Abstract
To determine the magnitude of Plasmodium vivax relapsing malaria in rural Amazonia, we carried out a study in four sites in northeastern Peru. Polymerase chain reaction-restriction fragment length polymorphism of PvMSP-3α and tandem repeat (TR) markers were compared for their ability to distinguish relapse versus reinfection. Of 1,507 subjects with P. vivax malaria, 354 developed > 1 episode during the study; 97 of 354 (27.5%) were defined as relapse using Pvmsp-3α alone. The addition of TR polymorphism analysis significantly reduced the number of definitively defined relapses to 26 of 354 (7.4%) (P < 0.05). Multivariate logistic regression modeling showed that the probability of having > 1 infection was associated with the following: subjects in Mazan (odds ratio [OR] = 2.56; 95% confidence interval [CI] 1.87, 3.51), 15–44 years of age (OR = 1.49; 95% CI 1.03, 2.15), traveling for job purposes (OR = 1.45; 95%CI 1.03, 2.06), and travel within past month (OR = 1.46; 95% CI 1.0, 2.14). The high discriminatory capacity of the molecular tools shown here is useful for understanding the micro-geography of malaria transmission.
Introduction
An estimated 2.6 billion people live in areas endemic for malaria caused by Plasmodium vivax1 where this parasite is responsible for a substantial burden of disease accounting for at least 70–80 million clinical cases yearly.2–5 Plasmodium vivax is the most common form of malaria in the Amazon region4–6; in Peru, 32.5% of the population is at risk for malaria and P. vivax is responsible for 70–90% of cases in the Peruvian Amazon, with Plasmodium falciparum accounting for the remainder.7 Plasmodium vivax and P. falciparum malaria transmission is characterized in the Peruvian Amazon region as low transmission. Older surveys have found 9.8/1,000 Anopheles spp. vector mosquitoes positive for Plasmodium species, with Anopheles darlingi being the primary vector in the Amazon region.8,9 Mixed infections with P. falciparum and P. vivax are uncommon,10,11 and asymptomatic malaria parasitemia is present in 3–5% of cross-sectional smears.12 Published entomological inoculation rates have been reported to range from 0.5 (0.2, 0.8) to 2.5 (1.0, 3.9).8,13
Control of vivax malaria at the public health level is complicated by the parasite's unique biological features: early gametocytogenesis, relapsing liver stages, and a wider geographic range caused by tolerance of different climatic conditions.2 Tools to delineate and quantify P. vivax relapses occurring in vivax-endemic regions are key to differentiating relapse from re-infection and to allow for the quantification of the burden of this phenomena, which is known to cause a cumulative lifetime malaria experience of 10–30 episodes.2 The biology of P. vivax relapse remains poorly understood and is an important obstacle to the public health control of P. vivax malaria.14–16
Genetic markers have been assessed to discriminate between P. vivax infecting strains. Some of the genes identified for this purpose include genes for the circumsporozoite protein (PvCSP) and merozoite surface protein-1 (PvMSP-1),17 apical membrane protein-1 (PvAMA-1), and merozoite surface protein-3α (PvMSP-3α)18; recently, tandem repeat (TR) polymorphism markers19 and microsatellite methods have been used for this purpose.20–22
The MSP-3α of P. vivax has a molecular weight ranging from 148 to 150 KD, an alanine-rich central domain, and a series of heptad repeats predicted to form a coiled-coil tertiary peptide structure. The TR markers are on a 100 kb chromosomal fragment that includes the P. vivax circumsporozoite gene, which is under selective immune pressure and thus is a highly dynamic area of the chromosome. In this study, we used polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of PvMSP-3α (enzymes Hha1 and Alu1) and PCR of nine TR markers. Because of extensive polymorphisms in these markers, it is unlikely that a parasite causing a new infection would possess an identical genotype; the probability of this being the product of individual allele frequencies of each allele of the genes.
This study was designed to test the hypothesis that recurrences caused by relapse are less common than recurrences caused by reinfection. In addition, we explore if a higher level of resolution of P. vivax infecting strains during initial and subsequent infections could be carried out by comparing PvMSP-3α PCR-RFLP versus TR polymorphism analysis in parasites from an initial versus a subsequent P. vivax infection, hence allowing relapses to be distinguished from new infections. Such analysis is the key to understanding the transmission dynamics and role of human movement in the maintenance and spread of P. vivax in endemic regions.
Materials and Methods
Human subject approvals.
All patients provided written informed consent to be enrolled in this study, which was approved by the Ethical Committees of Universidad Peruana Cayetano Heredia, Lima, Peru; Asociación Benéfica PRISMA, Lima, Peru; the Directorate of Health of Loreto-Peru; and the Institutional Review Boards of the University of California at San Diego and the Johns Hopkins Bloomberg School of Public Health.
Study sites and duration of follow-up.
The field activities of this study were carried out from 2005 to 2008 in northeastern Peru, in the area near the city of Iquitos in the province of Maynas, the capital of the Amazon Department of Loreto. Considering the region's geographical isolation from the rest of Peru, health services within the surrounding areas of Iquitos are relatively good and accessible. Four health posts provide medical services to the study villages (Figure 1) described as follows. The Santo Tomas health post is located 16 km from Iquitos with a catchment area of 2,650. The San Jose de Lupuna health post is located 10 km from Iquitos and accessible only by the Nanay River, has a catchment area of 1,250. The Padrecocha health post is located 6 km from Iquitos, is accessible only by the Nanay River, and has a catchment area of 1,800. The Mazan health center is located 50 km northeastern from Iquitos and has a catchment area of 8,000. The duration of follow-up was as follows: 16 months in Padrecocha (from January 2005 to May 2006), 29 months in Santo Tomas and Santo de Lupuna (from February 2005 to July 2007), and 20 months in Mazan (from June 29, 2006 to February 5, 2008).
Figure 1.
Location of study sites in surrounding areas of Iquitos, Peruvian Amazon (*). Axes of Malaria transmission in Loreto, Peru 2007. Sources: Loreto Health Regional Directorate, PAMAFRO.
Blood sampling.
Blood samples were collected from patients diagnosed by conventional light microscopy, on site, at the Peruvian Ministry of Health posts according to national norms. Patients identified as infected with only P. vivax (microscopy only) were invited to participate in the study. At each enrollment site, venous blood was collected in EDTA Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ) and taken to the field project laboratory where samples were aliquoted and frozen at −20°C. Samples were shipped on dry ice to Universidad Peruana Cayetano Heredia, in Lima, where they were stored frozen at −20°C until used for molecular analysis.
Molecular confirmation of P. vivax infection.
The DNA was extracted from 200 μL of thawed anticoagulated whole blood using the Qiagen Blood Kit (Qiagen, Valencia, CA). Fourteen PCR assays numbered from PCR1 to PCR14 were carried out (Table 1). The diagnosis of non-mixed P. vivax infections was confirmed in all patients using genus and species-specific nested PCR assays12; samples with mixed infections were omitted from the analysis. The PCR 1, part of a nested PCR (an 18S ribosomal RNA gene fragment specific for genus Plasmodium was used, and a 1,200 basepairs (bp) fragment, allowed for Plasmodium genus-level identification. Identification of P. vivax, and P. falciparum species was done using PCR2 (a 120 bp fragment specific for P. vivax), and PCR3 (a 205 bp fragment specific for P. falciparum), respectively.
Table 1.
Primer, polymerase chain reaction (PCR) mix and cycling conditions used for Plasmodium vivax confirmation and genotyping using Merozoite Surface Protein 3α (MSP3α) and tandem repeat (TR) markers
| Plasmodium vivax confirmatory nested PCR (PCR1: Genus PCR; PCR2 and 3: Species PCR) | ||
|---|---|---|
| Primer Sequence* (forward/reverse) | PCR-Mix† | PCR-Cycling‡ |
| PCR1: P1/P2: CTTGTTGTTGCCTTAAACTTC/TTAAAATTGTTGCAGTTAAAACG | 2 mM MgCl2, 200 μM dNTPs, 0.25 μM P1/P2, 0.3 U Taq-p§, 3 μL DNA. 1 μL product of PCR1, 0.5 μM V1/V2 or F1/F2 was used for species-specific PCR 2. | 94° for 3′; then 19 cycles of: 94° for 30″, annealing at 58° for 90″, extension at 72° for 90″; and finally 72° for 7′. 34 cycles were performed for species-specific subsequent PCR2, as above. |
| PCR2: V1/V2: CGCTTCTAGCTTAATCCACATAACTGATAC/ACTTCCAAGCCGAAGCAAAGAAAGTCCTTA | ||
| PCR3: F1/F2: TTAAACTGGTTTGGGAAAACCAAATATATT/ACACAATGAACTCAATCATGACTACCCGTC | ||
| MSP3-alpha nested PCR (PCR 4 and 5) | ||
| Primer sequence* (forward/reverse) | PCR-Mix† | PCR-Cycling‡ |
| PCR 4: M1/M2: CAGCAGACACCATTTAAGG/CCGTTTGTTGATTAGTTGC | PCR3: 2.5 mM MgCl2, 150 μM dNTP, 0.15 μM M1/M2 or N1/N2, 0.5 U Taq-p, 3μL DNA. PCR4: Idem but 1μL DNA product of PCR3. | 94° for 3′; then 34 cycles of: 94° for 30″, annealing at 50° for 30″, extension at 68° for 150″; and finally 68° for 4′. |
| PCR 5: N1/N2: GACCAGTGTGATACCATTAACC/ATACTGGTTCTTCGTCTTCAGG | ||
| Tandem repeat PCR (PCR 6 to 14) | ||
| Primer Sequence* (forward/Reverse) | PCR-Mix† | PCR-Cycling‡ |
| MN3: GAGAGTGACTAGCAATGG/AGATGCCTTTTTCTGCGTTT | 2 mM MgCl2, 150 μM dNTPs, 0.25 μM of each one of the pair of primers for the nine TR primer pairs (MN3, MN4, MN12, MN17, MN23, MN24, MN25, MN26, MN29), 0.5 U Taq-p, 3 μL DNA. | A single cycling protocol for all TR-PCR was used: 94°C for 2′; 35 cycles of: 94°C for 20″, 55°C for 10″, 65°C for 2′, and an extension step of 65°C for 5′. |
| MN4: TCACTTGGTTCTTCCCCG/AGTGTGAACATGGGTGCA | ||
| MN12: TCTGTTTCTGTTTCTGCGTT/AGGGTTGTTTAACATCTGCT | ||
| MN17: AACCGACTGGCTTACAACCA/GATGTGGACGTCTGCATAGT | ||
| MN23: AGCACAAGGTGACTCAAAAA/AGCTGCGTTTTGATGGAGAA | ||
| MN24: AATTTGACCTCCGGCACTTC/TTCGTCACTTCGTCGTTTCG | ||
| MN25: GGGGAAAAACAATGGCAA/AGGCAGTCGATTTGAAACT | ||
| MN26: TGAGCTGCGTAGAAGCCGTT/TTGTTCAGCATGTTGCGTTG | ||
| MN29: TGAAAAGAGCTGCTCGAACA/CACTTGTAGAGAGAGGCGAG | ||
Sequences are described 5′ to 3′.
All PCR mixes had 0.25 μL at final volume.
PCR reactions were performed using a PTC100, MJ Research Inc. thermal cycler (Bio-Rad).
Taq-p: Taq polymerase (Invitrogen).
Molecular genotyping assays.
Genotyping of P. vivax isolates was performed using two methods: 1) PCR-RFLP analysis of the P. vivax merozoite surface protein-3alpha (PvMSP3α) gene17,18,23; and 2) a PCR assay based on nine previously published tandem repeat (TR) polymorphism markers19,23 and recently validated in Peru.23
P. vivax Merozoite Surface Protein-3α.
To assess allelic types of the PvMSP-3α gene, a published PCR-RFLP method was used18 consisting of a nested PCR (PCR4 and PCR5) where a polymorphic fragment of the PvMSP-3α gene was amplified followed by PCR-RFLP (Alu1 and Hha1) as previously described (Table 1).23
TR polymorphism analysis.
We used nine primer pairs previously designed from a 200 kb contiguous DNA segment of 5 P. vivax strains from different parts of the world,19 syntenic to P. falciparum chromosome 3. We selected these nine primer pairs based on preliminary studies of seven different samples from different areas of the Peruvian Amazon; the other 24 primer pairs tested did not yield at least two alleles.23 No TR primer pair amplified DNA from either human or P. falciparum.19 The 5 μL of DNA extracted from 200 μL of whole blood and 0.5 μL of primer (Invitrogen, Carlsbad, CA) was added to the PCR mix. The TR markers used in this study were as follows: MN3, MN4, MN12, MN17, MN23, MN24, MN25, MN26, and MN29 (Table 1). The PCR electrophoresis and size quantification was done as previously described.23
Case definition and data analysis.
Recurrence was defined as any subsequent infection regardless of any genotyping pattern. To determine the relapse or reinfection status of all recurrent episodes, the following steps were followed: First, all PvMSP3-α PCR-RFLP patterns of the reference and subsequent infections were identified. Second, the identical patterns from the previous step were analyzed through TR markers using a tolerance of 5% on band sizes as previously described.23 Relapse was then defined as identical patterns after applying steps 1 and 2 in that order. Reinfection was defined as a difference in reference versus subsequent infection parasites after applying steps 1 and 2. No comparisons were done when mixed or composite infections.
The reference was the primary infection for all subjects with 2 episodes. In the case of > 2 episodes, we compared all episodes with the previous ones and if an episode met the criteria then a relapse was considered. On the other hand, because most of the subjects had a past history of vivax malaria, this is true that there would be a relapse of a parasite population that has not been typed because the primary infection has occurred before the study started.
Statistical analysis.
Data were analyzed using SAS v.9.1 (SAS, Cary, NC). The significance level for statistical tests was set at P ≤ 0.05. The risk of recurrence, relapse, and re-infection were estimated using Kaplan-Meier survival analysis. The Cox proportional hazard model was to compare survival functions when assumptions were met.
Results
Enrollment, baseline characteristics, and P. vivax recurrence rates.
After a baseline health assessment, 1,507 subjects with P. vivax malaria were enrolled in the study. Of these, 575 (38.2%) were enrolled in Santo Tomas, 198 (13.1%) in San Jose de Lupuna, 386 (25.6%) in Padrecocha, and 348 (23.1%) in Mazan. Eight hundred and thirty-one (831) (55.1%) were male, 676 (44.8%) female (Table 2).
Table 2.
Baseline characteristics of study subjects: total enrollees, enrollees who developed one episode, and enrollees who developed > 1 episode, by study sites (proportion of each cell per row are in italics)*
| Variable | Total (N = 1507) | 1 infection (N = 1153) | > 1 infection (N = 354) | P value | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Health Post (N = 1507) | < 0.0001 | ||||||
| San Jose de Lupuna (SJL) | 198 | (13.1) | 150 | (13.3) (75.8) | 48 | (12.8) (24.2) | |
| Santo Tomas (STO) | 575 | (38.2) | 446 | (38.5) (77.6) | 129 | (37.1) (22.4) | |
| Mazan (MZ) | 348 | (23.1) | 215 | (18.6) (61.8) | 133 | (37.7) (38.2) | |
| Padrecocha (PAD) | 386 | (25.6) | 342 | (29.6) (88.6) | 44 | (12.5) (11.4) | |
| Health Post (N = 1507) | < 0.0001 | ||||||
| STO, SJL, and PAD | 1,159 | (76.9) | 938 | (81.4) (80.9) | 221 | (62.3) (19.1) | |
| Mazan | 348 | (23.1) | 215 | (18.6) (61.8) | 133 | (37.7) (38.2) | |
| Time living in community (years) (N = 1507) | 0.2143 | ||||||
| Median (IQR) | 11.0 | (4–22) | 12.0 | (4–22) | 10.0 | (4–20) | |
| Age (years) (N = 1507) | 0.6573 | ||||||
| Mean (SD) | 27.1 | (17.0) | 27.0 | (17.2) | 27.4 | (16.2) | |
| 0.1568 | |||||||
| ≤ 4 | 46 | (3.1) | 33 | (2.9) (71.7) | 13 | (3.7) (28.3) | |
| 5–14 | 373 | (24.8) | 299 | (25.9) (80.2) | 74 | (20.9) (19.8) | |
| 15–44 | 844 | (56.1) | 630 | (54.6) (74.6) | 214 | (60.5) (25.4) | |
| ≥ 45 | 244 | (16.2) | 191 | (16.6) (78.3) | 53 | (14.9) (21.7) | |
| Gender (N = 1507) | 0.1653 | ||||||
| Male | 831 | (55.1) | 624 | (54.1) (75.1) | 207 | (58.5) (24.9) | |
| Female | 676 | (44.9) | 529 | (45.9) (78.3) | 147 | (41.5) (21.7) | |
| Education (in years) (N = 1477) | 0.8068 | ||||||
| Illiterate or < 6 years of age | 53 | (3.6) | 39 | (3.4) (73.6) | 14 | (4.1) (26.4) | |
| 0–6 | 748 | (50.6) | 582 | (51.1) (77.8) | 166 | (49.0) (22.2) | |
| 7–11 | 632 | (42.8) | 482 | (42.4) (76.3) | 150 | (44.2) (23.7) | |
| 12–19 | 44 | (3.0) | 35 | (3.1) (79.5) | 9 | (2.7) (20.5) | |
| Job (N = 1490) | < 0.0001 | ||||||
| Requires travel out of village | 286 | (19.2) | 188 | (16.5) (65.7) | 98 | (28.0) (34.3) | |
| Does not require travel out of village | 1204 | (80.8) | 952 | (83.5) (79.1) | 252 | (72.0) (20.9) | |
| Total number of malaria cases in lifetime (N = 1507) | < 0.0001 | ||||||
| Median (IQR) | 3.0 | (2–6) | 3.0 | (1-–5) | 4.0 | (2–8) | |
| Traveled before any of the last 4 episodes (N = 1446) | < 0.0001 | ||||||
| Yes | 349 | (24.1) | 216 | (19.8) (61.9) | 133 | (37.6) (38,1) | |
| No | 1097 | (75.9) | 876 | (80.2) (79.9) | 221 | (62.4) (20.1) | |
| Travel in past month (N = 1489) | < 0.0001 | ||||||
| No | 1305 | (87.6) | 1032 | (90.5) | 273 | (78.5) | |
| Yes | 184 | (12.4) | 109 | (9.5) | 75 | (21.5) | |
| Of those who traveled past month, number of trips (N = 184) | 0.4623 | ||||||
| Median (IQR) | 1,0 | (1–1.5) | 1.0 | (1–1) | 1.0 | (1–2) | |
| Traveled > 10 km, > 3days, not to Iquitos in the past month (N = 1490) | < 0.0001 | ||||||
| Yes | 159 | (10.7) | 95 | (8.3) (59.7) | 64 | (18.3) (40.3) | |
| No | 1331 | (89.3) | 1046 | (91.7) (78.6) | 285 | (81.7) (21.4) | |
IQR = interquartile range.
Recurrences of P. vivax infections after chloroquine (CQ)-primaquine (PQ) treatment were relatively common in the Peruvian Amazon. Despite the provision of standard therapy for P. vivax including CQ and PQ (0.5 mg/kg/day for 7 days) for all patients according to Peruvian Ministry of Health policy, during the study period, 354 out of 1,507 subjects (23.5%) developed at least one subsequent vivax malaria episode. Of these, 274 developed 1 subsequent episode, 60 developed 2 subsequent episodes, 15 developed 3 subsequent episodes, 4 developed 4 subsequent episodes, and 1 subject developed 5 subsequent episodes. The proportion of subjects who remained free of recurrence at the end of the follow up were 75.8% in San Jose Lupuna, 77.6% in Santa Tomas, 61.8% in Mazan, and 88.6% in Padrecocha (Table 2). Overall, there was a significantly higher risk of recurrences in Mazan compared with other villages (Santo Tomas, San Jose de Lupuna, and Padrecocha) (χ2 = 54.6, 1 degrees of freedom [df], P < 0.0001). Such differences were caused by differences in reinfection rates (χ2 = 18.95, 1 df, P < 0.0001) rather than relapse rates (χ2 = 0.17, 1 df, P = 0.68), which is expected provided that treatment is identical and that directly observed therapy was deployed in study sites.
PCR-RFLP and TR analysis of the Pvmsp-3α gene.
The Hha I and Alu I PCR-RFLP assays showed easily distinguishable restriction patterns in all samples. The PCR-RFLP patterns showed 500–600 bp for Alu I digests and the largest fragments between 950–1,100 bp for Hha I digests. These fragments and the smaller ones from 150–750 bp were analyzed by RFLP.
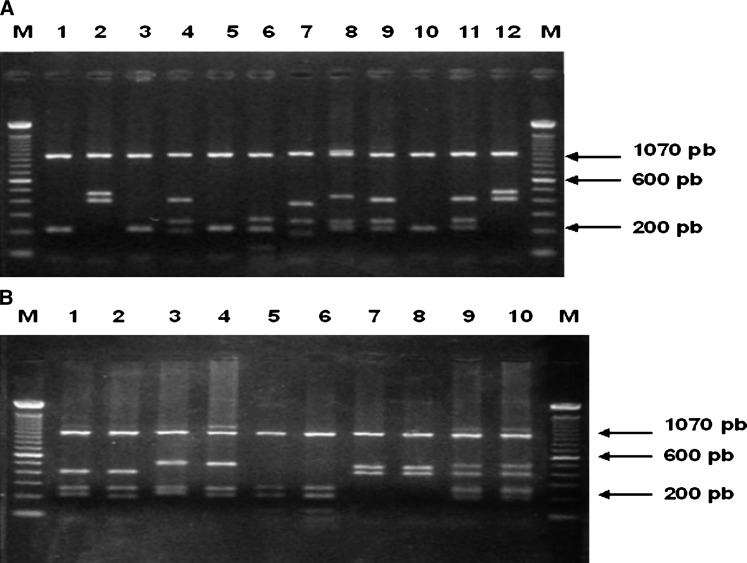
The RFLP analysis of MSP3-α PCR fragment produced 15 patterns after Hha I digestion (Table 3). From these 15 patterns, patterns 5, 11, 12, 14, and 15 were complex patterns; therefore, considered as mixed or composite P. vivax parasite samples, not unique patterns, because the sum of the restriction fragments exceeded the size of the primary product. Therefore, based on restriction patterns from digestion of PCR products with Hha I, 10 allele variants have been detected among the 354 patient-isolates (Figure 2 , Table 3).
Table 3.
MSP3α-HhaI PCR-restriction fragment length polymorphism alleles
| Hha I | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pattern | Allele | |||||||
| 1 | Allele 1 | 1070 | 431 | 275 | 207 | |||
| 2 | Allele 2 | 1070 | 530 | 275 | 236 | |||
| 3 | Allele 3 | 1070 | 275 | 223 | ||||
| 4 | Allele 4 | 1070 | 500 | 431 | ||||
| 5 | Complex | 1070 | 530 | 431 | 275 | 223 | ||
| 6 | Allele 5 | 1070 | 223 | |||||
| 7 | Allele 6 | 1070 | 431 | 275 | 223 | |||
| 8 | Allele 7 | 1070 | 385 | 275 | 188 | |||
| 9 | Allele 8 | 1070 | 475 | 275 | 204 | |||
| 10 | Allele 9 | 1070 | 343 | 275 | 223 | |||
| 11 | Complex | 1070 | 500 | 431 | 275 | 236 | ||
| 12 | Complex | 1070 | 603 | 501 | 426 | 275 | 223 | |
| 13 | Allele 10 | 1070 | 510 | 275 | 226 | |||
| 14 | Complex | 1070 | 460 | 431 | 275 | 188 | ||
| 15 | Complex | 1070 | 500 | 475 | 431 | 343 | 275 | 223 |
Figure 2.
Patterns after cutting with enzyme HhaI the gen coding MSP3α. from P. vivax (M: 100 bp DNA ladder) (A) Lines 1, 3, 5, and 10, pattern 1; line 6 pattern 2; line 7 pattern 3; line 8 pattern 4; line 4, 9 and 11 pattern 5; line 2 and 12 pattern 7. (B) Lines 1 and 2 pattern 6; lines 7 and 8 pattern 7; lines 3 and 4 pattern 8; lines 9 and 10 pattern 9; lines 5 and 6 pattern 2.
The RFLP analysis of PvMSP3-α PCR fragment produced 14 patterns after Alu I digestion. From these 14 patterns, patterns 6 and 10 were complex patterns; therefore, considered as mixed or composite P. vivax parasite samples, because the sum of the restriction fragments exceeded the size of the primary product. Therefore, based on restriction patterns from digestion of PCR products with Alu I, 12 allele variants have been detected among the 354 patient-isolates (Table 4).
Table 4.
MSP3α-AluI PCR-restriction fragment length polymorphism alleles
| Alu I | ||||||||
|---|---|---|---|---|---|---|---|---|
| Pattern | Allele | |||||||
| 1 | Allele 1 | 551 | 412 | 354 | 259 | 205 | 153 | |
| 2 | Allele 2 | 551 | 467 | 259 | 185 | 153 | ||
| 3 | Allele 3 | 551 | 398 | 349 | 259 | 192 | 173 | 153 |
| 4 | Allele 4 | 551 | 205 | 153 | ||||
| 5 | Allele 5 | 551 | 441 | 259 | 205 | 180 | 153 | |
| 6 | Complex | 551 | 464 | 412 | 293 | 259 | 192 | 153 |
| 7 | Allele 6 | 551 | 447 | 205 | 175 | 153 | ||
| 8 | Allele 7 | 551 | 405 | 205 | 175 | 153 | ||
| 9 | Allele 8 | 551 | 398 | 259 | 205 | 175 | 144 | |
| 10 | Complex | 551 | 441 | 354 | 293 | 259 | 205 | 153 |
| 11 | Allele 11 | 440 | 354 | 293 | 165 | 153 | ||
| 12 | Allele 12 | 551 | 354 | 259 | 205 | 173 | 153 | |
| 13 | Allele 13 | 523 | 253 | 198 | 144 | 130 | ||
| 14 | Allele 14 | 500 | 398 | 205 | 175 | 153 |
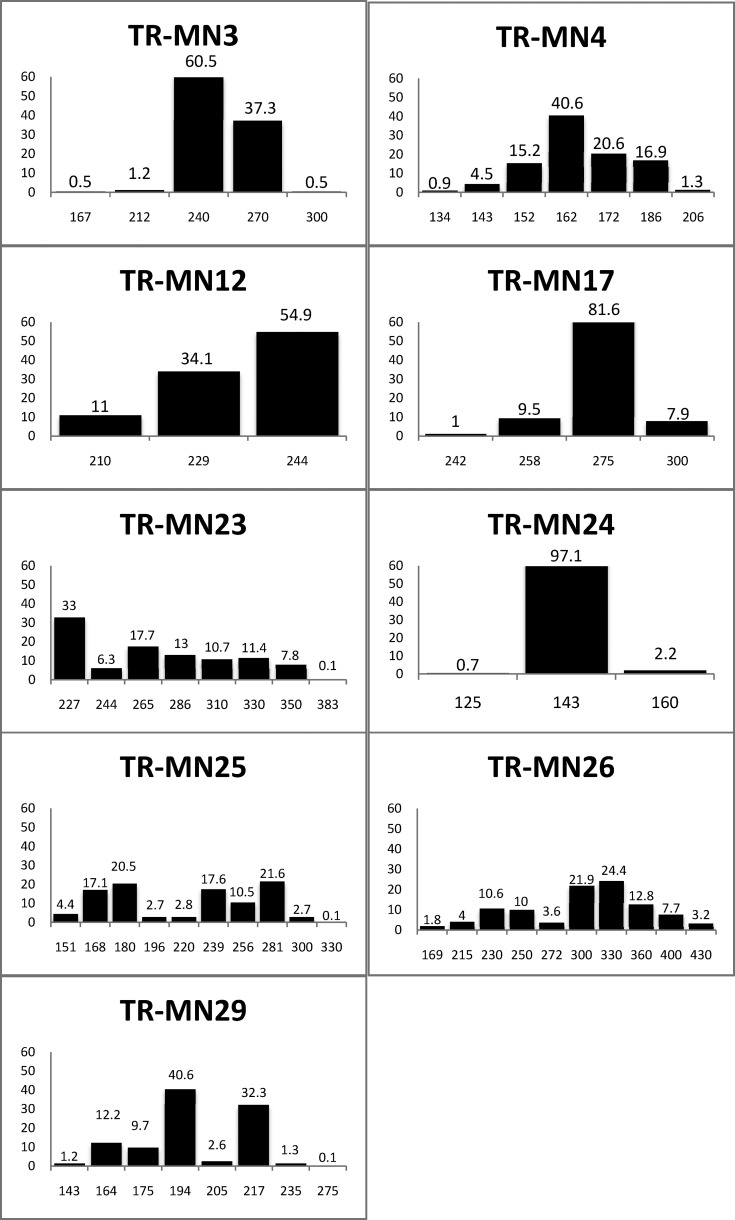
The overall diversity of P. vivax was determined on the basis of TR genotyping for the primary samples obtained from all study villages. The genetic diversity of each TR locus was 0.54 (MN3), 0.58 (MN24), 0.60 (MN17), 0.70 (MN4), 0.73 (MN12), 0.73 (MN19), 0.87 (MN23), 0.88 (MN25), and 0.88 (MN26) (Table 5, Figure 3 ). The allelic frequencies at each TR locus are shown in Figure 4 .
Table 5.
Schematic table for all nine tandem repeat markers used, allelic types found in the study, and gene diversity (heterozygosity)*
| Locus | MN3 | MN4 | MN12 | MN17 | MN23 | MN24 | MN25 | MN26 | MN29 |
|---|---|---|---|---|---|---|---|---|---|
| Alleles at each locus | 167 | 134 | 210 | 242 | 227 | 125 | 151 | 169 | 143 |
| 212 | 143 | 229 | 258 | 244 | 143† | 168 | 215 | 164 | |
| 240† | 152 | 244† | 275† | 265† | 160 | 180 | 230 | 175 | |
| 270 | 162†* | 300 | 286 | 196 | 250 | 194† | |||
| 300 | 172 | 310 | 220 | 272 | 205 | ||||
| 186 | 330 | 239 | 300 | 217 | |||||
| 206 | 350 | 256 | 330† | 235 | |||||
| 383 | 281† | 360 | 275 | ||||||
| 300 | 400 | ||||||||
| 330 | 430 | ||||||||
| %‡ | 57 | 41 | 34 | 51 | 26 | 47 | 21 | 21 | 42 |
| He§ | 0.54 | 0.70 | 0.73 | 0.60 | 0.87 | 0.58 | 0.88 | 0.88 | 0.73 |
Alleles were considered the same if molecular weights were within 5% of the band size.
Indicates the most common allele for that locus.
Frequency of most common allele for that locus.
Genetic diversity was assessed by measuring expected heterozygosity (He: chance of drawing 2 different alleles from a population sample) by use of the formula H = n/(n-1)(1-Σpi2), where p is the frequency of the ith allele and n is the sample size.
Figure 3.
MN23, this tandem repeat (TR) marker has 13 allelic types, e.g., allelic types: 1(227), 2(244), 345(227), 6(244), 7(227), 8(227), 9(315), 10(227), 11(265/227), 12(330/227), 13(310), 14(227), 15(227), 16(244), 17(315), and 18(350). Band sizes for TR markers were estimated by automated computational analysis using a 100 bp ladder (M) on each side of the gel as standard.
Figure 4.
Proportion of alleles of line tandem repeat (TR) markers in 814 case samples with Plasmodium vivax malaria.
Recurrences of P. vivax.
Of the 1,507 subjects enrolled with P. vivax malaria infection, 354 (23.5%) developed more than one vivax malaria episode; 1,153 subjects did not have another vivax malaria episode during the period of the study. Regarding the ability to differentiate between P. vivax genotypes, TR polymorphism analysis differentiated a higher proportion of subsequent infections than PCR-RFLP analysis alone (P < 0.05).
The odds of those enrolled with > 1 episode of P. vivax malaria, having increased lifetime malaria cases, compared with those who were enrolled with only one episode of P. vivax malaria, were 1.1 times higher for subjects within 5–14 years of age (P < 0.05) (95% CI 1.03, 1.19), and 1.05 times higher for subjects within 15–45 years of age (P < 0.05) (95% CI 1.01, 1.08) (Tables 6 and 7). The odds of developing > 1 episode of P. vivax malaria whether caused by relapse or reinfection were 2.6 times higher in the more remote village of Mazan than in villages closer to Iquitos (P < 0.001) (odds ratio [OR] = 2.6, 95% confidence interval [CI] 2.0, 3.4). There was no difference in age, gender, level of education, and time living in the village with respect to the odds of developing a subsequent vivax episode (Table 2). Significantly, occupation-related travel from a village (self-reported by the subject) was associated with a 2-fold elevated odds of developing more than one vivax malaria episode (whether caused by relapse or reinfection) during the study period (OR = 2.0, 95% CI 1.5, 2.6; P < 0.05) (Tables 2 and 8). People who developed a recurrent vivax malaria episode during the period of the study had a significantly higher median number of cumulative lifetime total of self-reported malaria cases than people who only developed one vivax episode in the study period (P < 0.001) (Table 2).
Table 6.
Comparison between relapse and reinfection subjects*
| Variable | Total (N = 336) | Relapse (N = 92) | Reinfection (N = 244) | P value | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Health Post (N = 336) | 0.2209 | ||||||
| SJL | 43 | (12.8) | 12 | (13.0) | 31 | (12.7) | |
| STO | 126 | (37.5) | 28 | (30.4) | 98 | (40.2) | |
| Mazan | 132 | (39.3) | 44 | (47.8) | 88 | (39.3) | |
| PAD | 35 | (10.4) | 8 | (8.7) | 27 | (10.4) | |
| Health Post (N = 336) | 0.0490 | ||||||
| STO, SJL, and PAD | 204 | (60.7) | 48 | (52.2) | 156 | (63.9) | |
| Mazan | 132 | (39.3) | 44 | (47.8) | 88 | (36.1) | |
| Time living in community (years) (N = 353) | 0.2014 | ||||||
| Median (IQR) | 10.0 | (4–20) | 9.0 | (3–18) | 10.0 | (4–20) | |
| Age (years) (N = 352) | 0.2301 | ||||||
| Mean (SD) | 27.4 | (16.2) | 29.0 | (17.3) | 26.6 | (15.6) | |
| 0.3264 | |||||||
| ≤ 4 | 13 | (3.9) | 5 | (5.4) | 8 | (3.3) | |
| 5–14 | 72 | (21.4) | 15 | (16.3) | 57 | (23.4) | |
| 15–44 | 201 | (59.8) | 55 | (59.8) | 146 | (59.8) | |
| ≥ 45 | 50 | (14.9) | 17 | (18.5) | 33 | (13.5) | |
| Gender (N = 336) | 0.5181 | ||||||
| Male | 195 | (58.0) | 56 | (60.9) | 139 | (57.0) | |
| Female | 141 | (42.0) | 36 | (39.1) | 105 | (43.0) | |
| Education (in years) (N = 322) | 0.1343 | ||||||
| Illiterate or < 6 years old | 14 | (4.4) | 6 | (6.9) | 8 | (3.4) | |
| 0–6 | 159 | (49.4) | 47 | (54.0) | 112 | (47.7) | |
| 7–11 | 142 | (44.1) | 34 | (39.1) | 108 | (45.7) | |
| 12–19 | 7 | (2.2) | 0 | (0.0) | 7 | (2.3) | |
| Job (N = 333) | 0.3877 | ||||||
| Requires travel out of village | 91 | (27.3) | 22 | (23.9) | 69 | (28.6) | |
| Does not require travel out of village | 242 | (72.7) | 70 | (76.1) | 172 | (71.4) | |
| Total number of malaria cases in lifetime (N = 353) | 0.3716 | ||||||
| Median (IQR) | 4.0 | (2–8) | 4.0 | (2–6.5) | 4.0 | (2–8) | |
| Traveled prior to any of the last 4 episodes (N = 336) | 0.5004 | ||||||
| Yes | 129 | (38.4) | 38 | (41.3) | 91 | (37.3) | |
| No | 207 | (61.6) | 54 | (58.7) | 153 | (62.7) | |
| Traveled in past month (N = 332) | 0.0272 | ||||||
| No | 258 | (77.7) | 64 | (69.6) | 194 | (80.8) | |
| Yes | 74 | (22.3) | 28 | (30.4) | 46 | (19.2) | |
| Of those who traveled in past month, number of trips (N = 76) | 0.3200 | ||||||
| Median (IQR) | 1.0 | (1–2) | 1.0 | (1–2) | 1.0 | (1–1) | |
| Traveled > 10 km, > 3 days, not to Iquitos in the past month (N = 332) | 0.3750 | ||||||
| Yes | 62 | (18.7) | 20 | (21.7) | 42 | (17.5) | |
| No | 270 | (81.3) | 72 | (78.3) | 198 | (82.5) | |
SLJ = San Jose de Lupuna; STO = Santa Tomas; PAD = Padrecocha; IQR = interquartile range.
Table 7.
Lifetime malaria cases for those enrolled with > 1 episode versus those enrolled with only 1 episode, stratified by age
| Age | (n) mean (IQR)* | OR† | 95% CI | P |
|---|---|---|---|---|
| < 5 | (46) 2 (0–3) | 1.035 | 0.803–1.335 | 0.7896 |
| 5–14 | (346) 2.9 (1–4) | 1.105 | 1.027–1.188 | 0.0072 |
| 15–44 | (844) 4.58 (2–6) | 1.046 | 1.013–1.080 | 0.0055 |
| > 45 | (244) 7.14 (3–10) | 1.019 | 0.970–1.070 | 0.4569 |
The total number of lifetime malaria cases stratified by age.
The odds of those enrolled with > 1 episode having increased lifetime malaria cases compared with those who were enrolled with only one episode.
Table 8.
Bivariate logistic regression modeling probability of > 1 infection, relapse episodes using ms3p α, and relapse episodes using tandem repeats*
| Variable | 1 infection vs. > 1 infection | Reinfection vs. relapse (Msp3 α) | Reinfection vs. relapse (tandem repeat) | |||
|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| Health Post | ||||||
| STO | 1.00 | 1.00 | 1.00 | |||
| SJL | 0.927 | 0.636–1.350 | 0.980 | 0.443–2.167 | 2.236 | 0.574–8.705 |
| Mazan | 2.054 | 1.532–2.753 | 1.603 | 0.923–2.785 | 3.131 | 1.092–8.973 |
| Padrecocha | 0.427 | 0.295–1.350 | 0.950 | 0.389–2.319 | 1.171 | 0.219–6.265 |
| Health Post | ||||||
| STO, SJL, and PAD | 1.00 | 1.00 | 1.00 | |||
| Mazan | 2.640 | 2.033–3.429 | 1.625 | 1.00–2.640 | 2.418 | 1.062–5.506 |
| Time living in community (years) | 0.994 | 0.986–1.003 | 0.991 | 0.974–1.009 | 0.983 | 0.951–1.017 |
| Age (years) | 1.002 | 0.995–1.009 | 1.009 | 0.994–1.024 | 0.993 | 0.967–1.019 |
| < 5 | 1.420 | 0.698–2.889 | 1.214 | 0.344–4.283 | 1.800 | 0.304–10.644 |
| 5–14 | 0.892 | 0.600–1.326 | 0.511 | 0.226–1.155 | 0.548 | 0.184–1.634 |
| 15–44 | 1.216 | 0.864–1.712 | 0.731 | 0.377–1.418 | 0.806 | 0.232–2.800 |
| ≥ 45 | 1.00 | 1.00 | 1.00 | |||
| Male Gender | 1.186 | 0.932–1.510 | 1.175 | 0.720–1.917 | 0.989 | 0.395–2.041 |
| Education (in years) (not enough subjects in 12–19) | ||||||
| N/A Under 6 years old | 1.154 | 0.610–2.182 | 2.382 | 0.772–7.349 | 6.129 | 1.372–27.377 |
| 0–6 | 0.917 | 0.713–1.179 | 1.333 | 0.797–2.230 | 1.959 | 0.768–4.995 |
| 7–11 | 1.00 | 1.00 | 1.00 | |||
| 12–19 | 0.826 | 0.388–2.182 | – | – | – | – |
| Job requires travel out of the village | 1.969 | 1.487–2.608 | 0.783 | 0.450–1.364 | 0.977 | 0.394–2.418 |
| Total number of malaria cases in lifetime | 1.042 | 1.018–1.066 | 0.985 | 0.930–1.043 | 0.970 | 0.879–1.071 |
| Traveled before any of the last 4 episodes | 2.411 | 1.856–3.133 | 1.183 | 0.725–1.930 | 1.375 | 0.604–3.128 |
| Times traveled in past month | 1.635 | 1.337–1.998 | 1.461 | 1.064–2.004 | 1.230 | 0.690–2.193 |
| Travel in past month | 2.601 | 1.883–3.592 | 1.845 | 1.067–3.193 | 1.882 | 0.777–4.559 |
| Of those who traveled in past month, times | 1.042 | 0.730–1.488 | 1.402 | 0.813–2.417 | 0.433 | 0.067–2.787 |
| Traveled > 10 km and > 3 days in past month | 2.473 | 1.755–3.484 | 1.310 | 0.721–2.379 | 2.225 | 0.914–5.413 |
| Household variables | ||||||
| Exterior wall materials | ||||||
| Brick/Cement | 1.00 | 1.00 | 1.00 | |||
| Wood | 1.121 | 0.761–1.653 | 2.231 | 0.935–5.324 | 1.202 | 0.334–4.329 |
| Other | 0.714 | 0.440–1.160 | 1.109 | 0.361–3.406 | 0.634 | 0.101–4.001 |
| Interior wall materials | ||||||
| Brick/Cement | 1.00 | 1.00 | 1.00 | |||
| Wood | 1.031 | 0.580–1.834 | 2.818 | 0.613–12.963 | – | – |
| Other | 0.722 | 0.401–1.302 | 2.257 | 0.476–10.704 | – | – |
| Location of kitchen | ||||||
| Exterior | 1.00 | 1.00 | 1.00 | |||
| Interior | 1.033 | 0.783–1.362 | 0.817 | 0.473–1.411 | 0.760 | 0.297–1.947 |
| Roof material | ||||||
| Calamine | 1.00 | 1.00 | 1.00 | |||
| Palm tree | 1.225 | 0.916–1.639 | 1.475 | 0.805–2.703 | 1.969 | 0.642–6.036 |
| Number bed nets/per bed | 0.998 | 0.777–1.282 | 1.758 | 0.943–3.277 | 1.642 | 0.995–2.710 |
| Possessions | ||||||
| Radio | 0.814 | 0.614–1.078 | 0.654 | 0.367–1.164 | 0.082 | 0.011–0.622 |
| Black and white TV | 0.785 | 0.531–1.160 | 1.081 | 0.514–2.276 | 1.633 | 0.366–7.300 |
| Color TV | 1.240 | 0.887–1.732 | 1.070 | 0.544–2.104 | 1.507 | 0.428–5.311 |
| VHS/DVD | 1.191 | 0.700–2.026 | 0.743 | 0.269–2.056 | 1.404 | 0.178–11.093 |
| Audio stereo | 0.743 | 0.404–1.367 | 0.746 | 0.247–2.257 | 0.971 | 0.120–7.853 |
STO = Santa Tomas; SJL = San Jose de Lupuna; PAD = Padrecocha.
The odds of developing > 1 episode of P. vivax malaria, whether caused by relapse or reinfection, were 2.6 times higher for subjects who had traveled in the prior month to any of their last four malaria episodes than in subjects without such travel (P < 0.05) (95% CI 1.9, 3.6) (Tables 2 and 8). The odds of developing > 1 episode of P. vivax malaria, whether caused by relapse or reinfection, were 2.5 times higher for subjects who had traveled > 10 Km and > 3 days in the prior month to episode than in subjects without such travel (P < 0.05) (95% CI 1.8, 3.5) (Tables 2 and 8). Household variables (exterior/interior wall materials, locations of kitchen, roof material, number of bed nets per bed, possessions) were found not to be different in any comparison between subjects with only 1 episode, subjects with > 1 episode, subjects with relapse, and subjects with reinfection) (Table 8). Multivariate logistic regression modeling probability of > 1 infection during study period showed that odds for subjects in Mazan (OR = 2.56), 15–44 years of age (OR = 1.5), traveling for job purposes (OR = 1.5), with increased lifetime malaria cases (OR = 1.1), and who traveled in the month before the malaria episode (OR = 1.5), were significantly higher than subjects without such profile (95% CIs 1.9, 3.5; 1.03, 2.2; 1.03, 2.1; 1.04, 1.1; 1.0, 2.1, respectively) (Table 9).
Table 9.
Multivariate logistic regression modeling probability of > 1 infection during study period*
| Variable | OR | 95% CI |
|---|---|---|
| Health Post | ||
| STO, SJL, and PAD | 1.00 | |
| Mazan | 2.559 | 1.866–3.510 |
| Age | ||
| < 5 | 1.683 | 0.749–3.780 |
| 5–14 | 1.488 | 0.949–2.333 |
| 15–44 | 1.489 | 1.031–2.152 |
| ≥ 45 | 1.00 | |
| Male gender | 0.891 | 0.679–1.168 |
| Job requires travel out of the village | 1.453 | 1.027–2.056 |
| Total number of malaria cases lifetime | 1.064 | 1.037–1.092 |
| Travel in past month | 1.464 | 1.001–2.140 |
OR = odds ratio; CI = confidence interval; STO = Santa Tomas; SJL = San Jose de Lupuna; PAD = Padrecocha.
Timing of P. vivax recurrences.
The median time interval between laboratory-confirmed P. vivax infections was 258.6 days in Santa Tomas, 244.5 days in San Jose Lupuna, 230.7, and 131.7 days in Mazan. More than a half of the recurrences were diagnosed up to 240 days: 54% in Santa Tomas, 60% in San Jose Lupuna, 56% in Padrecocha, and 85% in Mazan. Most of the recurrences were diagnosed up to 365 days after drug treatment: 72% in Santa Tomas, 75% in San Jose Lupuna, 74% in Padrecocha, and 97% in Mazan. Only five recurrences (two of them in Mazan, and one in each of the other sites) were diagnosed up to 28 days after treatment, and only one out of these five recurrences was found to be a relapse.
Comparison of relapse versus re-infection using PCR-RFLP versus TR markers.
The median number of times having traveled in the month previous to vivax malaria diagnosis was significantly higher in subjects who developed re-infections compared with the ones who relapsed with an identical genotype (P < 0.05) (Table 6). Regarding developing a subsequent relapse or re-infection, there was no difference between people who reported traveling further than 10 Km and for ≥ 3 days in the month before diagnosis and people who did not travel (OR = 2.2, 95% CI 0.9, 5.4; P = 0.38) (Table 6). Regarding developing a subsequent relapse or re-infection, there was no difference between people who travel out of village for work and people who do not (OR = 1.0, 95% CI 0.4, 2.5; P = 0.39) (Table 6). Of 814 infections, the different rates of multiple P. vivax genotypes were: 131 (16.1%) by TR-PCR, 37 (4.5%) by PvMSP-3α PCR-RFLP, and 153 (18.8%) if both typing schemes were used together.
Comparison of time intervals to relapse and to re-infection in all villages together.
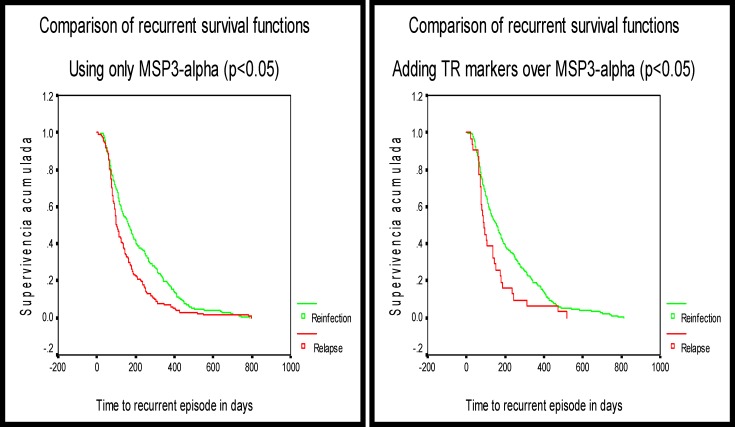
Using only PvMSP-3α markers, the risk of relapse was 1.5 higher than the risk of re-infection (rate ratio [RR] = 1.5, 95% CI 1.2, 2.0; P < 0.05). Adding TR markers, the risk of relapsing was 1.7 higher than the risk of a re-infection (RR = 1.7, 95% CI 1.2, 2.5; P < 0.05) (Figure 5).
Figure 5.
Comparison of recurrent survival functions for relapses (green solid line) and re-infections (red solid line) genotyping outcomes. Survival analysis techniques were used to compare the time interval for recurrent infections in relapsing and re-infection episodes, (A) Based on Merozoite Surface Protein-3α (MSP-3α) only and (B) adding the tandem repeat (TR) markers. In both cases the difference was statistically significant (P < 0.05) meaning there is a significant difference in the two survival recurrent functions when compared using either MSP-3α only or both MSP-3α/TR markers. Cox proportional hazard regression model.
Comparison of time intervals to relapse and to re-infection in Mazan.
In Mazan, using PvMSP-3α markers, the time to relapse versus re-infection showed a strong but not significant trend and was borderline not different (RR = 1.4, 95% CI 1.0, 2.0; P = 0.06). Adding TR markers, similar results were found (RR = 1.6, 95% CI 1.0, 2.7; P = 0.07).
Comparison of time intervals to relapse and to re-infection in the other villages.
In the non-Mazan villages, which are demographically different than Mazan, PvMSP-3α markers indicated that time intervals between relapse and re-infection were borderline significantly different. The relative risk of relapse was 1.47 times higher than re-infection (RR = 1.47, 95% CI 1.11, 1.94; P < 0.05). Adding TR markers, the risk of relapse was not different than the risk of a re-infection (P = 0.18).
Comparison of time intervals to relapse in Mazan versus the other villages.
People in Mazan were 2.4 times more likely to develop a recurrent vivax malaria caused by relapse (not caused by re-infection) than people in the other villages of the study (RR = 2.4, 95% CI 1.1, 5.5; P = 0.03).
Discussion
In a large, prospective study of P. vivax infection in diverse transmission settings in the Peruvian Amazon, the use of new molecular tools identified a high diversity of P. vivax parasites. Furthermore, combining the use of these molecular tools with socio-demographic analysis delineated a differential risk of relapse versus re-infection in distinct microgeographical settings within the Loreto region. These data are important to guide future surveillance and malaria intervention and control efforts not only in Peru but globally.
Molecular comparison of P. vivax parasites associated with relapses and those associated with the respective primary infections has been reported previously in a small number of patients: 6 patients in Canada who had acquired the infection in different areas of endemicity,16 10 patients in Brazil,24 and five patients in Thailand.25 Parasite diversity was assessed using molecular markers in the first two studies (pvcs and pvmsp1, and pvmsp-1 alone, respectively), whereas a panel of monoclonal antibodies was used in the third study. Although it was concluded that parasites associated with primary infection and those associated with relapse are usually of a similar genetic composition, evidence for novel genotypes in P. vivax associated with relapses was obtained for 24% of the patients (one of six patients in Canada,18 2 of 10 patients in Brazil, and 2 of 5 patients in Thailand.25 Another study showed relapses in 107 subjects (28 [78%] in Thailand, 55 [73%] in Myanmar, and 24 [63%] in India) with recurrent infections harboring none of the parasites that caused the primary infection; and concluded that activation of heterologous hypnozoite populations is the most common cause of first relapse in vivax malaria.14
This study was carried out to assess relapse versus reinfection in the Amazonian hypoendemic malaria setting by combining two molecular tools to characterize multiple episodes of vivax malaria. Given the high degree of internal mobility of people in the region, well-validated tools to determine the microgeographic characteristics of P. vivax geospatial transmission dynamics within the region will be essential for future malaria control efforts.
Previous studies have used molecular markers to distinguish relapse from reinfection but failed to take into account the field context of travel, comparison of villages, etc. Low transmission intensity but with high diversity is also an important finding of our study. Although some recrudescences could result from inadequate drug absorption or unusual disposition kinetics, recurrent infections in this study were newly acquired infections or relapses.26,27
The Pvmsp-3α polymorphism analysis has been an important molecular epidemiological tool for distinguishing P. vivax infections in malaria-endemic areas.18,28,29 This study used Pvmsp-3α PCR-RFLP and TR-PCR analysis as tools for differentiating primary from subsequent P. vivax infections. We found that the TR markers alone and in combination with the independent Pvmsp-3α locus were more highly discriminatory.
The TR markers used in this study are on a highly dynamic 100 kb chromosomal region where selective pressure is thought to be exerted because of the presence of the Pvcsp gene. Despite vivax malaria being hypoendemic at the population level in the Peruvian Amazon, considering that our study was done in a limited geographic area and time frame, in contrast to previous reports,21,30,31 our findings are consistent with a relatively high degree of P. vivax diversity in the Peruvian Amazon villages near to Iquitos city.
By Pvmsp-3α PCR-RFLP analysis, 12 different allelic variants were detected using the AluI PCR-RFLP and 11 allelic variants using HhaI. Using TR polymorphisms to analyze the same samples, a much greater degree of diversity was found allowing for greater discrimination within the patient groups. These data indicate that even within a region of hypoendemicity where little parasite outbreeding might be present, TRs are more useful and sensitive than the Pvmsp-3α markers. Previous studies have concluded that in some regions P. vivax populations emerging from hypnozoites commonly differ from the populations that caused the acute episode and that activation of heterologous hypnozoite populations is the most common cause of first relapse in patients with vivax malaria.16 Our data are consistent with this finding, because some of the apparent re-infections we found could be caused by relapse of a different clone acquired at the time of a primary infection or even afterward. Therefore, there is an inherent limitation of molecular epidemiological methods to differentiate infecting strains of P. vivax, highlighting the need of further understanding of the nature of relapses32 and the development of alternatives to PQ for radical treatment of hypnozoites.33
We have shown that by adding more polymorphic markers into the analysis, the proportion if identical genotypes may be even lower than what we found with msp3-alpha. For example, a similar study in Brazil34 has found very few 14-marker haplotypes in recurrent P. vivax infections using microsatellite typing. In this study, we considered a 5% tolerance in band sizes for TR markers to consider the possibility that relapsing parasites may be a minor, genetically different parasite clone that could have missed when typing parasites at the reference infection. Moreover, as shown in previous studies, we found 101 distinct molecular profiles among 110 patients2 meaning that here is a high degree of P. vivax diversity as demonstrated by our pool of TR markers,23 therefore, this is very unlikely that non-related parasites may have been considered identical after applying our 2-step tool of genotyping.
Relapse versus re-infection depending on socio-demographic context has important policy implications.
An important finding arising from this study is that people in a rural area of the Peruvian Amazon remote from the main city of Iquitos (Mazan) were significantly more likely to develop a subsequent episode caused by relapse (not to re-infection) than people in other villages where the socio-demographic context is different. Peruvian Ministry of Health policy indicates that anti-malarial drug therapy is to be directly observed. We suspect that the higher mobility of the more rural population (i.e., travel away from village for occupational reasons such as logging and forest extraction activities) make it likely both that patients might not receive the standard anti-P. vivax treatments compliantly, and also that this group of people might more likely to be re-infected. Other possible explanations of such phenomena could also be that relapse is stimulated by some activities or mechanisms related to the places where people travel or by mosquito bites.
In 2001, adherence problems to PQ led the Peruvian National Malaria Control Program to shorten the length of the treatment from a 14- to 7-day PQ course, but increasing the daily dose from 0.25 to 0.5 mg/kg/day. It was reported at the ASTMH 58th Annual Meeting that 7 days of PQ plus CQ proved as effective as the usual 14-day regimen. Another study by Solari and others35 during 1998–1999 found there was no difference between 7 days (10%) and 14 days (6.6%) of PQ. In this study, we found only 5 out of 354 subjects (1.4%) who experienced a recurrence during 28 days after treatment and no clinical resistance to malaria; therefore, we think resistance is not a problem with current treatment but drug resistance surveillance and research for new drugs are needed.
The data presented here showed an unexpectedly high proportion of infections caused by more than one parasite genotype. This is particularly remarkable given our current knowledge of transmission intensity and entomological inoculation rates in the Loreto region, and suggests that we lack critical knowledge of the micro-geography of malaria transmission at the actual places where people acquire their P. vivax infections. When two or more different genotypes or clones co-infect an individual or mosquito, cross-strain fertilization and genetic recombination are more likely to occur; therefore, it is quite possible that there is a higher risk subpopulation that makes a disproportionate contribution to the generation of high P. vivax genetic diversity. The present molecular epidemiology study and its development of high resolution tools provide new insights into the relationship of socio-demographic characteristics of malaria transmission dynamics in a low transmission/malaria hypoendemic region.
ACKNOWLEDGMENTS
We thank Paula Maguina for her research contributions that were essential to assure regulatory compliance, ethics, and logistical aspects of this project, and Amanda Rondinelli and Shira Abeles for their insightful discussions.
Footnotes
Financial support: This work was supported by the following grants from the United States Public Health Service, National Institutes of Health/National Institute of Allergy and Infectious Diseases: D43TW007120, K24AI068903, R01AI067727, U19AI089681. In addition, we are grateful for guidance and inspiration from Stephanie Brodine, John Weeks, and Richard Garfein, members of Dr. Chuquiyauri's thesis committee in the San Diego State University-University of California San Diego Doctoral Program in Global Health (supported by NIH grant R25TW007500).
Authors' addresses: Raul Chuquiyauri, ‘Alexander von Humboldt’ Tropical Medicine Institute, Cayetano Heredia Peruvian University, Lima, Peru, E-mail: rachuqui@ucsd.edu. Pablo Peñataro, Margaret Kosek, and Robert H. Gilman, Johns Hopkins Bloomberg School of Public Health, Department of International Health, Division of Global Disease Epidemiology and Control, Baltimore, MD, E-mails: pyori@jhsph.edu, mkosek@jhsph.edu, and rgilman@jhsph.edu. Manuel Fasabi, Maritza Calderon, and Sonia Torres, AB Prisma, Lima, Peru, E-mails: manuelmani1@gmail.com, mmcalderons@yahoo.es, and soniatadimi@yahoo.com. Kimberly C. Brouwer, Division of Global Public Health, School of Medicine, University of California San Diego, La Jolla, CA, E-mail: kbrouwer@ucsd.edu. Joseph M. Vinetz, Division of Infectious Diseases, Department of Medicine, University of California San Diego, La Jolla, CA, E-mail: jvinetz@ucsd.edu.
References
- 1.Guerra CA, Snow RW, Hay SI. Mapping the global extent of malaria in 2005. Trends Parasitol. 2006;22:353–358. doi: 10.1016/j.pt.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 3.Baird JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007;23:533–539. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 5.Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duarte EC, Gyorkos TW, Pang L, Abrahamowicz M. Epidemiology of malaria in a hypoendemic Brazilian Amazon migrant population: a cohort study. Am J Trop Med Hyg. 2004;70:229–237. [PubMed] [Google Scholar]
- 7.Pan American Health Organization, Health Information and Analysis . Health Situation in the Americas: Basic Indicators 2008. Washington, DC: 2008. [Google Scholar]
- 8.Flores-Mendoza C, Fernandez R, Escobedo-Vargas KS, Vela-Perez Q, Schoeler GB. Natural Plasmodium infections in Anopheles darlingi and Anopheles benarrochi (Diptera: Culicidae) from eastern Peru. J Med Entomol. 2004;41:489–494. doi: 10.1603/0022-2585-41.3.489. [DOI] [PubMed] [Google Scholar]
- 9.Schoeler GB, Flores-Mendoza C, Fernandez R, Davila JR, Zyzak M. Geographical distribution of Anopheles darlingi in the Amazon Basin region of Peru. J Am Mosq Control Assoc. 2003;19:286–296. [PubMed] [Google Scholar]
- 10.Branch O, Casapia WM, Gamboa DV, Hernandez JN, Alava FF, Ronca N, Alvarez E, Perez EJ, Gotuzzo E. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J. 2005;4:27. doi: 10.1186/1475-2875-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branch OH, Takala S, Kariuki S, Nahlen BL, Kolczak M, Hawley W, Lal AA. Plasmodium falciparum genotypes, low complexity of infection, and resistance to subsequent malaria in participants in the Asembo Bay Cohort Project. Infect Immun. 2001;69:7783–7792. doi: 10.1128/IAI.69.12.7783-7792.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roshanravan B, Kari E, Gilman RH, Cabrera L, Lee E, Metcalfe J, Calderon M, Lescano AG, Montenegro SH, Calampa C, Vinetz JM. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg. 2003;69:45–52. [PubMed] [Google Scholar]
- 13.Vittor AY, Gilman RH, Tielsch J, Glass G, Shields T, Lozano WS, Pinedo-Cancino V, Patz JA. The effect of deforestation on the human-biting rate of Anopheles darlingi, the primary vector of falciparum malaria in the Peruvian Amazon. Am J Trop Med Hyg. 2006;74:3–11. [PubMed] [Google Scholar]
- 14.Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, Nandy A, Guthmann JP, Nosten F, Carlton J, Looareesuwan S, Nair S, Sudimack D, Day NP, Anderson TJ, White NJ. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 15.Srivastava HC, Sharma SK, Bhatt RM, Sharma VP. Studies on Plasmodium vivax relapse pattern in Kheda district, Gujarat. Indian J Malariol. 1996;33:173–179. [PubMed] [Google Scholar]
- 16.Craig AA, Kain KC. Molecular analysis of strains of Plasmodium vivax from paired primary and relapse infections. J Infect Dis. 1996;174:373–379. doi: 10.1093/infdis/174.2.373. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, Escalante AA, Imwong M, Snounou G. The genetic diversity of Plasmodium vivax populations. Trends Parasitol. 2003;19:220–226. doi: 10.1016/s1471-4922(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 18.Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3alpha locus of Plasmodium vivax: global and local diversity. Am J Trop Med Hyg. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- 19.Feng X, Carlton JM, Joy DA, Mu J, Furuya T, Suh BB, Wang Y, Barnwell JW, Su XZ. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc Natl Acad Sci USA. 2003;100:8502–8507. doi: 10.1073/pnas.1232502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez JC, McNamara DT, Bockarie MJ, Baird JK, Carlton JM, Zimmerman PA. Identification of a polymorphic Plasmodium vivax microsatellite marker. Am J Trop Med Hyg. 2003;69:377–379. [PMC free article] [PubMed] [Google Scholar]
- 21.Karunaweera ND, Ferreira MU, Munasinghe A, Barnwell JW, Collins WE, King CL, Kawamoto F, Hartl DL, Wirth DF. Extensive microsatellite diversity in the human malaria parasite Plasmodium vivax. Gene. 2008;410:105–112. doi: 10.1016/j.gene.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 22.Nyachieo A, VAN Overmeir C, Laurent T, Dujardin JC, D'Alessandro U. Plasmodium falciparum genotyping by microsatellites as a method to distinguish between recrudescent and new infections. Am J Trop Med Hyg. 2005;73:210–213. [PubMed] [Google Scholar]
- 23.Kosek M, Yori P, Gilman RH, Calderon M, Zimic M, Chuquiyauri R, Jeri C, Pinedo V, Matthias MA, Llanos-Cuentas A, Vinetz JM. High degree of Plasmodium vivax diversity in the Peruvian Amazon demonstrated by tandem repeat polymorphism analysis. Am J Trop Med Hyg. 2012;86:580–586. doi: 10.4269/ajtmh.2012.11-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchgatter K, del Portillo HA. Molecular analysis of Plasmodium vivax relapses using the MSP1 molecule as a genetic marker. J Infect Dis. 1998;177:511–515. doi: 10.1086/517389. [DOI] [PubMed] [Google Scholar]
- 25.Khusmith S, Tharavanij S, Bunnag D. Antigenic disparity of Plasmodium vivax causing initial symptoms and causing relapse. Southeast Asian J Trop Med Public Health. 1998;29:519–524. [PubMed] [Google Scholar]
- 26.Pukrittayakamee S, Chantra A, Simpson JA, Vanijanonta S, Clemens R, Looareesuwan S, White NJ. Therapeutic responses to different antimalarial drugs in vivax malaria. Antimicrob Agents Chemother. 2000;44:1680–1685. doi: 10.1128/aac.44.6.1680-1685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pukrittayakamee S, Imwong M, Looareesuwan S, White NJ. Therapeutic responses to antimalarial and antibacterial drugs in vivax malaria. Acta Trop. 2004;89:351–356. doi: 10.1016/j.actatropica.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Zakeri S, Barjesteh H, Djadid ND. Merozoite surface protein-3alpha is a reliable marker for population genetic analysis of Plasmodium vivax. Malar J. 2006;5:53. doi: 10.1186/1475-2875-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruce MC, Galinski MR, Barnwell JW, Donnelly CA, Walmsley M, Alpers MP, Walliker D, Day KP. Genetic diversity and dynamics of Plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000;121:257–272. doi: 10.1017/s0031182099006356. [DOI] [PubMed] [Google Scholar]
- 30.Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, Menegon M, Severini C, Ayala FJ, Renaud F. Meager genetic variability of the human malaria agent Plasmodium vivax. Proc Natl Acad Sci USA. 2004;101:14455–14460. doi: 10.1073/pnas.0405186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen N, Auliff A, Rieckmann K, Gatton M, Cheng Q. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J Infect Dis. 2007;195:934–941. doi: 10.1086/512242. [DOI] [PubMed] [Google Scholar]
- 32.Collins WE. Further understanding the nature of relapse of Plasmodium vivax infection. J Infect Dis. 2007;195:919–920. doi: 10.1086/512246. [DOI] [PubMed] [Google Scholar]
- 33.Galappaththy GN, Omari AA, Tharyan P. Primaquine for preventing relapses in people with Plasmodium vivax malaria. Cochrane Database Syst Rev. 2007:CD004389. doi: 10.1002/14651858.CD004389.pub2. [DOI] [PubMed] [Google Scholar]
- 34.Orjuela-Sanchez P, da Silva NS, da Silva-Nunes M, Ferreira MU. Recurrent parasitemias and population dynamics of Plasmodium vivax polymorphisms in rural Amazonia. Am J Trop Med Hyg. 2009;81:961–968. doi: 10.4269/ajtmh.2009.09-0337. [DOI] [PubMed] [Google Scholar]
- 35.Lely Solari-Soto, Alonso Soto-Tarazona, Daniel Mendoza-Requena, Alejandro Llanos-Cuentas. Ensayo clinico del tratamiento de la malaria vivax con esquema acortado de primaquina comparado con el esquema tradicional. Rev Per Soc Med Intern. 2002;15:4. [Google Scholar]