Summary
The role of the trans-membrane receptor Notch in the adult brain is poorly understood. Here, we provide evidence that bunched, a negative regulator of Notch is involved in sleep homeostasis. Genetic evidence indicates that interfering with bunched activity in the mushroom bodies (MBs) abolishes sleep homeostasis. Combining bunched and Delta loss-of-function mutations rescued normal homeostasis, suggesting that Notch signaling may be involved in regulating sensitivity to sleep loss. Preventing the down regulation of Delta by over-expressing a wild-type transgene in MBs reduces sleep homeostasis and, importantly, prevents learning impairments induced by sleep deprivation. Similar resistance to sleep loss is observed with the Notchspl-1 gain-of-function mutants. Immunohistochemistry reveals that the Notch receptor is expressed in glia, whereas Delta is localized in neurons. Importantly the expression of the intracellular domain of Notch, a dominant activated form of the receptor, in glia is sufficient to prevent learning deficits after sleep deprivation. Together these results identify a novel neuronal-glia signalling pathway dependent on Notch and regulated by bunched. These data highlight the emerging role of neuron-glia interactions in regulating both sleep and learning impairments associated with sleep loss.
Results and discussion
Mutations in bunched, a regulator of Notch signaling, affect sleep homeostasis
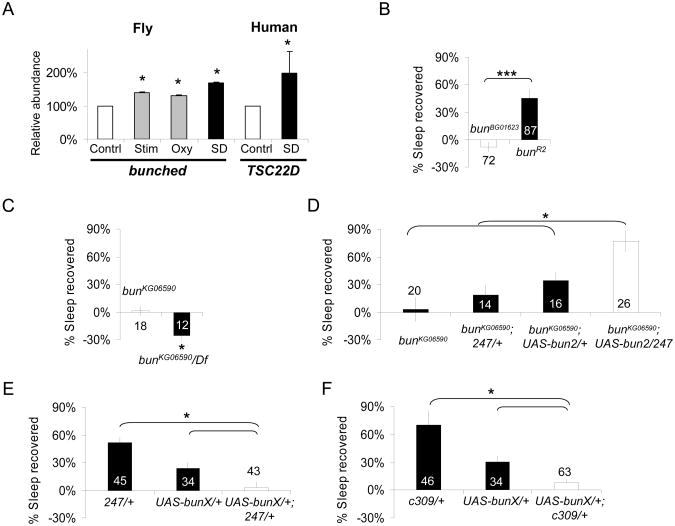
We, and others, have used microarrays to identify genes that are induced following sleep deprivation [1, 2]. Among the genes that have been identified, we found that bunched, a transcription factor regulating Notch activity in follicle cells [3], is up-regulated after sleep deprivation in fly heads (Figure 1A, left). bunched mRNA was also elevated in response to mechanical stimulation and exposure to oxidative stress suggesting that it is sensitive to physiological challenges in general (Figure 1A, left). Interestingly, mRNA for the bunched human homolog TSC-22D is detected in human saliva and is also up-regulated in sleep deprived healthy subjects (Figure 1A, right). In addition, TSC22D3 transcripts have been found to be up-regulated in the brain of sleep deprived mice [4]. Together, these results suggest that bunched function in response to sleep deprivation may be phylogenetically conserved. Nevertheless, it is important to note that gene profiling experiments are correlative in nature and do not necessarily reflect a change in gene activity. To determine whether bunched might, in turn, influence the response to sleep loss, we evaluated sleep homeostasis in several viable P element lines inserted in the bunched locus: bunBG01623, bunKG06590, bunKG00456 and bunKG00392. Sleep homeostasis is defined as a compensatory increase in sleep above baseline that occurs in the days following sleep loss [5-8]. The increase in sleep is highest immediately following sleep deprivation and progressively returns to pre-deprivation levels [5-8]. For comparisons across genotypes, individual flies are required to lose >90% of their nighttime sleep quota and the percentage of sleep recovered is calculated by dividing the minutes of sleep reclaimed during 48 h of recovery by the minutes of sleep lost; this metric represents a sensitive and reliable mean to assess sleep homeostasis [5, 8-10]. As seen in Supplemental Figure S1, all the bunched mutant lines showed a reduced homeostatic response to sleep deprivation. Excision of the P element in one of these lines, bunBG01623, restored normal homeostasis, indicating that the phenotype is due to the P element insertion (Figure 1B).
Figure 1. bunched is induced by sleep deprivation and regulates sleep homeostasis.
(A) bunched (left) mRNA levels are increased following 12hSD (SD, dark bar), compared to untreated controls (Contrl., white bar). bunched mRNA levels are also increased following a mechanical stimulation that reduces sleep disruption (Stim., grey bar), or following a 12h of exposure to oxidative stress (Oxy., grey bar). For the stimulation control, flies received the same number of mechanical stimuli provided during sleep deprivation by exposing them to the SNAP at twice the speed for 30min every two hours for 24 hours. For oxydative stress, flies were fed 20 μM paraquat dissolved in 1% agar/5% sucrose overnight (12h). mRNA for the bunched homolog TSC22D is elevated in saliva following 28h of waking in humans (n=9; Wilcoxon signed rank test, p=.038) Each saliva sample collected after sleep deprivation was compared to a circadian matched baseline sample from the same subject. Levels are expressed as % of baseline expression. *p<.05.
(B) bunBG01623 mutant flies do not display a sleep rebound following 12h of sleep deprivation, but excision of the P element in the bunR2 flies restores sleep rebound. *: p<0.05; Student's t-test.
(C) bunKG06590 flies (left) show no sleep rebound after sleep deprivation. bunKG06590 crossed to the Df(2L)prd1.7(Df) covering the bunched locus also failed to show a rebound after sleep deprivation (bunKG06590/Df, black bar, left graph). *p<.05, Student's t-test.
(D) Expression of a UAS-bun2 construct in the MB, using the 247-GAL4 driver is sufficient to increase sleep rebound in the bunKG06590 mutant background. ANOVA F(3,73)=8.4; p=7.18E-5.
(E) and (F) Expressing a dominant negative bunched construct (UAS-bunX) in the MBs using either the 247 (E) or c309 (F) GAL4 drivers (white bars in both graphs) is sufficient to reduce sleep homeostasis compared to genetic background controls (black bars). F(2,121)=12.12; p=1.6E-05 and F(2,142)=12.65; p=8.89E-06, respectively; *p<.05 modified Bonferroni test.
n is indicated in or beside each bar. mean±s.e.m is shown. See also Supplemental Figure S2 and Supplemental Table S1 for additional sleep data.
The MBs have been shown to regulate sleep [11, 12], Thus, to determine if bunched is required in the MB for the regulation of sleep homeostasis, we used the 247-GAL4 driver to express a wild-type copy of the bunched coding sequence in a bunched mutant background. bunBG01623 bears a GAL4 driver and could not be used. The bunKG06590 allele was used instead. Similarly to bunBG01623, bunKG06590 homozygous flies display a total lack of sleep homeostasis (Figure 1C, Supplemental Figure S1). The phenotype persists and was even enhanced if the allele is placed over a deficiency covering bunched (Figure 1C). This enhanced phenotype may be consequence of the inactivation of other genes deleted in the deficiency or may reflect the hypomorphic nature of bunKG06590 which results from a P element insertion in the last intron of bun (Figure S1). As shown in Figure 1D, bunKG06590; 247/+ and bunKG06590; UAS-bun2/+ had an increased homeostatic response compared to the bunKG06590 strain. However, bunKG06590;247/UAS-bun2 flies displayed a dramatically increased homeostatic response compared to both bunKG06590; 247/+ and bunKG06590; UAS-bun2/+ control lines, suggesting that activating bunched function in the MB is sufficient to increase the sleep homeostatic response. To confirm the requirement of bunched function in the MB, we expressed a dominant negative bunched construct, UAS-bunX [13], in the MB. Driving UAS-bunX in MB with two different GAL4 drivers resulted in a reduction in the homeostatic response to sleep loss compared to genetic controls, as anticipated (Figure 1E-F). Normal sleep rebound was observed when bunched was disrupted in the adult by feeding MB-Switch/UAS-bunX flies RU486 (not shown, [14]). The normal sleep rebound could reflect either an insufficient level of induction using this driver, or that disrupting bunched during development disrupts circuits that influence sleep regulation.
Sleep homeostasis and learning impairments after sleep loss in mutations affecting the Notch signaling
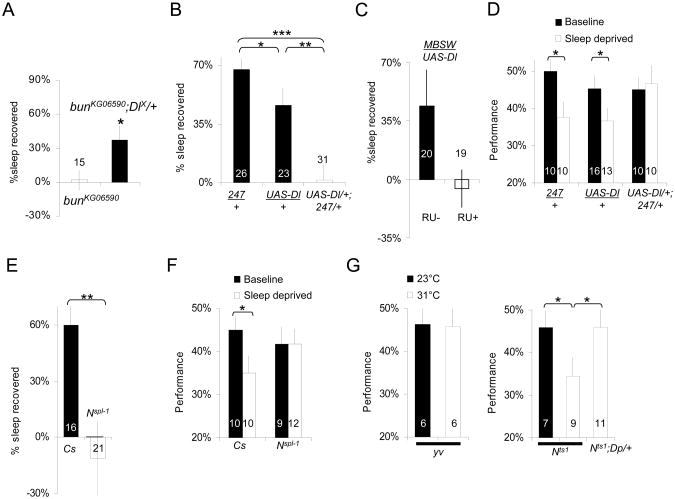
During oogenesis, bunched regulates Notch signaling [3], such that a loss of bunched function results in an up-regulation of the Notch pathway. Consistent with this idea, combining bunKG06590 with the amorphic DeltaX mutant rescued normal homeostasis (Figure 2A), suggesting that the bunched sleep homeostasis phenotype is linked to increased activation of Delta. Since expressing a dominant negative allele of bunched in the MBs reduces sleep homeostasis, we asked whether the activation of the Notch pathway in the MBs could protect against the negative effects of sleep loss. To begin, we first over-expressed the ligand Delta locally in the MB. Interestingly, over-expressing Delta in the MBs using the MB specific 247-GAL4 driver and a previously validated UAS-Delta construct (M.H insertion [15]) was sufficient to block the homeostatic response to sleep loss while both parental lines (247/+ and UAS-Delta/+) displayed a wild-type sleep rebound (Figure 2B). Similar results were obtained with another UAS-Delta construct (T.J insertion on the X chromosome [16], data not shown). Similar results were obtained when UAS-Delta was induced only at the adult stage using the MB-Switch GAL4 driver ([14], Figure 2C).
Figure 2. Notch regulates learning and sleep homeostasis after sleep deprivation.
(A) bunKG06590 flies (white bar, left) show no sleep rebound after sleep deprivation. Combining bunKG06590 to DeltaX(DlX) rescues normal homeostatic response (bunKG06590; DlX/+, black bar). *p<.05, Student's t-test.
(B) Over-expression of Delta using the MB specific driver 247 abolishes the sleep rebound (right, white bar). 247/+ and UAS-Dl/+ parental controls show a wild-type sleep homeostatic responses (black bars); F(2,77)=15.9; p=1,69E-06; *<.05, **<.005, ***<.0005 modified Bonferroni test, n is indicated in each bar. Sleep homeostasis is calculated for each individual as a ratio of the minutes of sleep gained above baseline during the 48 h of recovery divided by the total min of sleep lost during 12 h of sleep deprivation.
(C) Over-expression of Delta only at the adult stage using the MB-Switch GAL4 driver. Flies were fed RU 486 (RU+) or control food (RU-) for 48h before sleep deprivation (see methods). *p<.05 Student's t-test.
(D) Sleep deprivation does not disrupt learning in UAS-Dl/+;247/+ flies tested in the APS. In contrast both parental lines (247/+ and UAS-Dl/+) show learning impairments following 12 h of sleep deprivation; *<.05 modified Bonferroni test.
(E) Flies bearing the gain of function Notchspl-1(Nspl-1) allele do not show a sleep homeostatic response after 12 h of sleep deprivation compared to Cs controls; *<.05 t-test.
(F) Nspl-1 mutant flies (right) do not show learning impairments following 12h of sleep deprivation while Cs flies show significant impairments; *p<.05 Bonferroni test.
(G) Learning is not impaired by temperature in yv control flies (left graph); 23°C (black) vs. 31°C (white) p>.05, t-test. y Notchts1v flies (Nts1, right graph) learn normally at 23°C (permissive temperature), and are impaired at 31°C (non permissive temperature). A duplication covering the Notch locus, Dp (1;2) w+51b, rescues normal learning at 31°C (Nts1; Dp/+, right bar); F(2,24)=5.9; p=0.008; *p<.05 modified Bonferroni test.
n is indicated in or beside each bar. mean±s.e.m is shown. See also Supplemental Figure S2 for sleep in min/h graphs and Supplemental Table S2 for control metrics.
The lack of a homeostatic response may indicate that the animal is better able to withstand the negative effects of waking, or it may simply reflect a physiological impairment that globally disrupts sleep regulatory processes. To distinguish between these two possibilities we evaluated learning using Aversive Phototaxic Suppression (APS) [17]. We have recently shown that APS is sensitive to both sleep loss and sleep fragmentation [18]. Although both parental lines were impaired following 12 h of sleep deprivation, UAS-Delta/+; 247/+ flies maintained their ability to learn (Figure 2D). The Phototaxis Index (PI) and Quinine Sensitivity Index (QSI) for all lines were in the normal range of wild-type flies previously reported indicating that differences in performance cannot be attributed to changes in sensory thresholds (Supplemental Table S2) [1, 18, 19].
To confirm that activation of the Notch pathway regulates sensitivity to sleep deprivation and sleep homeostasis we evaluated sleep homeostasis in Notch gain-of-function mutants (NAx59b, Nspl-1). NAx59b flies were tested as heterozygous and exhibited a wild-type homeostatic response to sleep deprivation (Supplemental Table S1) presumably because the presence of wild type Notch reduces the dominant phenotype of this allele [20, 21]. However, the Nspl-1 allele did not compensate for lost sleep by initiating a homeostatic response (Figure 2E). Moreover, Nspl-1 flies maintained their ability to learn after sleep deprivation suggesting that the Nspl-1 mutation is protecting flies from the effects of sleep loss (Figure 2F). PI and QSI were in the normal range for Nspl-1 flies indicating that the changes in performance were not due to alterations in sensory thresholds (Supplemental Table S1). Nspl-1 encodes a Notch receptor with a single amino acid substitution in the extracellular domain, introducing a new O-fucosylation site [22, 23]. The effect of the Nspl-1 is context specific [21, 23]. A study of the effect of Nspl-1 on R8 photoreceptor development suggested that the mutation is increasing Notch sensitivity to the ligand Delta, leading to an ectopic activation of Notch in the R8 precursors cells [23].
We next tested several hypomorph/loss of function alleles (Nnd2, Nnd1, N5419, NCo). Except Nnd1, these alleles are homozygous lethal and were tested as heterozygotes. Surprisingly, all of these alleles showed a wild-type homeostatic response to sleep loss (Supplemental Table S2). The presence of a wild type Notch allele may have been sufficient to compensate for the effect of the mutation. Alternatively, the observed phenotypes could be attributed to developmental defects. Thus, we evaluated learning using the temperature-sensitive Notch allele Nts1 [24]. Nts1 flies showed normal APS performance at 23°C (Figure 2G, right graph). In contrast siblings that were tested at 31°C showed severe learning impairments. Genetic control flies (y v) showed normal performance both at 23°C and 31°C (Figure 2G, left graph). Importantly, Nts1 learning impairments at 31°C were not due to deficits in PI and QSI, which were in the normal range (Supplemental table S2), and could be rescued by introducing a duplication of the Notch locus in the genetic background (Figure 2G, right graph). In addition, similar learning impairments were observed with another thermo-sensitive allele: Nts2 (Supplemental Figure S3A). These results indicate that Notch is required for learning in the adult fly, in the absence of developmental defects. Given that temperature alters sleep homeostasis [10], we did not evaluate sleep rebound in Nts1 mutants.
Notch and Delta immuno-localization in the adult brain
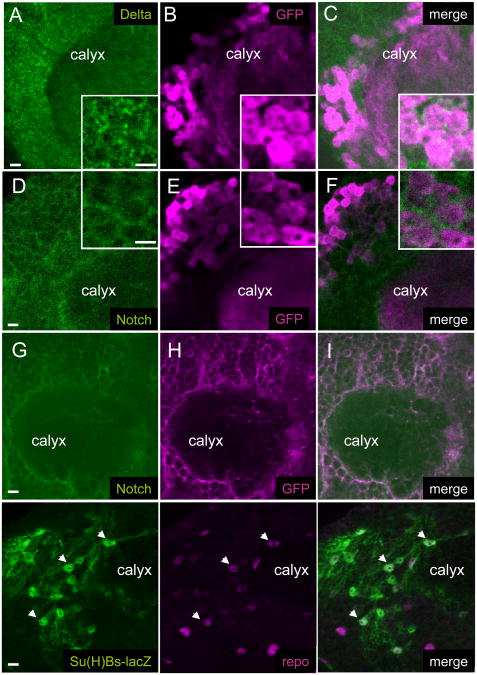
Whole brain immuno-histochemistry revealed different patterns of localization for Notch and Delta proteins (Figure 3A-I). Delta is expressed in a punctuate pattern throughout the brain cortex (Figure 3A, Supplemental Figure S3B shows an overall view) and is clearly detected in the cell bodies of Kenyon cells (Figure 3B-C). On the other hand, the Notch intracellular domain is predominantly detected in membranes surrounding the brain neuropiles and the cell bodies in the cortex, a pattern overlapping with glial cell membranes (Figure 3D-F, Supplemental Figure S3B shows an overall view). These results indicate that Notch and Delta could mediate neuron-glia signalling through cell-cell contacts. Such a possibility had already been suggested for the larval brain [25-27]. To further localize Notch activity in the adult brain, we have evaluated the expression of a Notch-reporter construct: Su(H)Bs-lacZ [28]. In this construct, lacZ is under the control of a promoter containing several Suppressor of Hairless (Su(H)) binding sites, and is induced following Notch activation. As shown in Figure 3J-L, Su(H)Bs-lacZ is specifically expressed in a subset of repo-positive glial cells, thus indicating that the Notch receptor is activated in glia. Similar results were obtained with a related Notch reporter construct driving EGFP expression ([29]; not shown).
Figure 3. Notch and Delta immuno-localization in the adult brain.
(A) to (I), confocal images showing the calyx, input neuropile of the MB and the surrounding neuronal cell bodies.
(A) to (F) Co-labelling of Delta (A,C, green) or Notch (D,F, green) and of GFP (B,C,E,F, magenta) in 247>UAS-GFP brains to identify a subset of Kenyon cells. Insets show high magnification views of the cell body region. Delta is localized in punctae within neuronal cell bodies whereas Notch is localized primarily in the surrounding membranes. Both Notch and Delta are only weakly expressed in the calyx neuropile itself.
(G) to (I) Co-labelling of Notch (green) and CD8-GFP (magenta) in a repo-GAL4>UAS-CD8-GFP brain to reveal co-localization of Notch with glial membranes.
(J) to (L) Co-labelling for the Notch reporter Su(H)Bs-lacZ (green) and the glial-specific repo (localized in glial nuclei, magenta). Arrows show examples of co-localization. All Su(H)Bs-lacZ positive cells were labeled with repo.
Bar: 5μm
See also Supplemental Figure S3B for an overall view of the brain.
Activating the Notch in glia abolishes sleep homeostasis and learning impairments after sleep loss
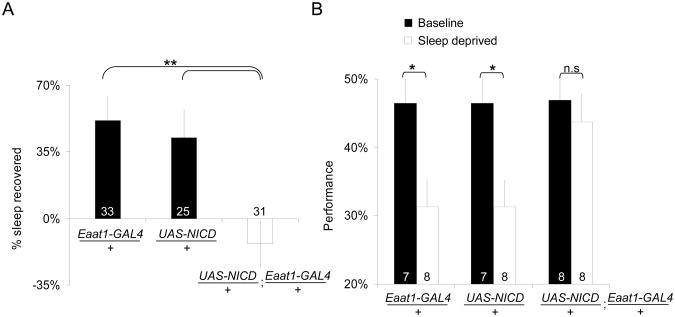
Upon binding to its trans-membrane ligand, Notch undergoes three rounds of cleavages leading to the shedding of the extracellular domain and the release of the intracellular domain into the cytoplasm. The intracellular domain associates with the transcription factor Su(H) and activates the transcription of targets genes such as members of the Enhancer of split complex [30]. Thus expressing the intracellular domain of Notch is sufficient to activate the pathway [31, 32]. We expressed the intracellular domain of Notch using a UAS-NICD (Notch Intra-Cellular Domain) and the glia-specific Eaat1-GAL4 driver [33]. As shown in Figure 4A, UAS-NICD/+; Eaat1-GAL4/+ flies did not show a homeostatic response to sleep deprivation. In addition, UAS-NICD/+; Eaat1-GAL4/+ flies showed normal learning after sleep deprivation (Figure 4B). In contrast, the genetic background controls showed a sleep rebound and learning impairments after sleep deprivation. Expressing UAS-NICD in neurons using elav-Switch did not alter sleep homeostasis (Supplemental Figure S4). Altogether, the data suggest that activating Notch signaling in glia can modulate the response to sleep loss as measured by sleep homeostasis and learning.
Figure 4. Expressing Notch intracellular domain in glial cells prevents impairments after sleep deprivation.
(A) Expression of Notch intracellular domain (NICD) using the glial specific driver Eaat1-GAL4 abolishes the sleep rebound (right, white bar). Eaat1-GAL4/+ and UAS-NICD/+ genetic controls show homeostatic responses comparable to wild type flies (black bars). F(2,86)=7.35; p=0.001; **p<.005 modified Bonferroni test; n is indicated in each bar.
(B) UAS-NICD/+; Eaat1-GAL4/+ flies do not show any significant impairment in learning following 12 h of sleep deprivation while performance is significantly impaired in both parental lines (Eaat1-GAL4/+ and UAS-NICD/+) after sleep loss (black vs. white); 2 way Genotype by Condition ANOVA shows main effect for condition F(1,46)=12.9; p=0.001; *p<.05 modified Bonferroni test.
n is indicated in or beside each bar. mean±s.e.m is shown. See also Supplemental Table S2 for control metrics and Supplemental Figure S2 for sleep in min/h graphs.
Conclusion
The evidence presented here suggests that Notch signaling controls factors that reduce the negative consequences of waking as measured by an attenuated sleep rebound and intact learning following 12 h of sleep deprivation. Although it is tempting to speculate that the intact learning seen following sleep loss is simply due to the flies not being sleepy, our previous studies have shown that sleepiness does not result in performance impairments in the APS [18]. Thus, Notch activation may preserve learning by preventing neuronal over-stimulation during extended waking. Reducing neuronal stimulation may also prevent the build-up of sleep debt and thus explain the lack of sleep rebound. Canonical Notch signaling leads to Su(H)-dependent changes in transcription but several other downstream pathways have been identified [30, 34], thus, further work is required to determine which pathway downstream of the receptor is effectively involved in this context. Our results suggest that Notch is mediating a neuron-glia signaling mechanism. These data provide additional support to recent work showing an involvement of glia in sleep homeostasis and cognitive impairments [35]. In mammals, adenosine released by glia appears to play a critical role [35]. Given that mutants for the only known Drosophila adenosine receptor have normal sleep homeostasis [36] other factors are likely to be involved. It is interesting to note in this context that expression of the cell adhesion molecule klingon, required for long term memory and controlled by Notch in the adult brain, has been reported to be expressed in the glia [37]. It should be noted that Notch localization and activation in glia may seem at odds with reports showing a requirement for Notch as well as the downstream effector Su(H) in MB neurons for memory consolidation [24, 38]. Our data do not exclude a low level of Notch expression in neurons. In fact, it would not be surprising that Notch is expressed in both cell types and mediate a two-way signaling between adjacent cells, as it occurs commonly during developmental processes [39].
Supplementary Material
Supplemental Table S1, related to Figures 1, 2 and 4: sleep data for bunched, Delta and Notch mutant conditions
Supplemental Table S2, related to Figure 2 and Figure 4: Phototaxis Index and Quinine Sensitivity Index
Supplemental Figure S1: sleep homeostasis in bunched mutants.
Supplemental Figure S2, related to Figures 1, 2 and 4: sleep and activity data for bu5ched, Delta and Notch mutant conditions for the baseline day, sleep deprivation day, and recovery days. .
Supplemental Figure S3, related to Figure 3: Learning impairments in flies mutant for the thermo-sensitive allele Nts2. Whole brain view of Delta and Notch immuno-localization.
Supplemental Figure S4, related to Figure 4: expressing UAS-NICD in neurons does not reduce the homeostatic response to sleep loss.
Highlights.
Mutations in bunched, a regulator of Notch, affect sleep homeostasis
Notch signaling can alter sleep homeostasis and sleep loss induced learning deficits
Notch is present in glial processes while Delta is predominantly in neuronal cell bodies
Results suggest that Notch mediate a neuron-glia pathway involved in sleep and learning
Acknowledgments
We thank Matthew Thimgan for helpful comments. This study was funded in part by 1 R01 NS051305-01A1, 5 K07 AG21164-02 the McDonnell Center for Cellular and Molecular Neurobiology, and the NIH Neuroscience Blueprint Core Grant, #NS057105).
Footnotes
The supplemental information contains two Supplemental tables and four Supplemental Figures as well as Supplemental Experimental Procedures, and Supplemental References:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seugnet L, Suzuki Y, Thimgan M, Donlea J, Gimbel SI, Gottschalk L, Duntley SP, Shaw PJ. Identifying sleep regulatory genes using a Drosophila model of insomnia. J Neurosci. 2009;29:7148–7157. doi: 10.1523/JNEUROSCI.5629-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 3.Dobens L, Jaeger A, Peterson JS, Raftery LA. Bunched sets a boundary for Notch signaling to pattern anterior eggshell structures during Drosophila oogenesis. Dev Biol. 2005;287:425–437. doi: 10.1016/j.ydbio.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Maret S, Dorsaz S, Gurcel L, Pradervand S, Petit B, Pfister C, Hagenbuchle O, O'Hara BF, Franken P, Tafti M. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 6.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 7.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 8.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seugnet L, Boero J, Gottschalk L, Duntley SP, Shaw PJ. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci U S A. 2006;103:19913–19918. doi: 10.1073/pnas.0609463104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 11.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 12.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 13.Dobens LL, Peterson JS, Treisman J, Raftery LA. Drosophila bunched integrates opposing DPP and EGF signals to set the operculum boundary. Development. 2000;127:745–754. doi: 10.1242/dev.127.4.745. [DOI] [PubMed] [Google Scholar]
- 14.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci U S A. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seugnet L, Simpson P, Haenlin M. Transcriptional regulation of Notch and Delta: requirement for neuroblast segregation in Drosophila. Development. 1997;124:2015–2025. doi: 10.1242/dev.124.10.2015. [DOI] [PubMed] [Google Scholar]
- 16.Jacobsen TL, Brennan K, Arias AM, Muskavitch MA. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 1998;125:4531–4540. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- 17.Le Bourg E, Buecher C. Learned suppression of photopositive tendencies in Drosophila melanogaster. Anim Learn Behav. 2002;30:330–341. doi: 10.3758/bf03195958. [DOI] [PubMed] [Google Scholar]
- 18.Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. D1 Receptor Activation in the Mushroom Bodies Rescues Sleep-Loss-Induced Learning Impairments in Drosophila. Curr Biol. 2008;18:1110–1117. doi: 10.1016/j.cub.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seugnet L, Suzuki Y, Stidd R, Shaw PJ. Aversive Phototaxic Suppression: evaluation of a short-term memory assay in Drosophila melanogaster. Genes Brain Behav. 2009 doi: 10.1111/j.1601-183X.2009.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portin P. Allelic negative complementation at the Abruptex locus of Drosophila melanogaster. Genetics. 1975;81:121–133. doi: 10.1093/genetics/81.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitzler P, Simpson P. Altered epidermal growth factor-like sequences provide evidence for a role of Notch as a receptor in cell fate decisions. Development. 1993;117:1113–1123. doi: 10.1242/dev.117.3.1113. [DOI] [PubMed] [Google Scholar]
- 22.Hartley DA, Xu TA, Artavanis-Tsakonas S. The embryonic expression of the Notch locus of Drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein. Embo J. 1987;6:3407–3417. doi: 10.1002/j.1460-2075.1987.tb02664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Lei L, Irvine KD, Baker NE. Notch activity in neural cells triggered by a mutant allele with altered glycosylation. Development. 2003;130:2829–2840. doi: 10.1242/dev.00498. [DOI] [PubMed] [Google Scholar]
- 24.Presente A, Boyles RS, Serway CN, de Belle JS, Andres AJ. Notch is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A. 2004;101:1764–1768. doi: 10.1073/pnas.0308259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stacey SM, Muraro NI, Peco E, Labbe A, Thomas GB, Baines RA, van Meyel DJ. Drosophila glial glutamate transporter Eaat1 is regulated by fringe-mediated notch signaling and is essential for larval locomotion. J Neurosci. 30:14446–14457. doi: 10.1523/JNEUROSCI.1021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehon RG, Johansen K, Rebay I, Artavanis-Tsakonas S. Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J Cell Biol. 1991;113:657–669. doi: 10.1083/jcb.113.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornbrooks C, Bland C, Williams DW, Truman JW, Rand MD. Delta expression in post-mitotic neurons identifies distinct subsets of adult-specific lineages in Drosophila. Dev Neurobiol. 2007;67:23–38. doi: 10.1002/dneu.20308. [DOI] [PubMed] [Google Scholar]
- 28.Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- 29.Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T, Durrenberger M, Paro R, Boutros M, Merdes G. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev Cell. 2010;18:862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 31.Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 32.Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 33.Rival T, Soustelle L, Strambi C, Besson MT, Iche M, Birman S. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 34.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61:213–219. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu MN, Ho K, Crocker A, Yue Z, Koh K, Sehgal A. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci. 2009;29:11029–11037. doi: 10.1523/JNEUROSCI.1653-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuno M, Horiuchi J, Tully T, Saitoe M. The Drosophila cell adhesion molecule klingon is required for long-term memory formation and is regulated by Notch. Proc Natl Acad Sci U S A. 2009;106:310–315. doi: 10.1073/pnas.0807665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Q, Sun K, Shuai Y, Lin R, You W, Wang L, Zhong Y. Suppressor of Hairless is required for long-term memory formation in Drosophila. J Neurogenet. 2009;23:405–411. doi: 10.3109/01677060903096133. [DOI] [PubMed] [Google Scholar]
- 39.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1, related to Figures 1, 2 and 4: sleep data for bunched, Delta and Notch mutant conditions
Supplemental Table S2, related to Figure 2 and Figure 4: Phototaxis Index and Quinine Sensitivity Index
Supplemental Figure S1: sleep homeostasis in bunched mutants.
Supplemental Figure S2, related to Figures 1, 2 and 4: sleep and activity data for bu5ched, Delta and Notch mutant conditions for the baseline day, sleep deprivation day, and recovery days. .
Supplemental Figure S3, related to Figure 3: Learning impairments in flies mutant for the thermo-sensitive allele Nts2. Whole brain view of Delta and Notch immuno-localization.
Supplemental Figure S4, related to Figure 4: expressing UAS-NICD in neurons does not reduce the homeostatic response to sleep loss.