Abstract
In 2012, Texas experienced the largest outbreak of human West Nile encephalitis (WNE) since the introduction of West Nile virus (WNV) in 2002. Despite the large number of WNV infections, data indicated the rate of reported WNE among human cases was no higher than in previous years. To determine whether the increase in WNV human cases could have been caused by viral genetic changes, the complete genomes of 17 isolates made from mosquito pools in Dallas and Montgomery Counties in 2012 were sequenced. The 2012 Texas isolates were found to be composed of two distinct clades, both circulating in Dallas and Montgomery Counties despite a 5-fold higher disease incidence in the former. Although minor genetic differences existed between Dallas and Montgomery WNV populations, there was weak support for population subdivision or adaptive changes. On the basis of these data, alternative explanations for increased WNV disease incidence in 2012 are proposed.
Introduction
West Nile virus (WNV) was first identified in North America in New York City in 1999.1 By 2002, WNV was detected in 44 states, including Texas.2 Epidemic levels of WNV human cases occurred in the United States in 2002 and 2003, and WNV has been endemic from 2004 to 2011 with a fairly stable annual incidence in humans.3–6 West Nile virus is maintained in an enzootic cycle between mosquitoes (predominantly Culex spp.) and passerine birds, with annual outbreaks in humans typically peaking in August.6 Previous studies have estimated that ∼20% of humans infected with WNV develop West Nile fever (WNF), and < 1% develop West Nile neuroinvasive disease (WNND).7
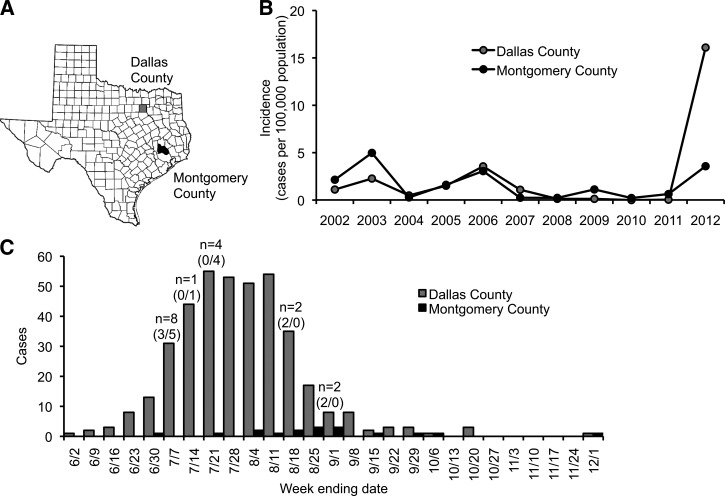
The largest outbreak of human WNF and WNND cases in the United States since 2003 occurred during 2012. As of December 11, 2012, 48 states and the District of Columbia reported a total of 5,387 confirmed or probable human WNF and WNND cases to the Centers for Disease Control and Prevention (CDC) through ArboNET. Approximately one-third of all United States WNF and WNND cases were reported from Texas. Within Texas, the incidence of WNV disease varied between counties. For example, the incidence of WNF and WNND was 16.1 cases/100,000 population in Dallas County and 3.6 cases/100,000 population in Montgomery County (Figure 1A and B). The reason for the increased incidence of WNV disease in 2012 is unknown, although climate, ecological changes in WNV vectors, and viral genetics have been suggested.8
Figure 1.
West Nile virus (WNV) disease incidence in Dallas and Montgomery Counties in 2012. Dallas and Montgomery Counties are highlighted in gray and black, respectively. (A) Map of Texas counties. (B) Annual incidence, reported as total West Nile fever (WNF) and West Nile neuroinvasive disease (WNND) cases per 100,000 populations. (C) Epidemic curve for total WNF and WNND cases in 2012. Numbers above bars represent the combined number of mosquito pool collections made in both Dallas and Montgomery Counties for this study, with numbers in parentheses representing the number of mosquito pool collections made in each county (Dallas/Montgomery).
In a manner similar to other arboviruses, WNV evolution is predominantly subject to purifying selection,9,10 and limited phylogeographic structure has been identified for WNV isolates10–12; few adaptive changes have been identified during its evolutionary history. Notable examples are a valine to alanine mutation at position 159 of the envelope protein that has been reported to infer fitness benefits in mosquitoes13,14 and has swept across the United States,15,16 and a threonine to proline mutation at position 249 in NS3 that has fitness benefits in crows and has been identified to have evolved under the effects of positive selection.17 As dead-end hosts, humans are unlikely to affect the evolution of WNV on a large scale; however, mutations arising in bird and mosquito hosts could stochastically affect viral replication and/or pathogenesis in humans. Additionally, altered transmissibility between mosquitoes and avian hosts could be associated with increased amplification and transmission to humans.
To determine whether the 2012 WNV outbreak may have been driven by recent WNV evolution, we assessed the phylogenetic relationship between WNV isolates from Texas in 2012 and isolates from North America in previous years. We also measured the genetic diversity and divergence of WNV isolates from Dallas and Montgomery Counties with variable disease incidence rates in Texas. Together, these data indicate that WNV evolution in 2012 was similar to previous years and is unlikely to explain the increased incidence in humans.
Materials and Methods
Incidence calculations.
Annual WNV disease incidence (cases per 100,000 population) in Dallas and Montgomery counties were calculated using total WNF and WNND case numbers reported to the CDC and population estimates from the U.S. Census Bureau for July 1 of each year, except for 2012, which was calculated using U.S. Census Bureau population change estimates for 2010–2012.
Virus collection.
Mosquito pools were collected by city agencies within Dallas and Montgomery Counties in July and August of 2012. Mosquito pools were screened at the Texas Department of State Health Services in BHK-21 and Vero cells for the growth of arboviruses including WNV. Indirect immunofluorescence of infected cells using monoclonal antibodies confirmed the presence of WNV.
Viral sequencing.
The WNV-positive mosquito pools were passaged once on BHK-21 cells. Viral RNA was extracted from clarified cell culture supernatant using the Qiagen Viral RNA Mini kit. Viral genomes were amplified by one-step reverse transcription-polymerase chain reaction (RT-PCR) (Qiagen, Valencia, CA) using specific primers to produce six overlapping RT-PCR products. The RT-PCR products were sequenced directly. Complete 5′ and 3′ noncoding region sequences were obtained from RACE products (Invitrogen, Carlsbad, CA). Primer sequences are available upon request; GenBank accession nos.: KC736486–KC736502.
Phylogenetic analyses.
Sequences were aligned with 76 additional full-length WNV sequences available in GenBank using Clustal Omega18 and edited manually. No evidence of recombination was detected using GARD in the HyPhy package, which was accessed through Datamonkey.19 Model selection was performed using jModelTest220,21 to determine the most appropriate nucleotide substitution model. Based on these results, a general time reversible + I + G model was used in further analyses. Maximum likelihood phylogenies were constructed using PhyML.22 One thousand (1,000) bootstrap replicates were used to obtain support for branches. Phylogenies were constructed for full-length sequences (11,029 nucleotides) as well as the entire coding region (10,299 nucleotides) and coding regions corresponding to individual proteins.
Diversity calculations.
Genetic diversity was calculated as the mean pairwise distance (substitutions per site) using the Maximum Composite Likelihood method23 in MEGA524; the P values were calculated using an unpaired two-tailed Student's t test.
Tests for selection.
Maximum likelihood tests for selection acting on individual sites were performed using FEL,25 and Bayesian tests for selection acting on individual sites were performed using FUBAR26 in the HyPhy package, which were accessed through Datamonkey.19
Results
2012. WNV epidemic in Dallas and Montgomery Counties.
Before the 2012 epidemic, WNV disease incidence in Texas peaked in 2003. In subsequent years, incidence fluctuated annually, but the general trend was toward a decrease in incidence through 2011.27 A similar trend was observed in Dallas and Montgomery counties, where total WNV disease incidence peaked in 2003 (2.3 and 5.0 cases/100,000 population, respectively) and 2006 (3.6 and 3.1 cases/100,000 population, respectively) (Figure 1B). In 2012, 396 WNF and WNND cases were reported to the CDC from Dallas County, yielding a WNV disease incidence of 16.1 cases/100,000 population, and 17 WNF and WNND cases were reported from Montgomery County, for a disease incidence of 3.6 cases/100,000 population. Of the total reported WNF and WNND cases, 175 (44%) were reported from Dallas County as WNND cases, and 8 (47%) were reported from Montgomery County as WNND cases. The severity of WNV disease in Dallas and Montgomery counties in 2012, represented by the percentage of WNND cases, is lower than the Texas state average from 2002 to 2011 of 67%27 and the national average from 1999 to 2011 of 60%.3–6 In addition, most of the 2012 WNF and WNND cases in Dallas and Montgomery Counties occurred during July and August (Figure 1C). This is consistent with WNV outbreaks in Texas in previous years, which typically peaked in August.27 Together, these data indicate that the 2012 epidemic was driven by an increase in overall WNV transmission from mosquitoes to humans, rather than an increase in WNV neuroinvasiveness. Increased WNV transmission could have been caused by the confluence of favorable biotic or abiotic factors that promote virus amplification.
Phylogenetic analyses of 2012 Texas WNV isolates.
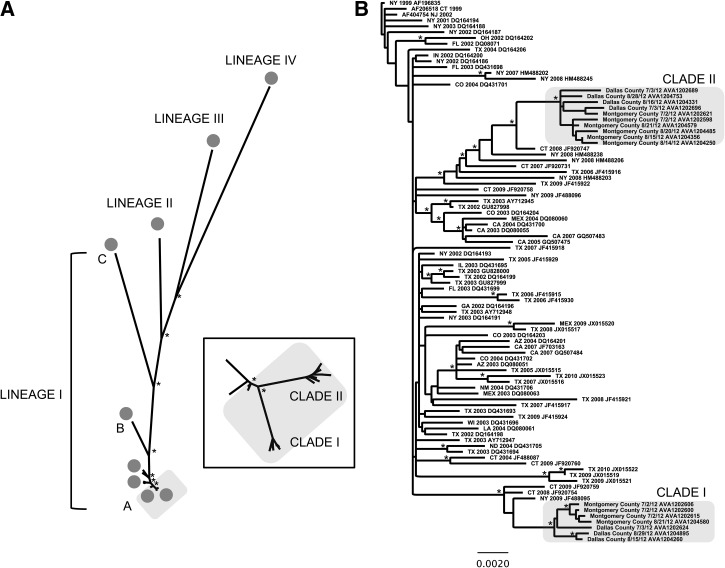
To determine whether the WNV strains circulating in Texas in 2012 were genetically different from previous outbreaks in the United States, we collected WNV isolates from mosquito pools before, during, and after the peak of the 2012 outbreak (Table 1 and Figure 1C). We sequenced seven full-length viral genomes from Dallas County and 10 full-length genomes from Montgomery County and aligned them with Lineage I–IV isolates of WNV. We constructed a maximum likelihood phylogeny and, as expected, identified that the 2012 Texas isolates clustered most closely with Lineage IA viruses from the United States (Figure 2A). Interestingly, the 2012 Texas isolates formed two distinct clades with high branch support (bootstrap value = 1,000; Figure 2A ), indicating that two strains of WNV were co-circulating in Texas in 2012.
Table 1.
West Nile virus (WNV) isolates made in Dallas and Montgomery Counties in 2012
| Isolate name | Date collected | County | City | Host species |
|---|---|---|---|---|
| AVA1202624 | 7/3/12 | Dallas | Grand Prairie | Culex quinquefasciatus |
| AVA1202689 | 7/3/12 | Dallas | Dallas | Cx. quinquefasciatus |
| AVA1202696 | 7/3/12 | Dallas | Dallas | Cx. quinquefasciatus |
| AVA1204260 | 8/15/12 | Dallas | Garland | Cx. quinquefasciatus |
| AVA1204331 | 8/16/12 | Dallas | Dallas | Cx. quinquefasciatus |
| AVA1204753 | 8/28/12 | Dallas | Garland | Cx. quinquefasciatus |
| AVA1204895 | 8/29/12 | Dallas | Garland | Cx. quinquefasciatus |
| AVA1202598 | 7/2/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1202600 | 7/2/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1202606 | 7/2/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1202615 | 7/2/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1202621 | 7/2/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1204250 | 8/14/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1204356 | 8/15/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1204485 | 8/20/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1204579 | 8/21/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
| AVA1204580 | 8/21/12 | Montgomery | The Woodlands | Cx. quinquefasciatus |
Figure 2.
Maximum-likelihood phylogeny of the coding region (nucleotides 97–10,398) of West Nile virus (WNV) isolates. Nodes supported by bootstraps over 90% are marked by asterisks. (A) WNV Lineage I–IV phylogeny. The North American Lineage IA is highlighted in gray. (Inset) Detailed view of Lineage IA. (B) WNV North American Lineage IA phylogeny. The 2012 Texas Clades I and II are highlighted in gray. Names are location and year of collection followed by GenBank accession nos. (described further in Supplemental Table 1).
Next, we aligned the 2012 Texas isolates with 76 representative full-length Lineage IA WNV genomes isolated between 1999 and 2010 from diverse geographic regions in North America (Supplemental Table 1). Using GARD, we found no evidence of recombination in the alignment. We constructed a maximum likelihood phylogeny of North American WNV isolates and found that Texas 2012 Clade I viruses clustered most closely with an isolate from New York in 2009, whereas the Texas 2012 Clade II viruses clustered most closely with an isolate from Connecticut in 2008. However, Clades I and II are phylogenetically similar groups containing isolates from both Dallas and Montgomery Counties, and weak branch support exists for separate Dallas and Montgomery populations within Clades I and II (bootstrap values < 500). This indicates that the Dallas County WNV isolates are not phylogenetically distinct from the Montgomery County WNV isolates.
We also calculated the average genetic diversity within Dallas and Montgomery populations and the average genetic divergence between Dallas and Montgomery populations and the most closely related isolates (NY 2009 and CT 2008). The mean pairwise diversity within the Dallas populations was higher than the mean pairwise diversity within the Montgomery populations (Table 2, Clade II: P < 0.05). However, the mean divergence between Dallas and Montgomery populations and the most closely related isolates are very similar between populations. Overall, the Dallas WNV populations contain greater genetic diversity compared with the Montgomery WNV populations, but they are derived from the same clades.
Table 2.
Genetic diversity of 2012 Texas isolates
Positive selection analyses of Texas WNV isolates.
Because single mutations can drastically affect WNV fitness, we next assessed whether individual amino acids are differentiated between 2012 Texas isolates and previous United States isolates, or between Dallas and Montgomery County WNV populations. Candidates for single sites that are correlated with high transmission would be derived non-synonymous mutations in at least one Dallas population. We found that five non-synonymous mutations are fixed in one 2012 Texas clade compared with the most closely related United States isolates (NY 2009 and CT 2008) (Table 3A). We also found that 14 population-specific non-synonymous mutations are polymorphic in one Dallas population (Table 3B). Two of these mutations are also polymorphic in Montgomery populations (E12 and NS2A52). In addition, 10 non-coding mutations in the 5′ and 3′ regions are polymorphic in one Dallas population, and one of these mutations is also polymorphic in a Montgomery population (Table 3C). Seven non-synonymous mutations and three non-coding mutations are polymorphic only in Montgomery populations (data not shown).
Table 3.
Variable nucleotides in 2012 Texas isolates
| (A) clade-specific non-synonymous mutations | ||||
|---|---|---|---|---|
| Residue | JF488095 | Clade I | JF920747 | Clade II |
| C119 | A | S | A | A |
| E123 | T | N | T | T |
| NS1308 | I | V | I | I |
| NS2A58 | V | V | V | I |
| NS4B240 | I | I | I | M |
| (B) population-specific non-synonymous mutations | ||||
|---|---|---|---|---|
| Clade I | Clade II | Clade I | Clade II | |
| Residue | Dallas | Dallas | Montgomery | Montgomery |
| E12 | L | L/V | L | L/V |
| E231 | T/A | T | T | T |
| E367 | A/V | A | A | A |
| E479 | R/L | R | R | R |
| NS1105* | T | T/A | T | T |
| NS1349 | Q | Q/R | Q | Q |
| NS2A52† | I | I/T | I/T | I/T |
| NS4A65 | M | M/T | M | M |
| NS4A85 | A | A/S | A | A |
| NS4B30 | G/R | G | G | G |
| NS4B99 | A | A/S | A | A |
| NS5587 | A | A/V | A | A |
| NS5706 | Y | Y/H | Y | Y |
| NS5866 | E/G | E | E | E |
| (C) population-specific non-coding mutations | ||||
|---|---|---|---|---|
| Clade I | Clade II | Clade I | Clade II | |
| Nucleotide | Dallas | Dallas | Montgomery | Montgomery |
| 19 | G | G/A | G/A | A |
| 10407 | T/G | T | T | T |
| 10408 | T | T/C | T | C |
| 10429 | A/G | A | A | A |
| 10435 | C | C/T | C | C |
| 10448 | T | T/G | T | T |
| 10450 | T | T/C | T | T |
| 10563 | A | A/G | A | A |
| 10734 | G | G/A | G | G |
| 10774 | C | C/T | C | C |
ω < 1, P < 0.05.
ω > 1, P < 0.05.
To determine whether these differences may be adaptive, we performed an analysis of selection pressures acting on the North America Lineage IA WNV dataset (Supplemental Table 1). Using two methods, we estimated the ratio of the non-synonymous and synonymous mutation rate (designated ω) for each codon across the WNV genome, where ω > 1 is indicative of positive selection and ω < 1 is indicative of purifying selection. Two codons contain signatures of positive selection: NS2A52 and NS5314 (ω = ∞, P < 0.05). The NS2A52 is mutated in all 2012 Texas populations (Table 3B). In contrast, although two loci showed evidence of positive selection across the WNV genome, 832 loci showed evidence of purifying selection (P < 0.05, data not shown). One of these negatively selected resides, NS1105, exhibited a variable amino acid in the Dallas Clade II population (Table 3B). Together, these data indicated that, although small genetic differences existed in Dallas and Montgomery WNV isolates compared with previous WNV isolates, most changes were unlikely to be adaptive. Rather, divergence and diversity of the 2012 Texas isolates was primarily driven by genetic drift.
Discussion
In this study, we investigated the evolution of WNV isolates made in Texas during the largest outbreak in recent United States history. To determine whether WNV genetics may have contributed to the increased incidence of human WNV disease in 2012, we sequenced full-length WNV genomes from mosquitoes collected in Texas during the 2012 epidemic. We found that the mutations in the 2012 Texas isolates were largely driven by genetic drift and viral population structure molded by purifying selection in a manner similar to previous years. Furthermore, despite differences in WNF and WNND incidence between Dallas and Montgomery Counties, the WNV isolates from these areas were phylogenetically indistinguishable. Together, these data indicate that a major role for WNV genetic determinants in the 2012 outbreak was unlikely.
The 2012 Texas WNV isolates clustered in two distinct clades, with the most closely related isolates from Connecticut in 2008 and New York in 2009 (Figure 2). This indicates that the 2012 WNV epidemic was derived from two WNV strains endemic to the United States; however, there may have been two recent reintroductions of WNV into Texas from the East coast. This is characteristic of the long-range movements of WNV across North America.11 However, there is likely short-distance viral gene flow between Dallas and Montgomery Counties because WNV isolates from these areas were phylogenetically similar. Even though WNF and WNND incidence in Dallas County was five times higher than in Montgomery County (Figure 1), similar WNV strains were found in both locations. This shows that a geographically isolated virulent strain of WNV did not cause the 2012 outbreak in Dallas. Thus, differences in WNV genetics are unlikely to explain the difference in WNF and WNND incidence between Dallas and Montgomery Counties.
Rapid evolution is a characteristic of genomes involved in antagonistic virus-host interactions28; therefore, identifying sites evolving under positive selection can pinpoint mutations with fitness consequences. In this study, we identified two residues that are rapidly evolving in the United States lineage of WNV: NS2A52 (Table 3) and NS5314. The NS5314 has previously been shown to have evolved under positive selection, making it a tempting candidate for an adaptive change29; however, NS5314 only showed variation in Montgomery County isolates and was therefore unlikely to have affected the 2012 outbreak. In a previous study of WNV pathogenesis in American crows, an alanine-to-threonine mutation at NS2A52 was shown to be one of 11 mutations potentially involved in the increased virulence in American crows of WNV NY99 compared with a Kenyan strain of WNV.30 However, most United States WNV strains since 1999 have contained a threonine at NS2A52, suggesting that this mutation is either unrelated to the 2012 outbreak or requires additional mutations to affect fitness. Of the 17 additional non-synonymous mutations in the 2012 Dallas isolates, one site was identified to be subject to purifying selection (Table 3), suggesting this mutation may be deleterious to viral fitness. The remaining 16 mutations, although not rapidly evolving or having a known functional consequence, may elicit an adaptive phenotype such as increased transmission efficiency in mosquitoes; however, vector competence studies would be required to fully assess this question.
Although WNV sequences from humans infected during the 2012 outbreak were not available for analyses, the mosquito isolates that were assayed should encompass the diversity of the strains circulating during the summer of 2012. Genetic diversity of WNV is maintained in mosquitoes,31,32 and studies of human and mosquito WNV isolates have not shown phylogenetic separation by host species.11 In addition, the level of divergence between the 2012 Texas isolates and other United States isolates was comparable to estimates from previous years in Texas and other geographic regions of the United States.9,10,33–35 Interestingly, significantly higher genetic diversity within the Clade II Dallas population compared with the Clade II Montgomery population was observed (Table 2), although the sampling area and timeframe of isolate collection were not matched between populations. The Dallas isolates were collected over 8 weeks from multiple cities, whereas the Montgomery isolates were collected over a 7-week span from one city (Table 1 and Figure 1). Thus, our population genetic diversity estimates could be skewed by mosquito sampling.
In addition to viral genetics, ecological factors have been shown to increase viral transmission in mosquitoes and birds. For example, temperature affects the WNV extrinsic incubation period of mosquitoes, such that increased temperature increases the rate of WNV transmission by mosquitoes.13,36 Previous outbreaks of WNV in 2002–2004 were associated with higher than average temperatures,36,37 and the average annual temperature in Texas in 2012 was 2° warmer than the average annual temperature in 2002–2011 (NOAA National Climatic Data Center). Furthermore, the average monthly temperature during the peak of the 2012 outbreak (June through September, Figure 1) was 1° to 6° warmer in Dallas than in Houston (NOAA National Climatic Data Center), which may help to explain the higher viral diversity in Dallas County mosquito isolates or the greater rates of WNV transmission in Dallas County. In addition, changes in mosquito or avian populations may have contributed to altered viral transmission rates. Based on the results presented here, rather than genetic changes in WNV leading to increased transmission efficiency to humans, it is more likely that a combination of other environmental and ecological factors could explain the magnitude of the 2012 WNV outbreak.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janae Stovall and Karen Boroughs for sequencing support, Jennifer Lehman for assistance with ArboNET data, and Bill Reisen and Marc Fisher for comments on the manuscript.
Footnotes
Financial support: N.K.D. was supported by an APHL Emerging Infectious Disease postdoctoral fellowship.
Authors' addresses: Nisha K. Duggal, Roger Nasci, and Aaron C. Brault, Division of Vector-Borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mails: wwd3@cdc.gov, rsn0@cdc.gov, and abrault@cdc.gov. Mary D'Anton, Jeannie Xiang, Robyn Seiferth, and Joanne Day, Texas Department of State Health Services, Laboratory Services Section Austin, TX, E-mails: Mary.D'Anton@dshs.state.tx.us, Jeannie.Xiang@dshs.state.tx.us, Robyn.Seiferth@dshs.state.tx.us, and Joanne.Day@dshs.state.tx.us..
References
- 1.CDC Outbreak of West Nile-like viral encephalitis–New York, 1999. MMWR Morb Mortal Wkly Rep. 1999;48:845–849. [PubMed] [Google Scholar]
- 2.CDC Provisional surveillance summary of the West Nile virus epidemic–United States, January–November 2002. MMWR Morb Mortal Wkly Rep. 2002;51:1129–1133. [PubMed] [Google Scholar]
- 3.CDC West Nile virus activity–United States, 2009. MMWR Morb Mortal Wkly Rep. 2010;59:769–772. [PubMed] [Google Scholar]
- 4.CDC West Nile virus disease and other arboviral diseases–United States, 2010. MMWR Morb Mortal Wkly Rep. 2011;60:1009–1013. [PubMed] [Google Scholar]
- 5.CDC West Nile virus disease and other arboviral diseases–United States, 2011. MMWR Morb Mortal Wkly Rep. 2012;61:510–514. [PubMed] [Google Scholar]
- 6.Lindsey NP, Staples JE, Lehman JA, Fischer M. Centers for Disease Control and Prevention (CDC) Surveillance for human West Nile virus disease–United States, 1999–2008. MMWR Surveill Summ. 2010;59:1–17. [PubMed] [Google Scholar]
- 7.Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, Busch MP. Estimated cumulative incidence of West Nile virus infection in US adults, 1999–2010. Epidemiol Infect. 2012;28:1–5. doi: 10.1017/S0950268812001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen L. Record heat may have contributed to a banner year for West Nile virus. Interview with Lyle Petersen. JAMA. 2012;308:1846–1848. doi: 10.1001/jama.2012.13495. [DOI] [PubMed] [Google Scholar]
- 9.Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertolotti L, Kitron U, Goldberg TL. Diversity and evolution of West Nile virus in Illinois and the United States, 2002–2005. Virology. 2007;360:143–149. doi: 10.1016/j.virol.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, Gray RR, Arinaminpathy N, Stramer SL, Busch MP, Delwart EL. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci USA. 2012;109:15066–15071. doi: 10.1073/pnas.1206598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray RR, Veras NM, Santos LA, Salemi M. Evolutionary characterization of the West Nile Virus complete genome. Mol Phylogenet Evol. 2010;56:195–200. doi: 10.1016/j.ympev.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Kilpatrick AM, Meola MA, Moudy RM, Kramer LD. Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathog. 2008;4:e1000092. doi: 10.1371/journal.ppat.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moudy RM, Meola MA, Morin LL, Ebel GD, Kramer LD. A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am J Trop Med Hyg. 2007;77:365–370. [PubMed] [Google Scholar]
- 15.Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DW, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett AD. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. Am J Trop Med Hyg. 2004;71:493–500. [PubMed] [Google Scholar]
- 17.Brault AC, Huang CY, Langevin SA, Kinney RM, Bowen RA, Ramey WN, Panella NA, Holmes EC, Powers AM, Miller BR. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 20.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 22.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 26.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolan MS, Schuermann J, Murray KO. West Nile virus infection among humans, Texas, USA, 2002–2011. Emerg Infect Dis. 2013;19:137–139. doi: 10.3201/eid1901.121135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMullen AR, May FJ, Li L, Guzman H, Bueno R, Jr, Dennett JA, Tesh RB, Barrett AD. Evolution of new genotype of West Nile virus in North America. Emerg Infect Dis. 2011;17:785–793. doi: 10.3201/eid1705.101707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brault AC, Langevin SA, Bowen RA, Panella NA, Biggerstaff BJ, Miller BR, Komar N. Differential virulence of West Nile strains for American crows. Emerg Infect Dis. 2004;10:2161–2168. doi: 10.3201/eid1012.040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brackney DE, Pesko KN, Brown IK, Deardorff ER, Kawatachi J, Ebel GD. West Nile virus genetic diversity is maintained during transmission by Culex pipiens quinquefasciatus mosquitoes. PLoS ONE. 2011;6:e24466. doi: 10.1371/journal.pone.0024466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amore G, Bertolotti L, Hamer GL, Kitron UD, Walker ED, Ruiz MO, Brawn JD, Goldberg TL. Multi-year evolutionary dynamics of West Nile virus in suburban Chicago, USA, 2005–2007. Philos Trans R Soc Lond B Biol Sci. 2010;365:1871–1878. doi: 10.1098/rstb.2010.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armstrong PM, Vossbrinck CR, Andreadis TG, Anderson JF, Pesko KN, Newman RM, Lennon NJ, Birren BW, Ebel GD, Henn MR. Molecular evolution of West Nile virus in a northern temperate region: Connecticut, USA 1999–2008. Virology. 2011;417:203–210. doi: 10.1016/j.virol.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann BR, McMullen AR, Guzman H, Tesh RB, Barrett AD. Dynamic transmission of West Nile virus across the United States-Mexican border. Virology. 2013;436:75–80. doi: 10.1016/j.virol.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reisen WK, Fang Y, Martinez VM. Effects of temperature on the transmission of West Nile virus by Culex tarsalis (Diptera: Culicidae) J Med Entomol. 2006;43:309–317. doi: 10.1603/0022-2585(2006)043[0309:EOTOTT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 37.Hartley DM, Barker CM, Le Menach A, Niu T, Gaff HD, Reisen WK. Effects of temperature on emergence and seasonality of West Nile virus in California. Am J Trop Med Hyg. 2012;86:884–894. doi: 10.4269/ajtmh.2012.11-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe KE, Crabtree MB, Scherret JH, Hall RA, MacKenzie JS, Cropp CB, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage HM, Stone W, McNamara T, Gubler DJ. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 39.Anderson JF, Andreadis TG, Vossbrinck CR, Tirrell S, Wakem EM, French RA, Garmendia AE, Van Kruiningen HJ. Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science. 1999;286:2331–2333. doi: 10.1126/science.286.5448.2331. [DOI] [PubMed] [Google Scholar]
- 40.Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, Bowen M, McKinney N, Morrill WE, Crabtree MB, Kramer LD, Roehrig JT. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology. 2002;298:96–105. doi: 10.1006/viro.2002.1449. [DOI] [PubMed] [Google Scholar]
- 41.Deardorff E, Estrada-Franco J, Brault AC, Navarro-Lopez R, Campomanes-Cortes A, Paz-Ramirez P, Solis-Hernandez M, Ramey WN, Davis CT, Beasley DW, Tesh RB, Barrett AD, Weaver SC. Introductions of West Nile virus strains to Mexico. Emerg Infect Dis. 2006;12:314–318. doi: 10.3201/eid1202.050871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herring BL, Bernardin F, Caglioti S, Stramer S, Tobler L, Andrews W, Cheng L, Rampersad S, Cameron C, Saldanha J, Busch MP, Delwart E. Phylogenetic analysis of WNV in North American blood donors during the 2003–2004 epidemic seasons. Virology. 2007;363:220–228. doi: 10.1016/j.virol.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 43.May FJ, Li L, Davis CT, Galbraith SE, Barrett AD. Multiple pathways to the attenuation of West Nile virus in south-east Texas in 2003. Virology. 2010;405:8–14. doi: 10.1016/j.virol.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrade CC, Maharaj PD, Reisen WK, Brault AC. North American West Nile virus genotype isolates demonstrate differential replicative capacities in response to temperature. J Gen Virol. 2011;92:2523–2533. doi: 10.1099/vir.0.032318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.