Summary
Localized cell shape change initiates epithelial folding, while neighboring cell invagination determines the final depth of an epithelial fold. The mechanism that controls the extent of invagination remains unknown. During Drosophila gastrulation, a higher number of cells undergo invagination to form the deep posterior dorsal fold, whereas far fewer cells become incorporated into the initially very similar anterior dorsal fold. We find that a decrease in α-Catenin activity causes the anterior fold to invaginate as extensively as the posterior fold. In contrast, constitutive activation of the small GTPase Rap1 restricts invagination of both dorsal folds in an α-Catenin-dependent manner. Rap1 activity appears spatially modulated by Rapgap1, whose expression levels are high in the cells that flank the posterior fold, but low in the anterior fold. We propose a model whereby distinct activity states of Rap1 modulate α-Catenin-dependent coupling between junctions and actin to control the extent of epithelial invagination.
Introduction
Epithelia are the most abundant tissue type in the animal kingdom. During animal development, epithelial tissues undergo a diverse array of morphogenetic processes to stretch, contract or deform (Fristrom, 1988). During early embryonic development, epithelial morphogenetic processes such as tissue invagination and cell delamination produce the initial internal tissue layers. In the later stages of development, morphogenetic changes of the epithelium produce vital organ structures and ultimately shape the form of the body. The mechanisms that underlie epithelial morphogenesis are thus fundamental to the understanding of a wide variety of developmental processes that occur during the entire lifetime of the animals.
One of the most basic processes of epithelial morphogenesis is epithelial folding, during which a sheet of two-dimensional epithelium undergoes dramatic cell shape changes and tissue reorganization to form a three-dimensional groove or a furrow, in some cases producing an enclosed tube and in others resulting in the internalization of cells. Epithelial folding is initiated by spatially restricted cell shape changes that deform the tissue. In most of the epithelial folding events that have been examined previously, the initial cell shape changes result from the accumulation and activation of actin-based molecular motor myosin that contracts the apical cell surface (Sawyer et al., 2010). Such apical constriction produces wedge-shaped cells, thereby deforming the tissue. Recently, however, we identified an alternative initiation mechanism during Drosophila gastrulation. This novel initiation process involves the repositioning of adherens junctions along the apical-basal axis of the initiating cells, but not spatially restricted activation of myosin contractility (Wang et al., 2012). This process occurs on the dorsal side of the early Drosophila gastrula that forms two epithelial folds called the anterior and posterior dorsal folds. Both dorsal folds undergo junctional repositioning that requires spatially restricted modulation of the epithelial apical-basal polarity. Specifically, the levels of the basal-lateral determinant Par-1 kinase decrease in the initiating cells, relative to a constant level of its substrate, the scaffolding protein Bazooka (Benton and St Johnston, 2003b). The resulting higher ratio of Bazooka/Par-1 in the initiating cells relative to that in the neighboring cells enables basal repositioning of adherens junctions, while the junctions in the neighboring cells remain in the subapical region. This junctional shift leads to the subsequent narrowing of cell apex and the ultimate shortening of the initiating cells, allowing the dorsal epithelium to deform.
Unlike epithelial folds (e.g. the ventral furrow that forms during Drosophila gastrulation) that are composed primarily of cells that display initial cell shape changes, dorsal fold formation involves the incorporation of neighboring cells adjacent to the initiating cells that do not display the junctional shift and apical narrowing during the initiation event, but become incorporated into the eventual tissue fold structure during the subsequent invagination process. Although the two dorsal folds display identical junctional shifts and cell shape changes (apical narrowing and the subsequent shortening) in their initiating cells (Wang et al., 2012), their ultimate morphology differs because their neighboring cells undergo distinct degrees of invagination. A higher number of neighboring cells become incorporated into the posterior fold, while far fewer cells do so in the anterior fold, producing a deep posterior fold and a shallow anterior fold (Figure 1). Previous work on epithelial folding generally assumed that cell shape changes that occur during initiation produce mechanical forces that are themselves sufficient to drive tissue rearrangement (Sawyer et al., 2010). However, it remains unclear whether additional cellular and mechanical processes control neighboring cell invagination to shape the final morphology of an epithelial fold. The dorsal fold system with its two epithelial folds exhibiting distinct degrees of invagination thus offers a unique opportunity to investigate this issue.
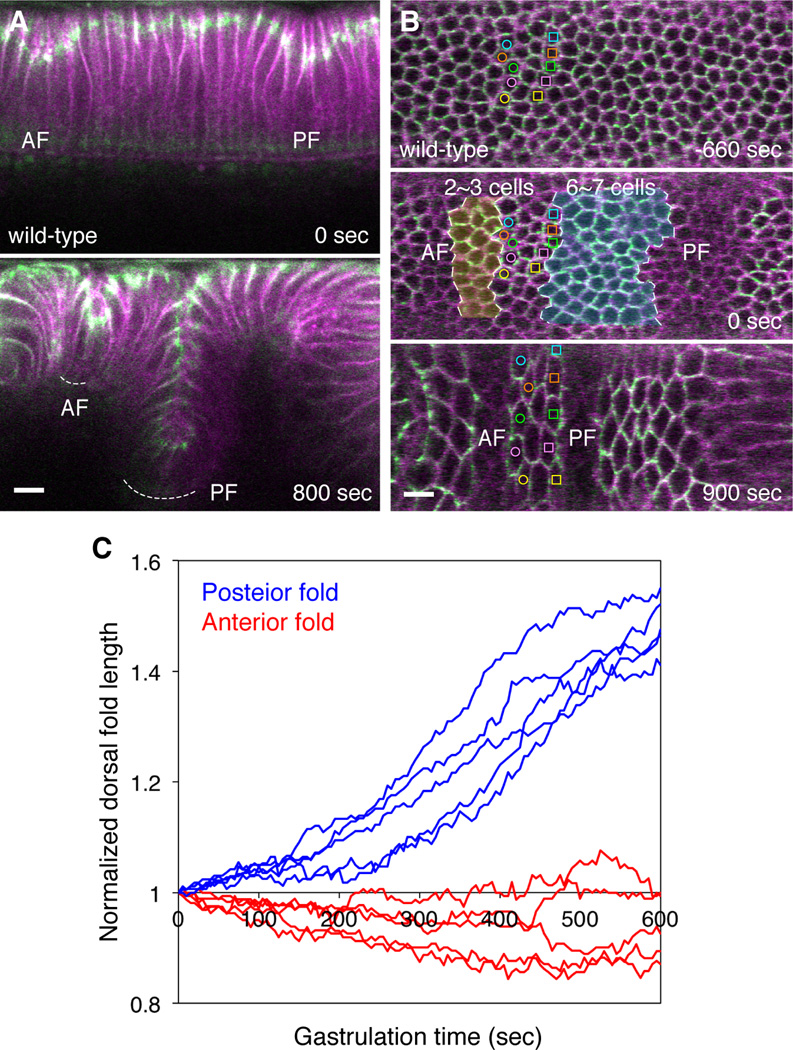
Figure 1. The two dorsal folds undergo distinct extent of invagination.
(A and B) Two-photon time-lapse images of the dorsal epithelium in gastrulating wild-type embryos expressing E-Cadherin-GFP (green) and membrane-mCherry (magenta). AF, anterior fold. PF, posterior fold. The mid-sagittal section view (A) shows the initial junctional shift at the onset of gastrulation (0 sec) and the eventual morphology of the dorsal folds (800 sec). Dashed curves indicate the bottom of the dorsal folds. The dorsal view (B) shows a single Z slice at 8 µm below the embryonic surface at three time points: prior to the junctional shift (−660 sec), onset of gastrulation (0 sec) and the completion of dorsal fold formation (900 sec). Cells shaded in light yellow and blue are neighboring cells that reside in the region between the anterior and posterior fold initiating cells and ultimately undergo invagination into the anterior and posterior folds as determined by tracing the anterior and posterior borders of the cells that remain on the embryonic surface at 900 sec time point (labeled with color-coded open circles and squares). Scale bar, 10 µm. (C) Total dorsal fold length as a function of time. Dorsal fold length was measured as the vertical distance between the embryonic surface and the bottom of the fold and normalized by the initial length at the onset of gastrulation (time = 0). Each trace represents measurements from one embryo (n = 5). See also Figure S1 and Movie S1.
Extensive invaginations such as those displayed by the posterior folds represent significant reorganization of the tissue architecture and likely require substantial restructuring of adherens junctions that hold the cells together within the epithelia. Adherens junctions are composed of transmembrane Cadherin and cytoplasmic catenins that linked the Cadherin molecules to the underlying actin cytoskeleton. In particular, α-Catenin, whose N- and C-terminal domains bind to the junctional core protein β-Catenin and the filamentous actin, respectively, has been thought of as the key molecule that couples the junctions to actin (Cavey et al., 2008; Costa et al., 1998; Gates and Peifer, 2005; Maiden and Hardin, 2011; Pokutta et al., 2008; Sarpal et al., 2012). Although interactions between the junctional complexes and the actin filaments are crucial for the stability and integrity of the junctional structure, recent experimental evidence suggests that that disruption of junction-actin coupling by reducing the activity of α-Catenin retains cell-cell adhesion at least temporarily, while increasing the mobility of junctional complexes (Cavey et al., 2008; Sarpal et al., 2012; Vasioukhin et al., 2001). We wondered whether uncoupling junctions from actin might enhance junctional restructuring when junctions are under stress and thus may increase the extent of dorsal fold invagination.
In the present study, we present genetic evidence that differing strength of junction-actin coupling underlies distinct degrees of dorsal fold invagination. Uncoupling junctions from actin by reducing α-Catenin activity causes extensive invagination of the anterior fold, mimicking the phenotype of the posterior fold. In the wild-type embryo, the physical strength of junction-actin coupling appears to depend on the activity states of the small GTPase Rap1, conferred by the spatially restricted expression of its GTPase activating protein Rapgap1 in the cells surrounding the posterior fold, but not in the anterior fold. These results suggest a model whereby differential modulation of junction-actin coupling controls the distinct extent of epithelial invagination.
Results
The two dorsal folds display distinct degrees of invagination
The dorsal epithelium of the early Drosophila gastrula forms a shallow anterior fold and a deep posterior fold (Wang et al., 2012). We used live imaging to monitor cellular and morphological changes during dorsal fold formation (Figure 1A, Movie S1 Part 1). During initiation, the junctional shift occurs in the initiating cells of the anterior and posterior folds in a similar fashion. The initiation is followed by invagination of two to three rows of flanking cells on each side of the initiating cells in the anterior fold, ultimately producing an epithelial fold that contains one row of cells at the bottom and two to three rows of cells on each side. In contrast, the invagination of the posterior fold involves several more cells. Six to seven rows of cells that flank each side of the initiating cells undergo invagination, producing an epithelial fold that contains one row of cells at the bottom and nine to ten rows of cells on each side. Upon completion of dorsal fold formation, only three rows of cells remain on the embryonic surface in the intervening region between the two dorsal folds, while other cells become invaginated (Figure 1B). Quantitation of the total depth of dorsal folds reveals that the anterior fold does not elongate, whereas the depth of the posterior fold increases by 40 to 50% over the course of ten minutes (Figure 1C). The quantitative measurement of the depth of the dorsal folds faithfully reflects the extent to which the neighboring cells become incorporated into the dorsal folds and thus represents a reliable readout for the degree of invagination.
The process of dorsal fold invagination coincides with the global gastrulation movement of germband extension during which the posterior midgut that forms at the posterior pole moves anteriorly as the lateral ectoderm undergoes tissue elongation. The movement of the posterior midgut appears to compress the dorsal epithelium. One possible explanation for the differing degrees of dorsal fold invagination is that the posterior fold experiences a greater degree of mechanical stress during germband extension and that the apparent compression pushes more cells into the eventual posterior fold than the anterior fold. This possibility, however, seems unlikely given that the differing depths of the dorsal folds remained intact in the torso-like mutant embryo in which the formation of posterior midgut was disrupted and the compression was substantially reduced. During the first 1000 seconds of gastrulation, the velocity of the dorsal tissue is severely decreased in the torso-like mutants (1.08 ± 0.48 µm/min moving anteriorly as compared to 4.15 ± 0.31 µm/min in the wild type). Despite this reduction in dorsal tissue compression, the posterior folds in the torso-like mutants undergo an identical degree of lengthening to that observed in the wild type (Figure S1, Movie S1 Part 2). Interestingly, however, in these mutant embryos the duration of posterior fold invagination prolongs and varies widely among individual embryos. It seems that the external compression ensures temporal robustness of posterior fold invagination, although it does not ultimately determine the depth of dorsal folds. Since the extent of invagination is not altered in situations in which the external compression is reduced, local processes, rather than global patterns of stress, may account for the differing degrees of dorsal fold invagination.
Reduction of α-Catenin activity dramatically enhances anterior fold invagination
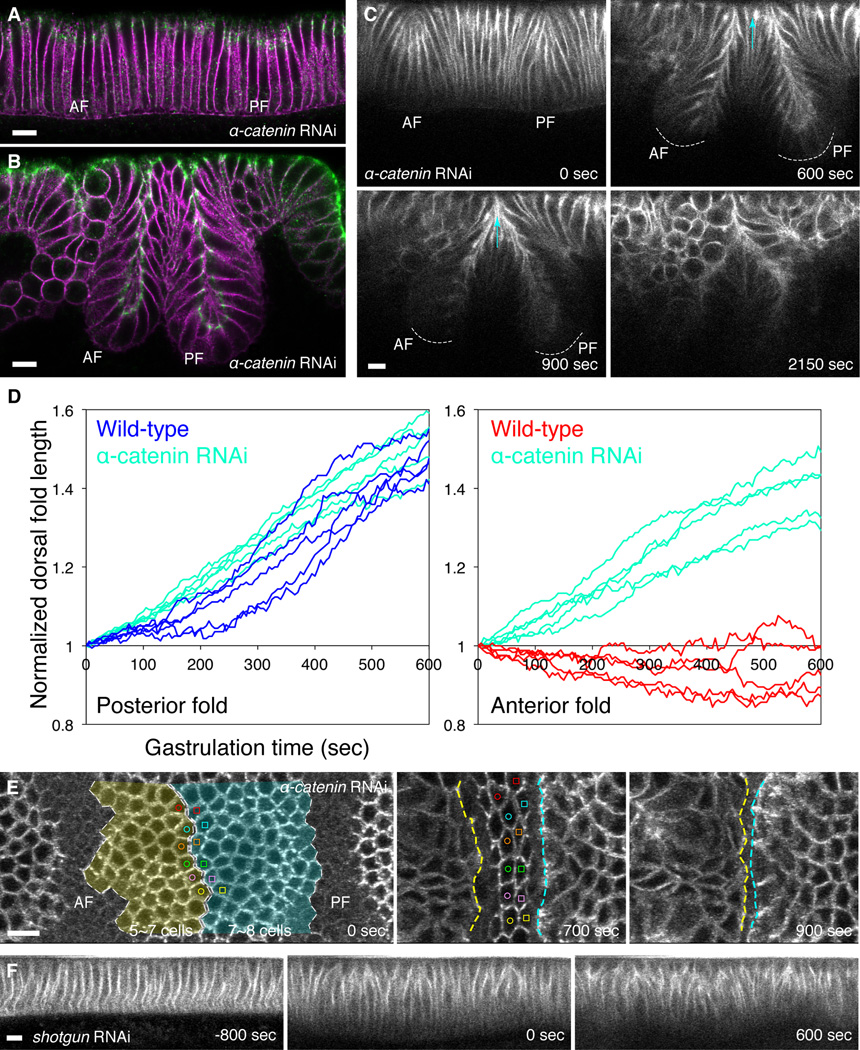
We wondered whether the physical coupling between junctions and actin represents the local event that is spatially modulated to confer distinct degrees of neighboring cell invagination. To explore this possibility, we asked whether uncoupling junctions from actin by reducing the levels of α-Catenin activity could facilitate invagination. We used a hairpin construct that mediates RNA interference (RNAi) of α-catenin (Ni et al., 2011) to knock down the activity of α-Catenin (Figure S2). In these α-catenin RNAi embryos, the assembly, the initial positioning and the basal shift of adherens junctions in the initiating cells all appear normal (Figure 2A), consistent with the retention of junctional function and integrity. Strikingly, however, the anterior folds that form in these embryos contain a higher number of cells (one cell at the bottom and six to seven cells on each side) and invaginate far deeper than those in the wild-type embryo (Figure 2B-2D, Movie S2 Part 1). In many cases, the anterior folds undergo such extensive invagination that the two dorsal folds are nearly identical in depth. Live imaging analysis indicates that a much greater number (five to seven rows) of neighboring cells become invaginated into the anterior fold, while the number (seven to eight rows) of cells that invaginate into the posterior fold is comparable to that in the wild type (Figure 2E). As a result, all intervening cells become invaginated, the gap between the dorsal folds closes, and no cell remains visible on the embryonic surface after the invagination. Thus, reducing α-Catenin activity, and thereby presumably weakening the physical coupling between junctions and actin, can promote epithelial invagination.
Figure 2. Abrogation of α-Catenin promotes epithelial invagination.
(A and B) Confocal mid-sagittal section of immunofluorescence of junctional core component Armadillo (Green) and membrane marker Neurotactin (magenta) in the dorsal epithelium of an α-catenin RNAi embryo at the onset of gastrulation (A) or at the end of dorsal fold formation (B). (C) Two-photon time-lapse images of the mid-sagittal section view of the membrane marker Resille-GFP in a gastrulating α-catenin RNAi embryo. Dashed curves indicate the bottom of the dorsal folds. Light blue arrow indicates invagination of all intervening cells. Many cells round up or become severely distorted after completion of dorsal fold formation (2150 sec). (D) Comparison of total dorsal fold length as a function of time between the wild-type and α-catenin RNAi (n = 5) embryos. (E) Two-photon time-lapse images of the dorsal view of E-Cadherin-GFP in a gastrulating α-catenin RNAi embryo. All neighboring cells in the intervening region between dorsal folds become invaginated into the anterior and posterior folds (shaded in light yellow and blue in D) as determined by cell tracing denoted with color-coded open circles and squares. This results in the closure of the gap between dorsal folds (the region between yellow and blue dashed lines). (F) Two-photon time-lapse images of the mid-sagittal section view of Resille-GFP in the dorsal epithelium of a shotgun RNAi embryo during cellularization (−800 sec) or gastrulation (0 and 600 sec). AF, anterior fold. PF, posterior fold. Scale bar, 10 µm. See also Figure S2 and Movie S2.
As described above, the α-catenin RNAi embryos retain the integrity of the epithelium, display normal localization of the junctional components and complete dorsal fold formation (Figure 2A and 2B). The cell adhesion and epithelial integrity are ultimately lost, however, as a large number of cells round up or become distorted, and the dorsal fold structure appears crumbled (Figure 2C, panel 2150 sec; see also Movie S2 Part 1). Importantly, the phenotype of α-catenin RNAi during dorsal fold formation is distinct from that of a complete loss of cell adhesion. RNAi against shotgun, which encodes E-Cadherin, the transmembrane protein that is a core component of the junctions, results in a widespread distortion of the columnar cell shape prior to the onset of gastrulation, indicative of a loss of epithelial integrity, and the dorsal folds do not form (Figure 2G, Movie S2 Part 2). These observations are in agreement with previous studies that cell adhesion could be maintained at least temporarily when α-Catenin is disrupted, while the long-term consequence of loss of α-Catenin is similar to that of the loss of junctional core components (Cavey et al., 2008; Sarpal et al., 2012; Vasioukhin et al., 2001). Our data thus unveil a transient and context-specific function for α-Catenin during epithelial invagination.
Increased Rap1 activity restricts epithelial invagination in an α-Catenin-dependent manner
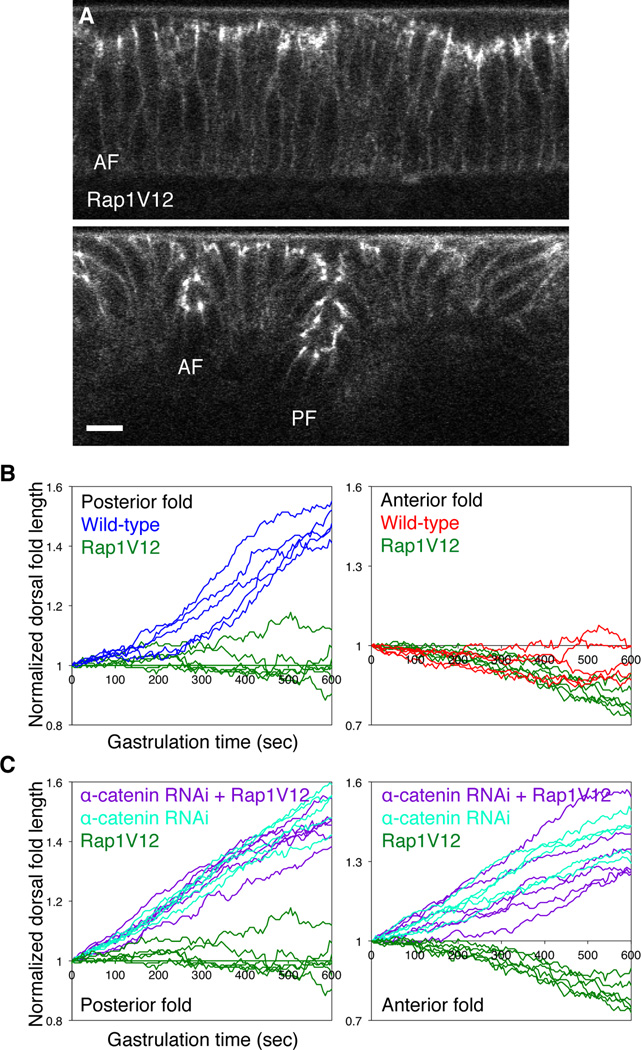
We next asked whether inhibition of junctional restructuring could restrict invagination. Recent work suggests that the small GTPase Rap1 promotes physical coupling between junctions and actin network (Noda et al., 2010; Sawyer et al., 2009; Spahn et al., 2012). Although Rap1 plays diverse roles in cell adhesion, migration and polarization (Boettner and Van Aelst, 2009; Bos, 2005), we considered the possibility that elevated Rap1 activity might strengthen junction-actin coupling in the dorsal epithelium, thereby suppressing junctional restructuring. Live imaging analysis of embryos overexpression of Rap1V12, a GTP-locked, constitutively active form of Rap1 (Boettner et al., 2003) shows that these embryos form two small dorsal folds (Figure 3A, Movie S3; compared to Figure 1A, Movie S1 Part 1). The depth of the posterior fold increases by less than 15%, in contrast to the normal 40 to 50% observed in the wild type (Figure 3B), while the anterior folds in these embryos still undergo restricted invaginations similar to those in the wild-type embryos. These results indicate that constitutive activation of Rap1 inhibits the invagination of neighboring cells that occurs after the initiation of the posterior fold.
Figure 3. Constitutive activation of Rap1 restricts epithelial invagination in an α-catenin-dependent manner.
(A) Two-photon time-lapse images of the mid-sagittal section view of E-Cadherin-GFP in the dorsal epithelium of a gastrulating embryo overexpressing Rap1V12. AF, anterior fold. PF, posterior fold. Scale bar, 10 µm. (B) Comparison of total dorsal fold length as a function of time between the wild-type embryos and embryos overexpressing Rap1V12 (n = 6). (C) Comparison of total posterior fold and anterior fold length as a function of time between the wild-type, α-catenin RNAi embryos and α-catenin RNAi embryos overexpressing RapV12 (n = 6). See also Movie S3.
If constitutively active Rap1 inhibits invagination by strengthening junctionactin coupling, Rap1-dependent restriction of epithelial invagination could depend on the same regulatory circuit that accounts for the opposing phenotype observed following knockdown of α-Catenin levels. We tested this hypothesis by performing RNAi knockdown of α-catenin in embryos that also overexpress Rap1V12. These conditions produced a phenotype very similar to that observed with the α-Catenin knockdown alone, namely extensive invagination in both the anterior and posterior folds, suggesting that active Rap1 depends on the presence of functional α-Catenin to confer restricted invagination (Figure 3C). The simplest interpretation of these results is that loss of α-Catenin is epistatic to the overexpression of constitutively active Rap1 and thus supports our hypothesis that active Rap1 promotes α-Catenin-dependent coupling between junctions and actin to restrict dorsal fold invagination.
The GTP activating protein Rapgap1 is expressed at high levels in the posterior fold region to promote invagination
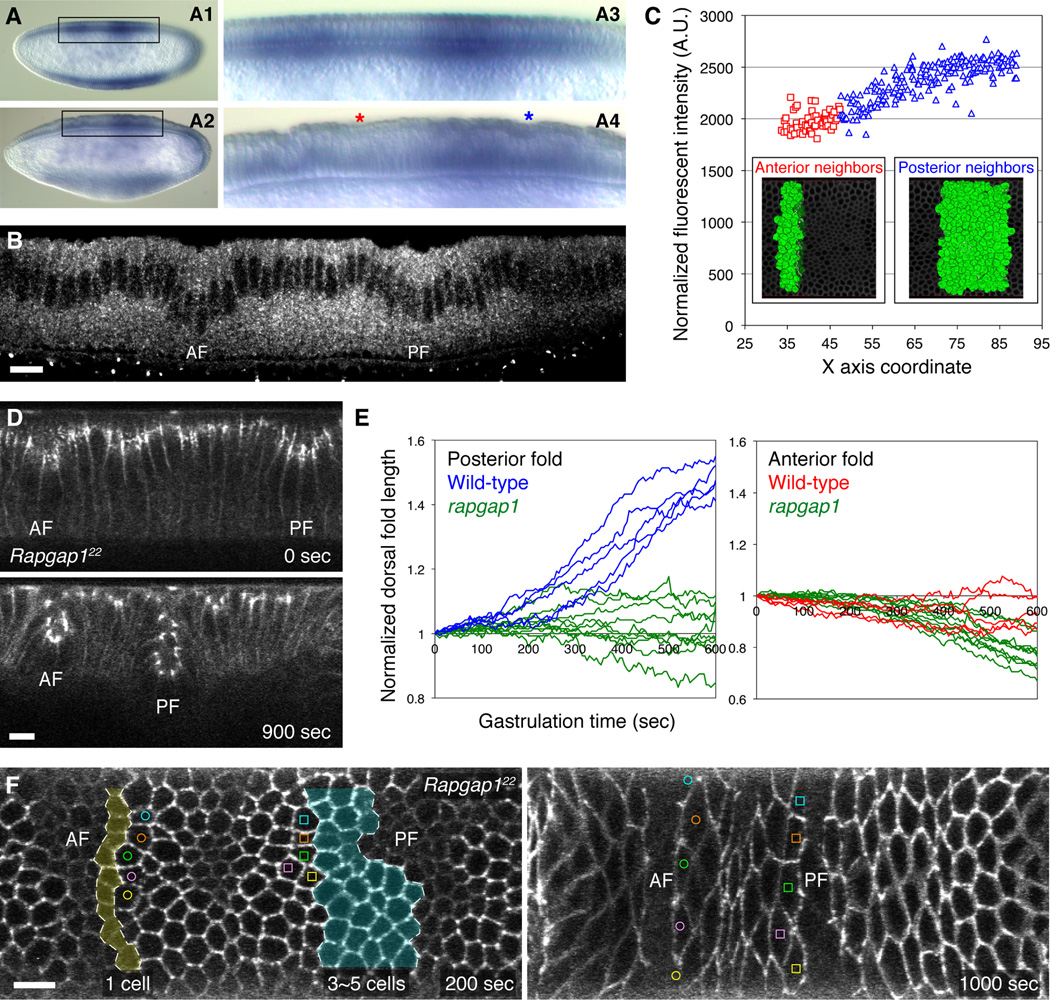
The differing degrees of dorsal fold invagination suggest that Rap1 activity is spatially modulated. Although Rap1 itself, and its activator PDZ-GEF homolog Dizzy, are maternally provided and uniformly distributed in the early embryo (Asha et al., 1999; Boettner and Van Aelst, 2007; Spahn et al., 2012), Rapgap1, a known GTPase activating protein for Rap1, provides a likely candidate that spatially modulates Rap1 activity (Chen et al., 1997). We used in situ hybridization to examine the expression pattern of Rapgap1. In the syncytial blastoderm, the maternal transcript of Rapgap1 is present uniformly in the somatic cells, which disappears prior to the onset of cellularization (data not shown and http://insitu.fruitfly.org/cgi-bin/ex/report.pl?ftype=3&ftext=GM01042-dg; Tomancak et al., 2007). During cellularization, the zygotic transcription of Rapgap1 displays a complex pattern that consists of four distinct, spatially restricted expression domains, overlapping the presumptive region of the cephalic furrow, the ventral furrow, the posterior midgut and the posterior dorsal fold, respectively (Figure 4A). This last expression domain centers on the initiating cells of the posterior fold and encompasses both the anterior and posterior flanking regions. Thus, across the dorsal epithelium, the levels of Rapgap1 zygotic transcription are high in the region surrounding the posterior fold, but low in the cells near the anterior fold. The distribution of Rapgap1 protein in the dorsal epithelium mirrors the spatial pattern of its transcript (Figure 4B). Quantitation of Rapgap1 concentration in individual cells demonstrates that among the cells that reside between the two dorsal folds, three to four rows of neighboring cells abutting the anterior fold has lowest levels of Rapgap1 expression (Figure 4C). These cells represent the cells that remain on the dorsal surface of the embryo upon completion of dorsal fold formation (Figure 1B). In contrast, the neighboring cells that will eventually become incorporated into the posterior fold express higher and graded levels of Rapgap1 that peak at the cells that immediately adjacent to the posterior fold initiating cells. Thus, the spatial distribution of Rapgap1 transcript and its protein product supports the model that Rapgap1 confers distinct activity states of Rap1 between the two dorsal folds.
Figure 4. The expression pattern and loss-of-function phenotype of Rapgap1.
(A) Whole mount in situ hybridization of Rapgap1 transcript visualized at the mid-sagittal view at two different stages: mid-cellularization (A1, boxed area enlarged in A3) and dorsal fold initiation (A2, boxed area enlarged in A4). Red and blue asterisks denote the anterior and posterior folds. (B) Confocal mid-sagittal section of Rapgap1 immunofluorescence in a wild-type embryo during dorsal fold initiation. AF, anterior fold. PF, posterior fold. Scale bar, 10 µm. (C) Normalized Rapgap1 immunofluorescence in individual cells in the region between the two dorsal folds plotted against the anterior-posterior position (represented by the X-axis coordinate) of these cells. The insets show 3D reconstruction models of the cells from which the measurements were derived. The red squares represent measurements from the 3 to 4 rows of cells that are immediately adjacent to the anterior fold, while the blue triangles represent the 9 to 10 rows of cells that are adjacent to the posterior fold. (D) Two-photon time-lapse images of the mid-sagittal section view of E-Cadherin-GFP in the dorsal epithelium of a Rapgap1 mutant embryo. (E) Comparison of total dorsal fold length as a function of time between the wild-type and Rapgap1 (n = 10) mutant embryos. (F) Two-photon time-lapse images of the dorsal view of E-Cadherin-GFP in a Rapgap1 mutant embryo. Cells at the anterior and posterior borders that remain on the embryonic surface are labeled with color-coded open circles and squares. Cells that invaginate into the anterior and posterior folds are shaded in light yellow and blue. AF, anterior fold. PF, posterior fold. See also Movie S4 and Movie S6.
To determine whether the expression pattern of Rapgap1 is relevant for the control of dorsal fold morphology, we examined the loss of function phenotype of Rapgap1. Live imaging analysis shows that Rapgap1 mutant embryos undergo normal dorsal fold initiation, but form two small dorsal folds (Figure 4D, Movie S4). Quantitative analysis indicates that the posterior folds in these embryos invaginate poorly, similar to the phenotype that was observed for embryos overexpressing the constitutively active Rap1V12 (Figure 4E). This defect stems from a decrease in the number of neighboring cells (three to five rows) that become incorporated into the posterior fold, leaving a higher number of cells – up to eight rows – remaining on the embryonic surface in the intervening region (Figure 4F). These results thus support the model whereby high levels of Rapgap1 expression in cells surrounding the posterior fold modulates Rap1 activity to promote invagination, whereas low levels of Rapgap1 expression allow accumulation of active Rap1 in the flanking cells of the anterior fold that restricts invagination.
Neighboring cell geometries correlate with the extent of invagination, the levels of α-Catenin function and the states of Rap1 activity
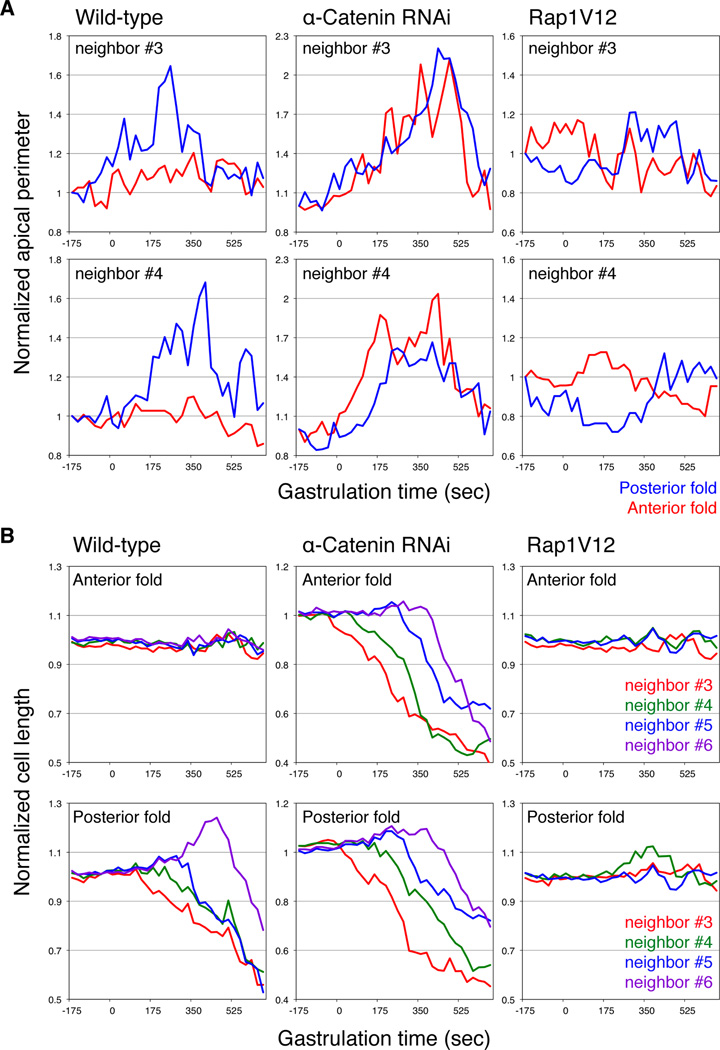
We next asked whether the cells that reside between the two dorsal folds behave differently according to the activity states of Rap1 given the differing levels of Rapgap1 expression. We measured the geometries of the neighboring cells that are at equivalent positions relative to the central cell of each dorsal fold (designated as cell #0). In neighboring cell #3 and #4 (the third and fourth neighbors anterior to cell #0) of the posterior fold in the wild-type embryo, we observed substantial expansion and subsequent contraction of the apical domain above the junctions as measured by the length of apical perimeter. In contrast, the #3 and #4 neighbors (the third and fourth neighbors posterior to cell #0) of the anterior fold do not undergo such dramatic changes (Figure 5A). In addition, the neighboring cells of the posterior fold undergo cell shortening sequentially, whereas those of the anterior fold maintain constant length relative to the dorsal cells that do not invaginate (Figure 5B). We found that RNAi knockdown of α-catenin causes the anterior fold neighboring cells to undergo dramatic apical expansion and contraction and to display sequential shortening similarly to the posterior fold neighboring cells, contrasting with the effect of Rap1V12 overexpression which causes the posterior fold neighbors to behave more similarly to the anterior fold neighbors in the wild-type in that their apical size displays only small fluctuations without substantial expansion and the total length remain constant. The dynamic expansion and contraction of the apical domain might suggest that junctions could undergo rapid restructuring and perhaps slide on the lateral surface of the cells, while cell shortening may reflect the ability for the neighboring cells to become incorporated into the tissue fold. Thus, these analyses establish that the behaviors of the neighboring cells correlate with the extent of invagination, the levels of α-Catenin activity and the activity states of Rap1.
Figure 5. Neighboring cell geometries correlate with extent of epithelial invagination.
(A) Normalized length of the apical domain perimeter as a function of time in the #3 and #4 neighboring cells in wild-type, α-catenin RNAi and Rap1V12- overexpressing embryos. The apical domain perimeter measures the cell outline apical to the junctions and represents a 2D approximation of the apical surface area. (B) Normalized cell length as a function of time in the #3 to #6 neighboring cells in wild-type, α-catenin RNAi and Rap1V12-overexpressing embryos.
Loss and gain of Rap1 function have similar effects on dorsal fold invagination
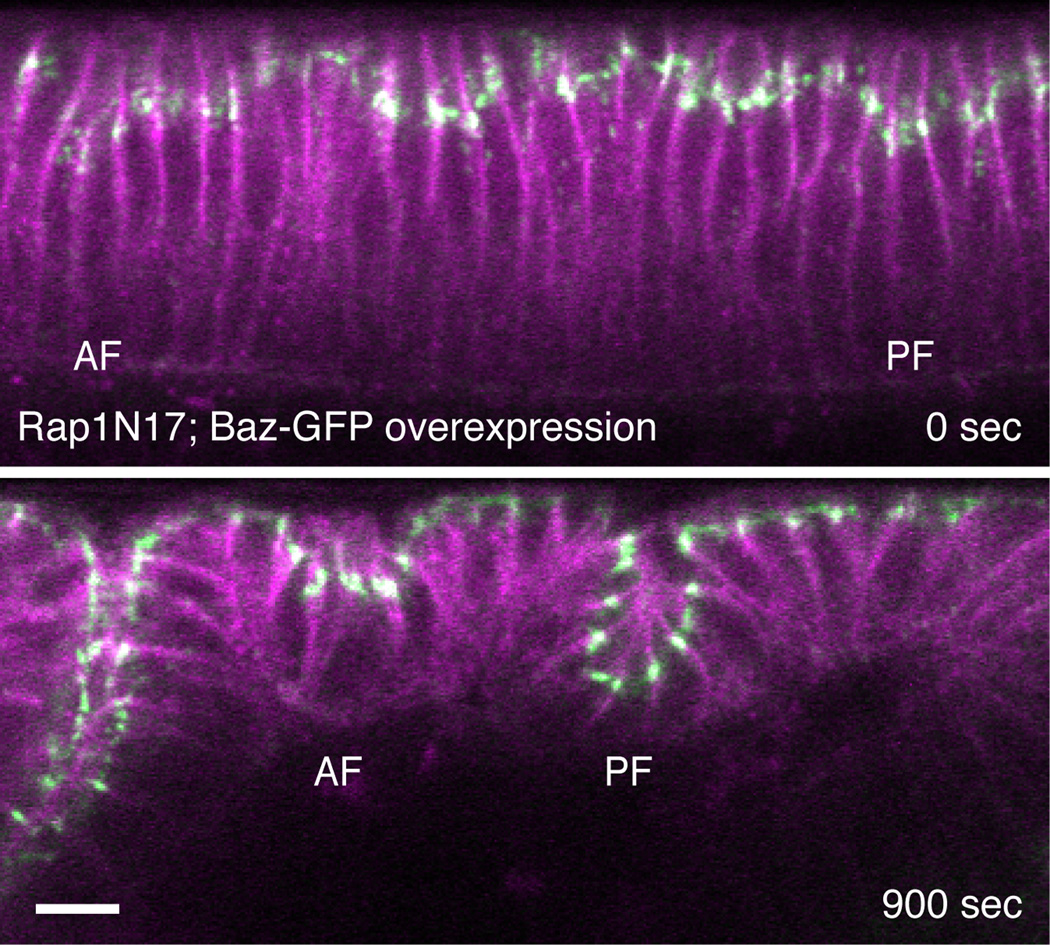
We next examined the phenotype of dorsal fold invagination in embryos that lack active Rap1. Loss of Rap1 activity results in an eventual elimination of the dorsal fold structures due to an earlier requirement of Rap1 function that is separate from its function during invagination (Figure S3, Movie S5 Part 1). We found that this initial requirement of Rap1 can be bypassed by overexpression of Bazooka, allowing us to determine the effect of loss of Rap1 function during dorsal fold invagination (see Discussion). A simple model might predict that loss of Rap1 might promote invagination, producing phenotypes complementary to overexpression of Rap1V12. Surprisingly, however, we find both the anterior and posterior folds display only limited invagination (Figure 6, Movie S5 Part 2). The similarity of this phenotype to that of Rap1V12 overexpression suggests that both the active and inactive forms of Rap1 are necessary for extensive invagination. It seems that the expression of Rapgap1 in the region of posterior fold promotes the formation of both forms, rather than simply reducing the levels of the active form. The presence of a GTPase activating protein could potentially increase the flux of the GTPase cycle by driving the rapid cycling of the small GTPase between its active and inactive forms. This regulatory principle, though seemingly counterintuitive, has been previously proposed and demonstrated for the Rho GTPase during cytokinesis (Bement et al., 2006; Miller and Bement, 2009; Miller et al., 2008). Thus, we propose that low levels of Rapgap1 expression in the anterior fold region allows for constitutive activation of Rap1, which inhibits invagination, whereas high levels of Rapgap1 expression in the region surrounding the posterior fold promotes Rap1 GTPase flux to facilitate invagination.
Figure 6. Loss of Rap1 function causes restricted invagination similar to constitutive activation of Rap1.
Two-photon time-lapse images of the mid-sagittal section view of E-Cadherin-GFP (green) and membrane-mCherry (magenta) in a gastrulating RapN17 embryo that overexpresses Bazooka-GFP. AF, anterior fold. PF, posterior fold. Scale bar, 10 µm. See also Figure S3 and Movie S5.
Discussion
In the current study we used the dorsal fold system to investigate whether specific cellular mechanisms actively regulate the extent of epithelial invagination. We showed that α-Catenin is required for the restricted invagination caused by constitutive activation of Rap1 and identified Rapgap1 as a locally expressed modulator of Rap1 that is required for the extensive invagination of the posterior fold. These data suggest a model whereby Rap1 regulates dorsal fold invagination through an α-Catenin-dependent process, and establish that differential regulation of an active, specific cellular mechanism confers distinct properties to the neighboring cells to control the extent of epithelial invagination.
Our genetic analysis identifies two separate functions of Rap1 during dorsal fold formation. The early function appears to be a general role required in all cells that is important for junctional positioning. This was established via examination of embryos that lack Rap1 activity, such as embryos that are produced by the germline clones of null alleles of Rap1 and dizzy, which encodes the Drosophila homolog of PDZ-GEF, a known guanine nucleotide exchange factor that activates Rap1 or embryos that overexpress a GDP-locked, dominant negative form of Rap1, Rap1N17 (Figure S3A–S3E, Movie S5 Part 1; Boettner and Van Aelst, 2007; de Rooij et al., 1999; Huelsmann et al., 2006; Spahn et al., 2012). These embryos display normal assembly of the adherens junctions, the initial basal shift of junction positioning in the initiating cells and attempt to form dorsal folds. Subsequently, however, the junctions re-localize to the apical surface in all dorsal cells, reversing these initial attempts of dorsal fold formation and eliminating all folding structures.
Rap1 appears to maintain the junctional positioning by maintaining the junctional levels of Bazooka. This notion is supported by the lower levels of junctional Bazooka in the Rap1 mutant embryos (Figure S3F and S3G) and by suppression of the loss-of-function phenotype of Rap1 by Bazooka overexpression, which restores the apical domain in the initiating cells and the dorsal fold structures in the Rap1 mutant embryos (Figure S3H, Figure 6, Movie S5 Part 2). Because Bazooka levels are uniform across the dorsal epithelium (Wang et al., 2012), this early function of Rap1 appears broadly required, independently of the levels of Rapgap1 expression, and operates in addition to Rap1’s later role during epithelial invagination. The effective suppression of Rap1 loss-of-function following Bazooka overexpression suggests that the two separate functions of Rap1 – the maintenance of Bazooka levels and the regulation of junction-actin connection during epithelial invagination – could be decoupled, allowing us to compare the effect of loss of Rap1 function to that of constitutively active Rap1V12.
The later, spatially regulated function of Rap1 is independent of Bazooka and is differentially modulated by the spatially restricted expression of Rapgap1. Since active Rap1 appears to act through α-Catenin to inhibit invagination, it seems plausible that distinct Rap1 activity states modulate the coupling strength between junctions and actin, thereby conferring distinct properties of junctional restructuring to the neighboring cells of the anterior and posterior folds. The geometric measurements of the neighboring cells suggest a model whereby constitutively active Rap1 inhibits junctional mobility so that the size of the apical domain remains constant in the cells surrounding the anterior fold where Rapgap1 levels are low. In contrast, Rapgap1 expression modulates Rap1 activity to promote junctional mobility in the neighboring cells of the posterior fold so that their apical domain expands. In this view, both the initiation and invagination processes require active remodeling of the junctions, but differ in their underlying cellular mechanisms (Figure 7). During initiation, the junctional shift is induced by a modification of the epithelial apical-basal polarity as a result of the downregulation of Par-1 in the initiating cells (Wang et al., 2012). During invagination, since Par-1 levels do not decrease, mechanical stress might be the dominant force that causes the junctions to move in the neighboring cells.
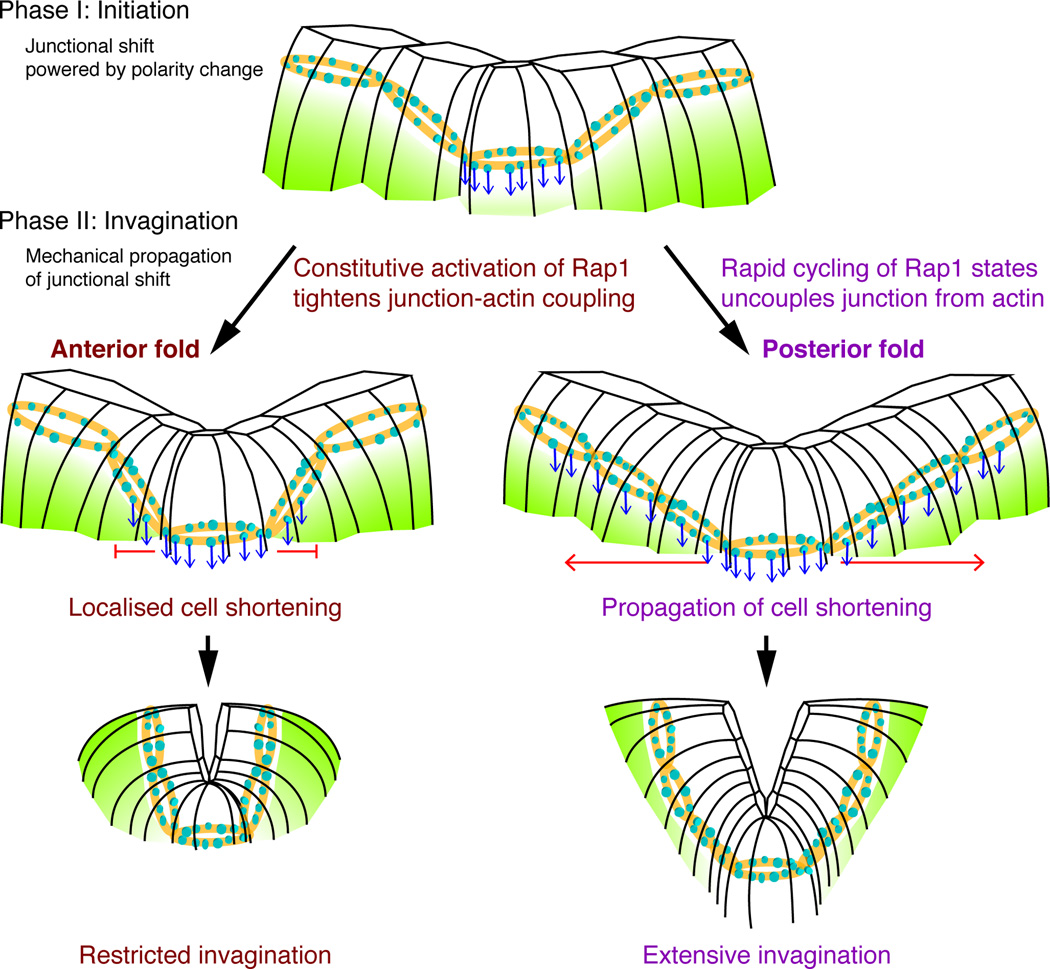
Figure 7. A model for the initiation and invagination of dorsal fold formation.
The anterior and posterior folds undergo a similar process of initiation during the phase I of dorsal fold formation during which polarity modification resulting from a decrease in the levels of Par-1 (light green) in the initiating cells causes a basal 0 shift of adherens junctions (blue arrows). In the phase II of dorsal fold formation, high levels of Rapgap1 expression in the neighboring cells of posterior fold likely drive rapid GTPase cycle of Rap1 to uncouple junctions from actin, allowing cell shortening to propagate across the tissue and the incorporation of these cells into a deep tissue fold. In contrast, low levels of Rapgap1 expression in the neighboring cells of anterior fold ensures constitutive activation of Rap1 that tightly couples junctions to actin to restrict neighboring cell shortening and invagination.
How Rap1 modulates α-Catenin-dependent junction-actin coupling remains unknown. We examined the intensities, localization and turnover kinetics (as measured by fluorescent recovery after photobleaching) of the core junctional components (E-Cadherin and Armadillo), α-Catenin, two junctional proteins that interact with both α-Catenin and actin (Canoe and Vinculin; Choi et al., 2012; Pokutta et al., 2008; Sawyer et al., 2009; Yonemura et al., 2010), but did not detect a difference between the neighboring cells of the anterior and posterior folds (data not shown). Recent work in mammalian tissue culture cells showed that the FRET (fluorescent resonance energy transfer) intensities of an E-Cadherin tension sensor correlate with the actin-coupling states of adherens junctions (Borghi et al., 2012). The use of such a sensor in the living Drosophila embryo might help revealing the difference in junction-actin coupling states between the anterior and posterior fold neighboring cells.
Recent work suggests that α-Catenin undergoes a conformational change upon mechanical stretch at the cell junctions (le Duc et al., 2010; Pokutta et al., 2008; Yonemura et al., 2010). Such conformational change could in principle relieve α-Catenin from an intramolecular inhibition on actin binding, thereby increasing its affinity to, or stabilizing its interaction with, the junctional actin (Choi et al., 2012). Changes in α-Catenin conformation thus may determine its ability to mediate the physical coupling between junctions and actin. It is of note that expression of a mutant form of α-Catenin that lacks the domain that modulates its conformational change can support static junctional function, but fails to effectively rescue the loss-of-α-Catenin phenotype in dynamic morphogenetic processes (Desai et al., 2013). It is possible that mechanical forces during morphogenesis dynamically modulate the conformational states of α-Catenin, the maintenance of which may require distinct Rap1 activity states. The dynamic changes of the α-Catenin conformations and the actin-coupling states of adherens junctions that they confer might be crucial for morphogenetic processes that involve extensive restructuring of cell-cell adhesion.
If Rapgap1 dictates the spatial extent of cell invagination, one simple model would envision that elevating the levels of Rapgap1 expression in the anterior region could promote anterior fold invagination. We explored this possibility using a UAS transgene to uniformly express Rapgap1 under the control of a maternal Gal4 driver. Two classes of phenotypes were observed: either a complete loss of dorsal fold formation (Movie S6 Part 1), or a limited degree of invagination similarly in both dorsal folds (Movie S6 Part 2). The former class suggests that the level of expression may be too high to permit the normal function of Rap1, while the latter class suggests that a reversal of the expression pattern – high in the anterior fold, but low in the posterior fold – might be necessary. We have attempted to express Rapgap1 in the cells anterior to the anterior fold using a Gal4 driver localized through the 3’UTR of the bicoid gene and have also used the enhancer of the Kr gene to direct the expression of Rapgap1 in cells that are posterior to the anterior fold. In neither case was there an effect on anterior fold invagination (data not shown). It is possible that driving extensive invagination for the anterior fold would require that Rapgap1 be expressed only in the surrounding cells of the anterior fold in a manner that mimics the endogenous pattern of Rapgap1 expression in the region of posterior fold. Currently, we know of no cis-regulatory element or a Gal4 driver that can drive gene expression in such a specific pattern. Thus, it remains unresolved whether ectopic expression in the anterior fold region would be sufficient to cause extensive invagination.
In summary, our data suggest an exciting conceptual framework in which regulated coupling between junctions and actin has a profound impact on the levels of tissue reorganization and on the cellular responses to mechanical stresses that arise during tissue reorganization. We define a specific molecular pathway that produces drastically different epithelial structures from a morphogenetic process whose initiation mechanism appears similar. The regulatory principles that we unveil for Rap1 and α-Catenin might be employed in other contexts of morphogenesis in which a tissue undergoes dramatic remodeling, while unperturbed tissue integrity and cell adhesion must be maintained.
Experimental Procedures
Drosophila Genetics
Drosophila stocks used for live imaging were: E-Cadherin-GFP (Oda and Tsukita, 2001), membrane-mCherry (Martin et al., 2010), Resille-GFP and Spider-GFP (Morin et al., 2001). Embryos that are mutant for torso-like were laid by torso-like1/torso-like4 females. Embryos that lack maternal activity of dizzy (also known as Gef26) or Rap1 (also known as Roughened) were produced by females containing germline clones of the dizzy null mutations (dizzyΔ1 or dizzyΔ8; Huelsmann et al., 2006) or Rap1 null mutations (Rap1rvB1 or Rap1CD3; Asha et al., 1999) using the FLP-DFS technique with the ovoD1 FRT40A or FRT2A ovoD1 chromosomes (Chou and Perrimon, 1996). The zygotic contribution for both of these genes is negligible as all embryos display the same phenotypes irrespective of their zygotic genotypes. Loss-of-function embryos for Rapgap1 were produced by a cross between males and females that are homozygous for the null allele Rapgap122 or Rapgap126 (Chen et al., 1997). For all of these genes, identical phenotypes were observed between the two alleles that were used and only data from the first of the two alleles were presented. Transgenic flies used in the overexpression experiments were: UASp-Bazooka-GFP (Benton and St Johnston, 2003a), UAS-Rap1N17, UAS-Rap1V12 (Boettner and Van Aelst, 2009), and UASp-Rapgap1. UASp-Rapgap1 was generated by cloning the coding region of the Rapgap1-RF transcript (http://flybase.org/reports/FBtr0112605.html) into pTIGER (Ferguson et al., 2012), which was then integrated into the attP2 landing site (Groth et al., 2004) at Genetic Services. The UAS transgenes were driven in the female germline using the maternal driver matαTub-Gal4VP1667C or matαTub-Gal4VP1667C and matαTub-Gal4VP1615.
RNAi Knockdown
RNAi knockdown of α-catenin was performed using the small hairpin construct P{TRiP.HMS00317}attP2 (Ni et al., 2011) driven by the maternal driver matαTub-Gal4VP1667C or both matαTub-Gal4VP1667C and matαTub-Gal4VP1615. RNAi knockdown of shotgun was performed by injecting double stranded RNA into the embryos at the syncytial blastoderm stage, typically 3–4 hours before imaging. The double stranded RNA against shotgun was synthesized using Megascript T7 kit (Ambion) from the PCR product that contains the T7 promotor sequence (5’-TAATACGACTCACTATAGGGTACT-3’) at each end. The PCR product used in the in vitro transcription reaction was amplified from 0–4 hours embryonic cDNA using the following primer pair: 5’-TGACTATCAGCGCCAGTGAC-3’, 5’-CGTGTGTATTCCGCACAATC-3’.
Live Imaging, Immunofluorescence and in situ Hybridization
Two-photon live embryo imaging was performed on a custom-made system built on an upright Olympus BX51 microscope that is equipped with a Ti:Sapphire tunable laser ranged from 720 to 1080 nm (Coherent). The laser was tuned at 920 nm for excitation of GFP or 960 nm for excitation of both GFP and mCherry. Immunofluorescence was performed on heat-methanol fixed embryos (Muller and Wieschaus, 1996) that were labeled with the following antibodies: anti-Neurotactin (BP106; 1:20; Developmental Studies Hybridoma Bank), rabbit anti-Armadillo (1:200), rat or guinea pig anti-Bazooka (1:500; Simoes Sde et al., 2010), rat anti-Rapgap1 (1:500, see below), and visualized with the Alexa 488, 568 and 647 conjugated secondary antibodies (Molecular Probes). Rat anti-Rapgap1 was generated at Panigen by immunizing rats with a full-length Rapgap1 recombinant protein produced at GenScript. Immunofluorescent images were taken on a Leica SP5 system. In situ hybridization was performed as described (Kosman et al., 2004) with a full-length Rapgap1 RNA probe labeled with digoxigenin. Images of in situ hybridization were captured on a Zeiss Axioplan microscope equipped with a Spot digital camera system (Diagnostics Instruments). Images were processed, assembled into figures and converted into movies using Image J, Adobe Photoshop and Adobe Illustrator.
Western Blot
Gastrulating embryos were devitellinized in bleach, washed and hand selected in 1XPBS with 0.1% Triton-X-100, and homogenized with plastic pestles in 1% Triton-X 100 buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 1.0% Triton-X 100) that contains 1mM DTT and Proteinase Inhibitor Cocktail (Sigma). Lysate of 40 embryos for each genotype was used for SDS-PAGE. Immunoblots were stained for rat anti-α-Catenin (DCAT-1; 1:20; Developmental Studies Hybridoma Bank,) or mouse anti-Tubulin (1:5000; Sigma).
Image Processing, Quantitation and Three-dimensional Reconstruction
Time course analyses of the dorsal fold depth (Figure 1C, Figure 2D, Figure 3B, Figure 3C, Figure 4E and Figure S1B) were performed by manually measuring the vertical distance between the embryonic surface and the bottom of the dorsal folds in Image J. These measurements were then plotted as a function of time. Time zero of gastrulation was defined as the end of cellularization, i.e. the time at which cells in the intervening region between dorsal folds reach their maximal cell length.
Time course analyses of the apical domain perimeter (Figure 5A) were performed by manually tracing the cell outline above the junctions, which were then measured in Image J. These measurements were then plotted as a function of time. Time zero of gastrulation was defined as described above. Neighboring cells were designated based on physical proximity to the central cell of each dorsal fold, which is the most basal cell at the end of dorsal fold formation and designated as cell #0.
Time course analyses of the neighboring cell length fold depth (Figure 5B) were performed by manually measuring the distance between the top and bottom of an individual cell in Image J. These measurements were then plotted as a function of time. Time zero of gastrulation was defined as described above.
Quantitation of immunofluorescent intensity of total Rapgap1 (Figure 4C) and junctional Bazooka (Figure S3G) was performed as described using the 3D reconstruction software that we developed previously (Wang et al., 2012). Briefly, Image stacks of fixed embryos doubly labeled for a junctional marker (Bazooka or Armadillo) and Neurotactin were processed for three-dimensional reconstruction based on the Neurotactin staining. For Figure 4C, cells that reside between the anterior and posterior fold initiating cells were analyzed. The total Rapgap1 immunofluorescent intensity in each cell was normalized by cell volume and plotted against the X-axis coordinate that represents the cell’s position along the anterior-posterior axis. For Figure S3G, 300 to 500 well-reconstructed dorsal cells of each embryo were used for quantitation. The immunofluorescent intensity of Bazooka was normalized by the junctional volume determined by the Bazooka staining and averaged within each embryo for plotting in Figure S3G. The average intensity among embryos was also plotted for comparison between the wild-type and Rap1N17-expressing embryos.
Time course analyses of the apical domain length (Figure S3H) were performed by manually measuring the vertical distance between the apex of the central initiating cell in the posterior fold and its junctions in Image J. These measurements were then plotted as a function of time. Time zero of gastrulation was defined as described above.
Supplementary Material
Acknowledgments
We thank, B. Boettner, S. Ferguson, U. Gaul, I. Hariharan, D. St Johnston, A. Martin, R. Reuter, L. Van Aelst, J. Zallen and Bloomington Stock Center for providing reagents; J. Goodhouse and S. Thiberge for assistance in microscopy; Shelby Blythe for help with Western blotting; G. Deshpande, members of the Wieschaus and Schüpbach labs for helpful comments on the manuscript and discussion. This work is supported by a postdoctoral fellowship from Helen Hay Whitney Foundation to Y.-C.W., and a National Institute of Child Health and Human Development grant (5R37HD15587) to E.F.W. E.F.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asha H, de Ruiter ND, Wang MG, Hariharan IK. The Rap1 GTPase functions as a regulator of morphogenesis in vivo. Embo J. 1999;18:605–615. doi: 10.1093/emboj/18.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Miller AL, von Dassow G. Rho GTPase activity zones and transient contractile arrays. Bioessays. 2006;28:983–993. doi: 10.1002/bies.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003a;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003b;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, Gaul U. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics. 2003;165:159–169. doi: 10.1093/genetics/165.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L. The Rap GTPase activator Drosophila PDZ-GEF regulates cell shape in epithelial migration and morphogenesis. Mol Cell Biol. 2007;27:7966–7980. doi: 10.1128/MCB.01275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21:684–693. doi: 10.1016/j.ceb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosingenerated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Cavey M, Rauzi M, Lenne PF, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453:751–756. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- Chen F, Barkett M, Ram KT, Quintanilla A, Hariharan IK. Biological characterization of Drosophila Rapgap1, a GTPase activating protein for Rap1. Proc Natl Acad Sci U S A. 1997;94:12485–12490. doi: 10.1073/pnas.94.23.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Pokutta S, Cadwell GW, Bobkov AA, Bankston LA, Liddington RC, Weis WI. alphaE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci U S A. 2012;109:8576–8581. doi: 10.1073/pnas.1203906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. The Journal of cell biology. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric alpha-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- Ferguson SB, Blundon MA, Klovstad MS, Schupbach T. Modulation of gurken translation by insulin and TOR signaling in Drosophila. J Cell Sci. 2012;125:1407–1419. doi: 10.1242/jcs.090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristrom D. The cellular basis of epithelial morphogenesis. A review. Tissue Cell. 1988;20:645–690. doi: 10.1016/0040-8166(88)90015-8. [DOI] [PubMed] [Google Scholar]
- Gates J, Peifer M. Can 1000 reviews be wrong? Actin, alpha-Catenin, and adherens junctions. Cell. 2005;123:769–772. doi: 10.1016/j.cell.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. The Journal of cell biology. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden SL, Hardin J. The secret life of alpha-catenin: moonlighting in morphogenesis. The Journal of cell biology. 2011;195:543–552. doi: 10.1083/jcb.201103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nat Cell Biol. 2009;11:71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, von Dassow G, Bement WM. Control of the cytokinetic apparatus by flux of the Rho GTPases. Biochem Soc Trans. 2008;36:378–380. doi: 10.1042/BST0360378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. The Journal of cell biology. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K, Zhang J, Fukuhara S, Kunimoto S, Yoshimura M, Mochizuki N. Vascular endothelial-cadherin stabilizes at cell-cell junctions by anchoring to circumferential actin bundles through alpha- and beta-catenins in cyclic AMP-Epac-Rap1 signal-activated endothelial cells. Mol Biol Cell. 2010;21:584–596. doi: 10.1091/mbc.E09-07-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J Cell Sci. 2001;114:493–501. doi: 10.1242/jcs.114.3.493. [DOI] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Yamada S, Nelson WJ, Weis WI. Biochemical and structural analysis of alpha-catenin in cell-cell contacts. Biochem Soc Trans. 2008;36:141–147. doi: 10.1042/BST0360141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal R, Pellikka M, Patel RR, Hui FY, Godt D, Tepass U. Mutational analysis supports a core role for Drosophila alpha-catenin in adherens junction function. J Cell Sci. 2012;125:233–245. doi: 10.1242/jcs.096644. [DOI] [PubMed] [Google Scholar]
- Sawyer JK, Harris NJ, Slep KC, Gaul U, Peifer M. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. The Journal of cell biology. 2009;186:57–73. doi: 10.1083/jcb.200904001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, Goldstein B. Apical constriction: a cell shape change that can drive morphogenesis. Dev Biol. 2010;341:5–19. doi: 10.1016/j.ydbio.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes Sde M, Blankenship JT, Weitz O, Farrell DL, Tamada M, Fernandez-Gonzalez R, Zallen JA. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev Cell. 2010;19:377–388. doi: 10.1016/j.devcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn P, Ott A, Reuter R. The PDZ-GEF Dizzy regulates the establishment of adherens junctions required for ventral furrow formation in Drosophila. J Cell Sci. 2012 doi: 10.1242/jcs.101196. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- Wang YC, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484:390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.