binding. Although PAD4 is able to bind five calcium ions, two ions that bind in the C-terminal domain are vital for the formation of the active site and are required for citrullination of BAEE (Ca1 and Ca2) [19]. Three other calcium ions (Ca3, Ca4, and Ca5) bind in a disordered region at the interface of the N- and C-terminal domains to create an alpha helix that is thought to be involved in protein-protein interactions and only minimally affect citrullination of BAEE [19]. It follows that PAD3/PAD4 cross-reactive antibodies may stabilize this region, in turn reducing the requirement for high calcium ion concentrations to maintain the structure needed for interaction with macromolecular substrates. Interestingly, Ca4 is coordinated by aspartic acid at position 388 (D388), and binding of this calcium ion contributes to stabilizing this ordinarily unstructured region of PAD4 [19]. D388 is located within the sequence MGPD388, a tetrapeptide which also satisfies the consensus for cleavage by the serine protease granzyme B (GrB) [20]. Since susceptibility to cleavage by GrB is a frequent feature of autoantigens [21], we addressed whether PAD4 was a GrB substrate. PAD4 was very efficiently cleaved by GrB (kcat/Km = 1.09 × 105 M−1s−1) generating two fragments, an N-terminal 45 kDa fragment and a C-terminal 30 kDa fragment (Figure 5A). Generation of these dominant fragments was abolished by an aspartic acid to alanine mutation at position 388 (MGPD388→MGPA388), confirming that GrB cleaves predominantly at this site.
Figure. 5. Cross-reactive antibodies modify accessibility of PAD4 to protease cleavage at D388.
(A) IVTT wild type or D388→A mutated PAD4 (filled arrows) was incubated with 0, 5, or 50 nM GrB for 1 hour at 37°C and dominant fragments were detected (open arrows). (B) IVTT PAD4 or the negative control IVTT B23 was pre-incubated with no IgG, PAD4-only IgG (P4), or cross-reactive IgG (XR) prior to incubating with 50 nM GrB. (C) Proteins were visualized by radiography and “% Protection from GrB cleavage” was determined by densitometry. Representative data from 3 independent experiments performed with either 5 nM or 50 nM protease is compiled and shown. A two-tailed t-test was performed and a p-value<0.05 was considered significant.
Since protection from and/or alterations in the pattern of digestion by proteases can be employed to interrogate changes in protein structure [22], GrB digestion was used to explore the effect of calcium and cross-reactive antibodies on the structure of PAD4. Interestingly, 2mM calcium inhibited the cleavage of PAD4, (Figure 5B (upper panel, lanes 1-4) and C), showing that the form of PAD4 exposed to these higher concentrations of calcium is resistant to cleavage, likely due to known calcium ion-induced structural changes around D388 [19]. Similar calcium concentrations had minimal effect on cleavage of the known GrB substrate B23 [22], suggesting that this effect is on the substrate and not the protease (Figure 5B (lower panel, lanes 1-4) and D). When PAD4 was pre-incubated with cross-reactive antibodies prior to exposure to GrB, PAD4 proteolysis was inhibited even at lower calcium concentrations (0 and 0.2 mM calcium), (Figure 5B (upper panel, lanes 8-13) and C). Cross-reactive antibodies provided little additional protective effect at 2 mM calcium suggesting that cross-reactive antibodies and calcium are operating through a similar mechanism. This protective effect was not observed with control or PAD4-only antibodies at 0 and 0.2 mM calcium (Figure 5B (upper panel, lanes 5-7) and C), strongly supporting the conclusion that cross-reactive autoantibodies affect GrB cleavage of PAD4 by inducing structural changes, rather than by steric hindrance alone. These results support the proposal that cross-reactive antibodies induce a PAD4 conformation similar to calcium ion binding that is necessary for optimal citrullination of macromolecular substrates, without the requirement for supraphysiologic calcium concentrations.
Discussion
Citrullinated proteins are highly specific targets of the immune response in RA [3] and are present at increased levels in the synovial tissue and fluid of patients [5], suggesting that dysregulated PAD activity may be present in this disease. Although anti-PAD4 antibodies are detected prior to disease onset [15] and are associated with severe erosive RA [13,15,16], a pathogenic role for these antibodies remains undefined. In this study, the discovery of a unique group of anti-PAD4 antibodies that cross-react with PAD3 has defined a RA subset with the most erosive and progressive arthritis. These PAD3/PAD4 cross-reactive antibodies have the striking ability to enhance the sensitivity of PAD4 to calcium, supporting protein citrullination at physiologically relevant calcium concentrations. We propose that this ability to activate PAD4 enzymatic activity has pathogenic consequences in RA.
The striking difference in affinity and calcium requirements for BAEE (Km= 960 uM; K0.5= 0.56 mM) [6] and histone H3 (Km= 0.256 uM; K0.5= 3.3 mM) catalysis suggests that citrullination of macromolecules requires extended substrate-enzyme interactions dependent upon binding of additional calcium ions. Since binding of Ca1 and Ca2 is sufficient for creation of the active site cleft and citrullination of BAEE [19], the low K0.5 for BAEE catalysis likely reflects binding of these two calcium ions. Binding of Ca3, Ca4, and Ca5 by residues remote from the active site, at the interface of the N- and C-terminal domains, likely requires higher calcium concentrations to induce the structural changes required for interaction with macromolecular substrates. The ability of cross-reactive antibodies to shift the calcium requirements for histone citrullination (K0.5= 0.5 ± 0.1 mM with RA45) to a range observed for small molecule substrates suggests that these antibodies can induce a PAD4 structure similar to that normally achieved via Ca3-Ca5 binding. This is further supported by the observation that cross-reactive antibodies can recapitulate the effect of 2 mM calcium and protect PAD4 from proteolysis by GrB at D388, a residue required for the coordination of Ca4 [19]. Thus, these studies suggest a model in which PAD3/PAD4 cross-reactive antibodies bind to the interface between the N- and C-terminal domains and lock PAD4 into a calcium-bound conformation favorable for the citrullination of macromolecular substrates at physiologic calcium concentrations (Figure S3).
Although the pathologic site of autoantigen citrullination by PAD enzymes in RA remains unclear, the detection of soluble PAD2, PAD4, and citrullinated autoantigens in RA synovial fluid suggests that PADs are active extracellularly [23]. Due to the supraphysiologic calcium requirements for PAD4 activity in vitro and the suboptimal free-calcium concentrations present in synovial fluid (0.49-0.98 mM) [9], it is likely that PAD3/PAD4 cross-reactive antibodies may have a major role in revealing PAD4 activity at this site. PADs are clearly also activated intracellularly by several stimuli [4,24]. The discovery of a protein factor that renders PAD4 active against macromolecular substrates in the setting of physiological Ca2+ concentrations suggests a novel mechanism by which PAD4 may be regulated intracellularly in vivo. The existence of endogenous regulatory PAD4-binding partners that perform a similar function during physiologic activation of PADs could explain the striking mismatch between the in vitro and in vivo PAD4 calcium requirements. Identification of such modulators is an important priority.
It is clear that the anti-PAD4 response is polyclonal with multiple epitopes being targeted by a single patient serum and heterogeneity in the epitopes targeted between individuals [7,13]. Autoantibodies binding to distinct PAD4 epitopes may have different effects on PAD4 enzymatic activity. Indeed, a recent study by Auger et al. suggested that anti-PAD4 antibodies may inhibit, enhance, or have no effect on PAD4 function [7]. Although that study has the limitation that saturating concentrations of calcium were used to characterize these antibodies, it is noteworthy that linear epitope mapping was not useful to predict the effect of different antibodies on PAD4 activity and clinical associations were not addressed. The PAD4-activating autoantibodies described in our study were initially defined by their cross-reactivity with PAD3, suggesting a specific conformational epitope on the PAD4 enzyme that modulates calcium sensitivity and enzymatic function. Performing citrullination assays at physiologic calcium concentrations will likely be critical for revealing previously undefined antibody subsets that modulate PAD catalytic function and potentially play a role in RA pathogenesis.
Although we have defined PAD4-activating autoantibodies that are associated with a worse disease outcome in RA, the studies have some limitations. Our study does not address the temporal development of PAD3 autoantibodies during the course of disease. Since antibodies to citrullinated proteins often precede clinical disease, and undergo epitope spreading just prior to disease onset, it would be of great interest to examine the appearance of anti-PAD3 in emerging RA. In addition, longitudinal data was not available to define the variation of these antibodies over time and possible associations with disease activity. Finally, since PAD-activating antibodies likely function in the extracellular environment, direct demonstration that PAD3/PAD4 cross-reactive antibodies activate PAD4 in RA joint cells and fluid in vivo is still needed.
Markers that effectively separate patients into informative disease subsets are often mechanism-based and are useful for prediction, monitoring, and guiding treatment [25]. These studies have identified such an antibody marker that is associated with more erosive disease and strikingly augments the function of a key pathogenic enzyme (PAD4) under physiological conditions. These cross-reactive antibodies may therefore identify RA patients in whom PAD-inhibition would be particularly beneficial therapeutically [26,27].
Materials and Methods
Rheumatoid arthritis subjects and controls
Sera were obtained from 36 healthy controls, 30 psoriatic arthritis patients, 44 RA patients from a convenience sample, and 194 patients from the “Evaluation of Subclinical Cardiovascular Disease and Predictors of Events in Rheumatoid Arthritis” (ESCAPE RA) longitudinal cohort [13]. In ESCAPE RA, disease activity and severity were assessed at baseline and at a final visit occurring an average of 39 ± 4 months post-baseline. Radiographs of the hands and feet were obtained at these visits and scored according to the Sharp-van der Heijde (SvdH) method. All individuals provided informed consent as approved by the Johns Hopkins Institutional Review Board and were followed at the Johns Hopkins Arthritis Center.
Recombinant proteins and antibodies
Recombinant PAD4 and PAD3 proteins were purified as previously described [28] and histone H3 (New England Biolabs, M2503S) was purchased. Immunoblotting was performed using rabbit anti-PAD4 (1:5000, raised against FEGIKKKKQQKIKN), rabbit anti-histone H3 (1:25000), rabbit anti-citrullinated H3 (1:2000, Abcam ab5103), or goat anti-human IgG (1:10000, Jackson 109-036-088).
Immunoprecipitation and blocking
cDNA encoding pCDNA3.1-PAD3 or pEFDEST51-PAD4 was in vitro transcribed and translated (IVTT) in the presence of S35-methionine (Promega, L1170). Radiolabeled proteins were immunoprecipitated with 1 μL of serum in NP40 lysis buffer/0.2%BSA for 1 hour at 4°C. Protein A beads were added and incubated for 30 minutes at 4°C. Beads were washed and boiled in SDS sample buffer. Samples were separated by gel electrophoresis and immunoprecipitated proteins were visualized by radiography. For blocking experiments, patient serum was incubated with 0 or 300 ng of recombinant PAD3 or PAD4 at 4°C for 30 minutes prior to use in IVTT PAD3 or PAD4 immunoprecipitations. Densitometry was performed on all films, values were normalized to a high titer anti-PAD3 serum, and antibody positivity was defined as a normalized densitometry value of >0.01. For blocking experiments, blocking efficiency was calculated as follows: (1- (blocked densitometry value/unblocked densitometry value)) × 100.
Depletion of anti-PAD3 antibodies by ELISA
High-binding ELISA plates were coated with 1000 ng/well of rPAD3 in PBS/0.02% azide overnight at 4°C. Plates were blocked with 3%BSA/PBS/0.02% azide for 2 hours then incubated with 5mM EDTA/PBS at 37°C for 1 hour to chelate free calcium. IgG was added and incubated overnight at 4°C prior to use in citrullination assays or immunoprecipitations.
Citrullination assay
IgG from PAD3/PAD4 cross-reactive, PAD4 only, or PAD-negative sera was purified using Melon Gel columns. 1 μM IgG was incubated with 10 nM recombinant PAD4 for 45 minutes at 4°C in 100 mM Tris-HCl pH 7.4. PAD4/IgG mixtures were mixed with 0-10 μM histone H3.1 in the presence of increasing amounts of CaCl2 and incubated at 37°C for 90-110 minutes. Proteins were boiled in SDS sample buffer, separated by electrophoresis, transferred to nitrocellulose, and immunoblotted as indicated. Molar equivalents of citrullinated H3 were calculated by normalizing to a densitometry value obtained from 1 μM H3 maximally citrullinated with 100 nM PAD4, 5 mM calcium, and DTT. Citrullination of 0-10 mM Benzoyl-Arginine Ethyl Ester (BAEE) (RDI, DR-AV009X) was performed with 50 nM PAD4 and 2 mM calcium and quantified as previously described [6].
Granzyme B cleavage assays
In order to map the GrB cleavage site, pEFDEST51-PAD4 was mutated by site directed mutagenesis (from MGPD388 to MGPA388). Wild-type or mutated IVTT PAD4 was incubated with 0, 5, or 50 nM recombinant human GrB for 1 hour at 37°C in either ICE assay buffer (10 mM Hepes pH 7.4/2 mM EDTA pH 7.4/1% NP-40). For experiments with IgG, IVTT PAD4 was pre-incubated with 5 μg of IgG for 1 hour at 4°C in the presence of 0, 0.2, or 2 mM calcium. The PAD4/IgG mixture was then incubated at 37°C for 1 hour with 50 nM GrB in 100 mM Tris-HCL pH 7.4. GrB cleavage was visualized by radiography and cleavage was quantified by densitometry.
Calculations
Data were fit to the Michaelis-Menten or Hill equations by nonlinear regression using Prism software (GraphPad). The Michaelis-Menten equation was expressed as v = Vmax*[S]/(Km +[S]). The Hill equation was expressed as v = Vmax*[Ca2+]n/(K + [Ca2+]n), with n representing the Hill coefficient. Values for K0.5, the calcium concentration producing half-maximal activation, were calculated from the relation K0.5 = nth root (K) [29]. Granzyme B cleavage efficiency was calculated using the equation, % substrate cleaved = 100 × (1 − e−kcat×[E]/Km×time).
Statistical Methods
The distributions of all variables were examined and compared according to autoantibody status using t-tests for normally distributed continuous variables, the Kruskal-Wallis test for non-normally distributed continuous variables, and the Chi-square or Fisher’s exact test (as appropriate) for categorical variables. Crude and adjusted associations of PAD antibody status with outcomes were explored using ordinary logistic regression or linear regression, appropriate to the outcome, in crude and adjusted models. Variables were normally transformed, as required. Non-contributory confounders were excluded based on the Likelihood Ratio Test for nested models or Akaike’s Information Criterion, as appropriate to the model. Throughout, a two-tailed alpha of 0.05 was used. All calculations were performed using Intercooled Stata 12 (College Station, TX).
Supplementary Material
Acknowledgments
We thank David Hines for providing technical support and Dr. Mark J. Soloski for providing psoriatic arthritis patient sera.
Funding: Supported by the Sibley Memorial Hospital grant, Donald B. and Dorothy L. Stabler Foundation, ACR-REF Within our Reach grant, NIH grant T32 AR048522, and NIH P30 AR053503.
Footnotes
Author contributions: ED discovered and characterized the effects of cross-reactive antibodies, and was involved in study design and data analysis. JTG provided ESCAPE RA sera samples, clinical data, and performed statistical analysis. MLO provided PAD4 cDNA constructs, determined the granzyme B cleavage site, and observed that calcium inhibits GrB cleavage of PAD4. FA provided PAD3 cDNA constructs and protein and was involved in study design. HB guided and interpreted enzyme kinetic and calcium-dependence studies. AR guided study design and data analysis/interpretation. All authors contributed to preparation of the manuscript.
Competing interests: Authors Antony Rosen, Erika Darrah, Felipe Andrade, and Jon T. Giles are authors on a provisional patent application entitled “Human Autoantibodies specific for PAD3 which are cross-reactive with PAD4 and their use in the diagnosis and treatment of rheumatoid arthritis” (P12072-01) that was filed with the USPTO. The other authors declare no competing interests.
References and Notes
- 1.Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat. Rev. Rheumatol. 2012;8:573–586. doi: 10.1038/nrrheum.2012.134. [DOI] [PubMed] [Google Scholar]
- 2.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessay. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 3.Willemze A, Trouw LA, Toes RE, Huizinga TW. The influence of ACPA status and characteristics on the course of RA. Nat. Rev. Rheumatol. 2012;8:144–152. doi: 10.1038/nrrheum.2011.204. [DOI] [PubMed] [Google Scholar]
- 4.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 5.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Mechin MC, Vincent C, Nachat R, Yamada M, Takahara H, Simon M, Guerrin M, Serre G. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 6.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 7.Auger I, Martin M, Balandraud N, Roudier J. Rheumatoid arthritis-specific autoantibodies to peptidyl arginine deiminase type 4 inhibit citrullination of fibrinogen. Arthritis Rheum. 2010;62:126–131. doi: 10.1002/art.27230. [DOI] [PubMed] [Google Scholar]
- 8.Pollmann S, Stensland M, Halvorsen EH, Sollid LM, Kvien TK, Fleckenstein B, Molberg O. Anti-PAD4 autoantibodies in rheumatoid arthritis: levels in serum over time and impact on PAD4 activity as measured with a small synthetic substrate. Rheumatol. Int. 2011 doi: 10.1007/s00296-010-1765-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson WG, Marshall RW. Ionized calcium in body fluids. Crit. Rev. Clin. Lab. Sci. 1981;15:85–125. doi: 10.3109/10408368109105869. [DOI] [PubMed] [Google Scholar]
- 10.Hill HR, Augustine NH, Jaffe HS. Human recombinant interferon gamma enhances neonatal polymorphonuclear leukocyte activation and movement, and increases free intracellular calcium. J. Exp. Med. 1991;173:767–770. doi: 10.1084/jem.173.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong K, Kwan-Yeung L. Sphingosine mobilizes intracellular calcium in human neutrophils. Cell Calcium. 1993;14:493–505. doi: 10.1016/0143-4160(93)90008-t. [DOI] [PubMed] [Google Scholar]
- 12.Halvorsen EH, Pollmann S, Gilboe IM, van der Heijde D, Landewe R, Odegard S, Kvien TK, Molberg O. Serum IgG antibodies to peptidylarginine deiminase 4 in rheumatoid arthritis and associations with disease severity. Ann. Rheum. Dis. 2008;67:414–417. doi: 10.1136/ard.2007.080267. [DOI] [PubMed] [Google Scholar]
- 13.Harris ML, Darrah E, Lam GK, Bartlett SJ, Giles JT, Grant AV, Gao P, Scott WW, Jr, El-Gabalawy H, Casciola-Rosen L, Barnes KC, Bathon JM, Rosen A. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum. 2008;58:1958–1967. doi: 10.1002/art.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Zhao Y, He J, Jia R, Li Z. Prevalence and significance of anti-peptidylarginine deiminase 4 antibodies in rheumatoid arthritis. J. Rheumatol. 2008;35:969–974. [PubMed] [Google Scholar]
- 15.Kolfenbach JR, Deane KD, Derber LA, O’Donnell CI, Gilliland WR, Edison JD, Rosen A, Darrah E, Norris JM, Holers VM. Autoimmunity to peptidyl arginine deiminase type 4 precedes clinical onset of rheumatoid arthritis. Arthritis Rheum. 2010;62:2633–2639. doi: 10.1002/art.27570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halvorsen EH, Haavardsholm EA, Pollmann S, Boonen A, van der Heijde D, Kvien TK, Molberg O. Serum IgG antibodies to peptidylarginine deiminase 4 predict radiographic progression in patients with rheumatoid arthritis treated with tumour necrosis factor-alpha blocking agents. Ann. Rheum. Dis. 2009;68:249–252. doi: 10.1136/ard.2008.094490. [DOI] [PubMed] [Google Scholar]
- 17.Darrah E, Rosen A, Giles JT, Andrade F. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann. Rheum. Dis. 2012;71:92–98. doi: 10.1136/ard.2011.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Heijde D. How to read radiographs according to the Sharp/van der Heijde method. J. Rheumatol. 1999;26:743–745. [PubMed] [Google Scholar]
- 19.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 20.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 21.Casciola-Rosen L, Andrade F, Ulanet D, Wong WB, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J. Exp. Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulanet DB, Torbenson M, Dang CV, Casciola-Rosen L, Rosen A. Unique conformation of cancer autoantigen B23 in hepatoma: a mechanism for specificity in the autoimmune response. Proc. Natl. Acad. Sci., U. S. A. 2003;100:12361–12366. doi: 10.1073/pnas.2035245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, Saxne T, Malmstrom V, Venables PJ. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum. 2008;58:2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 24.Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J. Immunol. 2008;180:1895–1902. doi: 10.4049/jimmunol.180.3.1895. [DOI] [PubMed] [Google Scholar]
- 25.Rosen A, Casciola-Rosen L. Autoantigens in systemic autoimmunity: critical partner in pathogenesis. J. Intern. Med. 2009;265:625–631. doi: 10.1111/j.1365-2796.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. N-alpha-benzoyl-N5-(2-chloro-1-iminoethyl)-L-ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J. Immunol. 2011;186:4396–4404. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X. Autocitrullination of human peptidylarginine deiminase 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010 doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segel IH. Enzyme Kinetics. John Wiley and Sons; New York: 1975. pp. 360–361. chapter. [Google Scholar]
- 30.Delano W. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.