Abstract
The flea-borne rickettsioses murine typhus (Rickettsia typhi) and flea-borne spotted fever (FBSF) (Rickettsia felis) are febrile diseases distributed among humans worldwide. Murine typhus has been known to be endemic to Kenya since the 1950s, but FBSF was only recently documented in northeastern (2010) and western (2012) Kenya. To characterize the potential exposure of humans in Kenya to flea-borne rickettsioses, a total of 330 fleas (134 pools) including 5 species (Xenopsylla cheopis, Ctenocephalides felis, Ctenocephalides canis, Pulex irritans, and Echidnophaga gallinacea) were collected from domestic and peridomestic animals and from human dwellings within Asembo, western Kenya. DNA was extracted from the 134 pooled flea samples and 89 (66.4%) pools tested positively for rickettsial DNA by 2 genus-specific quantitative real-time PCR (qPCR) assays based upon the citrate synthase (gltA) and 17-kD antigen genes and the Rfelis qPCR assay. Sequences from the 17-kD antigen gene, the outer membrane protein (omp)B, and 2 R. felis plasmid genes (pRF and pRFd) of 12 selected rickettsia-positive samples revealed a unique Rickettsia sp. (n=11) and R. felis (n=1). Depiction of the new rickettsia by multilocus sequence typing (MLST) targeting the 16S rRNA (rrs), 17-kD antigen gene, gltA, ompA, ompB, and surface cell antigen 4 (sca4), shows that it is most closely related to R. felis but genetically dissimilar enough to be considered a separate species provisionally named Candidatus Rickettsia asemboensis. Subsequently, 81 of the 134 (60.4%) flea pools tested positively for Candidatus Rickettsia asemboensis by a newly developed agent-specific qPCR assay, Rasemb. R. felis was identified in 9 of the 134 (6.7%) flea pools, and R. typhi the causative agent of murine typhus was not detected in any of 78 rickettsia-positive pools assessed using a species-specific qPCR assay, Rtyph. Two pools were found to contain both R. felis and Candidatus Rickettsia asemboensis DNA and 1 pool contained an agent, which is potentially new.
Key Words: Rickettsia, Fleas, PCR, Multilocus sequence typing, Surveillance
Introduction
Rickettsiae are obligate intracellular gram-negative bacteria that are associated with various arthropod vectors, including ticks, mites, lice, and fleas, and can cause mild to life-threatening human disease (Kelly et al. 2002). The first rickettsia associated with fleas was Rickettsia typhi, the causative agent of murine typhus. R. typhi, a typhus group (TG) rickettsia, has been most commonly associated with Xenopyslla cheopis (oriental rat flea), although at least 10 other species of fleas have been found to be naturally or experimentally infected with R. typhi. In addition, other arthropods (lice and mites) have been found to be infected with R. typhi (Azad 1990). Murine typhus is endemic to Kenya, but at a low level, especially in rural areas (Heisch and Harvey 1959, Maina et al. 2012). During the early 1960s it was reported that of batches of the fleas Ctenopthalmus cabirus, Dinopsyllus lypusus, and X. cheopis collected from wild rodents only X. cheopis were infected with R. typhi (Heisch et al. 1962), although in experimental infections both X. cheopis and Xenopsylla braziliensis could be infected equally well with R. typhi in the laboratory (Heisch 1969). More recently a serosurvey conducted in western Kenya showed that of a randomly selected subset of stored sera (n=357) collected from January, 2007, through October, 2008, from patients who presented to a rural health clinic with respiratory illness, jaundice, and acute febrile illness with or without a clinically apparent cause, 5 (1.4%) of the serum specimens contained immunoglobulin G (IgG) antibodies that reacted against R. typhi, as determined by an indirect fluoresence antibody assay (IFA) (Maina et al. 2012). This low prevalence of antibodies reactive to typhus group rickettsiae corroborates the earlier assertion by Heisch and Harvey (1959) that murine typhus is rare in Kenya, especially in rural areas.
A second rickettsia associated with fleas, Rickettsia felis, was discovered in Ctenocephalides felis during the 1990s (Adams et al. 1990, Azad et al. 1992). R. felis, initially characterized as a TG rickettsia because of its reactivity to guinea pig antiserum against R. typhi, was subsequently placed in the spotted fever group (SFG) of rickettsiae due to presence of ompA and phylogenetic studies. More recently, some have further separated R. felis from both the TG and SFG rickettsiae into a transition group of rickettsiae (Gillespie et al. 2007). It was successfully cultivated in the XTC-2 cell line derived from Xenopus laevis and fully described in 2001 (Raoult et al. 2001). Early on, R. felis was determined to be a human pathogen (Shriefer et al. 1994) causing flea-borne spotted fever (FBSF) that now is frequently diagnosed around the world, especially in sub-Saharan Africa (Parola 2011). Two recent reports from our group have shown FBSF due to infection with R. felis to be endemic to both the northeastern and western regions of Kenya (Richards et al. 2010, Maina et al. 2012). Risk factors of flea-borne rickettsioses in humans have not been fully characterized in those areas.
To identify the potential causative agent(s) of human flea-borne rickettsioses, we surveyed fleas collected from domestic and peridomestic animals and village homes for rickettsial agents. R. felis and a new rickettsia, Candidatus Rickettsia asemboensis, were identified among the fleas collected in Asembo, western Kenya.
Materials & Methods
Study site
The study was conducted in the Asembo division of Rarieda District, Nyanza Province in western Kenya. This rural site on the eastern shore of Lake Victoria falls within a health and demographic surveillance system (HDSS) run by the Kenya Medical Research Institute (KEMRI) and US Centers for Disease Control and Prevention (CDC) (Adazu et al. 2005). The Kenyan International Emerging Infections Program of KEMRI/CDC has conducted population-based infectious disease surveillance (PBIDS) of the human population in Asembo since late 2005 (Feikin et al. 2011). The PBIDS population is approximately 25,000 people living in 33 villages. The Asembo area is approximately 225 km2 and is culturally homogeneous, with 95% of people being ethnically Luo. It has a bimodal rainfall pattern, with rain seasons occurring from March to May and from September to November. The community lives in dispersed settlements, and houses are made of mud, cement, or brick, with roofs of iron sheet or thatch (Bigogo et al. 2010). The primary economic livelihood is subsistence farming and fishing. According to the demographic surveillance system (DSS) 2007/2008 census, the mean number of animals per compound was 2.6 cattle, 3.3 small ruminants, and 11 poultry (KEMRI/CDC, unpublished data).
Collection of fleas from domestic and peridomestic animals and human households
Three hundred livestock-owning compounds (LOC) were randomly selected from all known LOCs (n=4528) among the 33 PBIDS villages from January through May, 2009. The sampling frame of LOCs was compiled from livestock census data collected by the HDSS. A LOC was defined by ownership of 1 or more of the following animals—cattle, sheep, goats, and chickens. Where present, a maximum of 3 dogs and cats in each compound were surveyed for fleas. Fleas were also collected from 16 randomly selected houses from the 300 LOCs using light traps. The light traps were homemade and consisted of a hurricane lamp that was suspended a few inches above a tray containing water with dish soap and Vaseline smeared on the sides to prevent fleas from crawling out.
Additionally, peridomestic small mammal sampling was conducted in collaboration with the National Museums of Kenya using baited Sherman live traps over the period July 30 to August 7, 2009. Fifty compounds were randomly selected from the 300 LOCs. Informed consent for participation was obtained from household representatives prior to collection of animals. Five traps were placed within dwellings, in outbuildings, and outdoors in each compound for 4 consecutive nights, and the GPS locations of key points were recorded. Captured animals were euthanized using an overdose of halothane inhalant anesthesia (Rhodia Limited, Avonmouth, Bristol, BS119YF, UK). Fleas were collected from the animals by combing. Animal species identification was made on the basis of morphometric data at the National Museums of Kenya, where all specimens were submitted for archiving.
All fleas were preserved in 70% ethanol until delivery to the laboratory and then stored at −80°C. Fleas were identified using the entomological taxonomic keys of Segerman (1995), then pooled by individual host animal, method of collection, and flea species (49.3% of pools contained a single flea, the remainder contained 2–14 fleas per pool).
DNA extraction and rickettsiae detection from fleas
Fleas were washed in molecular-grade water and mechanically disrupted using a bead mill (QIAGEN TissueLyser LT, Valencia, CA). Genomic DNA was extracted using QIAamp blood and tissue kits (QIAGEN) according to the manufacturer's instructions, using a final elution volume of 50 μL.
A qPCR assay that amplifies and detects a 74-bp segment of the citrate synthase (gltA) gene (Stenos et al. 2005) was used to screen flea specimens for the presence of rickettsial DNA. A second Rickettsia genus-specific qPCR assay that amplifies and detects a 115-bp segment of the 17-kD antigen gene (Rick17b) (Jiang et al. 2012) was used to confirm the results.
Identification of rickettsiae in fleas by species-specific qPCR assays
To identify which rickettsiae were present in the rickettsia-positive flea pools, DNA samples were subsequently tested by 2 species-specific qPCR assays: (1) The Rfelis qPCR assay, in which a 129-bp fragment of the R. felis ompB was amplified (Henry et al. 2007), and (2) the Rtyph qPCR assay, in which a 122-bp fragment of the R. typhi ompB was amplified (Henry et al. 2007).
Confirmation of rickettsial species and identification of new rickettsiae by multilocus sequence typing
To confirm the identity of the flea-borne rickettsiae, PCR and sequencing were performed on a subset of selected samples using the 17-kD antigen gene, ompB and R. felis plasmid genes pRF and pRFδ. At least one of these positive samples was randomly selected from each of the different arthropod vectors, hosts, and method of collection.
To identify novel rickettsiae, we conducted multilocus sequence typing (MLST) using segments of 5 rickettsial genes, rrs, gltA, ompB, ompA, and sca4 (Fournier et al. 2003). Standard and nested PCR were carried out using primers and procedures previously described (Roux and Raoult 2000, Jiang et al. 2005) and new primers listed in Table 1. No positive controls were used in these PCR and nested PCR procedures to decrease chances of contamination; however, a negative control (molecular biology-grade water; Life Technologies) was run with the samples, and they were consistently negative in all runs.
Table 1.
New Primers Used for PCR and Sequencing
| Gene | Primer | Sequence (5′ to 3′) |
|---|---|---|
| rrs | 16SU17F | AGAGTTTGATCCTGGCTCAG |
| 16sR34F | CAGAACGAACGCTATCGGTA | |
| 16SU1592R | AGGAGGTRATCCAGCCGCA | |
| 16sOR1198R | TTCCTATAGTTCCCGGCATT | |
| 17 kD | R17k31F | GCTCTTGCAGCTTCTATGTTACA |
| R17k469R | ACTTGCCATTGTCCGTCAGGTTG | |
| Rr17k2608RN | CATTGTCCGTCAGGTTGGCG | |
| ompA | RompAM50F | TTGCGTTATAACACTTTTTAAGTGA |
| RompA642R | ATTACCTATTGTTCCGTTAATGGCA | |
| ompB | ompB1570R | TCGCCGGTAATTRTAGCACT |
| Rf1524R | CACACCCGCAGTATTACCGTT |
PCR products were purified using a QIAquick PCR purification kit (QIAGEN). Sequencing reactions were performed for both DNA strands using a Big Dye Terminator v3.1 Ready Reaction Cycle Sequencing Kit (Life Technologies) according to the manufacturer's instructions. After purification of sequenced products using Performa DTR Gel Filtration Cartridges (Edge BioSystems, Gaithersburg, MD), sequencing was run in an ABI Prism 3130xl Genetic Analyzer (Life Technologies). Sequences were assembled with Vector NTI Advance 11 software (Life Technologies), and BLAST searches were performed on the National Center for Biotechnology Information website (http://blast.ncbi.nlm.hih.gov/).
Phylogenetic analyses were performed using MEGA version 5 (Tamura et al. 2011) based on the multialignment of rrs, gltA, ompA, ompB, and sca4 sequences from the flea isolates and other rickettsial isolates from GenBank. The phylogenic trees were constructed using the neighbor-joining method, and the bootstrap analyses were performed with 1000 replications.
Development of the agent-specific qPCR assay (Rasemb) for the new flea-borne rickettsia, Candidatus R. asemboensis
A 4311-bp fragment of ompB amplified from Candidatus R. asemboensis was aligned with the ompB from 28 different Rickettsia species obtained from GenBank using MEGA version 5; a unique 24-bp sequence fragment was identified to be the target of the probe. To insure the specificity of the assay, the probe Rasem2893BP (6-FAM-CCGCAGCTCCAATAC CTTCGCCTAAGCCATATGCGG-BHQ-1) was designed as a molecular beacon with the first and the last 6-bp sequences as the stem. The primers Rasem2828F (5′-CACACTTAGCGG CGGTATTC) and Rasem2939R (5′-AAGTTGTTATAGTCTG TAGTAAACG) amplified a 112-bp fragment of ompB.
The concentrations of the primers, the probe, and MgCl2, were optimized, as well as the annealing temperature using Platinum Quantitative PCR SuperMix-UDG (Life Technologies), and run on a StepOnePlus Real-Time PCR platform (Life Technologies). Each 25-μL reaction contained 0.7 μM of each forward and reverse primer, 0.3 μM of probe, and 7 mM of MgCl2. Two microliters of template DNA was used in each reaction. The cycler parameters included incubation for 2 min at 50°C, initial denaturation for 2 min at 95°C and 45 cycles of denaturation for 5 s at 95°C, and annealing/elongation for 30 s at 59°C.
The specificity of Rasemb assay was confirmed by testing a panel of bacterial nucleic acid preparations (Jiang et al. 2012) representing genetically closely and distantly related bacteria, including R. felis URRWXCal2 and 19 other Rickettsia species and 12 nonrickettsial bacteria.
GenBank accession numbers
The sequences of the new rickettsial molecular isolates (F30, obtained from Ctenocephalides canis; F82, obtained from C. felis) have been deposited in GenBank with accession numbers JN315967 to JN315972 and JN315973 to JN315977 for rrs, gltA, 17-kD antigen, sca4, ompA, and ompB, respectively.
Ethical review
The collection of arthropod specimens from human dwellings and animals was approved by the KEMRI and CDC Animal Care and Use Committees (protocols nos. 1191 and 1562BRETBDX, respectively). Written informed consent was obtained from all animal owners before specimen collection.
Results
Vertebrate hosts and residences surveyed:
Of the 300 LOCs identified, 149 and 26 compounds owned at least 1 dog or a cat, respectively. Fleas were searched for on 299 domestic dogs and 26 cats from a total of 149 compounds. Thirty-nine (17%) dogs and 9 (34.6%) cats were found to be infested with fleas. Other domestic animals noted but not surveyed for fleas at the 300 LOC included cattle (n=463), goats (n=378), sheep (n=159), and chickens (n=760).
One or more peridomestic small mammals were trapped at 27 of the 50 compounds (54%). Fifty-five rodents and shrews were trapped from a total of 1142 trap placements (4.8%) and 18 (32.7%) were found to host fleas. The 55 peridomestic mammals were identified as Crocidura oliveiri (n=20), Rattus rattus (n=17), Mastomys natalensis (n=15), Lemniscomys striatus (n=2), and Mus minutoides (n=1).
All of the 16 randomly selected LOC dwellings were infested with fleas. A total of 88 fleas were trapped, with an average of 5.5 fleas/home (range 1–26 fleas) during a single night's collection using a single light trap/household.
Invertebrate hosts and rickettsia detection
A total of 330 fleas were collected and identified as 5 flea species: X. cheopis (n=77), C. canis (n=7), C. felis (n=193), Echidnophaga gallinacea (n=45), and Pulex irritans (n=8). DNA preparations were obtained from 134 flea pools, of which 66 (49.3%) consisted of a single individual flea. Overall 89 of 134 (66.4%) flea pools were positive for rickettsiae by using gltA, Rick17b, and Rfelis qPCR assays, which included all 4 (100%) C. canis pools, 75 (100%) C. felis pools, 8 (22.9%) X. cheopis pools, 1(20%) P. irritans pool, and 1(6.7%) E. gallinacea pool (Table 2). Seventy-eight of the 89 rickettsia-positive pools were tested by the R. typhi qPCR assay, and all of those pools were negative.
Table 2.
Rickettsia qPCR Results by Flea Species
| Number of pools | Number of positive gltA and Rick17 qPCR | Percent positive | |
|---|---|---|---|
| Dogs (n=39 dogs)a | |||
| Ctenocephalides felis | 58 | 58 | 100 |
| Echidnophaga gallinacea | 3 | 0 | 0 |
| Cats (n=9 cats) | |||
| Ctenocephalides canis | 1 | 1 | 100 |
| Ctenocephalides felis | 5 | 5 | 100 |
| Echidnophaga gallinacea | 8 | 1 | 12.5 |
| Rodents (n=18 rodents) | |||
| Echidnophaga gallinacea | 2 | 0 | 0 |
| Xenopsylla cheopis | 26 | 7 | 26.9 |
| Light trap (n=16 households) | |||
| Ctenocephalides canis | 3 | 3 | 100 |
| Ctenocephalides felis | 12 | 12 | 100 |
| Echidnophaga gallinacea | 2 | 0 | 0 |
| Pulex irritans | 5 | 1 | 20 |
| Xenopsylla cheopis | 9 | 1 | 11.1 |
| Total | 134 | 89 | 66.4 |
For each species, n represents the number of animals or houses.
Confirmation of rickettsial identification by 17-kD antigen gene, ompB, pRF, and pRFδ PCR sequencing
To confirm the identity of the rickettsiae, 12 of the 89 rickettsia-positive samples were selected and assessed by PCR and sequencing of 17-kD antigen gene and/or ompB, as well as the plasmid genes pRF and pRFδ. A 390-bp fragment of 17-kD antigen gene and/or a 599-bp fragment of ompB were amplified from all 12 samples. The sequences of 17 kD and ompB from 1 of the 12 samples was 100% identical to R. felis URRWXCal2 (GenBank accession no. CP000053). The R. felis-positive sample was further tested for the presence of plasmids. The plasmid pRF, but not the pRFδ, was detected by PCR in this sample. The 429-bp pRF amplicon was sequenced and was determined to be 100% identical to that of R. felis URRWXCal2. The sequences from the other 11 samples were identical to each other for both genes. BLAST searches revealed a unique sequence of ompB that was different from all rickettsial ompB sequences in GenBank. However, the 371-bp fragment of the 17-kD gene sequence was identical (100%) to Rickettsia sp. SE313 isolated from E. gallinacea in Egypt. The plasmids pRF and pRFδ were not detected in any of these 11 samples.
MLST results (rrs, gltA, ompB, ompA, sca4)
To determine the identification of and to more fully characterize the novel flea-borne rickettsial agent, sequences for rrs, gltA, ompA, ompB, and sca4, from 6 of the 11 novel rickettsia-positive samples were determined and compared to those available on GenBank. The sequences of the 1395-bp fragment of rrs and 1130-bp of gltA from the new isolate were closely related to R. felis URRWXCal2 (99.5% and 98.0%, respectively) and R. sp. RF2125 (99.6% for gltA only), indicating that this new isolate belongs within the genus Rickettsia. A 1517-bp fragment of ompA (3′ end) was amplified, providing evidence that the new molecular isolate is a member of the SFG of rickettsiae. The fragment of the 5′ end of ompA was not amplified by a SFG universal primer set, which suggests that the gene might be truncated or the sequence was not recognized by 1 or both of the primers. The sequence of the 1517-bp fragment of ompA was most similar to R. felis URRWXCal2 (92.3%). The 4311-bp fragment of ompB sequence from the new agent was most similar to R. felis URRWXCal2 (94.8%), although the sequence of a 790-bp fragment was most similar to an ompB sequence (P. Parola, unpubl. data; GenBank #JX183538) from Rickettsia RF2125 (99.7%), suggesting a very close relationship between these 2 new flea-borne rickettsiae. The sequence of a 1034-bp fragment of sca4 from the novel rickettsia was most similar to R. felis URRWXCal2 (95.5%). Because of the nucleotide dissimilarity between this new agent and the most homologous validated species R. felis, this novel agent should be considered for further studies to characterize it as a new Rickettsia species. Therefore, we propose this isolate be identified as Candidatus R. asemboenis until it is grown in culture and its biology is described more completely.
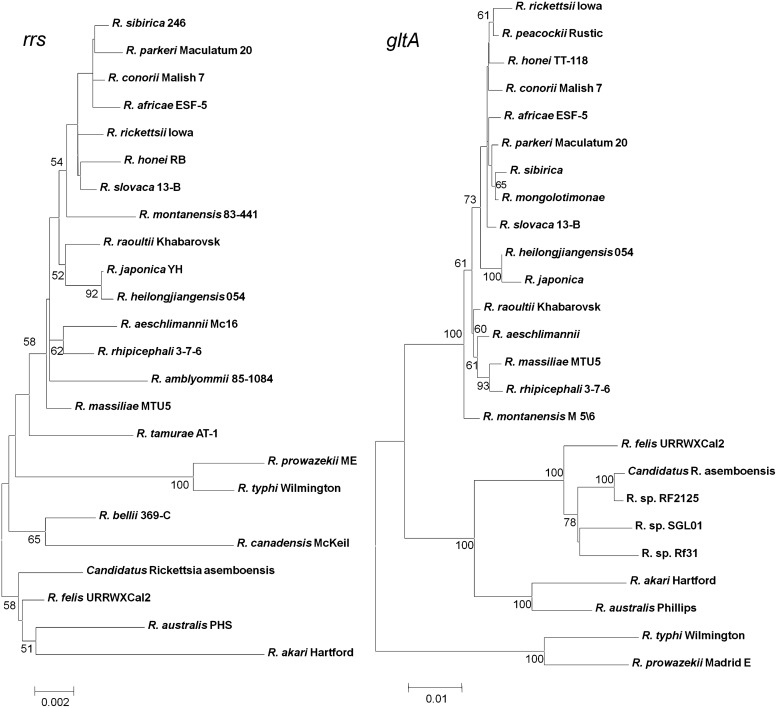
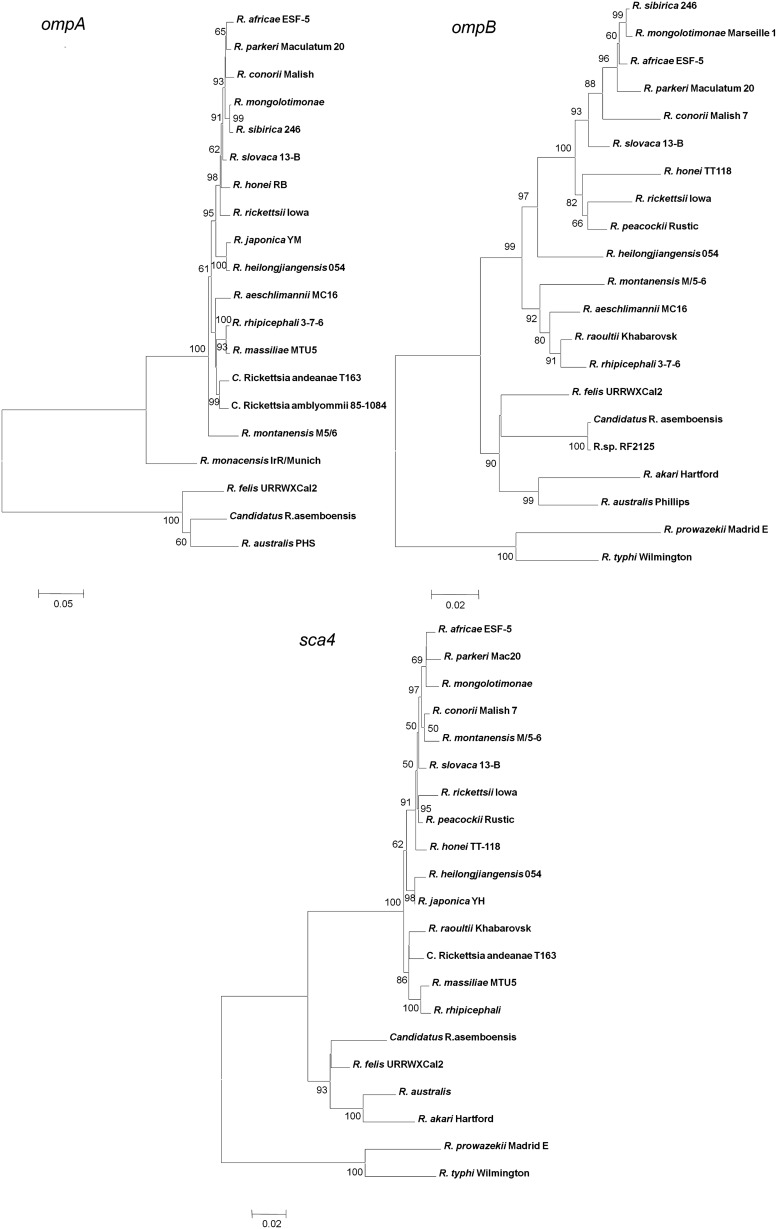
Phylogentic relationships between Candidatus R. asemboensis and other validated rickettsial species were analyzed by constructing phylogentic trees using rrs (1395 bp), gltA (1130 bp), ompA (1517 bp), ompB (1481 bp), and sca4 (1034 bp). Candidatus R. asemboensis was either clustered with or placed closely to R. felis URRWXCal2 for all 5 genes (Fig. 1). Candidatus R. asemboensis closely related to R. felis matched up with the results from the blast searches.
FIG. 1.
Phylogenetic trees from 5 genes. Phylogenetic relationships: Candidatus Rickettsia asemboensis with other rickettsiae; trees were constructed for rrs (1395 bp), gltA (1130 bp), ompB (1481bp), ompA (1517 bp), and sca4 (1034 bp) using maximum composite likelihood model, neighbor-joining method (MEGA software).
Development of the Rasemb qPCR assay
In determining the specificity of the Rasemb qPCR assay, it was found that only the 6 MLST-identified Candidatus R. asemboensis DNA preparations were positive, whereas R. felis and 19 near-neighbor rickettsial DNA preparations (Jiang et al. 2012) were negative.
Subsequently the Rasemb qPCR assay was used to assess all 134 DNA preparations. All 45 of the rickettsia-negative preparations were also negative for the Rasemb qPCR assay. Of the 89 rickettsia-positive samples, 81 samples (which include the 6 MLST-confirmed Candidatus R. asemboensis DNA preparations and 2 pools that had both Candidatus R. asemboensis and R. felis) were positive (Table 3) and 8 were negative. The identities of the 8 negative samples were determined by PCR and sequencing a 599-bp fragement of ompB. Seven of the 8 were determined to be 100% identical to R. felis. One sample's ompB sequence (from a DNA preparation of F28-a pool of 13 X. cheopis), was determined to be a unique rickettsial sequence that had only 98.3% similarity to R. felis and 92.0% similarity to Candidatus R. asemboensis (GenBank # JX183537). Thus, in addition to our near-neighbor panel of DNA preparations, the Rasemb qPCR assay was shown to be specific to Candidatus R. asemboensis using this study's sample of flea DNA preparations.
Table 3.
Rickettsia Detected in 5 Flea Species from Asembo
| |
Female |
Male |
|
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Flea species | No. pool | R. felis | Candidatus R. asemboensis | R. felis and R. asemb | No. pool | R. felis | Candidatus R. asemboensis | R. spp F28 | No. pool | SumPositive % |
| Ctenocephalides canis | 1 | 1 | 3 | 3 | 4 | 100 | ||||
| Ctenocephalides felis | 43 | 42 | 1 | 32 | 3 | 29 | 75 | 100 | ||
| Echidnophaga gallinacea | 10 | 1 | 5 | 1 | 6.7 | |||||
| Xenopsylla cheopis | 20 | 2 | 3 | 1 | 15 | 1 | 1 | 8 | 22.9 | |
| Pulex irritans | 3 | 2 | 1 | 1 | 20 | |||||
| Total | 77 | 3 | 46 | 2 | 57 | 4 | 33 | 1 | 89 | 66.4 |
Discussion
With the recently determined risk of FBSF to citizens of and travelers to western Kenya (Maina et al. 2012), we investigated the breadth, diversity, and prevalence of flea vectors containing R. felis in Asembo, because effective mitigation measures would certainly depend upon informed preventive medicine decisions. Of 300 LOCs selected, 149 had 1 or more dogs and cats, vertebrate hosts to C. felis, C. canis, and P. irritans, fleas known to be infected with R. felis (Venzal et al. 2006). Due to the ubiquitous nature of peridomestic rodents also known to harbor fleas infected with R. felis (Jiang et al. 2006), we included them in our investigation. Therefore, we surveyed from 300 randomly selected LOCs 50 compounds for fleas from domestic and peridomestic mammals that might harbor fleas infected with R. felis. From these locations 299 dogs, 26 cats, and 55 rodents were obtained and searched for fleas. In addition, 16 randomly selected residences were assessed for the presence of rickettsia-containing fleas by use of a light trap. Results showed that, 17% dogs (39/299), 34.6% cats (9/26), 32.7% rodents (18/55), and 100% homes (16/16) contained arthropod vectors known to transmit flea-borne rickettsiae.
The prevalence of rickettsial infections within the vectors was determined by 2 genus-specific qPCR assays and 1 presumed species-specific qPCR assay, Rfelis (Henry et al. 2007). Most notable was the high prevalence of rickettsiae found in dog (100%) and cat (97.3%) fleas (Ctenocephalides spp.). When combined with the frequent detection of these species on domestic dogs and cats, and within households, it is clear that the potential for human exposure to rickettsiae in these communities is very high.
When identities of the rickettsiae were confirmed by sequencing small fragments of the 17-kD antigen gene and ompB gene, it was determined that of the 12 samples assessed, only 1 was R. felis; the other 11 samples contained DNA of novel rickettsial agents. Further characterization by MLST (Fournier et al. 2003) of this unique agent determined that it was most closely related to the validated species R. felis. However, it did not have enough nucleotide homology to R. felis to be considered to be that species. Therefore, by molecular techniques, this agent was found to represent a new rickettsial species that has some nucleotide similarity to members of a recently described, but poorly characterized, rickettsial R. felis-like genotype.
The R. felis-like genotype group includes molecular isolates with partial sequences of a few conserved rickettsial genes (17-kD gene, rrs, and gltA) and have been found not only among various fleas species, but in mites, ladybird (Coccidula rufa), soft ticks (Ornithodoros moubata), and tsetse flies (Khaldi et al. 2012, Mediannikov et al. 2012, Roucher et al. 2012). The rickettsiae of this R. felis-like genotype group were initially identified in the 2000s and included Rickettsia RF2125 and Rickettsia RF31, which were detected in C. canis and C. felis (Parola et al. 2003) following a survey in Kanchanaburi Province, Thailand, near the Myanmar border. Rickettsia RF2125 and Rickettsia RF31 were distinct from other rickettsiae but most similar to R. felis (97.9 and 97.4%, respectively, for gltA). Subsequently, Reeves et al. (2005) identified, using molecular techniques, these 2 rickettsiae in C. felis collected from animals killed by automobiles in the Piedmont and foothills of South Carolina. A similar agent named Rickettsia sp. SE313, similar to RF2125, was also discovered in 12 of 12 Echidnophaga gallinacea (sticktight fleas) and in 7 of 120 (5.8%) pools of Ornithonyssus bacoti (tropical rat mites) obtained from rats live-trapped in Egypt (Loftis et al. 2006, Reeves et al. 2007). In a survey of ectoparasites and associated pathogens of free-roaming and captive animals in zoos of South Carolina, Rickettsia RF2125 was found in C. felis removed from a zookeeper and a grizzly bear (Nelder et al. 2009). In addition, Rickettsia RF2125 was identified in P. irritans (human flea) obtained from a dog in Hungary (Hornok et al. 2010). Most recently, an agent similar to RF2125 (99.7% identical sequence to a 728-bp portion of the conserved gltA gene) has been identified in Synosternus pallidus fleas from Dielmo village, Senegal (Roucher et al. 2012). Collectively, these references indicate that other flea-borne rickettsiae exist throughout the world and that a much more thorough characterization of these agents by molecular and biological means is needed, especially in regard to pathogenicity.
The high prevalence of Candidatus R. asemboensis among fleas known to bite humans and to transmit agents that cause human disease raises the potential that this newly identified rickettsia could be an important cause of human illness. However, in the same area of Asembo where 50 of 699 (7.2%) fever patients were determined to have FBSF, 21 of 21 molecular isolates from the 50 positive blood samples had sequence-confirmed R. felis DNA (Maina et al. 2012). Similarly, in a study of non-malaria fever patients, 6 of 163 (3.7%) patients had evidence of FBSF, and all 6 samples were sequence verified to have R. felis DNA in the serum samples (Richards et al. 2010). Candidatus R. asemboensis was not detected in any patients' samples from those areas. However, before concluding that Candidatus R. asemboensis is not pathogenic for humans, screening of specimens from additional febrile patients will be done while ensuring that illnesses from a distribution of age groups and during different months of the year and different years will be needed.
In contrast to the high prevalence of Candidatus R. asemboensis in arthropods obtained from animals and human dwellings was the low prevalence of R. felis (5.97%), a demonstrated human pathogen, among the same arthropods. This low prevalence of R. felis seems contrary to the 7.2% prevalence of FBSF among fever patients in Asembo (Maina et al. 2012) and the 4.4% and 3.7% prevalence among malaria-negative febrile patients in Senegal (Socolovschi et al. 2010) and northeastern Kenya (Richards et al. 2010), but more in line with the sporadic nature of this disease reported from non-sub-Saharan locations (only 68 cases from 20 countries on 6 continents; Parola 2011).
In conclusion, we report our findings that fleas collected from Asembo, Kenya, contain in addition to the rickettsial pathogen R. felis (the causative agent of flea-borne spotted fever), a new rickettsia, similar to the previously described Rickettsia RF2125 from Thailand, South Carolina, Egypt, Hungary, and Senegal. This new rickettsia, provisionally referred to as Candidatus R. asemboensis, is commonly found within fleas known to vector R. felis in western Kenya where flea-borne spotted fever is endemic. Although we did not find evidence of human infection with this new rickettsia, more testing on human specimens will be needed to determine whether the organism is pathogenic for humans.
Acknowledgments
We would like to acknowledge and thank the data team Allan Audi and Linus Ochieng; the lab team Terryson Yator, Sylvia Omul'o, and Fredrick Ade; and the entire CDC-Kenya Integrated Human and Animal Health Program (IHAHP) field team. We would also like to thank Bernard Risky Agwanda and his team at the National Museums of Kenya for their assistance with small mammal trapping.
This work is supported by the Global Emerging Infections Surveillance and Response System, a Division of the Armed Forces Health Surveillance Center; work unit number 0000188M.0931.001.A0074 and the Wellcome Trust (grant no. 81828).
Author Disclosure Statement
The views expressed in this article are those of the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government. As an employee of the US Government this work was prepared as part of my official duties and therefore under Title 17 USC paragraph 105 copyright protection is not available.
References
- Adams JR. Schmidtmann ET. Azad AF. Infection of colonized cat fleas, Ctenocephalides felis (Bouché), with a rickettsia-like microorganism. Am J Trop Med Hyg. 1990;43:400–409. doi: 10.4269/ajtmh.1990.43.400. [DOI] [PubMed] [Google Scholar]
- Adazu K. Lindblade KA. Rosen DH. Odhiambo F. Ofware P. Kwach J, et al. Health and demographic surveillance in rural western Kenya: A platform for evaluating interventions to reduce morbidity and mortality from infectious diseases. Am J Trop Med Hyg. 2005;73:1151–1158. [PubMed] [Google Scholar]
- Azad AF. Epidemiology of murine typhus. Ann Rev Entomol. 1990;35:553–569. doi: 10.1146/annurev.en.35.010190.003005. [DOI] [PubMed] [Google Scholar]
- Azad AF. Sacci JB., Jr Nelson WM. Dasch GA. Schmidtmann ET. Carl M. Genetic characterization and transovarial transmission of a typhus-like rickettsia in cat fleas. Proc Natl Acad Sci USA. 1992;89:43–46. doi: 10.1073/pnas.89.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigogo G. Audi A. Aura B. Aol G. Breiman RF. Feikin DR. Health seeking patterns among participants of population-based morbidity surveillance in rural western Kenya: Implications for calculating disease rates. Int J Infect Dis. 2010;14:967–973. doi: 10.1016/j.ijid.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Feikin DR. Olack B. Bigogo GM. Audi A. Cosmas L. Aura B, et al. The burden of common infectious disease syndromes at the clinic and household level from population-based surveillance in rural and urban Kenya. PLoS One. 2011;6:e16085. doi: 10.1371/journal.pone.0016085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE. Dumler S. Greub G. Zhang J. Wu Y. Raoult D. Gene sequence-based criteria for identification of new Rickettsia Isolates and description of Rickettsia heilongjiangensis sp. Nov. J Clin Microbiol. 2003;41:5456–5465. doi: 10.1128/JCM.41.12.5456-5465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ. Beier MS. Rahman MS. Ammerman NC. Shallom JM. Purkayastha A, et al. Plasmids and rickettsial evolution; insight from Rickettsia felis. PLoS One. 2007;2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisch RB. Urban Rattus as the main reservoir of murine typhus in Kenya. J Trop Med Hyg. 1969;72:195–196. [PubMed] [Google Scholar]
- Heisch RB. Harvey AEC. Rickettsioses in Kenya: Serological reactions of wild rodents and inoculated guinea pigs. East Afr Med. 1959;36:116–118. [PubMed] [Google Scholar]
- Heisch RB. Grainger WE. Harvey AEC. Lister G. Feral aspects of rickettsial infections in Kenya. Trans R Soc Trop Med Hyg. 1962;56:272–286. doi: 10.1016/0035-9203(62)90048-2. [DOI] [PubMed] [Google Scholar]
- Henry KM. Jiang J. Rozmajzl PJ. Azad AF. Macaluso KR. Richards AL. Development of quantitative real-time PCR assays to detect Rickettsia typhi and Rickettsia felis, the causative agents of murine typhus and flea-borne spotted fever. Mol Cell Probes. 2007;21:17–23. doi: 10.1016/j.mcp.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Hornok S. Meli ML. Perreten A. Farkas R. Willi B. Beugnet F, et al. Molecular investigation of hard ticks (Acari: Ixodidae) and fleas (Siphonaptera: Pulicidae) as potential vectors of rickettsial and mycoplasmal agents. Vet Microbiol. 2010;140:98–104. doi: 10.1016/j.vetmic.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Jiang J. Blair PJ. Felices V. Moron C. Cespedes M. Anaya E. Schoeler GB. Sumner JW. Olson JG. Richards AL. Phylogenetic analysis of a novel molecular isolate of spotted fever group rickettsiae from northern Peru: Candidatus Rickettsia andeanae. Ann NY Acad Sci. 2005;1063:337–342. doi: 10.1196/annals.1355.054. [DOI] [PubMed] [Google Scholar]
- Jiang J. Soeatmadji DW. Henry KM. Ratiwayanto S, Bangs MJ, Richards AL. Rickettsia felis in Xenopsylla cheopis, Java, Indonesia. Emerg Infect Dis. 2006;12:1281–1283. doi: 10.3201/eid1208.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. Stromdahl EY. Miller M. Richards AL. Detection of Rickettsia parkeri and Rickettsia andeanae in Amblyomma maculatum Gulf Coast ticks collected from humans, USA. Vector Borne Zoonotic Dis. 2012;12:175–182. doi: 10.1089/vbz.2011.0614. [DOI] [PubMed] [Google Scholar]
- Kelly DJ. Richards AL. Temenak JJ. Strickman D. Dasch GA. The past and present threat of rickettsial diseases to military medicine and international public health. Clin Infect Dis. 2002;34(Suppl 4):s145–s169. doi: 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- Khaldi M. Socolovschi C. Benyettou M. Barech G. Biche M. Kernif T. Raoult D. Parola P. Rickettsiae in arthropods collected from the North African Hedgehog (Atelerix algirus) and the desert hedgehog (Paraechinus aethiopicus) in Algeria. Comp Immunol Microbiol Infect Dis. 2012;35:117–122. doi: 10.1016/j.cimid.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Loftis AD. Reeves WK. Szumlas DE. Abbassy MM. Helmy IM. Moriarity JR, et al. Surveillance of Egyptian fleas for agents of public health significance: Anaplasma, Bartonella, Coxiella, Ehrlichia, Rickettsia, and Yersinia pestis. Am J Trop Med Hyg. 2006;75:41–48. [PubMed] [Google Scholar]
- Maina A. Knobel D. Jiang J. Halliday JEB. Feikin DR. Cleveland S, et al. Rickettsia felis infection in febrile patients western Kenya, 2007–2010. Emerg Infect Dis. 2012;18:328–331. doi: 10.3201/eid1802.111372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediannikov O. Audoly G. Diatta G. Trape JF. Raoult D. New Rickettsia sp. in tsetse flies from Senegal. Comp Immunol Microbiol Infect Dis. 2012;35:145–150. doi: 10.1016/j.cimid.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Nelder MP. Reeves WK. Adler PH. Wozniak A. Wills W. Ectoparasites and associated pathogens of free-roaming and captive animals in zoos of South Carolina. Vector Borne Zoonotic Dis. 2009;9:469–477. doi: 10.1089/vbz.2008.0008. [DOI] [PubMed] [Google Scholar]
- Parola P. Rickettsia felis: From a rare disease in the USA to a common cause of fever in sub-Saharan Africa. Clin Microbiol Infect. 2011;17:996–1000. doi: 10.1111/j.1469-0691.2011.03516.x. [DOI] [PubMed] [Google Scholar]
- Parola P. Sanogo OY. Lerdthusnee K. Zeaiter Z. Chauvancy G. Gonzalez JP, et al. Identification of Rickettsia spp. and Bartonella spp. in fleas from the Thai–Myanmar border. Ann NY Acad Sci. 2003;990:173–181. doi: 10.1111/j.1749-6632.2003.tb07359.x. [DOI] [PubMed] [Google Scholar]
- Raoult D. La Scola B. Enea M. Fournier PE. Roux V. Fenollar F, et al. A flea-associated rickettsia pathogenic for humans. Emerg Infect Dis. 2001;7:73–81. doi: 10.3201/eid0701.010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WK. Nelder MP. Korecki JA. Bartonella and Rickettsia in fleas and lice from mammals in South Carolina, USA. J Vector Ecol. 2005;30:310–315. [PubMed] [Google Scholar]
- Reeves WK. Loftis AD. Szumlas DE. Abbassy MM. Helmy IM. Hanafi HA, et al. Rickettsial pathogens in the tropical rat mite Ornithonyssus bacoti (Acari: Macronyssidae) from Egyptian rats (Rattus spp.) Exp Apply Acarol. 2007;41:101–107. doi: 10.1007/s10493-006-9040-3. [DOI] [PubMed] [Google Scholar]
- Richards AL. Jiang J. Omulo S. Dare R. Abdirahman K. Ali A, et al. Human infections with Rickettsia felis, Kenya. Emerg Infect Dis. 2010;16:1081–1086. doi: 10.3201/eid1607.091885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucher C. Mediannikov O. Diatta G. Trape JF. Raoult D. A new Rickettsia species found in fleas collected from human dwellings and from domestic cats and dogs in Senegal. Vector Borne Zoonotic Dis. 2012;12:360–365. doi: 10.1089/vbz.2011.0734. [DOI] [PubMed] [Google Scholar]
- Roux V. Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer membrane protein rompB (ompB) Int J Syst Evol Microbiol. 2000;50:1449–1455. doi: 10.1099/00207713-50-4-1449. [DOI] [PubMed] [Google Scholar]
- Schriefer ME. Sacci JB., Jr Dumler JS. Bullen MG. Azad AF. Identification of a novel rickettsial infection in a patient diagnosed with murine typhus. J Clin Microbiol. 1994;32:949–954. doi: 10.1128/jcm.32.4.949-954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerman J. Handbook for Identification of Fleas. Publication of the South African Institute of Medical Research. No 57. Johannesberg, South Africa: South African Institute for Medical Research; 1995. Siphonaptera of southern Africa. [Google Scholar]
- Socolovschi C. Mediannikov O. Sokhna C. Tall A. Diatta G. Bassene H, et al. Rickettsia felis- associated uneruptive fever, Senegal. Emerg Infect Dis. 2010;16:1140–1142. doi: 10.3201/eid1607.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenos J. Graves SR. Unsworth NB. A highly sensitive and specific real-time PCR assay for the detection of spotted fever and typhus group rickettsiae. Am J Trop Med Hyg. 2005;73:1083–1085. [PubMed] [Google Scholar]
- Tamura K. Peterson D. Peterson N. Stecher G. Nei M. Kumar S. Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011:MEGA5. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venzal JM. Perez-Martinez L. Felix ML. Portillo A. Blanco JR. Oteo JA. Prevalence of Rickettsia felis in Ctenocephalides felis and Ctenocephalides canis from Uruguay. Ann NY Acad Sci. 2006;1078:305–308. doi: 10.1196/annals.1374.056. [DOI] [PubMed] [Google Scholar]