Summary
Subcellular protein localization is thought to promote protein-protein interaction by increasing the effective concentration and enabling spatial coordination and proper segregation of proteins. We found that protein overexpression allowed the assembly of a productive polysaccharide biosynthesis-export-anchoring complex in the absence of polar localization in Caulobacter crescentus. Polar localization of the holdfast export protein, HfsD, depends on the presence of the other export proteins, HfsA, and HfsB, and on the polar scaffold protein PodJ. The holdfast deficiency of hfsB and podJ mutants is suppressed by the overexpression of export proteins. Restored holdfasts are randomly positioned and co-localize with a holdfast anchor protein in these strains, indicating that functional complexes can form at non-polar sites. Therefore, overexpression of export proteins surpasses a concentration threshold necessary for holdfast synthesis. Restoration of holdfast synthesis at non-polar sites reduces surface adhesion, consistent with the need to spatially coordinate the holdfast synthesis machinery with the flagellum and pili. These strains lack the cell-specific segregation of the holdfast, resulting in the presence of holdfasts in motile daughter cells. Our results highlight the fact that multiple facets of subcellular localization can be coupled to improve the phenotypic outcome of a protein assembly.
Keywords: Caulobacter, asymmetry, bacterial adhesion, holdfast, polysaccharide, protein localization
Introduction
The coordinated localization of proteins at specific subcellular sites provides multiple advantages. For example, asymmetric localization of proteins prior to cell division results in their differential inheritance into daughter cells. Protein localization results in high effective concentration, which can promote protein-protein interaction (Nooren & Thornton, 2003), as demonstrated in many multiprotein assemblies in eukaryotic cells (Good et al., 2011, Kuersten et al., 2002, Benton & Johnston, 2003). Bacteria often localize proteins at their poles (Rudner & Losick, 2010, Curtis & Brun, 2010). Polar localization can have regulatory consequences, for example in the control of cell differentiation in Caulobacter crescentus; impart topological constraints, for example the concentration of the FtsZ inhibitors MinCD at the poles to ensure division at the midcell; or are important for optimized function, for example the polar localization of Shigella IcsA to form a polymerized actin tail to propel the cell (Rudner & Losick, 2010, Curtis & Brun, 2010). Finally, subcellular localization is important for the assembly of multiprotein machineries, such as pili biosynthesis machinery or external structures such as the flagellum. For instance, in the absence of the FlhF protein found in Vibrio, Pseudomonas, Helicobacter, and Campylobacter, flagella are synthesized randomly instead of their normal polar localization, and the efficiency of flagellum synthesis is decreased (Kirkpatrick & Viollier, 2011).
In this paper, we investigate the link between subcellular localization and the assembly and activity of the polysaccharide biosynthesis machinery for the polar holdfast of C. crescentus. Localized extracellular polysaccharide (EPS) is common in several species of Alphaproteobacteria that use a polar polysaccharide for surface attachment (Chertkov et al., 2011, Brown et al., 2009, Tomlinson & Fuqua, 2009, Pate et al., 1973, Poindexter, 1964, Moore & Marshall, 1981, Laus et al., 2006). One such polysaccharide, the C. crescentus holdfast, mediates a cell-to-surface attachment of impressive strength (Tsang et al., 2006). Even in bacterial species whose capsular polysaccharide (CPS) covers the entire cell, several lines of evidence indicate that biosynthesis can take place in localized regions of the cell envelope. De novo capsule synthesis of both group I and group II has been observed as randomly located patches (Kroncke et al., 1990, Bayer & Thurow, 1977), the biosynthesis of E. coli K5 capsule occurs at both poles (McNulty et al., 2006), and a portion of S. pneumoniae capsule is exported at the cell division septum (Henriques et al., 2011).
The C. crescentus holdfast is composed, at least in part, of N-acetylglucosamine (GlcNAc) polymer (Merker & Smit, 1988). The holdfast is first synthesized at the flagellar pole of swarmer cells and is ultimately located at the tip of the stalk after stalk growth at the site of holdfast biogenesis (Fig. 1C). Holdfast synthesis is regulated by developmental and surface contact cues (Bodenmiller et al., 2004, Levi & Jenal, 2006, Li et al., 2012). The polar developmental regulatory pathway triggers holdfast synthesis late in the swarmer phase in cells that do not contact a surface (Levi & Jenal, 2006). In addition, holdfast synthesis is rapidly stimulated at any time after cell division in order to permanently adhere swarmer cells that transiently interact with a surface via a flagellum and pili (Li et al., 2012). The co-localization of the site of holdfast synthesis with pili and the flagellum is important for optimal adhesion of cells to surfaces (Li et al., 2012).
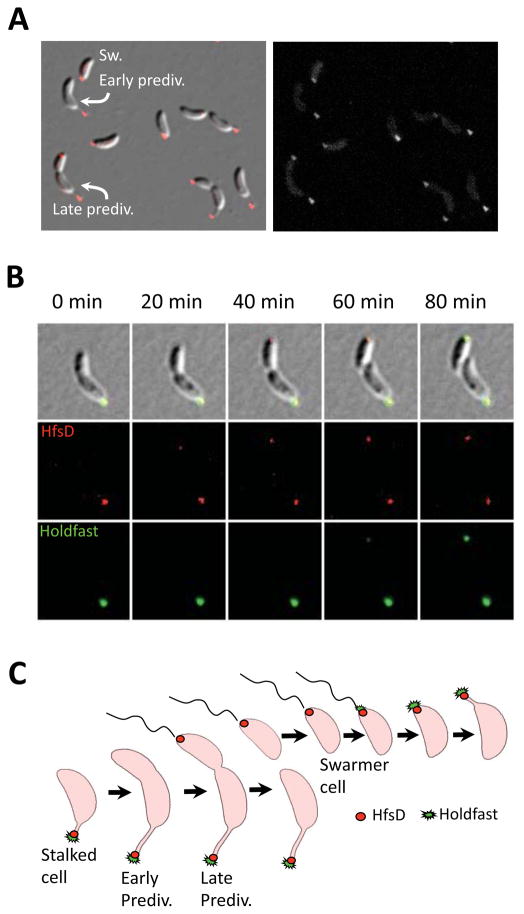
Figure 1.
HfsD-mCherry exhibits polar localization that is cell cycle dependent. (A) HfsD-mCherry localization in a mixed culture. Swarmer cells (Sw) have polar localization. HfsD-mCherry localizes at the tip of the stalk in stalked cells. Late predivisional cell (prediv) HfsD-mCherry localization is at the tip of the stalk and at the swarmer pole. (B) Time-lapse fluorescence microscopy of HfsD-mCherry (see Movie S1). At 0 min, the HfsD-mCherry focus (red) is at the tip of a short stalk on an elongating cell. As the predivisional cell starts to constrict, a second HfsD-mCherry focus appears at the flagellar pole. HfsD-mCherry remains at this pole after cell division, confirming that HfsD-mCherry localizes to the swarmer pole of swarmer cells. Holdfast (green) does not appear until after cell division (60 minute panel). (C) Model of HfsD cell-cycle localization pattern.
Holdfast biogenesis is mediated by the hfsEFGHDABC genes (Smith et al., 2003, Toh et al., 2008), which are homologous to the genes responsible for Wzy/Wzx-dependent biogenesis of lipopolysaccharide (LPS) O-antigen, CPS, and free EPS in a variety of Gram-negative and Gram-positive species (Paulsen et al., 1997). The hallmark of the Wzy/Wzx-dependent pathway is the synthesis of oligosacchride repeat units in the cytoplasm, which are transferred to the periplasm for polymerization into a large macromolecule (Whitfield, 2006). In Gram-negative bacteria, an additional trans-envelope complex is required to export CPS and EPS across the outer membrane (OM). Two families of proteins, OPX (OM polysaccharide export) and PCP (polysaccharide co-polymerase) make up this complex (Cuthbertson et al., 2009). OPX proteins, like E. coli Wza, are believed to form the OM secretion pore for the polysaccharide. Caulobacter HfsD is a homolog of Wza. PCPs, like E. coli Wzc, are inner membrane (IM) proteins with two transmembrane domains, a periplasmic loop that interacts with the OPX, and a cytoplasmic tyrosine autokinase domain (Cuthbertson et al., 2009). Caulobacter holdfast PCPs are among a handful of Gram negative PCPs encoded by two separate genes (Cuthbertson et al., 2009). HfsA is homologous to the IM periplasmic loop domain. HfsB is homologous to the autokinase domain, however, it lacks highly conserved ATP-binding residues and a conserved tyrosine-rich domain which serves as the autophosphorylation site. OPX and PCP mutants in the Wzx/Wzy-dependent pathway lose the ability to polymerize more than a few repeat units of the normally high molecular weight EPS (Cuthbertson et al., 2009). Cells lacking HfsD, HfsA, or HfsB do not produce holdfasts or attach to surfaces (Smith et al., 2003).
Once exported, the holdfast is anchored to the cell envelope by the holdfast anchor proteins, HfaA, HfaB, and HfaD (Cole et al., 2003, Hardy et al., 2010, Kurtz & Smith, 1992). Without these proteins, holdfast is shed into the medium. The anchor protein, HfaB, is homologous to CsgG, the predicted translocon for the E. coli fimbrial curlin, CsgA (Hardy et al., 2010). HfaA and HfaD anchor proteins have similarity to extracellular adhesins. All three anchor proteins localize to the flagellar pole of predivisional cells and persist at that pole after cell division and cell differentiation; during stalk synthesis, they are pushed out to the stalk tip, along with the holdfast. Polar localization of the three anchor proteins is lost in a ΔhfsDAB triple mutant as well as a deletion mutant of the polar development regulator gene podJ, which is necessary for holdfast synthesis (Hardy et al., 2010).
In this study, we show that polar localization is important for the assembly of active holdfast biosynthesis machinery. The rapidity of surface contact stimulation of holdfast synthesis suggested that the holdfast synthesis machinery is already localized to the flagellar pole in newborn swarmer cells, which is consistent with the polar localization of the holdfast anchor proteins at the flagellar pole of predivisional cells. We found that the Wza-like protein HfsD has the same localization pattern as the holdfast anchor proteins. Although this localization is lost in the holdfast-minus ΔpodJ and ΔhfsB mutants, holdfast synthesis can be restored when any holdfast export protein is overexpressed. Restored holdfasts were often randomly located, but were anchored to the cell, suggesting the formation of functional holdfast synthesis-export-anchoring complexes. Our model suggests that interactions between the holdfast anchor proteins and the holdfast export complex are destabilized in ΔpodJ or ΔhfsB mutants because these components are spread out across the entire cell body, instead of concentrated at a pole. We hypothesize that, in addition to the coordination of the flagellum, pili, and holdfast for surface adhesion, one important function of polar localization of holdfast synthesis components is to achieve the high effective concentration required for the assembly of functional holdfast synthesis machinery.
Results and Discussion
HfsD exhibits cell cycle dependent polar localization
We have recently found that the holdfast anchor proteins HfaA, HfaB, and HfaD, are first localized at the flagellar pole of predivisional cells and subsequently persist at the same pole after holdfast synthesis (Hardy et al., 2010). We hypothesized that the holdfast export proteins would exhibit a similar pattern given their likely participation in a multiprotein complex with anchor proteins and the fact that the localization of the holdfast anchor proteins depends on the holdfast export machinery (Hardy et al., 2010). HfsD localization was observed in vivo using a C-terminal mCherry fusion to HfsD created by integrating a C-terminal mcherry fusion at the 3′ end of hfsD into the genomic coding region of hfsD. Western blots confirmed the presence of a fusion protein of the correct size (Fig. S1A). The number of cells of the hfsD-mcherry fusion strain synthesizing holdfasts was ~40% compared to the wild-type (Fig. S1B). This might be explained by a comparable reduction in HfsD abundance as shown by Western blot (Fig. S1A). We were unable to obtain functional fluorescent protein fusions with HfsA and HfsB, nor were we able to localize them by immunofluorescence microscopy.
Epifluorescence microscopy showed that almost 97% of HfsD-mCherry exhibits polar localization across all cell types (Fig. 1A and 3D). We observed HfsD localization at the tip of the stalk, where the holdfast is found. Late predivisional cells exhibited bipolar localization, which suggests that HfsD localizes to the flagellar pole of swarmer cells and then gets pushed to the tip of the stalk as it elongates. This hypothesis was confirmed using time-lapse microscopy (Fig. 1B and Movie S1). HfsD first localized to the flagellar pole of late predivisional cells, where it remained in swarmer cells, culminating in its localization at the tip of the stalk after cell differentiation. The localization of HfsD to the swarmer pole before cell separation places it at the future pole of holdfast synthesis well in advance of holdfast export (Fig. 1C), which occurs at the swarmer pole late in the swarmer cell stage for planktonic cells (Levi & Jenal, 2006, Li et al., 2012). The localization pattern exhibited by HfsD is the same as that of the HfaB anchor protein (Hardy et al., 2010). From these data, we conclude that localization of HfsD is not the signal for the timing of holdfast synthesis. We hypothesize that the early localization of the holdfast anchor and synthesis/export complex allows new swarmer cells to start producing holdfast as soon as they come into contact with a surface.
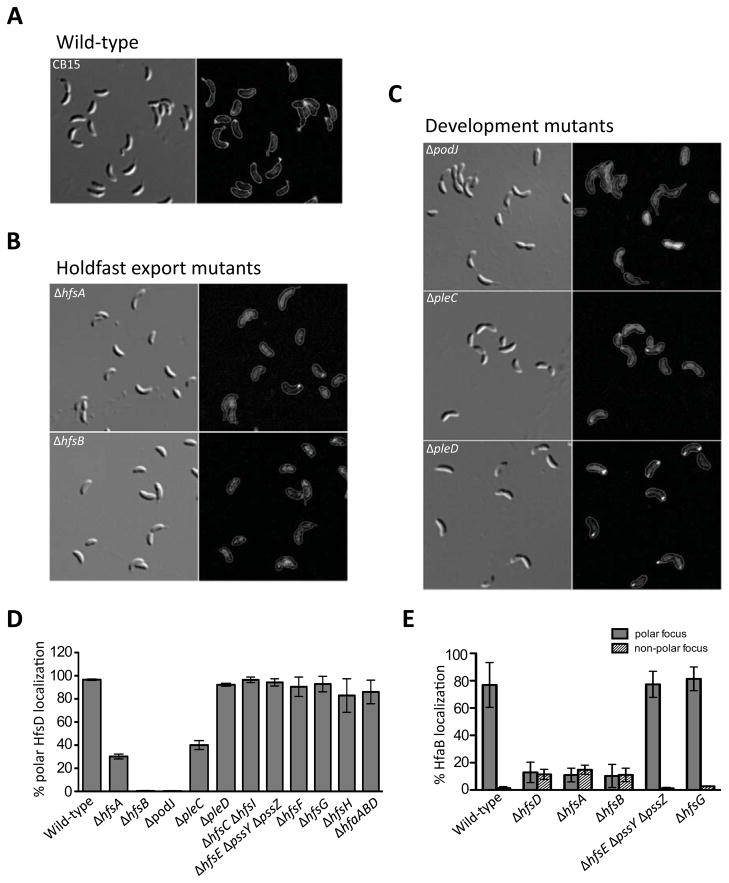
Figure 3.
Localization of HfsD-mCherry is dependent on holdfast export proteins and polar development proteins. DIC images (left-hand columns) and HfsD-mCherry fluorescence signal, with the outlines of the cell bodies overlaid (right-hand columns) are shown for the following mutants: (A) Wild-type CB15, (B) holdfast export mutants ΔhfsA and ΔhfsB, and (C) polar development mutants ΔpodJ, ΔpleC and ΔpleD. The contrast was adjusted similarly for each fluorescence image. (D) The percentage of cells that have a polar HfsD focus was calculated. Displayed are the average percentages from two to five independent cultures and the standard error of the mean. The number of cells counted for each culture ranged from 73 to 236. (E) The percentage of total cells that have a polar HfaB focus was calculated (gray bars). The percentage of total cells that have a non-polar HfaB focus was calculated (striped bars). Displayed are the average percentages from 3 independent cultures and the standard error of the mean. The total number of cells counted ranged from 120 to 500.
In order to determine if the appearance of HfsD at the swarmer pole of predivisional cells coincided with increased holdfast export synthesis, we measured their steady-state levels in samples of synchronized cells throughout the cell cycle (Fig. 2). HfsA, HfsB, and HfsD were present throughout the cell cycle and their level did not vary dramatically. The chemotaxis protein, McpA was used as a synchrony control. McpA is present in swarmer cells, undergoes proteolysis during the swarmer-to-stalked transition, then is re-synthesized at the pre-divisional stage (Alley et al., 1991, Tsai & Alley, 2001).
Figure 2.
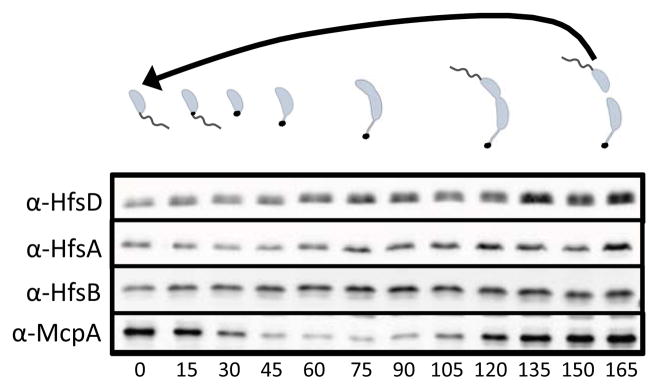
Abundance of holdfast export proteins during the cell cycle. Samples were taken every 15 min during a time course. The resulting Western blot was probed with α-HfsD, α-HfsA, α-HfsB, and α-McpA antibodies. The chemoreceptor protein McpA, which undergoes cell-cycle regulated proteolysis, was used as a synchrony control. The approximate developmental stage of the cell cycle is depicted above.
HfsD polar localization depends on HfsA and HfsB, but not on holdfast synthesis or anchor proteins
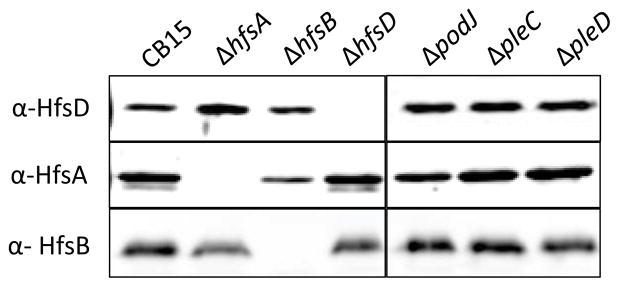
EPS export and synthesis proteins are predicted to work together in a large multiprotein complex (Whitfield, 2006). Furthermore, interactions have been demonstrated between Wza (HfsD homolog) and Wzc (HfsAB homolog) (Collins et al., 2007) as well as between homologs of HfsA and HfsB (Soulat et al., 2006). Thus, we reasoned that the HfsA or HfsB export proteins could play a role in HfsD localization. Indeed, HfsD polar localization was abolished in ΔhfsB and severely disrupted in ΔhfsA, albeit not completely (Fig. 3A and B). Around 30% of ΔhfsA cells counted had at least one HfsD focus (Fig. 3D), but many of these foci were much smaller and dimmer than those found in wild-type. The stronger HfsD localization defect in the ΔhfsB mutant was surprising, since, based on sequence similarity, HfsA is predicted to interact directly with HfsD, whereas HfsB is predicted to interact indirectly with HfsD through HfsA. Furthermore, there is a drop in HfsA abundance in a ΔhfsB mutant, suggesting that HfsA requires HfsB for stability (Fig. 4).
Figure 4.
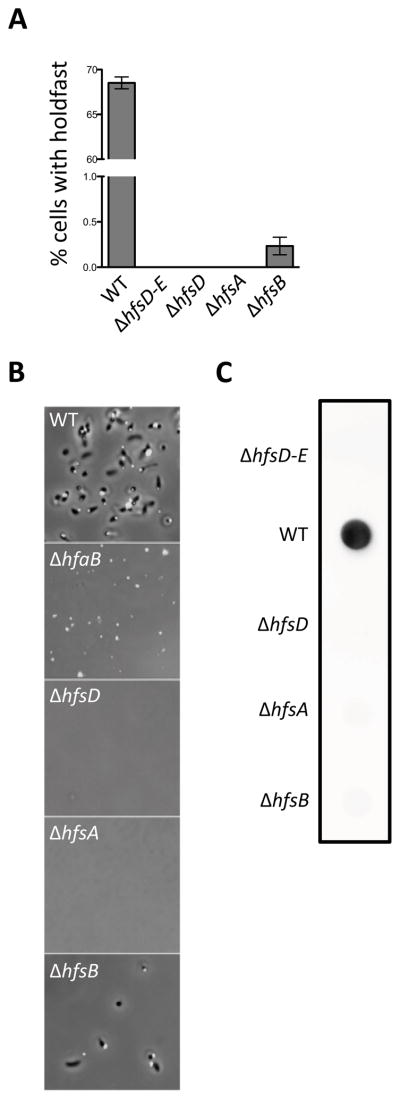
Deletion mutants of holdfast export genes are strongly deficient in holdfast synthesis. (A) Quantification of the number of planktonic cells that bind Alexa Fluor 488 conjugated WGA. Counts were taken from two to four independent cultures with between 1200 and 2800 cells counted for most samples. Data is expressed as a mean percent of the wild-type for each culture with the standard error of the mean. (B) Small volume cell attachment assay and holdfast labeling. DIC and fluorescence image overlays that are representative of the density of cells remaining attached to a glass slide after washing are shown. Additional panels can be found in Figure S4. (C) Total complement of holdfast polysaccharide in whole cell lysates as shown by HRP-WGA dot blot. A row of spots from the same blot representative of several trials is shown.
Since homologs of HfsA/B are predicted to have a role in CPS and EPS polymerization as well as its export, probably involving interaction with members of the biosynthesis machinery (Cuthbertson et al., 2009, Whitfield & Larue, 2008, Galván et al., 2013), we hypothesized that there might be a requirement of certain holdfast synthesis proteins for HfsD localization. We analyzed HfsD-mCherry localization in deletion mutants of each known holdfast synthesis gene and found that its subcellular localization pattern was unaffected (Fig. 3D and S2A), indicating that holdfast synthesis proteins (HfsCEFGHI) are not necessary for HfsD localization. Similarly, we showed that the polar localization of the OM holdfast anchor protein HfaB was not dependent on holdfast synthesis (Fig. 3E and S3A). Since polar localization of the HfaB anchor protein depends on HfsA, HfsB, and/or HfsD (Hardy et al., 2010; Fig. 3E and S3B), we tested if HfsD requires the holdfast anchor for polar localization, and found that HfsD polar localization was not altered in the triple anchor mutant, ΔhfaABD (Fig. 3D and S2B).
In each strain expressing HfsD-mCherry there is a weak background mCherry fluorescence dispersed throughout the entire cell body. This could be due to the formation of a non-functional HfsD degradation product still fused to functional mCherry, as suggested by Western blot (Fig. S1). Alternatively, a small pool of de-localized HfsD might persist at all cell cycle stages and only a portion becomes localized at the late pre-divisional stage.
Deletion of holdfast export proteins abolishes holdfast biogenesis
The loss of HfsD localization in mutants lacking HfsA and HfsB supports the hypothesis that these three proteins must interact to bring about holdfast export. We therefore decided to further investigate the individual contribution of each protein to holdfast synthesis. The holdfast is the only extracellular structure that binds wheat germ agglutinin (WGA), a lectin that binds specifically to GlcNAc polymers (Merker & Smit, 1988). Using fluorescent WGA to label and quantify external holdfast from a large number of cells, we found that the ΔhfsD and ΔhfsA mutants did not synthesize holdfasts (Fig. 5A), supporting earlier results (Smith et al., 2003). However, a very small percentage of ΔhfsB mutant cells synthesized holdfast. This difference between ΔhfsB and ΔhfsD and ΔhfsA became more apparent when observing cells attached to a glass slide after incubation of a small volume of cell culture on its surface. The small volume ensures that cells frequently come into contact with the surface, thereby maximizing adhesion. Therefore, mutants with very small percentages of holdfast-synthesizing cells can be “fished out” of the culture. The hfsA and hfsD mutants were unable to attach, whereas holdfast-bearing hfsB mutant cells were readily detectable at the glass surface, albeit at a much lower surface coverage than wild-type cells (Fig. 5B and S4).
Figure 5.
Abundance of holdfast export proteins in holdfast export and in polar development mutants as determined by Western blot using whole cell lysates.
In E. coli ABC-transporter-dependent systems, which have limited sequence similarity to Wzy/Wzy-dependent systems, mutating CPS export genes results the entrapment of high molecular weight polysaccharides in the periplasm (Silver et al., 1987, Bronner et al., 1993). On the other hand, when wza (hfsD homolog) or wzc (hfsA-hfsB homolog) are mutated in the Wzx/Wzy-dependent pathway, CPS repeat unit polymerization is completely abolished (Drummelsmith & Whitfield, 2000, Wugeditsch et al., 2001, Reuber & Walker, 1993, Vojnov et al., 1998). Even though HfsD, HfsA, and HfsB are homologous to Wzx/Wzy dependent EPS export proteins, the sequence similarity is low (Smith et al., 2003), which led us to question if holdfast material could be trapped inside the cell in holdfast export mutants.
Incubating cells with fluorescent WGA, as described in the previous section, only labels exported holdfast. We measured the entire complement of holdfast in whole cell lysates using a dot blot assay, probing with WGA conjugated to horseradish peroxidase. Wild-type CB15 cells exhibited a strong signal. In stark contrast, each of the holdfast export mutants had a signal that was similar to that of a strain with a deletion of the entire holdfast synthesis and export gene cluster (Fig. 5C). We suspect that the small percentage of ΔhfsB cells that make holdfast is under the detection limit of this assay.
These results suggest that each component of the holdfast export machinery is necessary for the synthesis of the holdfast polysaccharide, not just its export, perhaps as the result of feedback inhibition or by causing instability in a multiprotein complex. To evaluate the possibility that the link between export and synthesis is at the transcriptional level, we measured holdfast synthesis gene expression in a ΔhfsD mutant using a transcriptional lacZ fusion. There was no change in expression between the mutant and wild-type (Fig. S5), indicating that the holdfast synthesis defect of this mutant is not due to feedback inhibition of holdfast synthesis gene transcription.
We predict that HfsA, HfsB, and HfsD interact based on similarity to Wza and Wzc, the fact that HfsA and HfsB are necessary for HfsD subcellular localization, and the fact that single deletion mutants of each export gene has virtually the same phenotype. To further investigate the interrelationship of the holdfast export proteins, we determined if their abundance was affected by the lack of the other export proteins. The abundance of HfsA was reduced in a ΔhfsB mutant to about 22% (± 5, standard error) of wild-type as revealed by densitometry of three separate blots (Fig. 4). There was no substantial difference in HfsA abundance for the rest of the mutants tested. The abundance of HfsD and HfsB was similar in all mutants tested.
The effect of developmental regulators on holdfast export proteins
The polar development proteins, PodJ, PleC, and PleD are polarly localized and are necessary for optimal holdfast biogenesis. podJ and pleC mutants do not synthesize holdfasts (Wang et al., 1993, Smith et al., 2003) and pleD mutants exhibit delayed holdfast synthesis (Levi and Jenal, 2006). Furthermore, PodJ is required for polar localization of the HfaB anchor protein (Hardy et al., 2010). We hypothesized that any of these proteins could have a role in HfsD localization. Epifluorescence microscopy showed that wild-type HfsD-mCherry localization is abolished in a podJ mutant and severely impaired, albeit not totally abolished, in a pleC mutant (Fig. 3C and 3D). About 40% of the ΔpleC cells counted had at least one HfsD focus, but most of these foci were smaller and dimmer than those found in the wild-type. In contrast, HfsD-mCherry polar localization was normal in a pleD mutant.
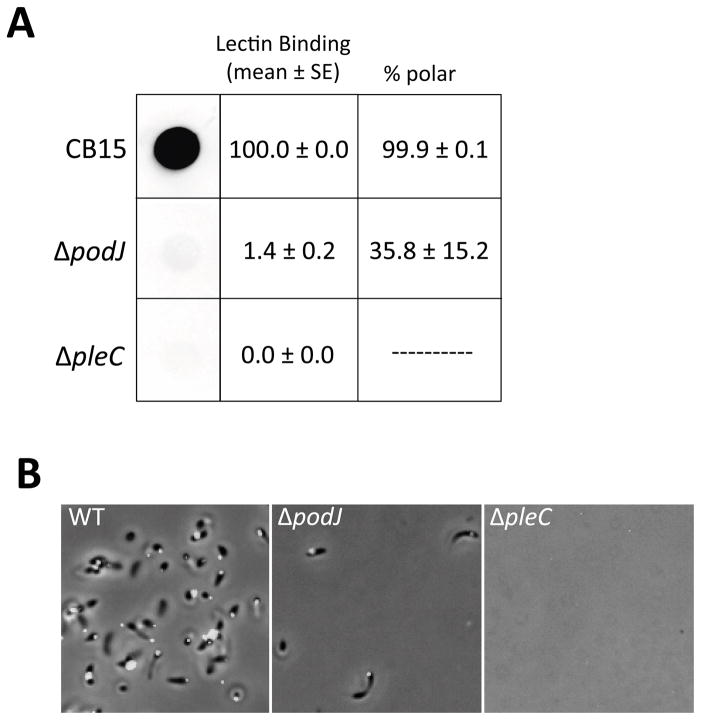
The difference in HfsD localization in the podJ and pleC mutants prompted us to analyze the holdfast deficiency in those mutants more closely. Lectin staining and dot blot analysis indicated that the podJ mutant synthesized holdfasts at a very low frequency and that only about one-third of these holdfasts were localized to the cell pole (Fig. 6). In contrast, the pleC mutant was completely deficient in holdfast synthesis. As with the ΔhfsB mutant, ΔpodJ cells could be readily “fished out” of a culture using the small volume attachment assay (Fig. 6B and S4). In contrast to attached ΔhfsB cells (Fig. 5B), attached ΔpodJ cells often had mis-localized holdfasts (Fig. 6 and S4). These results suggest that the holdfast synthesis machinery is still localized properly but much less active in a hfsB mutant, while it is mostly delocalized in a podJ mutant.
Figure 6.
A small proportion of ΔpodJ cells can synthesize holdfast whereas holdfast is never detected in a ΔpleC mutant. (A) Left panel: Total amount of holdfast in a whole culture as detected using a dot blot. Representative spots were cropped from the same blot. Middle panel: The percentage of individual planktonic cells that bind fluorescent WGA was quantified and normalized to wild-type as a reference strain. The mean and its standard error are shown. Cells (200 to 500 per sample) were counted from four independent cultures and then averaged after normalization. Right panel: The percentage of total polarly localized holdfasts was calculated. The mean and its standard error are shown. (B) Developmental mutants were tested using the small volume cell attachment assay and fluorescent holdfast labeling. DIC and fluorescent image overlays that are representative of the density of cells remaining attached to a glass slide after washing are shown. Additional panels can be found in Figure S4.
We have seen that PodJ plays a role in localizing the biosynthesis machineries of polar structures such as the pili and the holdfast (Viollier et al., 2002; and this work). PodJ also has a role in ensuring proper intracellular signaling through its function as a localization factor for many developmental regulators, such as PleC (Curtis et al., 2012, Hinz et al., 2003, Duerig et al., 2009). The fact that a ΔpodJ mutant still synthesizes a small amount of holdfast indicates that some, albeit low, PleC activity persists even when it is delocalized in a PodJ mutant. Although a small amount of HfsD is localized at the proper pole in a pleC mutant, holdfast synthesis is completely abolished. Therefore, PleC signaling is required for holdfast synthesis even if it is partially dispensable for the localization of the holdfast export complex.
Holdfast synthesis of podJ and hfsB mutants is restored by overexpression of holdfast export proteins
Our findings that ΔpodJ and ΔhfsB mutants produce a small but measurable amount of holdfast polysaccharide by comparison to ΔpleC, ΔhfsD, and ΔhfsA mutants (Fig. 5 and 6), suggest that the defects of the podJ and hfsB mutants may be regulatory or due to problems in the assembly or localization of the holdfast synthesis machinery. Further evidence to support this idea is that the abundance of HfsA is substantially reduced in a ΔhfsB mutant (Fig. 4). McNulty et al (2006) has suggested that polar localization serves to increase the effective concentration of CPS export proteins, which have low expression. We therefore hypothesized that overexpression of holdfast export proteins might restore holdfast synthesis.
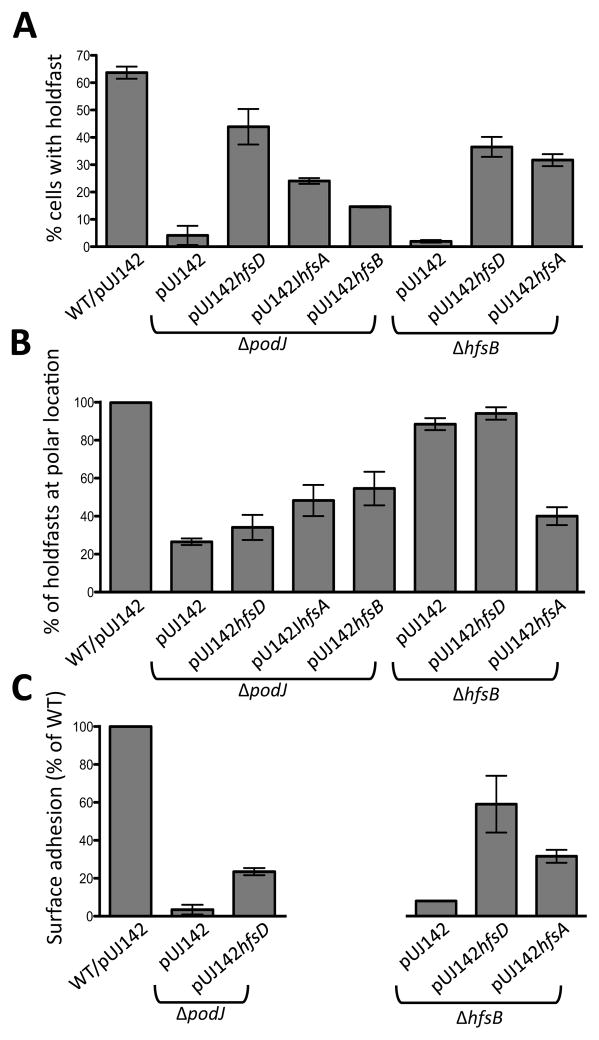
Overexpression of HfsD restored holdfast synthesis in ΔpodJ (Fig. 7A). Lectin staining demonstrated that the proportion of cells displaying holdfast was ~70% of wild-type, but only 34% of these had a polar holdfast (Fig. 7B). This was similar to the polar localization of holdfast in the ΔpodJ parent strain where 27% of holdfasts were polarly localized for the small number of cells that produced them. In contrast, over 99% of holdfasts were polarly localized in wild-type cells. Overexpression of HfsA and HfsB also restored holdfast synthesis to the ΔpodJ mutant, but to a lesser extent than overexpression of HfsD, and about half of the holdfasts were polar (Fig. 7B). The lack of polar localization of restored holdfast in these strains suggests a requirement of PodJ for holdfast export complex localization. There was no effect of overexpressing HfsD in a ΔpleC mutant, or a ΔpodJ ΔpleC double mutant, indicating that PleC is epistatic to PodJ in this pathway (Fig. 8). The respective overexpression strains had a significantly increased abundance of HfsD, HfsA, and HfsB (Fig. S6A). Overexpression of each holdfast export protein in the wild-type background did not impact the amount of WGA- staining holdfast material or the polar localization of holdfasts (Fig. S6B and S6C). Finally, the holdfast synthesis defect of the ΔhfsD, ΔhfsA, and ΔhfsB mutants could be complemented by overexpression of the corresponding protein (Fig. S6B). These results indicate that the increased abundance of holdfast export proteins in the overexpression strains does not have a dominant negative effect.
Figure 7.
Holdfast biogenesis is restored in ΔpodJ and ΔhfsB mutants when holdfast export proteins are overexpressed. (A) Percentage of cells binding Alexa Fluor 488-WGA normalized to wild-type (WT) as the reference strain. The mean of two to four independent cultures is shown with the standard error of the mean. Between 140 and 2100 cells were counted with most samples containing over 500 cells. (B) Quantification of the percentage of holdfasts from each sample from panel A that were polarly localized. (C) Surface attachment as measured by crystal violet staining. The A600 of the eluted dye for each sample was normalized to wild-type as a standard. Three samples of each independent culture were used in each experiment. Data from two to three experiments was averaged and the standard error of the mean is shown.
Figure 8.
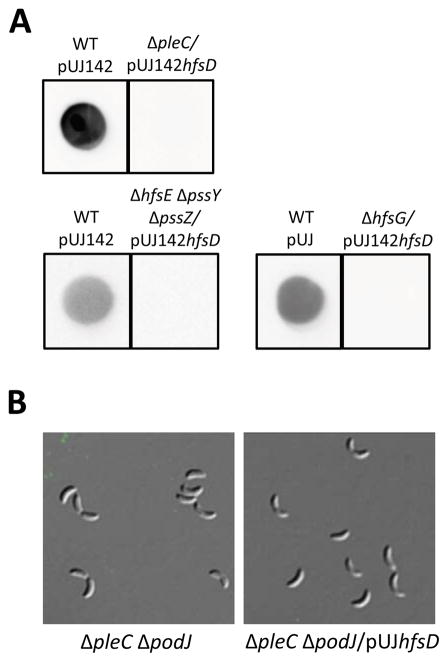
Overexpressing HfsD in holdfast synthesis mutants and ΔpleC does not restore holdfast synthesis. (A) Total holdfast in whole cultures of the developmental mutant ΔpleC overexpressing HfsD (top row) and two required synthesis glycosyl transferase mutants (ΔhfsE ΔpssY ΔpssZ and ΔhfsG) overexpressing HfsD (bottom row) was measured by dot blotting with HRP-WGA. Each sample set was cropped from a different blot (three total). (B) Overexpressing HfsD in a pleC podJ double mutant does not restore holdfast. Cells from these strains do not bind fluorescent lectin. DIC and fluorescence image overlays are shown for each strain.
Although the percentage of hfsB mutant cells that synthesized a holdfast was very low, these holdfasts were localized at the cell pole (Fig. 7A and B). Holdfast biogenesis could be restored when HfsD or HfsA were overexpressed in a ΔhfsB mutant with approximately 37% and 32 % of cells synthesizing a holdfast, respectively (Fig. 7A). Most holdfasts (94%) were correctly localized when HfsD was overexpressed in the hfsB mutant (Fig. 7B). Interestingly, only 40% of holdfasts restored by overexpressing HfsA in the hfsB mutant were polar. This suggests that HfsA is neither interacting with the localization factor, nor improving HfsD’s interaction with the localization factor upon overexpression.
The restoration of holdfast synthesis in podJ and hfsB mutants by export protein overexpression is similar, however, one key difference is that only when HfsD is overexpressed in a ΔhfsB mutant, are most of the holdfasts correctly localized at the pole. This suggests that HfsD is recruited to the pole by the holdfast export/synthesis complex localization factor, PodJ, perhaps via an intermediate. HfsA and HfsB probably have a role in stabilizing this interaction, since HfsD localization is perturbed in these mutants. Suppression of the ΔhfsD or ΔhfsA phenotypes could not be achieved by overexpressing holdfast export proteins (Table S1). It is not surprising that holdfast synthesis could not be restored to a hfsA mutant since HfsA is thought to stimulate the activity of the polysaccharide polymerase and bridge this activity to the export pore (Cuthbertson et al., 2009). In this context, it is interesting to note that it is not the overexpression of HfsA that resulted in the strongest restoration of holdfast synthesis in hfsB and podJ mutants, but the overexpression of the putative OM pore protein, HfsD. These results suggest that the export activity of the export-co-polymerase complex is limiting as compared to its polymerase-stimulation activity. Overexpression of any of the holdfast export proteins in a wild-type background did not increase holdfast synthesis (Fig. S6B and S6C). However recent studies of xanthan gum synthesis suggest that overexpressing HfsDA, HfsAB or HfsDAB may produce a level of polysaccharide that exceeds that of wild-type (Galván et al., 2013).
Strikingly, even though overexpression of HfsA and HfsD in the ΔhfsB mutant resulted in a similar percentage of cells with holdfasts (Fig. 7A), cells with HfsA overexpression had approximately 50% of the surface adhesion efficiency as compared to cells overexpressing HfsD (Fig. 7C, right graph), recapitulating the difference in the percentage of polar holdfasts (Fig. 7B). Overexpression of HfsD in the ΔpodJ developmental mutant, which produced mostly non-polar holdfasts, was even lower (Fig. 7C, left graph); this was expected since a podJ mutant does not synthesize pili, which are also important for adhesion. These results indicate that polar holdfasts are more efficient for surface adhesion than non-polar ones, consistent with the need to spatially coordinate the holdfast synthesis machinery with the flagellum and pili for the efficient transition from the reversible adhesion to the irreversible, holdfast-mediated adhesion. An additional consequence of holdfast mislocalization is that the asymmetry of holdfast inheritance is often perturbed, leading to the aberrant production of holdfast in early- stage swarmer cells that have not responded to surface contact signals (Fig. 9 and Movie S2).
Figure 9.

Asymmetry of holdfast inheritance is perturbed in cells that produce mis-localized holdfast. ΔpodJ/pUJ142hfsD cells were observed using time-lapse microscopy (see Movie S2). A pre-divisional cell is shown in the first panel. After cell division, both daughter cells will inherit holdfast. Top row: DIC images overlaid with fluorescence. Bottom row: fluorescence signal from AlexaFluor 488-conjugated WGA.
In order to verify that the increase in holdfast production upon holdfast export protein overexpression is due to the improved function of the holdfast export complex and not the synthesis of another GlcNAc-containing polysaccharide, we overexpressed HfsD in two holdfast synthesis mutants. ΔhfsE ΔpssY ΔpssZ is a mutant that lacks the redundant initiating glyosyl transferases of holdfast synthesis (Patel et al., 2012), and ΔhfsG is a mutant that lacks another glycosyl transferase essential for holdfast synthesis (Toh et al., 2008). No holdfast was restored as a result of overexpressing HfsD in either mutant, indicating that suppression of holdfast synthesis defects requires the biosynthesis of holdfast repeat units (Fig. 8).
The effect of hfsB and podJ suppression on HfsD subcellular localization
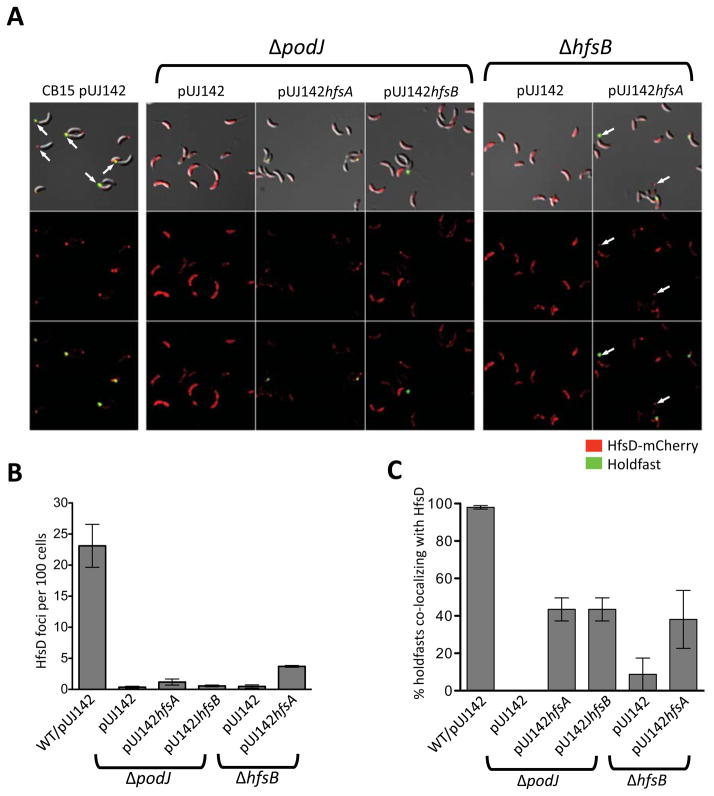
Since holdfasts of suppressed ΔhfsB and ΔpodJ mutants still localized as discrete punctae at lateral sites instead of being diffusely localized around the cell, we wondered if HfsD, whose localization was diffuse in ΔhfsB and ΔpodJ mutants, had re-gained punctate localization. Localization of HfsD-mCherry as punctae in strains that overexpress wild-type HfsD was rare (data not shown), likely due to abundant untagged HfsD outcompeting the HfsD-mCherry at the point of localization. Therefore, we focused our analysis on suppression by HfsA and HfsB overexpression, using ImageJ software (Rasband, 1997–2012) to pick out fluorescent HfsD-mCherry foci in these strains. We found that overexpressing HfsA in either a ΔpodJ or ΔhfsB mutant resulted in a slight increase of HfsD foci, but overexpressing HfsB did not change the number of HfsD foci in ΔpodJ (Fig. 10A and B). Overexpressing HfsA in a ΔhfsB mutant also resulted in frequent localization of HfsD at the tip of the stalk, when this was rarely, if ever, seen in the other holdfast-restored strains (Fig. 10A).
Figure 10.
HfsD-mCherry localization pattern in ΔpodJ and ΔhfsB mutants in which holdfast export proteins were overexpressed. (A) Strains expressing HfsD-mCherry as the only copy of HfsD. Top row: DIC and fluorescence overlay (both channels). Middle row: HfsD-mCherry fluorescence images. Bottom row: HfsD-mCherry and WGA-Alexa Fluor 488 fluorescence overlay. Contrast was adjusted similarly for all fluorescence images for best viewing of foci. Arrows point out HfsD foci located at the tip of the stalk. (B) Foci as detected and quantified using ImageJ. Foci were enumerated in 2 to 5 samples for each strain and compared to the number of cells estimated to be in the sample by ImageJ. The estimated number of cells ranged from about 175 to 2,800 cells (with most samples having over 400 cells). (C) The percentage of total holdfasts that co-localized with HfsD-mCherry foci for each strain.
When HfsA or HfsB was overexpressed in the ΔpodJ mutant or when HfsA was overexpressed in the ΔhfsB mutant, there was an increase in holdfast co-localization with HfsD-mCherry foci (Fig. 10C). These results further suggest that HfsA and HfsB have a role in stabilizing HfsD focal accumulation. These data also suggest that holdfast biogenesis is more likely to take place at sites of HfsD-mCherry focal localization than at any one random site around the cell.
That a very low percentage of ΔhfsB makes holdfasts suggests that an HfsDA complex may have low-level activity in the absence of an HfsB interaction with HfsA. The similar low percentage of holdfast bearing ΔpodJ mutant cells, combined with the holdfast and HfsD localization defects in these cells suggests that the absence PodJ destabilizes the HfsA-HfsD holdfast export complex found in the ΔhfsB mutant. In the wild-type, HfsB is the likely candidate to activate the export complex. Most homologs of HfsB have a regulatory role in that they are tyrosine autokinases in which cycling between phosphorylated and de-phosphorylated states is required for the biogenesis of CPS/EPS. HfsB belongs to an aberrant group of PCP proteins that lack both a key “non-variant” lysine in the Walker A ATP binding motif and a tyrosine rich domain that is phosphorylated in other systems, indicating that it is unlikely to regulate polysaccharide biosynthesis through tyrosine phosphorylation (Cuthbertson et al., 2009). But a regulatory role for these proteins may still be evolutionarily conserved, for instance if regulated interaction with an unknown protein takes the place of autokinase activity. For example, the presence of GelE, but not its ATP binding activity, is required for gellan gum synthesis in Sphingomonas elodea (Moreira et al., 2004).
Holdfast anchoring mechanism is in place for restored holdfasts
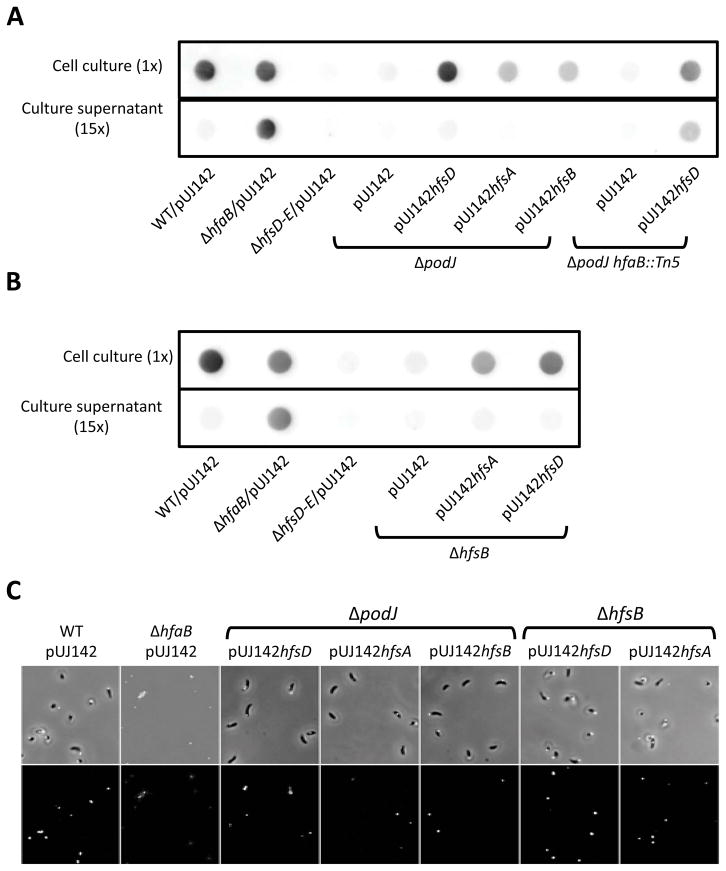
The three holdfast anchor proteins (HfaA, HfaB, and HfaD) are delocalized in the ΔpodJ and ΔhfsDAB mutants (Hardy et al., 2010) and we found that the anchor HfaB-mCherry protein is similarly delocalized in holdfast export mutants (Fig. 3E and S3B). This suggests that the holdfast produced in the ΔhfsB and ΔpodJ mutants restored for holdfast synthesis might not be anchored since they would be produced at a site devoid of sufficient anchor proteins, unless suppression restored co-localization of the export and anchoring complexes. We therefore measured the amount of shed holdfast in the suppressed hfsB and podJ mutants to determine if their holdfasts were anchored to the cell body. The ΔhfaB anchor mutant, which readily sheds holdfast into the medium (Hardy et al., 2010) was used as a shedding control. Strikingly, holdfast was virtually undetectable in the supernatants of the suppressed ΔhfsB and ΔpodJ mutants (Fig. 11A and B), suggesting that restored holdfasts were anchored to the cell envelope. In contrast, holdfast was readily detectable in the supernatant when HfsD was overexpressed in a ΔpodJ mutant with an hfaB anchor gene knockout, confirming that shed holdfasts are detectable in the suppressed strains by this method. Furthermore, all mutants suppressed by overexpression of holdfast export proteins were able to adhere to a glass slide after thorough washing in contrast to what is seen in a holdfast anchor mutant (Fig. 11C). Interestingly, the podJ/hfaB double mutant overexpressing HfsD produced less holdfast in the whole cell culture than when HfsD was overexpressed in a podJ single mutant (Fig. 11A), suggesting that the HfaB anchor protein could play a role in holdfast biogenesis in addition to its function as an anchor, perhaps through a stabilizing effect on the export complex.
Figure 11.
Holdfasts restored due to suppression by overexpression of holdfast export proteins are anchored to the cell body. Comparison of holdfast restored in (A) ΔpodJ and (B) ΔhfsB mutants by holdfast export protein overexpression. Holdfast material from whole cell lysates was detected on a nitrocellulose membrane using chemiluminescence from an HRP conjugated lectin (top rows). In the blots on the bottom rows, the amount of holdfast in cell culture supernatants is detected in a volume equal to 15 times that of the whole cell culture sample. Culture and corresponding supernatant samples were taken from the same culture and were from the same blot. Each set of samples (cell culture with corresponding culture supernatant for ΔpodJ or ΔhfsB) is representative of several independent blots, but is from the same blot. (C) ΔpodJ and ΔhfsB mutants in which holdfast export proteins were overexpressed remain attached to a glass slide after thorough washing with water.
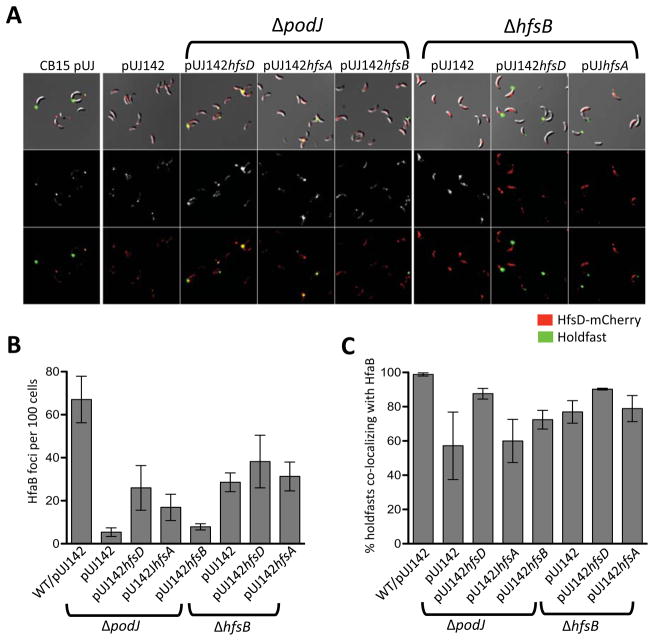
The above results suggest that a sufficient amount of holdfast anchoring proteins is present at the site of holdfast synthesis to anchor the holdfast. In the ΔpodJ, ΔhfsD, ΔhfsA, and ΔhfsD mutants, most of the signal from the anchor fusion protein, HfaB-mCherry, is diffuse and peripheral. However, many cells also have small randomly located foci (Hardy et al., 2010; Fig. 3E and S3B). We identified foci in mutants with restored holdfast synthesis by export protein overexpression using ImageJ and found that there were less HfaB-mCherry anchor foci in ΔpodJ and ΔhfsB strains compared to wild-type (Fig. 12A and B). Overexpression of HfsD or HfsA in a ΔpodJ mutant resulted in an increase in HfaB-mCherry anchor foci. Overexpressing HfsA or HfsD in a ΔhfsB mutant did not affect the amount of HfaB anchor foci, but it increased the amount of foci localized at the poles and the tip of the stalk (Fig. 12A and B). We also found that most holdfasts in ΔpodJ and ΔhfsB strains with restored holdfast synthesis co-localized with HfaB anchor foci, suggesting that holdfast synthesis is likely to occur where HfaB anchor protein has focally localized (Fig. 12C).
Figure 12.
HfaB-mCherry localization pattern in ΔpodJ and ΔhfsB overexpressing holdfast export proteins were overexpressed. (A) Strains expressing HfsD-mCherry as the only copy of HfsD. Top row: DIC and fluorescence overlay (both channels). Middle row: mCherry fluorescence images. Bottom row: mCherry and Alexa Fluor 488 fluorescence overlay. Contrast was adjusted similarly for all fluorescence images for best viewing of foci. (B) Foci as detected and quantified using ImageJ for each strain. Foci were enumerated in two to five samples for each strain and compared to the number of cells estimated to be in the sample by ImageJ. The estimated number of cells ranged from about 175 to 2,800 cells with most samples having over 400 cells). (C) The percentage of total holdfasts that co-localized with HfaB-mCherry foci for each strain.
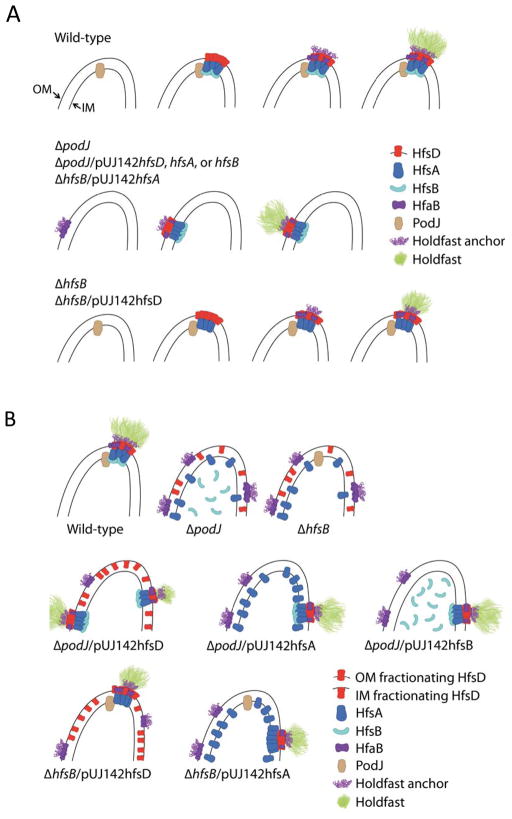
We hypothesize that, in wild-type cells PodJ either directly or indirectly recruits HfsD, or perhaps the whole export complex in concert, which in turn recruits the holdfast anchor proteins (Fig. 13A). The interaction would most likely be between one or more members of the export complex and the integral OM HfaB anchor protein. HfaA and HfaD anchor proteins, on the other hand, are extracellular proteins secreted by HfaB (Hardy et al., 2010). Our data suggest that HfaB anchor secretion pores have an intrinsic ability to self-assemble into small foci even in the absence of the target export complex, Furthermore, these HfaB anchor foci usually co-localize with holdfast (Fig. 12C), suggesting the presence of functional holdfast biogenesis machinery at these randomly located sites. We therefore hypothesize that HfaB anchor complexes recruit the holdfast export machinery in the absence of PodJ or HfsB (Fig. 13A). In wild-type cells, HfsD appears to lack the ability to form focal complexes in the absence of its target, PodJ or HfsB, which might enhance the association with HfsD’s target. Increasing the abundance of export complex components probably drives the interaction of export complexes with pre-existing anchor foci, resulting in the assembly of a functional export-anchor complex (Fig. 13A and B). This model is supported by the fact that HfsD overexpression does not restore holdfast synthesis as efficiently in a double mutant of podJ and the hfaB anchor gene as compared to a podJ single mutant.
Figure 13.
Proposed model for holdfast export and anchor complex localization. Depiction of a holdfast biogenesis complex at the pole indicates that more than 90% of holdfasts observed are polarly localized. (A) Sequence of localization events. For clarity, de-localized HfsD is not shown. (Top row) In wild-type cells, the polar development protein, PodJ, recruits the HfsDAB holdfast export complex to the cell pole, which in turn, recruits the holdfast anchor complex. The holdfast anchor/export complex is in place in advance of the holdfast synthesis. (Middle row) In ΔhfsB and ΔpodJ cells, HfaB assembles into randomly localized small foci. These small HfaB assemblies can recruit holdfast export complexes at these random sites in a small percentage of ΔpodJ cells, or when either HfsD, HfsA, or HfsB are overexpressed in ΔpodJ, or when HfsA is overexpressed in a ΔhfsB mutant. (Bottom row) In the rare holdfast-synthesizing ΔhfsB cells, the holdfast is usually correctly localized at the pole, presumably because the HfsDA complex can still be recruited by PodJ. Consequently, when HfsD is overexpressed in a ΔhfsB mutant, the restored holdfasts are polarly localized. The fact that holdfasts are non-polar when HfsA is overexpressed in a ΔhfsB mutant (middle row) suggests that HfsD, and not HfsA is interacting with PodJ, either directly, or via an intermediate. (B) Localization of holdfast export and anchor proteins. (Top row) In wild-type cells, holdfasts, holdfast export, and holdfast anchor complexes co-localize at the poles along with PodJ. When podJ or hfsB is deleted, holdfast export proteins are de-localized, and holdfast anchor proteins are localized in small foci at random locations around the cell body. Furthermore, HfsD no longer fractionates with the OM. (Middle and bottom row) Overexpression of HfsA, HfsB, or HfsD holdfast export proteins stabilizes interactions between the holdfast export and anchor complexes causing holdfast export proteins to co-localize with randomly localized holdfast anchor complexes. HfsD protein only fractionates with the OM when it is incorporated into a functional holdfast export/anchor complexes, suggesting that its stable interaction with the OM requires a functional complex. OM-fractionating HfsD is illustrated by the red protein traversing the OM in the diagram, whereas HfsD that does not fractionate with the OM is indicated by the red protein that just touches the OM.
Membrane fraction localization of holdfast export proteins
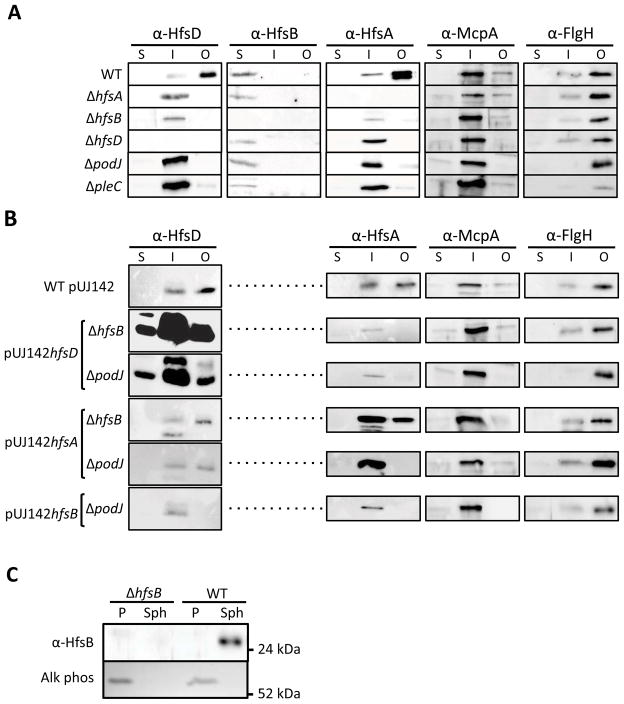
Our investigation thus far has supported the idea that the holdfast export proteins work together in a complex as predicted from sequence similarity to OPX and PCP proteins. This complex is stabilized by PodJ-directed localization and PleC-mediated developmental signaling. We hypothesized that membrane fractionation of any one holdfast export protein may be affected in the absence of other export proteins or the developmental regulators, PodJ or PleC. Whole cells were fractionated into soluble, inner membrane (IM), and outer membrane (OM) components using sarkosyl to solubilize the inner membrane from the total membrane fraction (Fig. 14A). The purity of the fractions was verified by probing the blot with antibodies to the integral IM protein, McpA, and the OM protein, FlgH (Alley et al., 1992, Jenal et al., 1994). As expected, McpA fractionated in the IM fraction and FlgH in the OM fraction.
Figure 14.
Cell fraction localization of holdfast export proteins. (A) Cell fractionation of HfsA, HfsB, HfsD, McpA (IM positive control), and FlgH (OM positive control) into soluble (S), inner membrane (I), and outer membranes (O) was determined by Western Blot of wild-type (WT), holdfast export and developmental mutants. (B) Fractionation of the same proteins was determined by Western blot of ΔhfsB and ΔpodJ mutants suppressed by overexpression of various holdfast export proteins (C) Extraction of periplasmic proteins shows that HfsB localizes to the cytoplasm. Equal volumes of periplasm (P) and the spheroplast fraction (Sph) were loaded in SDS-PAGE gels and blotted to nitrocelluose. A zymogram for alkaline phosphatase (Alk Phos) shows clean separation between periplasm and spheroplasts.
HfsD localized to the OM as would be expected based on similarity to Wza (Fig. 14A). HfsB fractionated in the soluble fraction. Based on similarity to the cytoplasmic C-terminal domain of typical PCP proteins, we expected HfsB to fraction with the cytoplasm. We confirmed this by separating the periplasmic fraction from the spheroplast fraction following chloroform extraction (Fig. 14C). HfsA fractionated predominantly with the OM fraction with a smaller portion in the IM fraction. Since Wzc (an HfsA homolog) interacts with the OM Wza (Collins et al., 2007), we hypothesized that the OM fractionation of HfsA is a result of its interaction with HfsD, since PCP proteins similar to HfsA are thought to reside in the inner membrane.(Cuthbertson et al., 2009). Indeed, HfsA localized to the IM fraction in ΔhfsB, ΔhfsD, ΔpodJ, and ΔpleC mutants. We also found that HfsD switched to the IM fraction in ΔhfsA, ΔhfsB and the developmental mutants. We attribute the altered membrane fractionation in these strains to decreased stability of the export complex due to the absence of key interacting partners. The HfsB protein always remained in the cytoplasmic fraction in all the mutants tested. Using whole cell lysates, we confirmed that the abundance of HfsD, HfsA, and HfsB was normal in the podJ and pleC mutants (Fig. 4).
There is no difference in fractionation pattern between the ΔhfsB, and ΔpodJ mutants, which make some holdfast, and the ΔhfsD, ΔhfsA, and ΔpleC mutants that make no holdfast. We hypothesize that in in the ΔhfsB and ΔpodJ mutants, a very small percentage of HfsD and HfsA has wild-type cell membrane fractionation – a percentage that is below the limit of detection of the Western blot.
Since HfsD has similarity to predicted EPS pore proteins, the lack of HfsD OM fractionation could be a contributing factor to the holdfast synthesis defect in ΔhfsB and ΔpodJ mutants. We therefore wondered if overexpressing export proteins restored HfsD and HfsA OM fractionation. Overexpressing HfsD in either mutant resulted in a large amount of HfsD being retained in the OM (Fig. 14B), but this increase is difficult to interpret since there was a high level of HfsD protein overall. More informative was the effect of overexpressing HfsA in the ΔpodJ and ΔhfsB mutants, which also restored OM targeting of HfsD. This suggests that HfsD is present in the OM, but that its association with the OM is weak when it is not forming critical interactions with other members of the protein complex. This causes HfsD to fractionate in the IM in holdfast export or developmental mutants. Overexpressing HfsB in ΔpodJ, however, did not restore detectable OM targeting of HfsD.
As for HfsA OM targeting: overexpressing HfsA restored its own OM targeting only in the hfsB mutant, and not in the podJ mutant. Neither did overexpressing HfsB or HfsD restore HfsA OM targeting in any of the mutants. Because these results fail to show a strict correlation between OM targeting of HfsD and HfsA and suppression of the holdfast synthesis defect, we hypothesize that the defect in HfsA and HfsD targeting to the OM is not the main reason for the deficiency in holdfast synthesis of the podJ and hfsB mutants
Conclusion
The holdfast of C. crescentus is synthesized at the flagellar pole of swarmer cells, and this synthesis can occur immediately after cell division for cells that contact a surface. In this paper, we investigate the role of polar localization in the control of holdfast synthesis. We show that the holdfast export protein, HfsD, localizes to the flagellar pole of late predivisional cells. HfsA and HfsB are required for the polar localization of HfsD and its retention in the OM, suggesting that the complete holdfast export complex is localized at the flagellar pole in late predivisional cells. The developmental polar localization factor PodJ is also required for HfsD localization and its proper membrane fractionation. Interestingly, a very small percentage of hfsB and podJ mutant cells can still synthesize holdfast, and overexpression of other holdfast export proteins in these strains can restore holdfast synthesis. Since we know that these mutants are not severely impacted in the level of holdfast export proteins, this suggests that these mutants have a defect in the regulation of holdfast synthesis protein activity. In agreement with this hypothesis, overexpression of holdfast export proteins results in a significant restoration of holdfast synthesis in hfsB and podJ mutants.
Restored holdfasts and HfsD are frequently randomly localized instead of polarly localized. Therefore, polar localization of HfsD is not required for holdfast synthesis when export components are at high concentration, implying that it is mostly the lack of focal accumulation of HfsD that prevents holdfast synthesis in ΔhfsB and ΔpodJ mutants. These results suggest that an important role of polar localization is to increase the effective concentration of holdfast export proteins in order to optimize formation of a productive holdfast export complex. Restored holdfasts also had no anchoring defects in either ΔhfsB or ΔpodJ mutants. These holdfasts also co-localized with small, randomly positioned HfaB anchor foci. In addition, small HfsD foci present in the suppressed mutants often co-localized with holdfast. These findings suggest that interaction between the holdfast anchoring and export complexes are involved in assembling a complete, productive holdfast biosynthesis machine.
Another role of localization is to spatially coordinate the holdfast synthesis machinery with the flagellum and pili for the efficient transition from the reversible to the irreversible, holdfast mediated adhesion. Restoration of holdfast synthesis at non-polar sites did not restore surface adhesion as well as restoration of polar holdfast synthesis, presumably eliminating the just-in-time production of holdfast upon surface contact that is thought to optimize the efficiency of adhesion and the ability of cells to modulate holdfast synthesis based on the quality of the environment (Li et al., 2012). Our results highlight the fact that multiple facets of subcellular localization can be coupled to improve the phenotypic outcome of a protein assembly.
Future work will need to focus on the mechanisms for the localization of the holdfast export-anchoring machinery and for its activation. While PodJ is required for the localization of a number of developmental signaling and organelle biogenesis proteins, direct interaction has only been demonstrated with the regulatory protein DivL (Curtis et al., 2012). PodJ has both a cytoplasmic and a periplasmic domain and it could therefore directly recruit any component of the holdfast export and/or anchoring machinery (Fig. 13). Our results indicate that the accumulation of a sufficient amount of HfsD at the pole, directed by PodJ, is important for the activation of holdfast biogenesis. Analogy to the Wza/Wzc complex and our results suggest that HfsA and HfsB localize at the same time as HfsD. This localization takes place at the flagellar pole of predivisonal cells before either the developmental or the surface contact activation signal takes place. Thus, the signals that activate holdfast biogenesis do not do so through localization of the holdfast export complex. Future work will have to determine if other proteins of the holdfast synthesis pathway localize to the proper pole contemporaneously with holdfast synthesis or earlier, with the holdfast export machinery. In addition, it remains to be seen how the holdfast biogenesis activation signal(s) activate the machinery. As mentioned above, HfsB is a likely target, but its mode of action cannot be through the canonical tyrosine phosphorylation of PCP proteins, which it lacks. A thorough analysis of the structure- function of HfsB and the identification of interacting proteins should provide insight into the mode of action of the aberrant group of PCP proteins that regulate polysaccharide biosynthesis by a means other than tyrosine phosphorylation (Cuthbertson et al., 2009). Finally, holdfast biogenesis is clearly subject to regulation by c-di-GMP (Abel et al., 2011), but the mechanism by which diguanylate cyclase activity activates holdfast biogenesis, perhaps through HfsB, remains to be determined.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed on Table S2. All Caulobacter strains were grown with peptone yeast extract (PYE) (Poindexter, 1964) liquid and solid agar media at 30°C. For cell synchrony, M2 minimal medium (Johnson & Ely, 1977) with glucose added to 0.2% was used (M2G). The following antibiotics were added when called for (solid, liquid): chloramphenicol (1 μg ml−1, 0.5 μg ml−1), nalidixic acid (20 μg ml−1, solid only) spectinomycin (100 μg ml−1, 25 μg ml−1), streptomycin (10 μg ml−1, solid only) and kanamycin (20 μg ml−1, 5 μg ml−1).. Strains carrying pUJ142 derivatives were grown overnight in PYE supplemented with 0.5 μg ml−1 chloramphenicol and 0.5% glucose (the repressing condition), and subcultured to OD600~0.1 in the same and grown for 4–5 h before the cells were collected and analyzed as described. Holdfast export proteins were sufficiently overexpressed with out adding the inducer (xylose) due to leaky expression.
All E. coli strains were grown with Luria-Bertani liquid and solid agar media at 37°C. The following antibiotics or supplements were added when called for (solid, liquid): chloramphenicol (30 μg ml−1, 20 μg ml−1), streptomycin (30 μg ml−1, 30 μg ml−1), and kanamycin (50 μg ml−1, 30 μg ml−1).
Genetic techniques
PCR primers were obtained from Operon (Huntsville, AL) and are listed in Table S3. Restriction digests and ligations using T4 DNA ligase were performed according to manufacturers suggestions (New England Biolabs, Beverly, MA). DNA was purified using the following kits according to manufacturer’s instructions: PCR products with the Qiaquick gel extraction kit (Qiagen, Valencia, CA), plasmids with the Zyppy Plasmid Miniprep Kit (Zymo Research, Orange, CA), and genomic DNA with the Bactozol Kit (Molecular Research Center, Cincinnati, OH) or with Promega Magic Miniprep columns (Promega, Madison, WI) and the protocol previously described in (Janakiraman & Brun, 1997). Purified plasmid or PCR fragments were sequenced at Indiana University’s Indiana Molecular Biology Institute with an Applied Biosystems 3730 automated fluorescence sequencing system using ABI Prism BigDye Terminator cycle sequencing version 3.1 (Applied Biosystems, Foster, CA).
To express HfsD-mCherry in CB15 and a variety of deletion strains, the last 219 base pairs of hfsD were amplified from genomic DNA using primers F_hfsD_3′NdeI and R_hfsD_endSacI and ligated into pCHYC-1. pCHYC-1hfsD3′ was introduced into chemically competent Bioline α-select silver efficiency cells for purification. pCHYC-1hfsD3′ was introduced into CB15 and a collection of in-frame deletion mutants via conjugation with chemically competent E. coli SM10. Integration in the hfsD open reading frame creates a C-terminal translational fusion of the complete hfsD gene with mCherry and a small fragment of with the displaced C-terminus of HfsD further downstream. Constructs using the pUJ142 plasmid were introduced into Caulobacter via electroporation: 1.5 kV, 100 Ohms, 25μF.
To create an in-frame deletion of the entire hfs cluster (hfsDABCHGFE; hfsD-E), two 500 bp fragments, one located just outside each end of the cluster were amplified. The FhfsDendHindIII/HfsDBamup primer pair is located downstream of HfsD (transcribed divergently from hfsA) and the FupEcoRIhfsE/RupHindIIIhfsE primer pair is located upstream of hfsE (transcribed divergently relative to hfsABC). The two fragments were digested with a combination of HindIII/BamHI or EcoRI/HindIII respectively and ligated into the BamHI and EcoRI sites of pNPTS138 and introduced into Bioline α-select silver efficiency cells for purification. This suicide vector was introduced into CB15 ΔhfsDAB via conjugation with chemically competent E. coli S17 and an in-frame deletion of hfsCHGFE resulted from two-step homologous recombination using sucrose sensitivity and kanamycin resistance as previously described (Gonin et al., 2000).
Western Blot Analysis
A volume of 50 μl of 2x Laemmli SDS sample buffer (Laemmli, 1970) was added to cell pellets collected from exponential phase and re-suspended in 50 μl 10 mM Tris, pH = 8. Samples were boiled for 5 min before being run on a 10% polyacrylamide gel, and transferred to nitrocellulose membranes. Nitrocellulose was blocked for 1 to 4 h in 3% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20. HfsA, HfsB and HfsD antibodies were used at a concentration of 1:1,000, 1:500 and 1:1,000 (or 1:2,000), respectively, and incubated overnight at 4°C. Nitrocellulose membranes incubated with anti-HfsA and anti-HfsD were developed with SuperSignal West Pico Substrate (Thermo Scientific, Rockford, IL). Nitrocellulose membranes incubated with anti-HfsB were incubated with SuperSignal West Dura Substrate.
Cell synchronization
A population of swarmer cells were isolated from a early exponential phase culture using the Ludox density centrifugation method (Evinger & Agabian, 1977). C. crescentus CB15 (ATCC 19089) cells from a 200 ml culture grown in M2G medium were re-suspended in 120 ml of 33.3% Ludox (DuPont) in M2G and centrifuged at 12,100 × g for 20 min. Purified swarmer cells (verified by microscopy) were washed three times in cold M2 salts via 5 min, 7,740 × g centrifugations. Cells were suspended in warm M2G and Western blot samples were collected every 15 min after a 10 min recovery. Since this procedure isolates newly divided swarmer cells, the cell number remains the same until cell division. Thus equal volumes of sample were used for Western blotting.
Lectin dot blot
One ml samples from mid-log phase cultures were either frozen directly at −20° C, or 0.5 ml of supernatant (from a 15 to 20 min centrifugation of 4,000 × g at 4 °C) was collected from the 1 ml sample and then frozen. The pellets and remaining supernatant were discarded. When thawed, Proteinase K (Bioline) was added to a final concentration of 0.5 mg ml−1 and incubated for 3 to 6 h at 55 °C, which cleared the peptidoglycan specific signal from the sample. Samples were re-frozen, and upon thawing, were incubated at 70°C for 10 min to deactivate Proteinase K, chilled briefly on ice, and 12.5 units of Benzonase endonuclease (Novagen) were added.
Samples were adsorbed to a nitrocellulose membrane using a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories, Hercules, CA). The volume used for each sample was normalized to be equivalent to 25 μl and 10 μl of culture with an OD600 of 0.6. Twenty-five or 150 μl of supernatant samples was used. Each volume was loaded in duplicate or triplicate. Samples were diluted with PBS, pH = 7.4 (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4), to a final volume of 300 μl before being loaded in the apparatus wells. Each identical sample was loaded at least in duplicate. Membrane was blocked with 3% BSA in PBS, 0.1% Tween 20 (PBST) for 1 to 3 h, then wheat germ agglutinin (WGA) conjugated to horseradish peroxidase (HRP) (Sigma, St. Louis, MO) was added to a dilution of 1:20,000 to 1:18,000 for 1 h at room temperature. After developing with SuperSignal West Pico Substrate (Thermo Scientific, Rockford, IL), the blot was imaged using a Kodak ImageStation 4000 MM.
Fluorescent Lectin Binding
Alexa Fluor 488-conjugated WGA (Invitrogen Molecular Probes) was added directly to mid-log phase cultures to a 1:10,000 dilution. Phase and fluorescent images of cells were gathered on either glass slides or 1% agarose pads in PYE. The percentage of cells with attached holdfasts was determined by counting cells using MicrobeTracker (Sliusarenko et al., 2011) and counting lectin spots by hand.
Small volume attachment assay
Fifteen μl of cells in mid-log phase were applied to individual wells of a 12-well multitest slide (MP Biomedicals, Aurora, OH) that had been cleaned prior by soaking in 100% Micro-90 detergent. Cells were allowed to incubate for 45 min to 1 h. Slides were rinsed thoroughly with water from a squirt bottle for 2 min and then gently rinsed with 7 ml of PYE with a 1:10,000 dilution of Alexa Fluor 488 conjugated-WGA (Invitrogen Molecular Probes). Phase and fluorescent images were recorded by microscopy.
Surface attachment assay
Polystyrene adhering cells were identified by crystal violet staining similar to (Bodenmiller et al., 2004). Aliquots of 500 μL of mid-log phase culture were added to wells of a 24-well plate in triplicate and incubated while shaking for 30 min at room temperature. The contents of the plate were decanted and plates were gently washed in a bucket of deionized water. Approximately 1 mL of 0.1% crystal violet was added to each well. After a 30 min incubation. the wells were rinsed in deionized water as above, and eluted with 500 μL of 10% acetic acid once dry. The A600 of each elution was read. All samples were normalized to a wild-type sample included in every experiment.
Microscopy
Cells binding fluorescent lectin or expressing HfsD-mCherry were observed with a Nikon Eclipse E800 or 90i microscope using 100x Plan Apo oil immersion objectives (NA 1.4). DIC, phase and fluorescent images were captured using a Photometrics Cascade 1K EMCCD camera. Cells imaged overnight in a time-lapse experiment were immobilized on a 1% agarose-PYE pad and images taken every 10 min at room temperature. Images were processed and compiled using ImageJ software (Rasband, 1997–2012)
Image analysis
Fluorescent HfsD-mCherry and HfaB-mCherry foci were calculated using ImageJ software v. 1.47a (Rasband, 1997–2012). For HfaB-mCherry foci expressed in various mutants (Fig. 3E), fluorescence images were first normalized to correct for background fluctuations over time or over different fields. The background intensity of phase contrast images was subtracted to optimize auto-thresholding operations. Cell boundary, centerline, and pole position were detected using a specifically developed plug-in for ImageJ. Briefly, cells were detected using an auto-thresholding function and sub-pixel resolution refined cell contours were obtained using a cubic spline fitting algorithm. Cells touching each other were resolved using an iterative auto-thresholding function. The centerline of each cell was deducted from the skeleton and expanded in both directions to the most probable point maximizing the cell-boundary curvature and minimizing the angle between the centerline and the cell boundary. The small fluorescent foci were detected using a local and sub-pixel resolution maxima detection algorithm. For each focus, the relative position was determined along the center line of the cell and used to differentiate polar foci from non-polar foci.
For cells overexpressing HfsD, HfsA, or HfsB, the background was subtracted from each fluorescent image and converted into a binary image using the auto-thresholding function. Foci were counted using the particle analyzer. The number of cells in the corresponding DIC images was estimated using the particle analyzer to count cell shapes in binary images generated using auto-thresholding, after using the Find Edges and Smooth commands. The number of cells was estimated by applying the total area of the objects in an image to the linear regression equation resulting from the comparison of a selection of images with the actual number of hand-calculated cells.
Cell fractionation
Cell fractionation using sarkosyl insolubility was performed as previously described (Hardy et al., 2010). This method has been shown to be effective at isolating OM proteins in C. crescentus (Cao et al., 2012). Cell pellets from 20 ml of exponentially growing cells normalized to OD600 = 0.6 were suspended in 1 ml of 20 mM Tris buffer pH = 8 and lysed by FastPrep®-24 Instrument (MP Biomedicals LLC) in 2.0 ml Lysing Matrix tube containing specialized Lysing Matrix beads for 45 sec. Unbroken cells were removed by centrifugation at 16,000 × g at 4°C for 2 min. The supernatant was removed and centrifuged at 100,000 × g at 4°C for 30 min. The supernatant containing the soluble protein was stored at −80°C. The pellet was suspended in 500 μl 20 mM Tris, pH 8.0 and 1% sodium lauryl sarcosine, rocked at RT for 45 min and centrifuged at 100,000 × g for 30 min. The pellet, which contains the OM, was washed briefly with 20 mM Tris, pH 7.8 and then suspended in 100 μl 20 mM Tris, pH 8.0 and stored at −80°C. The sarkosyl soluble inner membrane fraction, which was the supernatant, was isolated by ethanol precipitation by adding ethanol to a final concentration of 75%, incubating overnight at −20°C and centrifuging at 16,000 × g for 15 min at 4°C. Protein pellets were suspended in 100 μl of 20 mM Tris, pH 7.8 and stored at −80°C.
Periplasmic extraction
Periplasm was isolated from 3 ml of an exponentially grown culture by chloroform extraction as previously described (Ames et al., 1984). Extracts were stored at −20°C. Sample volumes were normalized and equal volumes were loaded for SDS-PAGE. Separation of periplasm was evaluated by alkaline phosphatse zymograms according to previously described methods (Michel and Baratti,1989; Pond et al., 1989). After electrophoresis, polyacrylamide gels were washed, 2 × 20 min, in 0.5M Tris-HCl pH8.5, 1mM ZnCl2, 2mM MgCl2, 2% Triton x-100 (wash 1 buffer). This was followed by 2 × 20 min. washes in 0.5M Tris-HCl pH8.5, 0.1mM ZnCl2, 1mM MgCl2 and 2% Triton x-100 (wash 2 buffer). Gel was developed by adding a final concentration of 9.8 mM CaCl2, to fresh wash 2 buffer, as well as a final concentration of 16.7 μg ml−1 5-Bromo-4-chloro-3-indolyl-phosphate (BCIP) and 32.3 μg ml−1 4-Nitro blue tetrazolium chloride (NBT).
Supplementary Material
Acknowledgments
We thank David Kysela for his help with microscopy and the use of ImageJ and MicrobeTracker, Adrien Ducret for writing and explaining the ImageJ plug–in, Christopher Smith for performing some preliminary experiments, and members of our laboratory for critical reading of the manuscript. This work was supported by grants GM51986 and GM102841 from the National Institutes of Health to YVB. J.J. was supported by the Indiana University Genetics, Molecular and Cellular Sciences Training Grant T32-GM007757 and the Ruth L. Kirschstein National Research Service Award Predoctoral Fellowship Award for Minority Students, F31 GM81903 (to J.J.) from the National Institute of General Medical Sciences.
References
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub M, et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MR, Gomes SL, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–341. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MR, Maddock JR, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Bayer ME, Thurow H. Polysaccharide capsule of Escherichia coli: microscope study of its size, structure, and sites of synthesis. J Bacteriol. 1977;130:911–936. doi: 10.1128/jb.130.2.911-936.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Johnston DS. A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. J Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner D, Sieberth V, Pazzani C, Roberts IS, Boulnois GJ, Jann B, Jann K. Expression of the capsular K5 polysaccharide of Escherichia coli: biochemical and electron microscopic analyses of mutants with defects in region 1 of the K5 gene cluster. J Bacteriol. 1993;175:5984–5992. doi: 10.1128/jb.175.18.5984-5992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJB, Hardy GG, Trimble MJ, Brun YV. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. In: Poole RK, editor. Advances in Microbial Physiology. Vol. 54. London: Academic Press Ltd-Elsevier Science Ltd; 2009. pp. 1–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertkov O, Brown PJB, Kysela DT, De Pedro MA, Lucas S, Copeland A, et al. Complete genome sequence of Hirschia baltica type strain (IFAM 1418(T)) Stand Genomic Sci. 2011;5:287–297. doi: 10.4056/sigs.2205004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JL, Hardy GG, Bodenmiller D, Toh E, Hinz A, Brun YV. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol Microbiol. 2003;49:1671–1683. doi: 10.1046/j.1365-2958.2003.03664.x. [DOI] [PubMed] [Google Scholar]
- Collins RF, Beis K, Dong C, Botting CH, McDonnell C, Ford RC, et al. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Quardokus EM, Lawler ML, Guo X, Klein D, Chen JC, et al. The scaffolding and signaling functions of a localization factor impact polar development. Mol Microbiol. 2012;84:712–735. doi: 10.1111/j.1365-2958.2012.08055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, I, Mainprize L, Naismith JH, Whitfield C. pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in Gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván EM, Ielmini MV, Patel YN, Bianco MI, Franceschini EA, Schneider JC, Ielpi L. Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiology. 2013;23:259–272. doi: 10.1093/glycob/cws146. [DOI] [PubMed] [Google Scholar]
- Gonin M, Quardokus EM, O’Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy GG, Allen RC, Toh E, Long M, Brown PJB, Cole-Tobian JL, Brun YV. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques MX, Rodrigues T, Carido M, Ferreira L, Filipe SR. Synthesis of capsular polysaccharide at the division septum of Streptococcus pneumoniae is dependent on a bacterial tyrosine kinase. Mol Microbiol. 2011;82:515–534. doi: 10.1111/j.1365-2958.2011.07828.x. [DOI] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Janakiraman RS, Brun YV. Transcriptional and mutational analyses of the rpoN operon in Caulobacter crescentus. J Bacteriol. 1997;179:5138–5147. doi: 10.1128/jb.179.16.5138-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, White J, Shapiro L. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. Poles apart: prokaryotic polar organelles and their spatial regulation. Cold Spring Harbor Perspectives in Biology. 2011:3. doi: 10.1101/cshperspect.a006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroncke KD, Golecki JR, Jann K. Further electron microscopic studies on the expression of Escherichia coli group II capsules. J Bacteriol. 1990;172:3469–3472. doi: 10.1128/jb.172.6.3469-3472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Arts GJ, Walther TC, Englmeier L, Mattaj IW. Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol Cell Biol. 2002;22:5708–5720. doi: 10.1128/MCB.22.16.5708-5720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz HD, Smith J. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J Bacteriol. 1992;174:687–694. doi: 10.1128/jb.174.3.687-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Laus MC, Logman TJ, Lamers GE, Van Brussel AAN, Carlson RW, Kijne JW. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol. 2006;59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- Levi A, Jenal U. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty C, Thompson J, Barrett B, Lord L, Andersen C, Roberts IS. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: the role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol Microbiol. 2006;59:907–922. doi: 10.1111/j.1365-2958.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- Merker RI, Smit J. Characterization of the adhesive holdfast of marine and freshwater Caulobacters. Appl Environ Microbiol. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel GPF, Baratti JC. Phosphateirrepressible alkaline phosphatase of Zymomonas mobilis. J Gen Microbiol. 1989;135:453–460. [Google Scholar]
- Moore RL, Marshall KC. Attachment and rosette formation by hyphomicrobia. Appl Environ Microbiol. 1981;42:751–757. doi: 10.1128/aem.42.5.751-757.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LM, Hoffmann K, Albano H, Becker A, Niehaus K, Sá-Correia I. The gellan gum biosynthetic genes gelC and gelE encode two separate polypeptides homologous to the activator and the kinase domains of tyrosine autokinases. J Mol Microbiol Biotechnol. 2004;8:43–57. doi: 10.1159/000082080. [DOI] [PubMed] [Google Scholar]
- Nooren IMA, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate J, Porter J, Jordan T. Asticcacaulis biprosthecum sp. nov. Life cycle, morphology and cultural characteristics. Antonie Van Leeuwenhoek. 1973;39:569–583. doi: 10.1007/BF02578901. [DOI] [PubMed] [Google Scholar]
- Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, Brun YV, et al. Functional characterization of UDP-glucose: undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J Bacteriol. 2012;194:2646–2657. doi: 10.1128/JB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Beness AM, Saier MH., Jr Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- Pond JL, Eddy CK, Mackenzie KF, Conway T, Borecky DJ, Ingram LO. Cloning, sequencing, and characterization of the principal acid phosphatase, the phoC+ product, from Zymomonas mobilis. J Bacteriol. 1989;171:767–774. doi: 10.1128/jb.171.2.767-774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Biological properties and classification of Caulobacter group. Bacteriol Rev. 1964;28:231. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, MD: U.S. National Instituites of Health; 1997–2012. [Google Scholar]
- Reuber TL, Walker GC. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Protein Subcellular Localization in Bacteria. Cold Spring Harbor Perspectives in Biology. 2010:2. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RP, Aaronson W, Vann WF. Translocation of capsular polysaccharides in pathogenic strains of Escherichia coli requires a 60-kilodalton periplasmic protein. J Bacteriol. 1987;169:5489–5495. doi: 10.1128/jb.169.12.5489-5495.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol. 2003;185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulat D, Jault JM, Duclos B, Geourjon C, Cozzone AJ, Grangeasse C. Staphylococcus aureus operates protein-tyrosine phosphorylation through a specific mechanism. J Biol Chem. 2006;281:14048–14056. doi: 10.1074/jbc.M513600200. [DOI] [PubMed] [Google Scholar]
- Toh E, Kurtz HD, Jr, Brun YV. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol. 2008;190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson AD, Fuqua C. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol. 2009;12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang PH, Li G, Brun YV, Freund LB, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci USA. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J-W, Alley MRK. Proteolysis of the Caulobacter McpA Chemoreceptor Is Cell Cycle Regulated by a ClpX-Dependent Pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojnov AnA, Zorreguieta A, Dow JM, Daniels MJ, Dankert MA. Evidence for a role for the gumB and gumC gene products in the formation of xanthan from its pentasaccharide repeating unit by Xanthomonas campestris. Microbiology. 1998;144:1487–1493. doi: 10.1099/00221287-144-6-1487. [DOI] [PubMed] [Google Scholar]
- Wang SP, Sharma PL, Schoenlein PV, Ely B. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci USA. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Larue K. Stop and go: regulation of chain length in the biosynthesis of bacterial polysaccharides. Nat Struct Mol Biol. 2008;15:121–123. doi: 10.1038/nsmb0208-121. [DOI] [PubMed] [Google Scholar]
- Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whitfield C. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.