Abstract
The microtubule (MT) “plus end” constitutes the platform for the accumulation of a structurally and functionally diverse group of proteins, collectively called “MT plus-end tracking proteins” (+TIPs). +TIPs control MT dynamics and link MTs to diverse sub-cellular structures. Neurospora crassa MicroTubule Binding protein-3 (MTB-3) is the homolog of yeast EB1, a highly conserved +TIP. To address the function of MTB-3, we examined strains with mtb-3 deletions, and we tagged MTB-3 with GFP to assess its dynamic behavior. MTB-3-GFP was present as comet-like structures distributed more or less homogeneously within the hyphal cytoplasm, and moving mainly towards the apex at speeds up to 4× faster than the normal hyphal elongation rates. MTB-3-GFP comets were present in all developmental stages, but were most abundant in mature hyphae. MTB-3-GFP comets were observed moving in anterograde and retrograde direction along the hypha. Retrograde movement was also observed as originating from the apical dome. The integrity of the microtubular cytoskeleton affects the presence and dynamics of MTB-3-GFP comets, while actin does not seem to play a role. The size of MTB-3-GFP comets is affected by the absence of dynactin and conventional kinesin. We detected no obvious morphological phenotypes in Δmtb-3 mutants but there were fewer MTs in Δmtb-3, MTs were less bundled and less organized. Compared to WT, both MT polymerization and depolymerization rates were significantly decreased in Δmtb-3. In summary, the lack of MTB-3 affects overall growth and morphological phenotypes of N. crassa only slightly, but deletion of mtb-3 has strong effect on MT dynamics.
Introduction
Microtubules (MTs) are part of the cytoskeletal system of all eukaryotic cells and are essential for many vital cellular activities, including maintenance of cell shape, division, migration and intracellular transport [1]–[3]. MTs are “polarized” because they have two distinct ends: a fast-growing, or “plus” end and a slow-growing, or “minus” end. In living cells, the minus-ends are often anchored, whereas the plus-ends are highly dynamic and stochastically switch between phases of growth and shrinkage by addition and loss of α/β tubulin heterodimers, respectively. This process is termed “dynamic instability” and is driven by GTP hydrolysis of the β-tubulin subunits, which is necessary for switching between “catastrophe” (MT depolymerization) and “rescue” (MT polymerization) [4]–[5]. The MT plus-end therefore constitutes a platform for the accumulation of a structurally and functionally diverse group of proteins, collectively called “MT plus-end tracking proteins” (+TIPs). +TIPs control MT dynamics and link MTs to various sub-cellular structures, such as the cell cortex and kinetochores.
“End-binding proteins” (EBs) are core components of +TIP networks due to their autonomous tracking to growing MT ends independently of any known binding partners [6]–[8]. EBs constitute a highly conserved family of proteins, and were initially described as interacting with APC, the adenomatous polyposis coli tumor suppressor protein, which had been implicated in colon cancer [9]. Homologues of EB1 (i.e., EB2, EB3) have been described not only for vertebrates [10]–[12], but also for plants, e.g. AtEB1a in Arabidopsis thaliana [13], insects, e.g. DmEB1 in Drosophila melanogaster [14], and fungi, e.g. BIM1 in Saccharomyces cerevisiae [15], MAL3 in Schizosaccharomyces pombe [16], PEB1 in Ustilago maydis [17] and AN2862.3 in Aspergillus nidulans [18]. EB1 localizes to the mitotic spindle and cytoplasmic MTs, at their distal tips, and regulates MT dynamics promoting anti-catastrophe activity (i.e., MT growth), cell polarization, and chromosome stability [2], [3], [19]. Hence, EB1 has been described as a good marker for the dynamics at the growing tips of MTs because it directly dictates the behavior of the MT plus-ends [20]. EB1 binds autonomously to growing MT ends, where it forms comet-like accumulations and interacts directly with the majority of other known +TIPs, stimulating MTs assembly and consequently their growth [1]–[3], [7], [21]–[25].
Previous studies in N. crassa have shown that cytoplasmic MTs are arranged generally longitudinal along the hyphal tube and that they move as a unit as hyphae are extending, although anterograde and retrograde motility have also been described in small MTs [5], [26]–[27]. We expected that the Neurospora EB1 homologue can play a significant role in orchestrating MT dynamics. Consequently, the main goal of this work was to characterize the localization and behavior of the N. crassa EB1 homologue, MicroTubule Binding protein-3 (MTB-3), during the dynamic instability of MTs, polarized growth and development, as well as the characterization of the MTB-3 deletion mutant.
Materials and Methods
Strains and culture conditions
Strains used in this study are listed in Table 1. Strains were maintained on Vogel's minimal medium (VMM) with 2% sucrose. All manipulations were according to standard techniques [28]. We used a ro-3 mutant with an UV-induced point mutation in p150glued, the largest subunit of dynactin [29]–[30], and a Δkin-1 deletion mutant that lacks conventional kinesin (“nkin”; [31]).
Table 1. Strains used in this study.
| Organism | Genotype | Reference |
| NC4200 | mat a | FGSC |
| NMF39 | mat A | FGSC2489 |
| N2928 | mat A his-3; Δmus-51::bar + | FGSC9717 |
| FGSC43 | mat a; ro-3 | FGSC43 |
| N2281-3A | mat A his-3 + ::Pccg-1-hH1 + -sgfp + | Freitag et al., 2004 |
| N2524 | ridRIP1 mat a his-3 + ::Pccg-1-Bml + -sgfp + | Freitag et al., 2004 |
| NMF138 | mat A his-3 + ::Pccg-1-hH1 + -tdimerRed + | Freitag et al., 2004 |
| FGSC11851 | mat a; Δmtb-3 | Colot et al., 2006 |
| FGSC11852 | mat A; Δmtb-3 | Colot et al., 2006 |
| XRM-10-OC | mat a his-3; Δkin-1 | This study |
| XTL2-1 | mat A his-3 + ::Pccg-1-mtb-3 + -sgfp + | This study |
| XTL2-2 | mat a his-3 + ::Pccg-1-mtb-3 + -sgfp + | This study |
| XRM-01-AR | his-3 + ::Pccg-1-Bml + -sgfp + ;Δmtb3 | This study |
| XRM-02-AR | his-3 + ::Pccg-1-hH1 + -sgfp + ; Δmtb3 | This study |
| XRM-03-AR | his-3 + ::Pccg-1- mtb3 + -sgfp + ; ro3 | This study |
| TRM-01-AR | Δkin-1; MTB-3 + ::sgfp + | This study |
| TRM-02-AR | mat A his-3 + ::Pccg-1-mtb3 + -mChfp | This study |
Construction of fps-containing plasmids
Standard PCR and cloning procedures were used to fuse the sgfp (green fluorescent protein; GFP) or mchfp (mCherry fluorescent protein; mChFP) gene to the carboxyl terminus of mtb-3. The mtb-3 gene (1,019 nt) was amplified by PCR from N. crassa genomic DNA isolated from a wild type (WT) strain (NMF39; FGSC2489) with primers that included XbaI and BamHI or PacI restriction endonuclease sites at their respective 5′ termini, the reverse primer eliminated the stop codon of mtb-3 to generate a translational fusion to sgfp or mChfp (OMF1, 5′-CCGTCTAGAATGGGTGAATCACGGTGTGTA-3′ and OMF2, 5′- GGCGGATCCTAAAGGTCTCCTGATCGTCGGC-3′) for pTL1; and for pRM43-AR01. PCR products were digested with XbaI and BamHI and cloned into XbaI- and BamHI-digested plasmid pMF272 (GFP, GenBank accession no. AY598428; [26]), or digested with XbaI and PacI and cloned into XbaI- and PacI-digested pJV15-2 (mChFP; [32]). This yielded pTL1 and pRM43-AR01, respectively. Inserts were verified by sequencing.
Transformation protocols, transformant selection and crosses
Transformation of N. crassa strain N2928 (FGSC9717) conidia with linearized plasmids (Table 1) was carried out by electroporation as previously described [33]–[34]. Prototrophic His+ transformants were screened for the expression of GFP or mChFP by epifluorescence microscopy as described previously [26]. Transformants showing fluorescence were selected. Heterokaryotic transformants were backcrossed to mat a strains to isolate homokaryotic progeny, following standard protocols [28]. For each cross, recipient strains were grown on medium for 5 days at 25°C and fertilized by adding conidia from the donor strain. After ∼14 days of incubation at 25°C, ascospores from the developed perithecia were collected from Petri dish covers. Ascospores were spread on VMM, heat-shocked at 60°C for 60 min and incubated overnight at room temperature or 32°C. Colonies were transferred to slants with VMM and incubated at 28°C. Strains were screened under an epifluorescence microscope to select progeny that expressed GFP or mChFP.
Complementation of Δmtb-3 by MTB-3-GFP
Strains carrying deletion alleles of mtb-3 (FGSC11851 and FGSC11852) were obtained from the Neurospora Functional Genomics project from the Fungal Genetics Stock Center (FGSC, Kansas City, MO). They were crossed to XTL2-1 and XTL2-2 to generate homokaryotic strains that carried MTB-3-GFP in the mtb-3 deletion background, resulting in XRM-AR-01and XRM-AR-02, respectively (Table 1).
Actin and MT depolymerization assays
Stock solutions of the anti-actin drug cytochalasin A (CytA; CAT# 14110-64-6, Sigma–Aldrich, St. Louis, Mo), and the anti-microtubule drug benomyl (Ben; CAT# PS222, Sigma–Aldrich) at 10 mg ml−1 were prepared in 100% ethanol. For further studies the concentration that inhibited the hyphal growth rate by 50% was selected based on our previous work [35]. To study the effect of CytA on the distribution of MTB-3 in N. crassa, we exposed the cells to 1 µg ml−1 of CytA in VMM plates and incubated at 28°C until the cells reached a young mycelium stage (∼16 h). For Ben experiments, we inoculated VMM plates amended with 2.5 µg ml−1 and incubated at 28°C. For both treatments, mycelia were observed following the procedure described for laser scanning confocal microscopy.
Laser scanning confocal microscopy of living cells
To record the behavior of all the strains under study, we used the “inverted agar block” method [36] for live-cell imaging with an inverted laser scanning microscope (LSM-510 Meta, Carl Zeiss, Göttingen, Germany) equipped with an argon ion laser for excitation at 488 nm wavelength and GFP filters for emission at 515–530 nm, and a He-Ne laser for excitation at 543 nm and mChFP filters for emission at 580–700 nm. An oil immersion objective was used: 100× (PH3)/1.3 N.A. planneofluar. Laser intensity was kept to a minimum (1.5%) to reduce photobleaching and phototoxic effects. Time-lapse imaging was performed at scan intervals of 0.5 to 4.5 s for periods up to 40 min. Image resolution was 512×512 pixels and 300 dpi. Confocal images were captured using LSM-510 software (version 3.2; Carl Zeiss) and evaluated with an LSM 510 Image Examiner. Some of the image series were converted into AVI movies using the same software. Time-lapse images of hyphae were recorded simultaneously by phase contrast microscopy and fluorescent confocal microscopy. Phase contrast images were captured with a photomultiplier for transmitted light using the same laser illumination for fluorescence [27]. Final images were processed, and figures were created using Adobe Photoshop CS3 Extended (Adobe Systems Inc, San Jose, CA). For fluorescence recovery after photobleaching (FRAP), a selected rectangular area of a hypha was overexposed at 20% intensity for 30 s (argon ion laser 488 nm wavelength). After photobleaching, images were scanned at intervals of 3.5 to 4.5 s for up to 3 min [27].
Total Internal Reflection Fluorescence Microscopy (TIRFM) of living cells
For TIRFM, an IX-70 inverted microscope equipped with a 60×/1.45 N. A. Apochromat objective lens (Olympus Corp., Melville, NY) and a krypton/argon laser (Melles Griot, Carlsbad, CA) (488 nm) was used [37]. Images were recorded with a Cascade 512B EMCCD camera (Photometrics, Tucson, AZ) for durations of 2–3 min at 512×512 resolution and frame rates of 50–100 msec. MetaMorph 6.0/6.1 software (Universal Imaging, Downingtown, PA) was used to control the camera and capture images. Final images were processed, and figures were created using Adobe Photoshop CS3 Extended (Adobe Systems Inc, San Jose, CA).
Membrane staining with FM4-64
Using the “inverted agar block method” of preparing and staining samples [36], hyphae of GFP-expressing strains were double-labeled with 2.5 µM FM4-64 (Invitrogen, Eugene, OR), which stains the plasma membrane and organelle membranes [26], [38].
Results
Localization and dynamics of MTB-3-GFP in N. crassa
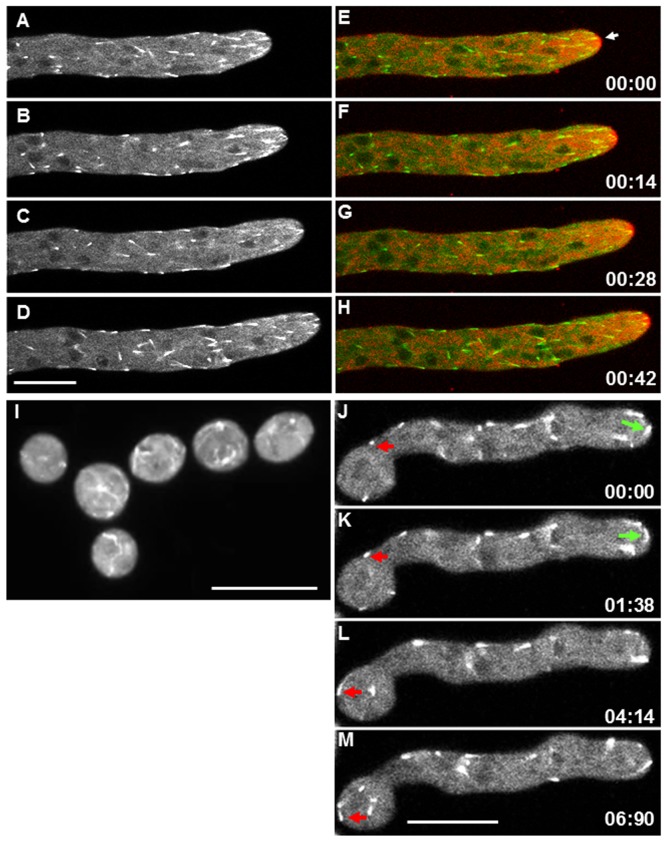
MTB-3-GFP was present as comet-like structures that appeared evenly distributed along the hypha (Fig. 1A–H; Movie S1). The comets had a length of 1.44±0.03 µm (mean ± standard error; n = 150). MTB-3-GFP comets moved mainly towards the apex with a speed of 0.64±0.02 µm s−1, about fourfold faster than the hyphal elongation rate (0.15±0.01 µm s−1) (p<0.05). We also observed instances of comets moving in retrograde direction (Movie S1). In distal regions, comets of MTB-3-GFP moved in both anterograde and retrograde directions along the hyphae and accumulated in regions around the nuclei. MTB-3-GFP comets were present in all developmental stages, but were less prevalent in spores (Fig. 1I) and germlings (Fig. 1J–M) than mature hyphae. MTB-3-GFP comets were seen moving in anterograde and retrograde direction along germ tubes and the germinal spore (Fig. 1J–M).
Figure 1. Distribution of MTB-3-GFP comets in different developmental stages of N. crassa.
(A–D) Time lapse images of MTB-3-GFP in mature hyphae. (E–H) merged images of MTB-3-GFP and membranes stained with 5 µM FM4-64 in mature hypha. The Spitzenkörper is indicated by a white arrow. (I) Conidia expressing MTB-3-GFP. (M–P) Time lapse images of MTB-3-GFP in germlings. Red arrows point to MTB-3-GFP comets that move in retrograde direction and green arrows point to comets that move in anterograde direction. Time scale is in min∶sec. Scale bars = 10 µm.
FRAP experiments, corroborated the faster speed of MTB-3-GFP comets relative to the cytoplasmic bulk flow through the growth axis of hyphae. The comets moved faster than the cytoplasm or the elongating tip, thus it was not possible to observe defined bleached areas; after application of the laser, the bleached area was immediately re-populated by fluorescent comets.
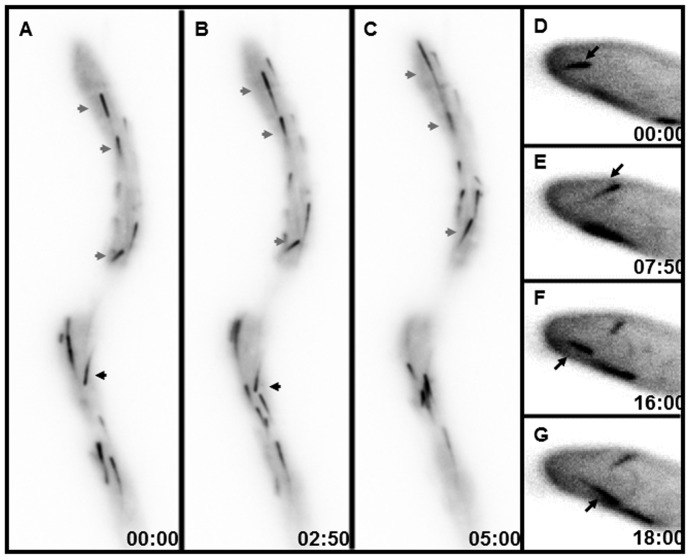
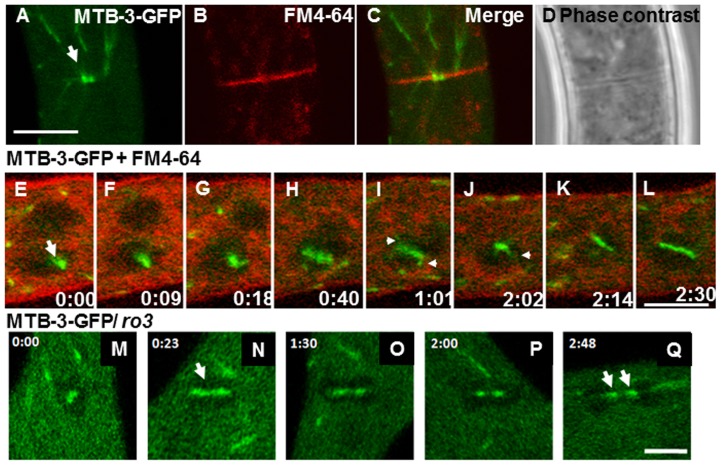
Additionally, both anterograde and retrograde motility of MTB-3 comets were detected in apical and subapical regions of the hyphal cortex when TIRFM was used (Fig. 2; Movie S2). With TIRFM, we were able to observe instances of comets moving from apical to basal regions of the hypha, suggesting the plus-end of MTs were growing in retrograde direction (Fig. 2D–2G). In other regions, MTB-3-GFP accumulations were observed in the septal pore where the MTs are trapped during septation (Fig. 3 A–D). The accumulation of MTB-3-GFP in the mature septa lasted throughout the whole observation period. MTB-3-GFP was also localized along the mitotic spindle during nuclear division. Although it is not possible to determine whether the fluorescence is associated to certain part of the spindle, it seems that MTB-3-GFP is decorating the whole spindle and also part of what seems to be astral MTs (Fig. 3 E–L). The dynamics of MTB-3-GFP in the mitotic spindle was not affected in the dynactin p150Glued subunit, the fluorescence of MTB-3-GFP was present in all mitotic stages and also after karyokinesis (Fig. 3M–3Q).
Figure 2. TIRFM of MTB-3-GFP in the cortex of mature hyphae.
(A–C) Apical and subapical regions. (D–G) Enlargement of apical regions. Grey arrows point to MTB-3-GFP comets moving in anterograde direction, and black arrows point to comets that move in retrograde direction. Time scale is in sec∶msec. Scale bars = 10 µm.
Figure 3. Localization of MTB-3-GFP in different hyphal regions of N. crassa.
(A–D) MTB-3-GFP remains close to the septal pore. (A). Septum of a mature hypha expressing MTB-3-GFP. (B) Membranes stained FM4-64 (C) Merged image of (A), (B) and (D) Phase contrast image. Arrow points to MTB-3-GFP accumulation in the completely formed septal pore when it is already formed. Scale bar = 10 µm. (E–L) Confocal time lapse images of a nucleus during mitosis in mature hypha of a strain expressing MTB-3-GFP, merged with images of membranes stained with FM4-64. The arrow points to accumulation of MTB-3-GFP on the spindle of a nucleus in mitosis. Arrowheads indicate the presence of MTB-3-GFP in what seem to be astral MTs. Time scale is in min∶sec. Scale bar = 5 µm. (M–Q) Confocal time lapse images of a nucleus during mitosis in a strain expressing MTB-3-GFP in the ro-3 mutant. Arrows show MTB-3-GFP in the spindle, even when karyokinesis has been completed. Time scale is in min∶sec. Scale bar = 5 µm.
MTB-3 during depolymerization of MTs and actin microfilaments
When MT cytoskeleton was depolymerized with 2.5 µg ml−1 Ben [35], all MTB-3-GFP comets disappeared, resulting in a punctuated and dispersal distribution of the fluorescence throughout the hypha (Fig. 4B). Only a few disorganized fluorescent accumulations were present along the hypha and some of them were concentrated around nuclei (Fig. 4B, arrowhead). As MTB-3 has a calponin domain, a motif that can regulate interactions of proteins with actin filaments, we also performed an experiment with the anti-actin drug CytA (1 µg ml−1) to observe potential effects of actin filament depolymerization on localization and integrity of MTB-3-GFP comets. We used a concentration of CytA that ensures disruption of the actin cytoskeleton without compromising cell survival [39]. Fluorescent comets were seen in treated hyphae, but they were fewer and smaller than WT strain (Fig. 4C).
Figure 4. Effect of drugs anti MTs and actin and mutation of motor proteins in MTB-3-GFP comets.
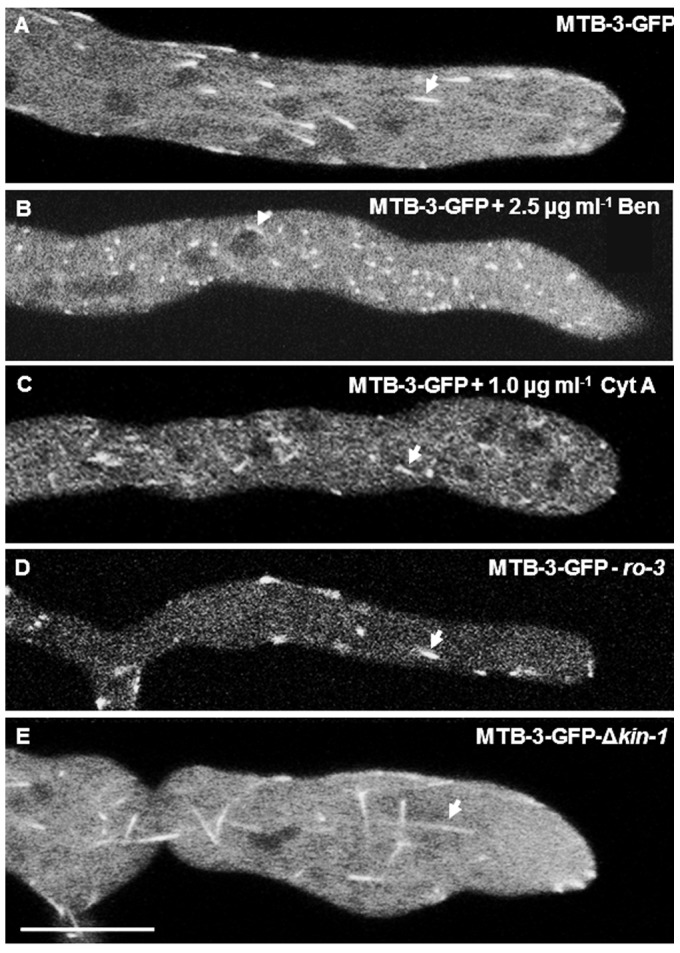
Effect of benomyl (Ben), cytochalasin A (CytA) or mutation of motor proteins (ro-3 and Δkin-1) on localization and dynamics of MTB-3-GFP comets. (A) MTB-3-GFP in the WT strain in untreated mature hypha, (B) MTB-3-GFP in mature hypha treated with 2.5 µg ml−1 of Ben to depolymerize MTs. (C) MTB-3-GFP in mature hypha treated with 1.0 µg ml−1 Cyt A, to disturb the actin cytoskeleton. (D) MTB-3-GFP in the dynactin mutant (p150glued subunit, ro-3) and (E) MTB-3-GFP in the conventional kinesin deletion mutant (Δkin-1). Arrows point to the MTB-3-GFP comets in each treatment and mutant. Scale bar = 10 µm.
The lack of MT associated motor proteins and MTB-3 dynamics
Irregular sizes of MTB-3-GFP comets were distributed evenly along the hypha of dynactin (ro-3) and conventional kinesin (Δkin-1) mutants (Fig. 5D–E). Slightly longer MTB-3-GFP comets were observed in ro-3 (1.83±0.05 µm; n = 100) relative to WT strains (Table 2) (p<0.05). Also, the abundance of fluorescent comets were reduced and motility was slower (0.42±0.02 μ s−1) than in WT (p<0.05) (Fig. 5D, Movie S3), resulting in altered shapes (blotchy instead of needlelike). In Δkin-1 mutant, MTB-3-GFP comets were two-times longer (3.42±0.10 µm; n = 100) than in WT (p<0.05), but they moved at similar speeds (0.68±0.03 µm s−1) to MTB-3-GFP comets as in WT (p>0.05) (Fig. 5E; Table 2; Movie S4).
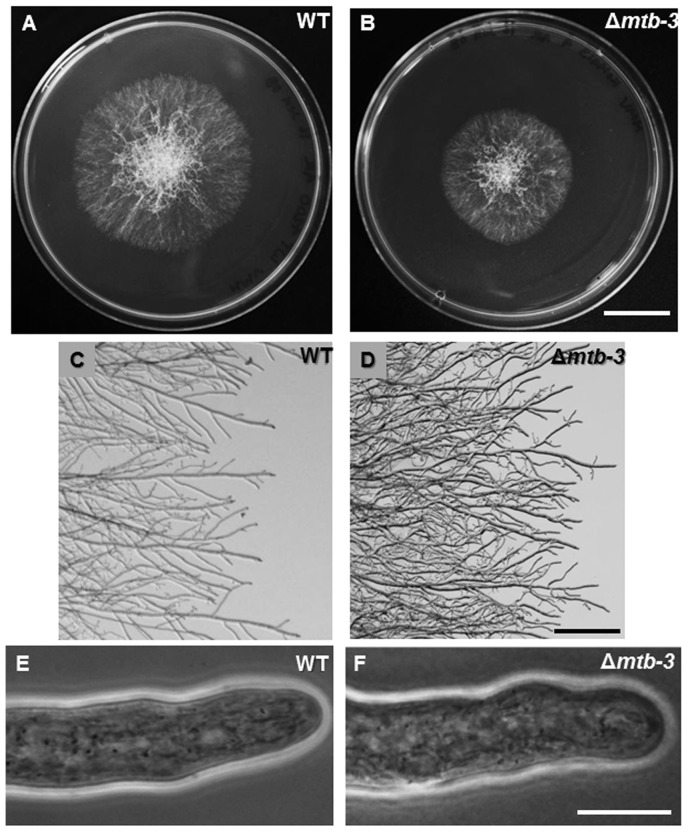
Figure 5. Phenotype of the Δmtb-3 mutant.
Macroscopic growth phenotype of colonies of (A) WT and (B) Δmtb-3, showing slightly reduced linear growth of the mutant. Stereomicroscopic image of (C) WT and (D) Δmtb-3, showing more compact colony margins of the mutant. Hyphal phenotype of (E) WT and (F) Δmtb-3, showing slightly more rounded appearance of the apex in the mutant. Scale bar (A–B) = 2 cm, (C–D) = 100 µm and (E–D) = 10 µm.
Table 2. Average (± standard error) of length and speed of MTB-3 for WT, Δkin-1 and ro-3 strains.
| WT | Δkin-1 | ro-3 | |
| Length (µm) | 1.44±0.03 | 3.42*±0.10 | 1.83*±0.05 |
| n = | 150 | 100 | 100 |
| Speed (μ s−1) | 0.64±0.02 | 0.68±0.03 | 0.42*±0.02 |
| n = | 50 | 50 | 50 |
The number of observations is indicated by n.
p<0.05.
Characterization of mtb-3 deletion strains
We found no obvious, morphological phenotypes associated with deletion of the mtb-3 gene. We found, however, slightly reduced linear growth rates and slightly increased conidiation rates compared to WT (Fig. 6, Table 3). The linear elongation rate in Δmtb-3 mutant was 34.1±1.1 µm min−1 compared to 41.9±0.4 µm min−1 in WT (p>0.05). Images obtained by stereomicroscopy showed somewhat denser mycelium in Δmtb-3 mutant. However, no differences in branching rate or biomass accumulation were observed (Fig. 6 C–D, Table 3).
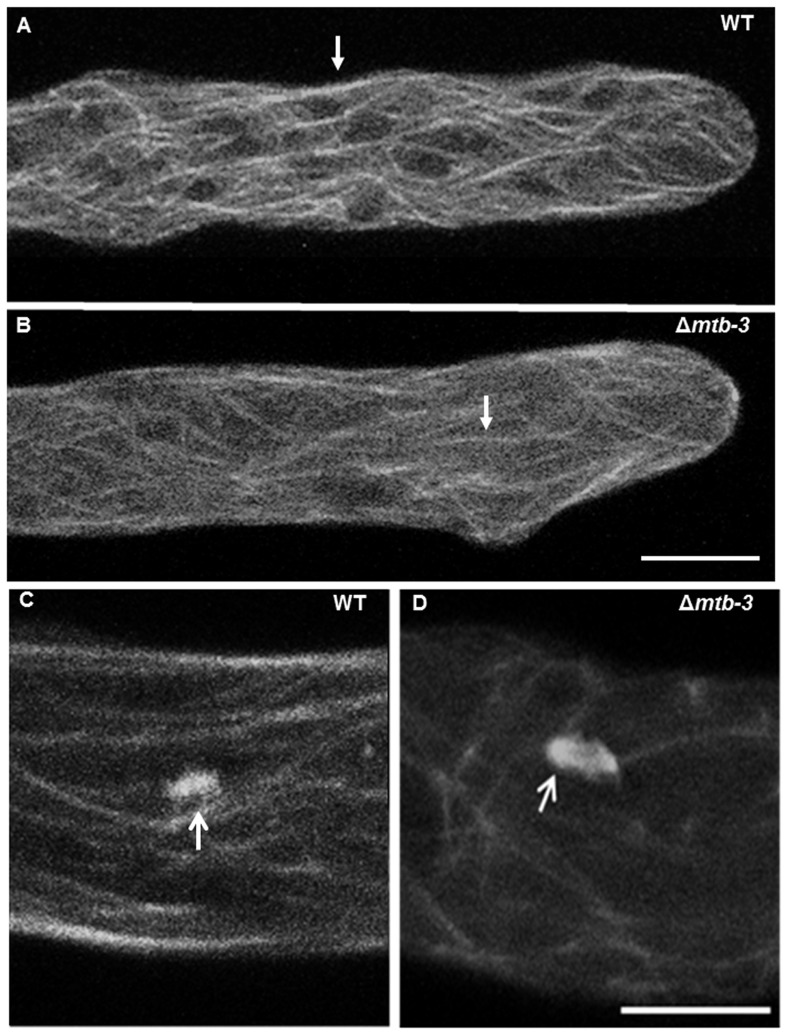
Figure 6. MTs in the Δmtb-3 mutant.
MT distribution, measured by imaging of β-tubulin-GFP, and dynamics in mature hyphae of (A) WT and (B) Δmtb-3 (also see Movies S5 and S6). The white arrow indicates a thicker and brighter arrangement of MTs, while the red arrow points to thinner appearance of MTs in Δmtb-3. Mitotic spindle in (C) WT and (D) Δmtb-3. It was easily observed how MTs are not compacted in mitotic spindles in Δmtb-3 (white arrow) in (D). Scale bar = 5 µm.
Table 3. Average (± standard error) of elongation, branching, conidiation and biomass rates for WT and Δmtb-3.
| WT | Δmtb-3 | |
| Elongation rate (µm s−1) | 41.9±0.4 | 34.1±1.1 |
| Branching in 500 lineal-µm | 2.0±0.2 | 2.0±0.2 |
| Total number of hyphae measured | 45 | 45 |
| Total number of branches | 111 | 84 |
| Total number of bilateral branches | 20 | 14 |
| % Bilateral branching | 18 | 17 |
| Conidiation rate (conidia ×107 per ml) | 86.7±2.5 | 117.2±5.6 |
| Biomass (mg of dry weight per day) | 193.2±2.7 | 192.9±5.8 |
Microtubules dynamics in Δmtb-3 mutant
MTs tagged with GFP (β-tubulin-GFP) were observed in Δmtb-3 by confocal microscopy compared with WT strain (Fig. 7). MTs in Δmtb-3 appeared thinner and less organized in bundles than in WT (Fig. 7B). MTs close to the apex were slightly less organized in Δmtb-3 mutant; fewer MTs reached the apical dome relative to WT (Fig. 7; Movie S5). Additionally, spindle MTs were less compact during nuclear division in Δmtb-3 compared to WT strain (Fig. 7 C+D). Dynamic instability of MTs appeared to be modified in Δmtb-3 mutant, polymerization (21.91±15.72 µm s−1) and depolymerization (38.90±26.98 µm s−1) rates in Δmtb-3 mutant were reduced by 21% and 28%, respectively, relative to WT (Fig. 7, Table 4).
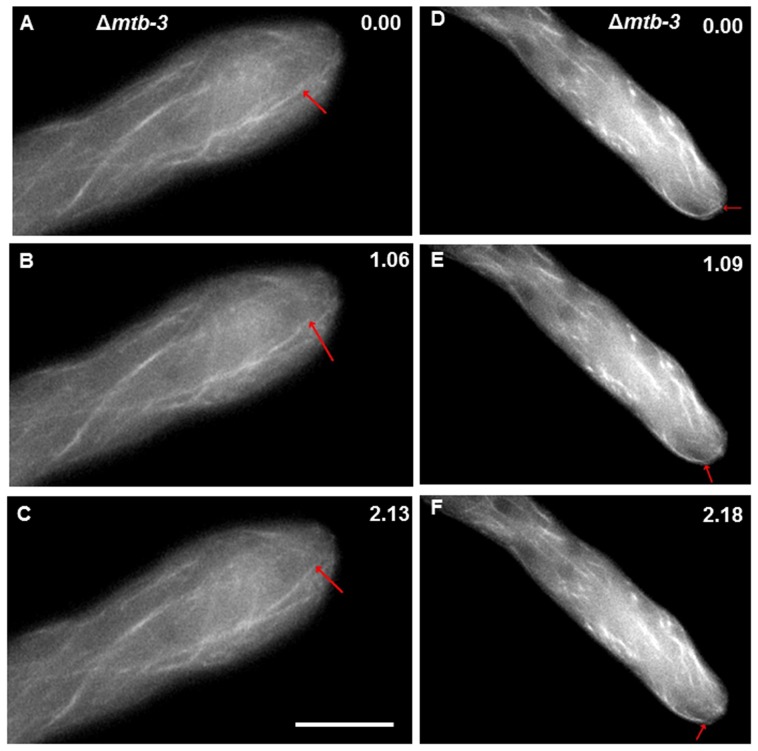
Figure 7. Dynamic instability of MTs in the Δmtb-3.
Instances of (A–C) a MT polymerizing and (D–F) depolymerizing in mature hyphae of the Δmtb-3. The MT plus ends are indicated with red arrows. Time scale in min∶sec. Scale bar = 10 µm.
Table 4. Average (± standard error) rates of dynamic instability of apical microtubules in mature hyphae of WT and Δmtb-3.
| WT | Δmtb-3 | |
| Polymerization rate | 27.96±6.34 | 21.91*±2.49 |
| (µm min−1) | n = 36 | n = 40 |
| Depolymerization rate | 58.24±6.42 | 38.90*±4.93 |
| (µm min−1) | n = 34 | n = 30 |
The number of observations is indicated by n.
p<0.05.
Discussion
MTB-3-GFP in N. crassa
A large variety of +TIPs are localized to the MTs plus-end. These proteins have different domains, structures and functional properties. EB1 homologues are core components of +TIP networks because of their autonomous tracking to growing MT plus ends, which is apparently independent of any known binding partners [7]–[8]. MTB-3 is the single EB1 homologue found in the N. crassa genome. MTB-3 tagged with GFP was localized at the plus-end of cytoplasmic and mitotic microtubules as comet-like structures. We also observed MTB-3-GFP in septal rings, where MTs are trapped during septation [26]. MTB-3-GFP comets were also observed in germlings and conidia, but in lower abundance than in mature hyphae.
The fluorescent comets of MTB-3 have been visualized moving not only toward apical but also basal regions, similar to the anterograde and retrograde motility of MTs and nuclei observed previously in N. crassa [5], [27]. Observing MT dynamics by autonomous tracking of MTB-3 with two independent microscopic techniques (confocal and TIRF), suggests that the MT plus-ends are growing in both directions along mature hyphae and germlings with speeds about four-times higher than the typical hyphal elongation rate. The direct binding of MTB-3 to the plus-end of polymerizing MTs was also corroborated, when only fluorescent yet stationary spots labeled by MTB-3-GFP were observed after exposure to benomyl at concentrations that depolymerized the microtubular cytoskeleton and altered hyphal shape and growth [35].
Conversely, even though we found homology to a calponin domain in Neurospora MTB-3, no evidence of strong disturbance in the MTB-3 dynamics along hyphae was found after cytochalasin A exposure. This was unexpected as calponin domains are often found in actin-binding proteins [40]. Only a slight reduction in the number and size of MTB-3 comets was observed, likely as an indirect consequence of the actin depolymerization that affected MTs at the apical and subapical regions [35]. Our observations of MTs plus-ends growing in both anterograde- and retrograde directions including the apical region, suggested an antiparallel growth of MTs from different nucleation sites distributed within the cytoplasm and likely very close to the apex of N. crassa [27], [41]. This behavior has been previously seen in interphase MTs of U. maydis; however, in those studies there was no indication for mixed polarities of MTs in the growing region [17], [42].
In spite of the relationship between motor proteins and the dynamic instability of MTs in N. crassa [5], we found no evidence for direct interactions of MTB-3 with dynactin and conventional kinesin, suggesting an autonomous behavior of this +TIP in this filamentous fungus. This may be accomplished by binding of MTB-3 to the MT plus-end through recognition of specific plus-end structure, thus stimulating MT assembly (polymerization) and consequently aiding MT growth [2]–[3]. In fact, the model of the interaction of MTB-3 with a certain nucleotide state of β-tubulin (GTP) [43] helps to explain that a lack of conventional kinesin (Δkin-1) in N. crassa, which likely caused an increase of GTP-β-tubulin in the microtubular structure, generated MTB-3 comets twice as long as those in WT. However, the displacement dynamic of the MTB-3 comets seemed unaltered by the lack of this motor protein, as shown by the similar speeds of comets in Δkin-1 and WT strains. This suggests that MTB-3 can reach MT tips independently. The lack of dynactin in ro-3 mutant only slightly increased the length of MTB-3 comet while causing appreciable changes in comet shape. We also observed a considerable decrease in the abundance and motility of MTB-3 comets along the mature hyphae in ro-3, caused by a reduction in the MT abundance and dynamic instability rates as were previously observed (reduction of 50% and 53% of polymerization and depolymerization rates; [5]). A decrease in the polymerization rate in ro-3 may cause distortions in “treadmilling”, which will affect directly how much MTB-3 is accumulated on MT tips and how rapidly it is exchanged with the cytoplasmic pool. This can cause an illusion of comets moving along the hypha, as suggested elsewhere [3]. Thus, it is possible that slower treadmilling was observed in ro-3 due to a reduced association-dissociation rate of MTB-3 with the binding sites at the growing MT, either by a decrease in the structural features at the MTs plus-end or by a low recognition signal of those features.
Function of MTB-3 in N. crassa
Previous work has suggested that EB1-like proteins specifically decorate freshly polymerized MT plus ends and that this fulfills a conserved role in promoting MT polymerization by increasing MT rescue frequencies and decreasing MT catastrophe or pausing [2], [10], [12], [15]–[16], [19]. Here, we observed a decrease in polymerization and depolymerization rates, an alteration in the form and size of MTs, thus resulting in thinner or less associated MT bundles, and reduced hyphal elongation rates in Δmtb-3 strains. Our results suggest an important role of MTB-3 in maintaining integrity of the microtubular cytoskeleton, and therefore in normal growth and development of N. crassa. Variations in MT morphology were also observed in Mal3 mutants of the fission yeast, S. pombe. Lack of mal3 resulted in short and often weak cytoplasmic MTs [16]. Maintenance of MT structural integrity by this class of +TIP was also observed in the budding yeast Bim1 homologue of the human pathogen Cryptococcus neoformans [44]. Abnormally short and relatively unstructured MTs were observed in bim1 mutants in filaments during sexual development and during diploid filamentation, consequently changing its normal filamentous growth [44]. Similarly, MT dynamics, specifically during G1 phase of cell division, has been shown on studies of S. cerevisiae Bim1 [15].
We also show a role for MTB-3 in keeping the structural integrity of the MT cytoskeleton in mitotic MTs of N. crassa. Lack of MTB-3 caused less compact mitotic spindles during nuclear division, probably by a reduction of structural stability of interpolar MTs as part of mitotic spindles. Indeed in previous work, deletion of S. cerevisiae bim1 showed shortening of the MTs that generated a reduction in the length of the anaphase antiparallel overlap zone, which is important for the efficient antiparallel cross-linking of motor proteins that maintain the forces required to stabilize the elongating anaphase spindle [45]. Additionally, MTs at the start of mitosis switch from polymerization to depolymerization at a rate that is 20 times faster than that of interphase MTs [46]. Therefore, the reduction in the dynamic instability rates by >20% that we found when Neurospora MTB-3 was lacking likely altered the initial assembling of the mitotic spindle, generating a more relaxed structure in Δmtb-3 than in WT.
The distal segment of MTs has been described as a “plus-end raft” that allows a cascade of proteins interactions that control the dynamics and function of the microtubular cytoskeleton [47]. Particularly, the interaction between EB1 with other proteins to stabilize the MTs during their growth has been reported. For example, interactions between EB1 and dynactin suggest that they function as a plus-end complex. While the p150Glued subunit of dynactin has potent activity for MT nucleation, EB1 has the ability to elongate MTs, thus the two proteins together would have large effects on overall MT dynamics [48]. In our work, deletion of the N. cassa mtb-3 gene, likely caused a partial loss of this plus-end complex functionality and altered the balance between individual effects of MTB-3 and the network of all other +TIPs on overall microtubule dynamics, suggested by the reduced instability dynamics and hyphae elongation rates observed. Consequently, our findings highlight the importance of MTB-3 not only in maintaining the integrity of the microtubular cytoskeleton structure but also, to interact with other +TIPs at the MTs plus-end, in order to guarantee optimal polarized growth and development of the filamentous fungus N. crassa.
Supporting Information
MTB-3-GFP comets dynamics in mature hypha of N. crassa . Comets are distributed homogeneously along the hypha and moved in anterograde and retrograde direction.
(AVI)
TIRFM of MTB-3-GFP cortical comets dynamics in mature hypha of N. crassa .
(AVI)
Dynamics of MTB-3-GFP in the UV mutant of the p150glued subunit of dynactin ( ro-3 ).
(MPG)
Dynamics of MTB-3-GFP in the deletion mutant of kin-1 gene (Δ kin-1 ).
(MPG)
Microtubules dynamics (β-tubulin-GFP) in the mtb-3 gene deletion mutant.
(MPG)
Microtubules dynamics (β-tubulin-GFP) in the WT.
(MPG)
Acknowledgments
We thank the Neurospora Functional Genomics project (NIH P01GM068087) and the Fungal Genetics Stock Center (FGSC, Kansas City, MO) for strains. We gratefully acknowledge Douglas Daniels for the training on and use of the TIRFM at ASU.
Funding Statement
This work was supported by grants from Consejo Nacional de Ciencia y Tecnología Ciencia Básica (133518) to RRMP and the American Cancer Society (RSG-08-030-01-CCG) to MF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Akhmanova A, Hoogenraad CC (2005) Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol 17: 47–54. [DOI] [PubMed] [Google Scholar]
- 2. Akhmanova A, Hoogenraad MO (2010) Microtubule +TIPs at a glance. J Cell Sci 123: 3415–3419. [DOI] [PubMed] [Google Scholar]
- 3. Akhmanova A, Steinmetz MO (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9: 309–322. [DOI] [PubMed] [Google Scholar]
- 4. Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13: 83–117. [DOI] [PubMed] [Google Scholar]
- 5. Uchida M, Mouriño-Pérez RR, Freitag M, Bartnicki-Garcia S, Roberson RW (2008) Microtubule dynamics and the role of molecular motors in Neurospora crassa. Fungal Genet Bio 45: 683–692. [DOI] [PubMed] [Google Scholar]
- 6. Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, et al. (2007) Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450: 1100–1105. [DOI] [PubMed] [Google Scholar]
- 7. Bieling P, Kandels-Lewis S, Telley IA, Van Dijk J, Janke C, et al. (2008) CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol 183: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, et al. (2009) Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci USA 106: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su LK, Burrell M, Hill DE, Gyuris J, Brent R, et al. (1995) APC binds to the novel protein EB1. Cancer Res 55: 2972–2977. [PubMed] [Google Scholar]
- 10. Morrison EE, Wardleworth BN, Askham JM, Markham AF, Meredith DM (1998) EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene 17: 3471–3477. [DOI] [PubMed] [Google Scholar]
- 11. Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, et al. (2003) Visualization of Microtubule Growth in Cultured Neurons via the Use of EB3-GFP (End-Binding Protein 3-Green Fluorescent Protein). J Neurosc 23: 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, et al. (2005) EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell 16: 5334–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan J, Calder G, Fox S, Lloyd C (2005) Localization of the Microtubule End Binding Protein EB1 Reveals Alternative Pathways of Spindle Development in Arabidopsis Suspension. Cells Plant Cell 17: 1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rogers SL, Rogers GC, Sharp DJ, Vale RD (2002) Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol 158: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tirnauer JS, O'Toole E, Berrueta L, Bierer B, Pellman D (1999) Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol 145: 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beinhauer JD, Hagan IM, Hegemann JH, Fleig U (1997) Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol 139: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Straube A, Brill M, Oakley BR, Horio T, Steinberg G (2003) Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol Biol Cell 14: 642–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fischer R, Zekert N, Takeshita N (2008) Polarized growth in fungi–interplay between the cytoskeleton, positional markers and membrane domains. Mol Biol 68 4: 813–826. [DOI] [PubMed] [Google Scholar]
- 19. Tirnauer JS, Bierer BE (2000) EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol 149: 761–766.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vaughan KT (2005) TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol 171 2: 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakamura N, Zhou XZ, Lu KP (2001) Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr Biol 11 13: 1062–1067. [DOI] [PubMed] [Google Scholar]
- 22. Hayashi I, Wilde A, Mal TK, Ikura M (2005) Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell 19: 449–460. [DOI] [PubMed] [Google Scholar]
- 23. Lansbergen G, Akhmanova A (2006) Microtubule plus end: a hub of cellular activities. Traffic 7: 499–507. [DOI] [PubMed] [Google Scholar]
- 24. Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, et al. (2009) An EB1-binding motif acts as a microtubule tip localization signal. Cell 138: 366–376. [DOI] [PubMed] [Google Scholar]
- 25. Komarova Y, De Groot CO, Grigoriev I, Montenegro S, Munteanu EL, et al. (2009) Mammalian end binding proteins control persistent microtubule growth. J Cell Biol 184: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND (2004) GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa . Fungal Genet Biol 41: 897–910. [DOI] [PubMed] [Google Scholar]
- 27. Mouriño-Pérez RR, Roberson RW, Bartnicki-García S (2006) Microtubule dynamics and organization during hyphal branching in Neurospora crassa . Fungal Genet Biol 43: 389–400. [DOI] [PubMed] [Google Scholar]
- 28.Davis R (2000) Neurospora: contributions of a model organism. Oxford University Press.
- 29. Courtright JB (1975) Growth of ropy mutants on glycerol and acetate. Neurospora News Let 22: 5. [Google Scholar]
- 30. Tinsley JH, Minke PF, Bruno KS, Plamann M (1996) p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol Biol Cell 7 5: 731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seiler S, Nargang FE, Steinberg G, Schliwa M (1997) Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa . EMBO J 16: 3025–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdín J, Bartnicki-García S, Riquelme M (2008) Neurospora crassa GS-1, a protein involved in beta-1,3-glucan synthesis, accumulates in the outer region of the Spitzenkörper, in: 9th European Conference on Fungal Genetics ECFG9. Edinburgh, United Kingdom.
- 33. Margolin BS, Freitag M, Selker EU (1997) Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newslett 44 34–36(and correction in FGN 47: 112).0. [Google Scholar]
- 34. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramos-García SL, Roberson RW, Freitag M, Bartnicki-Garcia S, Mouriño-Pérez RR (2009) Cytoplasmic bulk flow propels nuclei in mature hyphae of Neurospora crassa . Eukaryot Cell 8: 1880–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hickey PC, Jacobson D, Read ND, Glass NL (2002) Live-cell imaging of vegetative hyphal fusion in Neurospora crassa . Fungal Genet Bio 37: 109–119. [DOI] [PubMed] [Google Scholar]
- 37. Uchida M, Mouriño-Pérez RR, Roberson RW (2011) Total internal recflection microscopy of fungi. Fungal Biol Rev 24 3–4: 132–136. [Google Scholar]
- 38. Fischer-Parton S, Parton RM, Hickey PC, Dijksterhuis J, Atkinso HA (2000) Confocal microscopy of FM4-64 as a tool for analyzing endocytosis and vesicle trafficking in living fungal hyphae. J Microscopy 198: 246–259. [DOI] [PubMed] [Google Scholar]
- 39. Delgado-Alvarez DL, Callejas-Negrete OA, Gómez N, Freitag M, Roberson RW, et al. (2010) Visualization of F-actin localization and dynamics with live cell markers in Neurospora crassa . Fungal Genet Biol 47: 573–586. [DOI] [PubMed] [Google Scholar]
- 40. Hayashi I, Ikura M (2003) Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J Biol Chem 278: 36430–36434. [DOI] [PubMed] [Google Scholar]
- 41. Plamann M, Minke PF, Tinsley JH, Bruno KS (1994) Cytoplamic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol 127: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fink G, Steinberg G (2006) Dynein-dependent motility of microtubules and nucleation sites supports polarization of the tubulin array in the fungus Ustilago maydis . Mol Biol Cell 17: 3242–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Akhmanova A, Hoogenraad MO (2011) Microtubule End Binding: EBs Sense the Guanine Nucleotide State. Curr Biol 21: R283–R285. [DOI] [PubMed] [Google Scholar]
- 44. Staudt MW, Kruzel EK, Shimizu K, Hull CM (2010) Characterizing the role of the microtubule binding protein Bim1 in Cryptococcus neoformans . Fungal Genet Biol 47: 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gardner MK, Haase J, Mythreye K, Molk JN, Anderson M, et al. (2008) The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol 180: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, et al.. (2009) Essential Cell Biology, fourth ed. Garland, New York.
- 47. Galjart N, Perez F (2003) A plus-end raft to control microtubule dynamics and function. Curr Opin Cell Biol 15: 48–53. [DOI] [PubMed] [Google Scholar]
- 48. Ligon LA, Shelly SS, Tokito M, Holzbaur EL (2003) The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol Biol Cell 14: 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MTB-3-GFP comets dynamics in mature hypha of N. crassa . Comets are distributed homogeneously along the hypha and moved in anterograde and retrograde direction.
(AVI)
TIRFM of MTB-3-GFP cortical comets dynamics in mature hypha of N. crassa .
(AVI)
Dynamics of MTB-3-GFP in the UV mutant of the p150glued subunit of dynactin ( ro-3 ).
(MPG)
Dynamics of MTB-3-GFP in the deletion mutant of kin-1 gene (Δ kin-1 ).
(MPG)
Microtubules dynamics (β-tubulin-GFP) in the mtb-3 gene deletion mutant.
(MPG)
Microtubules dynamics (β-tubulin-GFP) in the WT.
(MPG)