Abstract
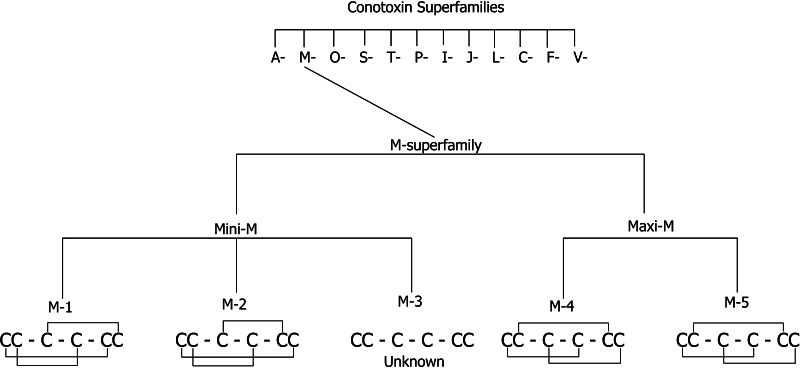
The focus of this review is the M-superfamily of Conus venom peptides. Disulfide rich peptides belonging to the M-superfamily have three loop regions and the cysteine arrangement: CC–C–C–CC, where the dashes represent loops one, two, and three, respectively. Characterization of M-superfamily peptides has demonstrated that diversity in cystine connectivity occurs between different branches of peptides even though the cysteine pattern remains consistent. This superfamily is subdivided into five branches, M-1 through M-5, based on the number of residues in the third loop region, between the fourth and fifth cysteine residues. M-superfamily peptides appear to be ubiquitous in Conus venom. They are largely unexplained in indigenous biological function, and they represent an active area of research within the scientific community.
Keywords: M-superfamily, Conotoxin, Review, Marine snail, Venom, Peptide
Introduction
Throughout the world there exist both predator and prey. This distinction is apparent though sometimes misleading. Take, for example, marine snails of the genus Conus that are present across the oceans of the southern hemisphere [1]. These snails are slow-moving animals that appear more prey than predator. However, they have evolved into effective predators through the development of venom consisting of biologically active peptides. The venom is loaded into a hollow harpoon that the snail injects into the intended prey: fish, worms, or other snails [2]. The categories of cone snails based on prey preference are piscivorous (fish eating), molluscivorous (mollusk eating), and vermivorous (worm eating) [3]. The cone snail venom contains myriad peptide components significant to the survival of the organism with respect to hunting and defense against being eaten [4].
Interest by researchers in snails of the genus Conus began in the early 1970s as evidence of their involvement in numerous human fatalities mounted [5]. Dr. Alan Kohn, an early pioneer in the study of hunter/prey relationship of cone snails, recognized that the venom of cone snails may possess therapeutic components [6]. During that time, Dr. Robert Endean and coworkers in Australia demonstrated that the venom of dissimilar species of cone snail contained a diversity of biologically active components. Dr. Baldomero (Toto) Olivera and coworkers at the University of Utah became the primary innovators of successful laboratory techniques in the study of venom components extracted from cone snails [7]. Foremost among these innovations was an avant-garde method of bio-assay using intracranial rather than intraperitoneal injection of toxin into subject mice. The intracranial delivery method revealed greater sensitivity to individual peptides in fish and mouse studies than those from traditional intraperitoneal injections [8]. This early research revealed the disulfide rich nature of the majority of peptide components from Conus snail venom. The disulfide rich peptides became broadly defined as conotoxins [9].
From conotoxins to drugs
The intracranial injection method of conotoxin delivery into fish and mouse subjects allowed researchers to begin unraveling the complicated chemistry of the neurotoxic peptides and paving the path for therapeutic applications. The venom of any individual cone snail contains upwards of 100 different peptides, each with a distinctive role when injected into the target subject. It is the cumulative effect of the individual peptides that causes the venom to be deadly to the prey. Researchers called the collective effect of the venom a “cabal”, after unspecified covert groups organized to overthrow equally unspecified governments [10]. With different snails, researchers noted different cabals: the ‘lightning-strike cabal’, the ‘motor cabal’, and the ‘nirvana cabal’, named after the general set of reactions elicited by the overall effect of the venom on the test subject. The lightning-strike cabal inhibits muscular contraction in prey due to a combination of paralytic peptides that act to block voltage-gated Na+ and K+ channels; this has an effect similar to electrocution. The motor cabal effectively inhibits the pre-synaptic Ca2+ channels, post-synaptic nicotinic receptors and Na+ channels. The combination achieves total inhibition of neuromuscular transmission [11]. The lightning-strike and motor cabals often work in conjunction. The nirvana cabal diminishes the sensory circuitry of the prey by producing a euphoric effect [5]. This allows the snail to capture its prey in a net-like extendable stomach for consumption.
Conotoxin components in the venom target myriad receptor sites in the prey, a predatory tactic that has been adopted across mammalia, from “Caenorhabditis elegans to humans” [12]. Individual conotoxins range between 10 and 100 amino acids in length [2]. The genus Conus contains over 700 species, representing a peptide library on the order of 70,000 sequences. Conotoxins represent extremely specific biological probes that offer researchers a tool to understand and differentiate between closely related receptors [13]. The simplicity of conotoxins has made them valuable in the advancement of neuroscience research and consequent drug development [14]. Active research in the field of conotoxins involves the isolation, identification, and assessment of biological activity for individual peptides that possess the potential to be made into potent and selective therapeutic drugs. Scientists either use small amounts of purified native conotoxin or they use automated synthesis techniques to prepare milligram quantities of material for their investigations. Currently, only about 0.2% of the conotoxin peptide library has been cataloged [9, 15].
Many diseases, such as epilepsy, schizophrenia, Tourette’s syndrome, Parkinson’s disease, and sclerosis, are associated with improper functioning of ligand- and voltage-gated channels. Conotoxin-based therapeutics have demonstrated great promise because they are relatively small, potent, selective antagonists and agonists of specific cell membrane channel proteins [16]. Examples of pharmaceutical companies that have been involved in developing and testing drugs based on conotoxins and/or conotoxin molecular scaffolds include Metabolic Pharmaceuticals and Xenome Ltd. from Australia, and Elan Corporation in Ireland. The Technology Commercialization Office of the University of Utah has recently released α-conotoxin peptides and derivatives U-2902, and U-4079 as potential therapeutics for neuropathic pain [17]. At this time, the only conotoxin drug approved by the United States Food and Drug Administration for public use is Prialt™ [15, 18–23]. Prialt™ is a synthetic conopeptide derived from Conus magus used to treat chronic pain and is one of the most powerful analgesics available [15]. Prialt™ is the trade name for ψ-conotoxin MVIIA, an N-type calcium channel blocker. This drug provides a non-addictive means to block pain in subject patients by inhibiting the source of pain transmission in nerve cells of the spinal cord. For this reason, it is a desirable alternative to traditional opiate derivatives like morphine or codeine [23].
An introduction into the M-superfamily of conotoxins
Members of the M-superfamily of conotoxins have been found in every Conus species tested to date [4]. Division into different superfamilies is based on signal sequences homology. Thus, each superfamily is unique [5]. Classification of this widely divergent superfamily is based on the general pattern derived from the number of residues contained in each of the three loop regions CC(X4–6)C(X4–5)C(X1–5)CC, where X4–6 represents four to six amino acids in the first loop, X4–5 represents four to five amino acids in the second loop, and X1–5 represents one to five amino acids in the third loop [24]. M-superfamily peptides are further divided into 5 branches, labeled M-1 to M-5, based on the number of residues that exist in the third cystine loop between the fourth and fifth cysteine residues. A further delineation separates the five branches into the Mini-M and Maxi-M conotoxins, where M-1 through M-3 are considered Mini-Ms and M-4/M-5 are Maxi-Ms (Fig. 1). This differentiation is based on the overall number of residues in the mature peptide. The Mini-M conotoxins all contain fewer than 22 residues, while the Maxi-M peptides contain more than 22 amino acids [13]. The Maxi-M (M-4 and M-5) peptides are sub-grouped into μ-, κM-, and ψ-conotoxins based on their biological targets. The μ-conotoxins block voltage-gated sodium channels, κM-conotoxins block voltage-gated potassium channels in muscle [2], and ψ-conotoxins block nicotinic acetylcholine receptors [24]. Members of the M-superfamily for which a target receptor is known are exclusively within the M-4 and M-5 branches with relatively little known about the biological receptors targeted by the Mini-M peptides. It is known that the target receptor for the Mini-M peptides is different from that of the Maxi-M peptides [24].
Fig. 1.
A flowchart of the organization of the M-superfamily with known disulfide connectivity shown for each branch (M-1–M-5)
Mini-Ms
Interestingly, the Mini-M peptides demonstrate unique disulfide bond connectivity patterns, loop size variability, and unique 3D folding structure even within the same branch. The M-superfamily is the only superfamily where such diverse structure and cysteine connectivity is present, especially in the Mini-Ms, M-1 through M-3.
M-1 branch
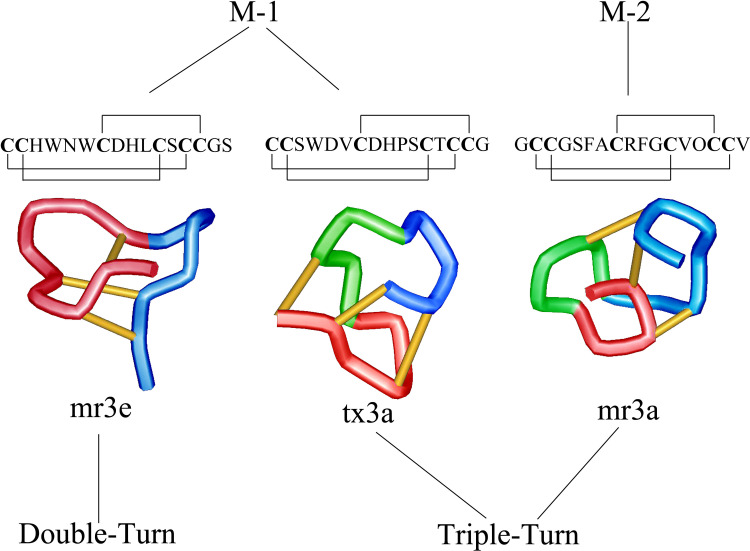
Although the presence of the Mini-M peptides is well known in laboratories that investigate Conus venom, little has been published due to the lack of definitive evidence pertaining to defined biological relevance. Much of what is presently known of the M-1 branch has been through the study of conotoxins mr3e (from C. marmoreus) and tx3a (from C. textile). Conotoxin mr3e has the disulfide connectivity Cys1-Cys5, Cys2-Cys4, and Cys3-Cys6, and its NMR-derived solution structure contains what authors described as a ‘double-turn’ motif [25, 26]. This is different than the ‘triple-turn’ motif described for M-1 branch peptide tx3a [4, 27]. Thus, even though the disulfide connectivity pattern is the same for these two peptides, their secondary structures vary considerably. Prior to this discovery, it was thought that once a structure was determined for a peptide family with the same disulfide pattern, the peptide scaffold was conserved for that family of peptides. The unique secondary structure characteristics of mr3e and tx3a are suspected to be responsible for the observed difference in response to intracranial injection in mice with conotoxin mr3e causing no observable effect and tx3a inducing excitatory behavior [28]. This excitatory behavior is also observed for the M-2 branch peptide, mr3a [26]. The mr3a structure consists of the triple-turn motif also observed for tx3a, but its backbone has a different cystine pattern. Figure 2 shows a comparison of the published structures, disulfide bond pattern, and sequences for mr3e, tx3a, and mr3a [25].
Fig. 2.
Structures of Mini-M peptides of the M-1 and M-2 branches. Depicted are the Double-turn motif of mr3e (M-1) and the Triple-turn of tx3a (M-1) and mr3a (M-2)
M-2 branch
There is limited published information describing members of the M-2 branch of the M-superfamily. The structure and sequence of conotoxin mr3a have been reported, but little is known about biological targets or action mechanisms for this and other M-2 peptides. Conotoxin mr3a was determined to have a cystine connectivity of Cys1-Cys6, Cys2-Cys4, and Cys3-Cys5 [26, 28] and a three-dimensional structure best described as a triple-turn motif [4]. To date, this connectivity belongs exclusively to the M-2 branch peptides.
Additionally, there appear to be similarities between the M-2 branch and the Maxi-M peptides. A resemblance between the prepropeptide sequences for the immature conotoxins has been reported. The M-2 and M-4/M-5 branches have a common-size signal peptide (25 residues) in addition to a high degree of homology in the first eight residues for that signal peptide, MMSKLGVL [28]. It has further been described that the M-2 peptide mr3a elicits a strong excitatory response in mice at nanomolar quantities upon intracranial injection [4]. This finding supports the notion that members of the M-2 branch peptides are significant and functional snail venom components with the potential to be used as a means of discovering the possible existence of uncharacterized receptors.
M-3 branch
The cystine pattern and structural characteristics of M-3 branch peptides have not been reported to date. Current knowledge of these peptides is limited to sequence comparisons with other branch families of the M-superfamily. A study exploring similarities to other families noted that both M-1 and M-3 peptides are rich in acidic residues, as opposed to basic residues in M-2 and M-4 [28]. cDNA studies were performed to further investigate the similarities between the M-1 and M-3 branches. From this, a new phylogenetic tree was constructed in which M-1 and M-3 conotoxins have “a closer relationship, falling in the same tree branch” [28]. The signal peptide and pro-peptide region of the immature toxins appear to be highly conserved; evidence that the M-1 and M-3 branch peptides are evolutionarily related. This close relationship does not appear between any other branch families in the M-superfamily [28]. A few recently reported examples of M-3 peptides are: reg12a (GCCOOQWCGODCTSOCC) [1], Tx3.5 (RCCKFPCPDSCRYLCC(nh2)) [24], and Qc3.1 (ACCDPDWCDAGCYDGCC) [25]. Most M-3 family peptides are discovered by PCR amplification of cDNA [25]. Peptide sequencing allows for characterization into the M-3 branch based on the cysteine pattern and three amino acid residues in the third loop.
Maxi-Ms
The Maxi-M peptides are divided into the M-4 and M-5 branches of the M-superfamily based on their primary sequence. These peptides are further separated into the κM-, ψ-, and μ-conotoxins based on their molecular targets consisting of voltage-gated potassium channels, nicotinic acetylcholine receptor, and voltage-gated sodium channels (VGSCs), respectively [4, 5]. The M-4 branch consists of all published κM- and ψ-conotoxins in addition to about one-third of the μ-conotoxins, (Fig. 3) while the M-5 branch consists of only μ-conotoxins to date. It is also interesting to note that the Maxi-M, κM-, ψ-, and μ-conotoxins are found mostly in fish-hunting Conus species, whereas the Mini-M peptides appear in the venom of the mollusk- and worm-hunting cone snails [28, 29]. The Maxi-M peptides induce paralysis in prey, while the M-1, M-2, and M-3 branch (Mini-M) peptides generally cause an excitatory physiological response [4]. This difference may be attributed to the feeding preferences of the snail. The fish-hunting cone snails paralyze their fast-moving prey before consumption, while worm- or snail-hunting cone snails rely on spasmodic response to drive the prey from its shelter.
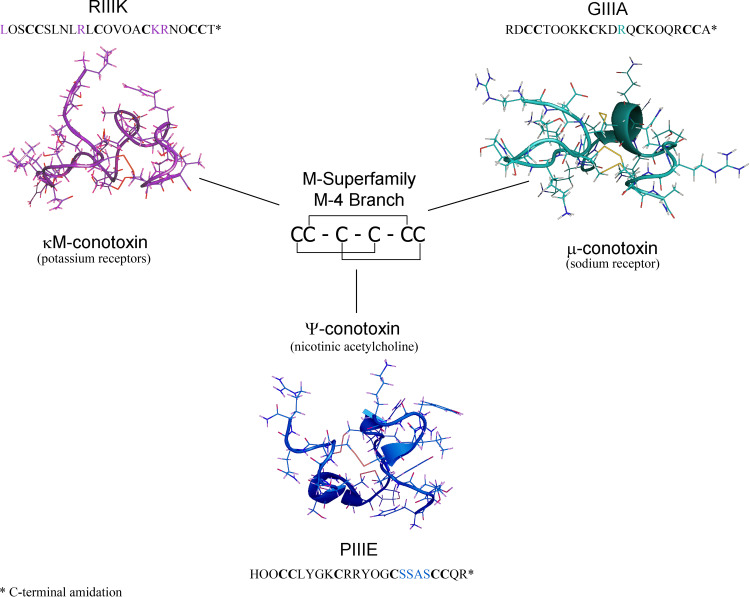
Fig. 3.
Structures of the M-4 branch sub-categories κM-, μ-, and ψ-conotoxins. Explanations of highlighted residues are described in the text
Characteristic differences for the Maxi-M peptides versus the Mini-Ms can be summarized as: a longer primary sequence (22–24 a.a. vs <20), a known molecular target (K+, nACh, Na+ receptors), and a common and distinctive cystine pattern (Cys1-Cys4, Cys2-Cys5, Cys3-Cys6) [25, 29–31]. Until the more recent characterization of the Mini-M peptides, it was believed that the Maxi-M disulfide bond pattern was the archetype of the entire M-superfamily; a commonality observed in other conotoxin superfamilies. It has recently been demonstrated that the disulfide bonds are significant structural and electrostatic elements involved in the interaction of conotoxins with the VGSC [32]. The electrostatic properties of the sulfur atoms in the disulfide bonds contribute to conotoxin agonist and antagonist activity toward a class of biological receptors that have recently been classified as the Cys-loop superfamily of receptors [33].
The Cys-loop superfamily receptors include nicotine acetylcholine receptors (nAChRs). nAChRs play a role in the transmission of nerve impulses, specifically in central and peripheral nervous tissue, and neuromuscular junctions, which control muscle contraction [34]. The structure of the Cys-loop receptors is largely comprised of β-sheets and α-helices, constituting a major class of receptor-coupled ion channels [33]. Regulation of these receptors is vital to the well-being of the organism; and it appears that conotoxins have evolved to target this class of receptors.
M-4 branch
The M-4 branch is defined by 4 residues in the third loop region of the primary sequence between Cys4 and Cys5 (CC(X4–6)C(X4–5)C(X 4)CC). The M-4 branch κM- and ψ-conotoxins were the first conotoxins for which a disulfide bond pattern was determined and ψ-PIIIE was the first M-superfamily peptide for which a three dimensional structure was determined by NMR spectroscopy. Thus, the M-4 peptides serve as a structure model for all ψ-conotoxins.
κM-conotoxins
κM-conotoxins are antagonists of K+ ion channels. In conotoxin nomenclature, the “κ“refers to activity toward K+ ion channels and the M indicates that the peptide belongs to the M-superfamily. The only published κM-conotoxin at this time is κM-RIIIK, with the primary sequence: LOSCCSLNLRLCOVOACKRNOCCT(nh2). This peptide was isolated and characterized from the venom of C. radiatus [29]. Its biological target was determined to be the “Shaker” channel and the similar mammalian Kv1.2 K+ channel [31]. The Shaker channel was first identified from Drosophila. Applied stimulus to induce rapid muscle contraction caused the fly to “shake” [35–37]. The inhibition of the channel protein responsible for the shaking affect was then selectively inhibited confirming the existence of what is now referred to as the Shaker channel. This channel was further classified as a potassium voltage-gated channel allowing for the identification of subsequent similar channels in numerous species.
κM-RIIIK has the highest demonstrated affinity for TSha1 (Trout Shaker Channel 1), a Shaker K+ channel from rainbow trout (Onchorychus mykiss) [29, 31, 38]. In comparison with the O-superfamily conotoxin, κ-PVIIA, [39, 40] and other venom components targeting the Shaker channel, κM-RIIIK appears to function uniquely; it does not have the characteristic κ-conotoxin functional dyad consisting of hydrophobic amino acids [30]. The proposed mechanism of κ-conotoxins is to use the hydrophobic dyad structure to plug the Shaker channel pore. In contrast, κM-RIIIK inhibits the Shaker channel by forming a ring that occludes the channel pore by acting as a surface lid rather than an intercalating plug in the channel [29–31, 40]. The amino acids essential for the K+ channel binding of κM-RIIIK were determined to be: Leu1, Arg10, Lys18, and Arg19 [30]. The structure activity relationship of κM-RIIIK provides a backbone template for drug development.
ψ-Conotoxins
The M-4 branch of the M-superfamily contains three examples of ψ-conotoxins that have been labeled ψ-PIIIE, ψ-PIIIF, and ψ-PrIIIE [41–43]. These ψ-conotoxins contain the same disulfide connectivity as the rest of the M-4 branch [41]. Similar to the κ- and κM- association, ψ-conotoxins function at the nicotinic acetylcholine receptors (nAChRs) that were previously determined to be the site of activity for the αA-conotoxins [41–45]. αA-Conotoxins are much smaller (< 20 amino acids) and only contain two disulfide bonds [42].
In 1997, ψ-PIIIE became the first ψ-conotoxin to be described in literature. This peptide was isolated from the venom of C. purpurascens and was reported to have the amino acid sequence: HOOCCLYGKCRRYOGCSSASCCQR(nh2) [41]. It was further discovered that C. purpurascens inhibits muscle nAChRs using a combination of ψ-PIIIE and αA-PIVA [41]. The binding of ψ-PIIIE to nAChRs in muscle was determined to occur at a ligand binding site complementary to that identified for αA-conotoxins [41, 44, 45]. Significant to the discovery of ψ-PIIIE was the identification of the novel ligand binding site on muscle nAChRs. Thus, ψ-PIIIE served as a useful tool to characterize a new target for therapeutic drug development, and to increase understanding of the Cys-loop superfamily of receptors.
ψ-PIIIF was reported in 2003, a component of the venom of C. purpurascens with the sequence: GOOCCLYGSCROFOGCYNALCCRK(nh2) [43]. Though also a ψ-conotoxin, PIIIF is ~50 times less potent than ψ-PIIIE in blocking both mice and elasmobranch nAChRs [43]. The difference in functionality is thought to be due to the amino acids of the third loop region. In ψ-PIIIE, the third loop consists primarily of small polar residues (SSAS), while in ψ-PIIIF the third loop consists of bulkier residues (YNAL) that collectively form a less polar region of the peptide. The variation in amino acid composition has led researchers to hypothesize that polarity and steric bulk in the third loop are the significant factors for the observed deviation in binding and activity of these peptides to nAChRs [43].
ψ-Conotoxin PrIIIE isolated from the venom of Conus parius has the sequence: AARCCTYHGSCLKEKCRRKYCC(nh2) [42]. ψ-PrIIIE is approximately 28 times more potent than ψ-PIIIE at inhibiting nAChRs (IC50 = ~ 250 nM for ψ-PrIIIE as compared to ~7,000 nM for ψ-PIIIE) [42]. Whereas ψ-PIIIE-F, and ψ-PrIIIE all inhibit fish nAChRs, only ψ-PrIIIE is active as a non-competitive inhibitor of mouse skeletal muscle nAChRs [42]. This is significant as the mouse model is used to predict therapeutic activity in humans.
M-4 μ-conotoxins
μ-Conotoxins belong to both the M-4 and M-5 branches of the M-superfamily. These conotoxins differ only in the number of residues in the third loop region and are similar in their common activity as antagonists of voltage-gated sodium channels (VGSCs). VGSCs sense the membrane potential across excitable cells plasma membrane and change their conformation into the open state if the potential results higher than a defined threshold, which allows firing of action potentials [46]. These VGSCs are Nav1.1–Nav1.9 based on their α-subunit sequence and further organized according to their sensitivity to tetrodotoxin (TTX). VGSCs are classified as either TTX-S, for sensitive, and TTX-R, for resistant, depending on their overall degree of inhibition by tetrodotoxin [47, 48]. All nine VGSCs are inhibited by TTX, but those with IC50 values in the nanomolar range constitute the TTX-S VGSCs (Nav1.1-4, Nav1.6, and Nav1.7), and the VGSCs that have IC50 values in the micromolar range are considered TTX-R (Nav1.5, Nav1.8 and Nav1.9) [32, 46, 47, 49–53]. The TTX-S VGSCs function in either neuronal communication (Nav1.1, Nav1.2-3, and Nav1.6-7) [47, 54] or skeletal muscle control (Nav1.4) [46, 47, 55]. Researchers have determined the connection between VGSCs and the symptoms caused by many diseases including neuropathic pain, arrhythmia, epilepsy, stroke, and bipolar disorder [46, 56]. For example, neuropathic pain originates from the TTX-R channels Nav1.8 and Nav1.9 [57–59].
μ-Conotoxins have been investigated as therapeutic drugs because of their potency and ability to differentiate between distinct VGSCs. The first μ-conotoxins isolated from the venom of Conus geographus were μ-GIIIA, B, and C. These peptides were shown to be active in blocking skeletal muscle VGSCs [52], specifically Nav1.4, while having a much lower affinity toward neuronal VGSCs. The activity of conotoxin μ-GIIIA toward the inhibition of Nav1.4 VGSCs was shown to be dependent on Arg13 in the primary sequence (R in bold): RDCCTOOKKCKDRQCKOQRCCA-(nh2) [52, 60]. Substitution of this Arg residue (Arg13) three amino acids removed from the third Cys15 residue (i.e., loop two) results in the inactivation of the peptide. This is a testament to the specificity and usefulness of conotoxins as tools to understand ligand to receptor activity.
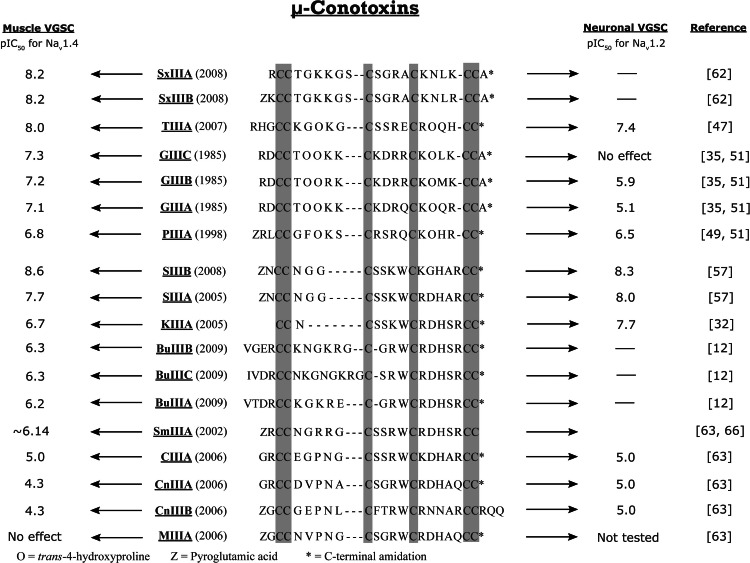
More recent discoveries have led to the identification, characterization, and publication of 18 μ-conotoxins to date. The 7 μ-conotoxins belonging to the M-4 branch are: GIIIA-C [35, 47, 61], PIIIA [47, 49], TIIIA [47], and SxIIIA-B [62]. The remaining 11 μ-conotoxin belong to the M-5 branch: SIIIA [46, 53], KIIIA [53], CIIIA, CnIIIA-B, MIIIA [63], SIIIB [57], SmIIIA [64], and BuIIIA-C [12].
μ-Conotoxin PIIIA blocks both muscle and neuronal TTX-S VGSCs [51, 54, 65]. μ-Conotoxin PIIIA was found to have an IC50 value of 2 μM in tests involving mammalian central nervous system (CNS) Type II muscle channels [51]. This represents an inhibition that is upwards of 50 times greater than that observed for μ-conotoxin GIIIA [49]. μ-PIIIA is also effective in blocking neuronal channels (Nav1.2) albeit with lower affinity than muscle VGSCs [63]. μ-PIIIA has a pIC50 of 6.8 (where pIC50 = −logIC50) in binding studies with Nav1.4 skeletal muscle VGSCs [32, 35, 50, 66, 67]. Also important for assays are the isoforms Nav1.1, expressed in the brain, and Nav1.6, expressed in the axons [68]. While Nav1.4 skeletal muscle VGSCs serve as the model system for muscle cell antagonist activity, the Nav1.2 VGSCs serve as the standard test receptor for neuronal VGSCs (Fig. 4).
Fig. 4.
Known binding efficacy for μ-conotoxins toward muscle voltage gated sodium channels (VGSCs) and neuronal VGSCs
More recently, μ-conotoxin TIIIA was isolated from the venom of Conus tulip. The primary sequence for this peptide contains the characteristic and functionally relevant positively charged basic amino acid Arg (or Lys), three amino acids removed from the third Cys residue (i.e., loop two), next to an acidic and negatively charged amino acid (Glu15) [47]. This is unusual as the sequences of previous μ-conotoxins contained the Arg (or Lys) neighbored by either Gln, Arg, or Trp [47]. μ-TIIIA inhibits VGSCs from both neuronal (Nav1.2) and skeletal muscle (Nav1.4), with a preference for Nav1.4 VGSCs. Mutation of Glu15 to Ala in the TIIIA peptide changed the biological target preference from skeletal muscle Nav1.4 VGSCs to neuronal Nav1.2 [47]. This is an example of the value of conotoxins as selective probes to develop structure activity relationship models for closely related receptors. Single-site mutation on the conotoxin scaffold provided a better understanding of the size and electrostatic topography of the ligand required to differentiate between receptor types.
μ-Conotoxins SxIIIA and SxIIIB demonstrate high affinity for muscle Nav1.4 VGSCs [62]. While little has been described in the literature regarding these recently reported μ-conotoxins, the process used to identify their disulfide connectivity made use of a novel technique referred to as rapid disulfide bridge mapping [64]. This procedure introduces isotope labeled cysteine residues during cloning, then uses nuclear magnetic resonance (NMR) spectroscopy to identify the connectivity of the labeled peptides and rapidly determine bridge order. When synthesizing SxIIIA, the first three cysteines were uniformly 15N and 13C enriched, while the remaining three cysteine residues were labeled with a 70:30 mix of (14N/12C):(15N/13C) [62]. The use of NMR to differentiate the cystine pattern is an efficient approach that may lead to the characterization of many additional conotoxins that are so far known only by sequence because of insufficient native sample quantities for confirmation of correct folding of synthetic peptide.
M-5 branch
The M-5 branch of the M-superfamily differs from those preceding it by containing five residues in the third cysteine loop between cysteines four and five. All reported M-5 conotoxins are μ-conotoxins [25].
M-5 μ-conotoxins
The μ-conotoxins within the M-5 branch are: SmIIIA [64], SIIIA [46, 53], KIIIA [53], CIIIA, CnIIIA-B, MIIIA [63], SIIIB [57] and BuIIIA-C [12]. In contrast to the M-4 branch μ-PIIIA and μ-GIIIA that inhibit TTX-S VGSCs, μ-SmIIIA is an antagonist of TTX-R VGSCs [64]. Although μ-SmIIIA inhibits TTX-R channels, it does not differentiate well between them (Nav1.5, Nav1.8, and Nav1.9) [66].
μ-Conotoxins SIIIA and KIIIA are antagonists of TTX-R channels similar to the M-4 peptide μ-SmIIIA. Interestingly, the size of the first loop varies between μ-SmIIIA (five amino acids) to μ-SIIIA (three residues) and finally μ-KIIIA (one residue) [46, 53]. The diversity in the number of amino acids in the first loop was an indication to researchers that the second and third loop regions contained the necessary structural and electrostatic characteristics for the peptides to be active toward inhibiting TTX-R VGSCs. This was confirmed in a study that removed the first loop of μ-KIIIA and showed that the remainder of the peptide retained efficacy toward the inhibition of TTX-R VGSCs. The modified μ-KIIIA provided a smaller yet equally effective model from which synthetic mimetics are being explored as pharmacological therapeutics [32].
Based on sequence comparison from the last two loop regions of μ-SmIIIA, μ-KIIIA, and μ-SIIIA, four new μ-conotoxins, CnIIIA-B, CIIIA, and MIIIA, were discovered and reported in 2006 [63]. As expected, these more recent μ-conotoxins inhibited TTX-R VGSCs. Despite their sequence similarity to one another, they were found to inhibit a divergent set of TTX-R channels, indicating the importance of the amino acid constituents within these peptides. Thus, it was determined that the secondary structures of the second and third loop regions of μ-SmIIIA, μ-KIIIA, and μ-SIIIA are significant for binding to TTX-R channels. This was tested by a comparison study of these three conotoxins with μ-CnIIIA, μ-CIIIA, and μ-MIIIA, which yielded a consensus sequence of  indicating the three significant amino acids present in the second and third loops that are responsible for differentiating between the TTX-R channels [63]. Common to these μ-conotoxins is the conservation of Arg in the second loop, usually in the same sequence position (third residue past Cys) [35, 46, 47, 49, 53, 57, 62–64], further validating the significance of Arg in VGSC inhibition first reported for μ-GIIIA.
indicating the three significant amino acids present in the second and third loops that are responsible for differentiating between the TTX-R channels [63]. Common to these μ-conotoxins is the conservation of Arg in the second loop, usually in the same sequence position (third residue past Cys) [35, 46, 47, 49, 53, 57, 62–64], further validating the significance of Arg in VGSC inhibition first reported for μ-GIIIA.
A similar effect, as reported with μ-TIIIA, was observed for μ-conotoxins SIIIA and SIIIB, where μ-SIIIA differs from μ-SIIIB by a single residue change in the third loop that results in μ-SIIIB preferentially targeting Nav1.4 (muscle) to Nav1.2 (neuron), while μ-SIIIA prefers neuronal VGSCs [57].
The M-superfamily peptides have advanced the methods used to identify novel peptides belonging to the M-5 branch based on exogene analysis. A study by Mandë Holford at the University of Utah presented the structure of a new sodium channel blocking conotoxin extracted from C. bullatus, μ-BuIIIA [12], which also shows an affinity for muscle VGSCs Nav1.4. μ-BuIIIA was identified and characterized by the isolation of exogenes which are the genes responsible for the mediation of biotic interactions between organisms [12]. Once isolated, these genes were compared with the genes of different cone snail species. The phylogenetic information resultant from this gene comparison approach was incorporated into a general strategy aimed at the discovery of new classes of peptides [12]. This technique led to the discovery of the C. bullatus μ-conotoxins, BuIIIA, BuIIIB, and BuIIIC. It is thought that this approach to conotoxin detection will expedite the characterization and reporting of novel peptides previously not identifiable by traditional means. After all, it was the sequence comparison of many M-superfamily peptides containing five amino acids in the third loop that resulted in the M-5 branch appearing in the literature in 2009.
Uncharacterized M-superfamily conotoxins
A new branch of the M-superfamily has been described in the literature that is homologous in signal sequence to other M-superfamily branches, but differs in cysteine order. The two conotoxins in this new category are Vx2 and Im6.1 [9, 15]. A study performed at the Shanghai Institute of Biological Sciences led to the publication of conotoxin Vx2. Their report demonstrated the commonality between cDNA and mRNA for conotoxin Vx2 and the M-4 branch peptides μ-GIIIA and μ-GIIIB. Out of the 25 amino acid residues comprising the signal sequences for Vx2 and the μ-GIIIA-C, there was only a difference in two residues: Leu9 was substituted by Val; and Met23 was replaced by Leu [9]. These differences are considered to be within an acceptable margin of variance to allow Vx2 to be characterized as an M-superfamily peptide. However, experts in the field disagree with this assignment because of the unique cysteine order in mature Vx2 (Table 1). M-superfamily peptides all have the characteristic cysteine arrangement of CC–C–C–CC while that of Vx2 is CCC–C–C–C [9].
Table 1.
A summary of known M-superfamily branches
O trans-4-hydroxyproline, Z Pyroglutamic acid, aC-terminal amidation
Conotoxin Im6.1, isolated from the venom of Conus imperialis, was placed in the same neoteric category of the M-superfamily as Vx2. Like Vx2, Im6.1 shows a similar cDNA and mRNA signal sequence consistent with other members of the M-superfamily. However, Im6.1 has the cysteine arrangement of C–C–CC–C–C with the disulfide connectivity C1–C4, C2–C5, and C3–C6 [15]. The biological targets of Vx2 and Im6.1 are yet to be identified.
Conclusion
The M-superfamily of conotoxins is arguably the most diverse of all the conotoxin superfamilies yet characterized. Five branches of this family have been definitively categorized with conotoxins Vx2 and Im6.1 demonstrating signal sequences consistent with the M-superfamily, yet with unique cysteine and cystine patterns. The study of the M-superfamily has been pivotal in increasing understanding of conotoxins and their specific receptors. Through study of the Maxi-M conotoxins, the μ-, κM-, and ψ-conotoxins have been characterized with their respective targets (Na+, K+, and nAChR). The Maxi-M peptides that constitute the M-4 and M-5 branches have defined disulfide connectivity and known target receptors.
The smaller Mini-M conotoxins of the M-1 and M-2 branches not only differ in cystine order from the Maxi-M peptides, but also from each other. This is unique to the M-superfamily and has led to the interesting discovery that conotoxins with different cystine pattern can have similar structures even when they belong to different branches of the superfamily. This is observed with the structures of the M-1 peptides mr3e and tx3a having dissimilar structure, while the M-2 peptide mr3a has a similar three dimensional structure to the M-1 peptide tx3a. This serves as another unique feature to the M-superfamily. The M-3 peptides can be said to have an uncharacterized disulfide pattern, unknown molecular target, and yet to be determined structural information.
Although much remains to be discovered regarding the M-superfamily of peptides, especially the Mini-M branches, the scientific advancement that has occurred through the study of these peptides has been significant. The study of Maxi-M conotoxins has involved the use of original techniques leading to the advancement in structure activity relationship comprehension for neuronal and muscular molecular targets. Research on the M-superfamily peptides has led to the development of new instruments, probes, and laboratory techniques that are broadly applicable to other fields of science. A specific example is the use of isotopic enrichment of cysteine side chains to characterize cystine arrangement by NMR. This technique can be applied to any disulfide rich peptide or protein to reduce the time required for structure determination and connectivity elucidation. Secondly, the use of exogene phylogeny comparison can be used across the genus Conus to identify and characterize peptides that would otherwise not be possible by traditional means.
The M-superfamily of conotoxins has displayed unique characteristics that have allowed for the development of therapeutic treatments and increased knowledge of receptors responsible for pain and sensory transmission. In summary, there remains much work to be done in this field.
Acknowledgments
We would like to thank Drs. Greg Bulaj and Kenneth A. Cornell for their contributions to this manuscript. Additional thanks are owed to Seth Eidemiller, Steven D. Jacob, Matthew Turner, Luke Woodbury, and Aubrey Johnston for their editorial input. This publication was made possible by NIH Grant #P20 RR016454 from the INBRE Program of the National Center for Research Resources, Mountain States Tumor Medical Research Institute, and Research Corporation Cottrell College Scholars Program.
References
- 1.Franco A, Pisarewicz K, Moller C, Mora D, Fields GB, Mari F. Hyperhydroxylation: a new strategy for neuronal targeting by venomous marine molluscs. Prog Mol Subcell Biol. 2006;43:83–103. doi: 10.1007/978-3-540-30880-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Myers RA, Cruz LJ, Rivier JE, Olivera BM. Conus peptides as chemical probes for receptors and ion channels. Chem Rev. 1993;93(5):1923–1936. doi: 10.1021/cr00021a013. [DOI] [Google Scholar]
- 3.Kohn AJ, Nishi M, Pernet B. Snail spears and scimitars: a character analysis of Conus radular teeth. J Mollus Stud. 1999;65:461–481. doi: 10.1093/mollus/65.4.461. [DOI] [Google Scholar]
- 4.McDougal OM, Turner MW, Ormond AJ, Poulter CD. Three-dimensional structure of conotoxin tx3a: an m-1 branch peptide of the M-superfamily. Biochemistry. 2008;47:2826–2832. doi: 10.1021/bi702388b. [DOI] [PubMed] [Google Scholar]
- 5.Olivera BM, Cruz LJ. Conotoxins, in retrospect. Toxicon. 2001;39:7–14. doi: 10.1016/S0041-0101(00)00157-4. [DOI] [PubMed] [Google Scholar]
- 6.Kohn AJ. Piscivorous gastropods of the genus Conus . Proc Natl Acad Sci USA. 1956;42:168–171. doi: 10.1073/pnas.42.3.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spence I, Gillessen D, Gregson RP, Quinn RJ. Characterization of the neurotoxic constituents of Conus geographus (L) venom. Life Sci. 1997;21:1759–1770. doi: 10.1016/0024-3205(77)90156-4. [DOI] [PubMed] [Google Scholar]
- 8.Clark C, Olivera BM, Cruz LJ. A toxin from Conus geographus venom which acts on the vertebrate central nervous system. Toxicon. 1981;19:691–699. doi: 10.1016/0041-0101(81)90106-9. [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, Wang CZ, Xu CQ, Fan CX, Dai XD, Chen JS, Chi CW. A novel M-superfamily conotoxin with a unique motif from Conus vexillum . Peptides. 2006;27:682–689. doi: 10.1016/j.peptides.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Olivera BM. Conus venom peptides, receptor and ion channel targets and drug design: 50 million years of neuropharmacology. Mol Biol Cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–798. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Holford M, Zhang MM, Gowd KH, Azam L, Green BR, Watkins M, Ownby JP, Yoshikami D, Bulaj G, Olivera BM. Pruning nature: biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus . Toxicon. 2009;53:90–98. doi: 10.1016/j.toxicon.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDougal OM (1998) Conus peptides investigated by NMR spectroscopic methods. Doctoral dissertation, University of Utah
- 14.Yao S, Zhang MM, Yoshikami D, Azam L, Olivera BM, Bulaj G, Norton RS. Structure, dynamics, and selectivity of the sodium channel blocker (mu)-conotoxin SIIIA. Biochemistry. 2008;47:10940–10949. doi: 10.1021/bi801010u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kass Q, Westermann JC, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24(3):445–446. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- 16.Mehdiratta R, Saberwal G. Bio-business in brief: the case of conotoxins. Curr Sci. 2007;92(1):39–45. [Google Scholar]
- 17.Watkins M, Olivera BM, Hillyard DR, McIntosh JM, Jones RM. Alpha-conotoxin peptides. United States Patent. 2007;7:279–549. [Google Scholar]
- 18.Alonso D, Khalil Z, Stakunanthan N, Livett BG. Drugs from the sea: conotoxins as drug leads for neuropathic pain and other neurological conditions. Mini Rev Med Chem. 2003;3:785–787. doi: 10.2174/1389557033487746. [DOI] [PubMed] [Google Scholar]
- 19.Bowersox SS, Luther R. Pharmacotherapeutic potential of omega-conotoxin MVIIA (SNX-111), an N-type neuronal calcium channel blocker found in the venom of Conus magus . Toxicon. 1998;36(11):1651–1658. doi: 10.1016/S0041-0101(98)00158-5. [DOI] [PubMed] [Google Scholar]
- 20.Atanassoff PG, Hartmannsgruber MW, Thrasher J, Wermeling D, Longton W, Gaeta R, Singh T, Mayo M, McGuire D, Luther RR. Ziconotide a new N-type calcium channel blocker, administered intrathecally for acute postoperative pain. Reg Anesth Pain Med. 2000;25:274–278. doi: 10.1016/s1098-7339(00)90010-5. [DOI] [PubMed] [Google Scholar]
- 21.Penn RD, Paice JA. Adverse effects associated with the intrathecal administration of ziconotide. Pain. 2000;85(1–2):291–296. doi: 10.1016/S0304-3959(99)00254-7. [DOI] [PubMed] [Google Scholar]
- 22.Levin T, Petrides G, Weiner J, Saravay S, Multz AS, Bailine S. Intractable delirium associated with ziconotide successfully treated with electroconvulsive therapy. Psychosomatics. 2002;43:63–66. doi: 10.1176/appi.psy.43.1.63. [DOI] [PubMed] [Google Scholar]
- 23.Skov MJ, Beck JC, de Kater AW, Shopp GM. Nonclinical safety of ziconotide: an intrathecal analgesic of a new pharmaceutical class. Int J Toxicol. 2007;26(5):411–421. doi: 10.1080/10915810701582970. [DOI] [PubMed] [Google Scholar]
- 24.Corpuz GP, Jacobsen RB, Jimenez EC, Watkins M, Walker C, Colledge C, Garrett JE, McDougal O, Li W, Gray WR, Hillyard DR, Rivier J, McIntosh JM, Cruz LJ, Olivera BM. Definition of the M-conotoxin superfamily: characterization of novel peptides from molluscivorous Conus venoms. Biochemistry. 2005;44:8176–8186. doi: 10.1021/bi047541b. [DOI] [PubMed] [Google Scholar]
- 25.Han YH, Wang Q, Jiang H, Liu L, Xiao C, Yuan DD, Shao XX, Dai QY, Chemng JS, Chi CW. Characterization of novel M-superfamily conotoxins with new disulfide linkage. FEBS J. 2006;273:4972–4982. doi: 10.1111/j.1742-4658.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 26.Du WH, Han YH, Huang FJ, Li J, Chi CW, Fang WH. Solution structure of an M-1 conotoxin with a novel disulfide linkage. FEBS J. 2007;274:2596–2602. doi: 10.1111/j.1742-4658.2007.05795.x. [DOI] [PubMed] [Google Scholar]
- 27.McDougal O, Poulter CD. Three-dimensional structure of the Mini-M conotoxin mr3a. Biochemistry. 2004;43:425–429. doi: 10.1021/bi0353732. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Jiang H, Han YH, Yuan DD, Chi CW. Two different groups of signal sequence in M-superfamily conotoxins. Toxicon. 2008;51:813–822. doi: 10.1016/j.toxicon.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Ferber M, Sporning A, Jeserich G, DeLaCruz R, Watkins M, Olivera BM, Terlau H. A novel Conus peptide ligand for K + channels. J Biol Chem. 2003;278(4):2177–2183. doi: 10.1074/jbc.M205953200. [DOI] [PubMed] [Google Scholar]
- 30.Al-Sabi A, Lennartz D, Ferber M, Gulyas J, Rivier JEF, Olivera BM, Carlomagno T, Terlau H. kappaM-conotoxin RIIIK, structural and functional novelty in a K+ channel antagonist. Biochemistry. 2004;43(27):8625–8636. doi: 10.1021/bi0495681. [DOI] [PubMed] [Google Scholar]
- 31.Verdier L, Al-Sabi A, Rivier JEF, Olivera BM, Terlau H, Carlomagno T. Identification of a novel pharmacophore for peptide toxins interacting with K + channels. J Biol Chem. 2005;280(22):21246–21255. doi: 10.1074/jbc.M502376200. [DOI] [PubMed] [Google Scholar]
- 32.Khoo KK, Feng ZP, Smith BJ, Zhang MM, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structure of the analgesic (mu)-conotoxin KIIA and effects on structure and function of disulfide deletion. Biochemistry. 2009;48:1210–1219. doi: 10.1021/bi801998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 34.Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. Cys-loop receptors: new twists and turns. Trends Neurosci. 2004;27(6):229–336. doi: 10.1016/j.tins.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Cruz LJ, Gray WR, Olivera BM, Zeikus RD, Kerr L, Yoshikami D, Moczydlowski E. Conus geagraphus Toxins that discriminate between neuronal and muscle sodium channels. J Biol Chem. 1985;260(16):9280–9288. [PubMed] [Google Scholar]
- 36.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol (Lond) 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catterall WA. The molecular basis of neuronal excitability. Science. 1984;233:653–661. doi: 10.1126/science.6320365. [DOI] [PubMed] [Google Scholar]
- 38.Ferber M, Al-Sabi A, Stocker M, Olivera BM, Terlau H. Identification of a mammalian target of κM-conotoxin RIIIK. Toxicon. 2004;43:915–921. doi: 10.1016/j.toxicon.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Shon K-J, Stocker M, Terlau H, Stuhmer W, Jacobsen R, Walker C, Grilley M, Watkins M, Hillyard DR, Gray WR, Olivera BM. kappa-Conotoxin PVIIA is a peptide inhibiting the shaker K + channel. J Biol Chem. 1998;273(1):33–38. doi: 10.1074/jbc.273.1.33. [DOI] [PubMed] [Google Scholar]
- 40.Jacobsen RB, Koch ED, Lange-Malecki B, Stocker M, Verhey J, Van Wagoner RM, Vyazovkina A, Olivera BM, Terlau H. Single amino acid substitutions in kappa-conotoxin PVIIA disrupt interaction with the shaker K + channel. J Biol Chem. 2000;275(32):24639–24644. doi: 10.1074/jbc.C900990199. [DOI] [PubMed] [Google Scholar]
- 41.Shon KJ, Grilley RJ, Cartier GE, Hopkins C, Gray WR, Watkins M, Hillyard DR, Rivier J, Torrres J, Yoshikami D, Olivera BM. A noncompetitive peptide inhibitor of the nicotinic acetylcholine receptor from Conus purpurascens venom. Biochemistry. 1997;36(31):9581–9587. doi: 10.1021/bi970235w. [DOI] [PubMed] [Google Scholar]
- 42.Lluisma AO, Lopez-Vera E, Bulaj G, Watkins M, Olivera BM. Characterization of a novel psi-conotoxin from Conus parius . Toxicon. 2008;51:174–180. doi: 10.1016/j.toxicon.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Wagoner RM, Jacobsen RB, Olivera BM, Ireland CM. Characterization and three-dimensional structure determination of psi-conotoxin PIIIF, a novel noncompetitive antagonist of nicotinic acetylcholine receptors. Biochemistry. 2003;42(21):6353–6362. doi: 10.1021/bi0272757. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell SS, Shon KJ, Foster MP, Davis DR, Olivera BM, Ireland CM. Three-dimensional solution structure of conotoxin psi-PIIIE, and acetylcholine gated ion channel antagonist. Biochemistry. 1998;37(5):1215–1220. doi: 10.1021/bi972186t. [DOI] [PubMed] [Google Scholar]
- 45.Van Wagoner RM, Ireland CM. An improved solution structure for psi-conotoxin PIIIE. Biochemistry. 2003;43(21):6347–6352. doi: 10.1021/bi027274e. [DOI] [PubMed] [Google Scholar]
- 46.Wang C-Z, Zhang H, Jiung H, Lu W, Zhao Z-Q, Chi C-W. A novel conotoxin from Conus striatus, mu-SIIIA, selectively blocking rat tetrodotoxin-resistant sodium channels. Toxicon. 2006;47:122–132. doi: 10.1016/j.toxicon.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Lewis RJ, Schroeder CI, Ekberg J, Nielson KJ, Loughnan M, Thomas L, Adams DA, Drinkwater R, Adams DJ, Alewood PF. Isolation and structure-activity of mu-conotoxin TIIIA, a potent inhibitor of tetrodotoxin-sensitive voltage-gated sodium channels. Mol Pharmacol. 2007;71:676–685. doi: 10.1124/mol.106.028225. [DOI] [PubMed] [Google Scholar]
- 48.Catterall WA, Goldin AL, Waxman SG. International union of pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 49.Shon KJ, Olivera BM, Watkins M, Jacobsen RB, Gray WR, Floresca CZ, Cruz LJ, Hillyard DR, Brink A, Terlau H, Yoshikami D. mu-Conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J Neurosci. 1998;18(12):4473–4481. doi: 10.1523/JNEUROSCI.18-12-04473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Safo P, Rosenbaum T, Shcherbatko A, Choi DY, Han E, Toledo-Aral JJ, Olivera BM, Brehm P, Mandel G. Distinction among neuronal subtypes of voltage-activated sodium channels by mu-conotoxin PIIIA. J Neurosci. 2000;20(1):76–80. doi: 10.1523/JNEUROSCI.20-01-00076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen KJ, Watson M, Adams DJ, Hammarstrom AK, Gage PW, Hill JM, Craik DJ, Thomas L, Adams D, Alewood PF, Lewis RJ. Solution structure of mu-conotoxin PIIA, a preferential inhibitor of persistent TTX-sensitive sodium channels. J Biol Chem. 2002;277:27247–27255. doi: 10.1074/jbc.M201611200. [DOI] [PubMed] [Google Scholar]
- 52.Nakamura M, Niwa Y, Ishida Y, Kohno T, Sato K, Oba Y, Nakumura H. Modification of Arg-13 of μ-conotoxin GIIIA with piperidinyl-Arg analogs and their relation to the inhibition of sodium channels. FEBS Lett. 2001;503:107–110. doi: 10.1016/S0014-5793(01)02714-4. [DOI] [PubMed] [Google Scholar]
- 53.Bulaj G, West PJ, Garrett JE, Watkins M, Zhan M-M, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44(19):7259–7265. doi: 10.1021/bi0473408. [DOI] [PubMed] [Google Scholar]
- 54.Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Kallen RG, Mandel G, Meiseler MH, Netter YB. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. doi: 10.1016/S0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 55.Wada A, Wanke E, Gullo F, Schiavon E. Voltage-dependent Nav1.7 sodium channels: multiple roles in adrenal chromaffin cells and peripheral nervous system. Acta Physiol. 2008;192:221–231. doi: 10.1111/j.1748-1716.2007.01810.x. [DOI] [PubMed] [Google Scholar]
- 56.Clare JJ, Tate SN, Nobbs M, Romanos MA. Voltage-gated sodium channels as therapeutic targets. Drug Discov Today. 2000;5:506–520. doi: 10.1016/S1359-6446(00)01570-1. [DOI] [PubMed] [Google Scholar]
- 57.Schroeder CI, Ekberg J, Nielsen KJ, Adams D, Loughnan ML, Thomas L, Adams DJ, Alewood PF, Lewis RJ. Neuronally mu-conotoxins from Conus striatus utilize an alpha-helical motif to target mammalian sodium channels. J Biol Chem. 2008;283:21621–21628. doi: 10.1074/jbc.M802852200. [DOI] [PubMed] [Google Scholar]
- 58.Kalso E. Sodium channel blockers in neuropathic pain. Curr Pharm Des. 2005;11:3005–3011. doi: 10.2174/1381612054865028. [DOI] [PubMed] [Google Scholar]
- 59.Wood JN, Boorman JP, Okuse K, Baker MD. Voltage-gated sodium channels and pain pathways. J Neurobiol. 2004;61:55–71. doi: 10.1002/neu.20094. [DOI] [PubMed] [Google Scholar]
- 60.Sato K, Ishida Y, Wakamatsu K, Kato R, Honda H, Ohizumi Y, Nakamura H, Ohya M, Lancelin JM, Kohda D. Active site of mu-conotoxin GIIIA, a peptide blocker of muscle sodium channels. J Biol Chem. 1991;266:16989–16991. [PubMed] [Google Scholar]
- 61.Hill JM, Alewood PF, Craik DJ. Three-dimensional solution structure of μ-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels. Biochemistry. 1996;35:8824–8835. doi: 10.1021/bi960073o. [DOI] [PubMed] [Google Scholar]
- 62.Walewska A, Skalicky JJ, Davis DR, Zhang M-M, Lopez-Vera E, Watkins M, Han TS, Yoshikami D, Olivera BM, Bulaj G. NMR-based mapping of disulfide bridges in cysteine-rich peptides: application to the mu-conotoxin SxIIIA. J Am Chem Soc. 2008;130(43):14280–14286. doi: 10.1021/ja804303p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang MM, Fiedler B, Green BR, Catlin P, Watkins M, Garrett JE, Smith BJ, Yoshikami D, Olivera BM, Bulaj G. Structural and functional diversities among mu-conotoxins targeting TTX-resistant sodium channels. Biochemistry. 2006;45:3723–3732. doi: 10.1021/bi052162j. [DOI] [PubMed] [Google Scholar]
- 64.West PJ, Bulaj G, Garrett JE, Olivera BM, Yoshikami D. µ-Conotoxin SmIIIA, a potent inhibitor of tetrodotoxin-resistant sodium channels in amphibian sympathetic and sensory neurons. Biochemistry. 2002;41(51):15388–15393. doi: 10.1021/bi0265628. [DOI] [PubMed] [Google Scholar]
- 65.Catteral WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/S0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 66.Keizer DW, West PJ, Lee EF, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structural basis for tetrodotoxin-resistant sodium channel binding by µ-conotoxin SmIIIA. J Biol Chem. 2003;278(47):46805–46813. doi: 10.1074/jbc.M309222200. [DOI] [PubMed] [Google Scholar]
- 67.Moczydlowski E, Olivera BM, Gray WR, Strichartz GR. Discrimination of muscle and neuronal Na-channel subtypes by binding competition between saxitoxin and p-conotoxins. Proc Natl Acad Sci USA. 1986;83:5321–5325. doi: 10.1073/pnas.83.14.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliveira JS, Redaelli E, Zaharenko AJ, Cassulini RR, Konno K, Pimenta DC, Freitas JC, Clare JJ, Wanke E. Binding specificity of sea anemone toxins to Nav 1.1-1.6 sodium channels: unexpected contributions from differences in the IV/S3–S4 outer loop. J Biol Chem. 2004;279:33323–33335. doi: 10.1074/jbc.M404344200. [DOI] [PubMed] [Google Scholar]