Abstract
In patients with Duchenne muscular dystrophy (DMD), the absence of a functional dystrophin protein results in sarcolemmal instability, abnormal calcium signaling, cardiomyopathy, and skeletal muscle degeneration. Using the dystrophin-deficient sapje zebrafish model, we have identified microRNAs (miRNAs) that, in comparison to our previous findings in human DMD muscle biopsies, are uniquely dysregulated in dystrophic muscle across vertebrate species. MiR-199a-5p is dysregulated in dystrophin-deficient zebrafish, mdx5cv mice, and human muscle biopsies. MiR-199a-5p mature miRNA sequences are transcribed from stem loop precursor miRNAs that are found within the introns of the dynamin-2 and dynamin-3 loci. The miR-199a-2 stem loop precursor transcript that gives rise to the miR-199a-5p mature transcript was found to be elevated in human dystrophic muscle. The levels of expression of miR-199a-5p are regulated in a serum response factor (SRF)-dependent manner along with myocardin-related transcription factors. Inhibition of SRF-signaling reduces miR-199a-5p transcript levels during myogenic differentiation. Manipulation of miR-199a-5p expression in human primary myoblasts and myotubes resulted in dramatic changes in cellular size, proliferation, and differentiation. MiR-199a-5p targets several myogenic cell proliferation and differentiation regulatory factors within the WNT signaling pathway, including FZD4, JAG1, and WNT2. Overexpression of miR-199a-5p in the muscles of transgenic zebrafish resulted in abnormal myofiber disruption and sarcolemmal membrane detachment, pericardial edema, and lethality. Together, these studies identify miR-199a-5p as a potential regulator of myogenesis through suppression of WNT-signaling factors that act to balance myogenic cell proliferation and differentiation.
Keywords: microRNA, zebrafish, miR-199a, WNT signaling, dystrophin, skeletal muscle
Skeletal muscle is a remarkably complex organ comprised of many different cell types that have important functional roles in disease, injury, and regeneration. Duchenne muscular dystrophy (DMD) is caused by mutations in the X-linked dystrophin gene.1 Although the molecular cause of the disease is well documented, the secondary signaling pathways that are altered by the lack of a functional dystrophin protein remain understudied.2, 3 Dystrophic muscle is known to undergo cycles of regeneration to form new myofibers as a consequence of myofibers loss and myoblast cell dysregulation.4 WNT (wingless-type MMTV integration site family) signaling factors are known to regulate both the maintenance of muscle satellite cells (MSCs), and their ability to differentiate into myofibers.5, 6, 7 Conversely, both dystrophic and aged skeletal muscle display activated WNT signaling resulting in increased muscle fibrosis.8 In DMD, a similar dysregulation of the transcriptional-level WNT signaling components has been reported.9
MicroRNAs (miRNAs) have been shown to have an essential role in muscle development, differentiation, and disease.10, 11, 12, 13, 14, 15 Previously, we defined a miRNA biosignature of different muscle diseases and revealed dysregulated miRNAs that were either common or unique to each muscle disease.12 Additional studies using the dystrophic mdx mouse muscle identified dysregulated miRNAs as a result of nNOS destabilization from the muscle membrane.11 Nevertheless, studies in the mdx dystrophic mouse model have limitations in their clinical usefulness due to the lack of severity of the disease progression in the mouse muscles. The sapje dystrophin-mutant zebrafish model is an excellent tool for studying dystrophic-muscle and disease progression, as the sapje mutants show dystrophic disease severity and can be analyzed in large numbers in a short period of time.16, 17 The sapje dystrophic zebrafish has a more severe muscle phenotype in which their dorsal muscles progressively waste resulting in significant early lethality as compared with mdx mouse models.16, 17, 18 The sapje zebrafish have weakened and degenerating muscle and the majority of mutants (95%) expire by 10 dpf due to the inability to swim and oxygenate their muscles. Thus, we chose to utilize the sapje dystrophic zebrafish as a model for analyzing dysregulated dystrophic miRNAs and to expand our previous findings obtained from DMD muscle biopsies.12
Here we have identified miRNAs that are commonly dysregulated in both zebrafish and human muscle DMD biopsies. One of these dysregulated miRNAs, miR-199a-5p, was conservatively induced in expression in dystrophic zebrafish, mouse, and human muscle. Previous studies have shown that miR-199a transcript is increased in expression in both human DMD muscle biopsies and the quadriceps and diaphragm muscles of mdx mice.12, 19 Overexpression or knockdown of miR-199a-5p in primary human myoblasts (MB) resulted in dramatic changes in cell size, differentiation, and proliferative capacity. Manipulation of miR-199a-5p levels in primary human MB and myotubes (MT) results in dramatic changes in myogenic cell proliferation, shape, and differentiation. Overexpression of miR-199a-5p exclusively in zebrafish muscle, results in myofiber disruption, pericardial edema, and early lethality, which resembles some of the phenotypic features observed in sapje dystrophic zebrafish. Several of the downstream targets of miR-199a-5p expression in the muscle belong to the WNT signaling pathway, which has been shown to regulate muscle stem cell proliferation and differentiation. Together, these studies identify miR-199a-5p as a potent regulator of the progression of myogenic differentiation in normal and dystrophic muscle by potentially modulating the expression levels of WNT signaling components.
Results
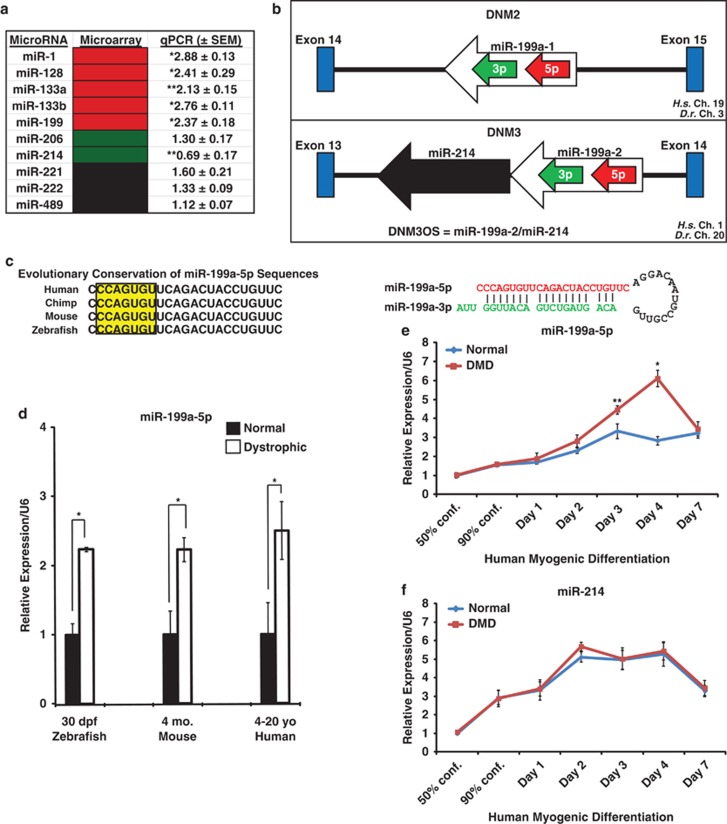
To identify miRNAs that were dysregulated in sapje (dystrophin exon 4 A/T transversion) mutant zebrafish, we performed miRNA microarray analysis on total mRNA isolated from unaffected and sapje zebrafish at 5 and 30 dpf. MiRNAs isolated at 5 and 30 dpf from unaffected and sapje zebrafish were compared using locally weighted scatterplot smoothing microarray analysis of dysregulated miRNAs. Comparisons revealed an altered miRNA biosignature in which several evolutionarily conserved miRNAs were dysregulated in the absence of dystrophin (Figure 1a). Additionally, sapje zebrafish that survived to 5 dpf exhibited uniquely dysregulated miRNAs earlier than zebrafish that survived to 30 dpf (Supplementary Figure S1A). Several muscle-enriched miRNAs were expressed at higher levels in sapje zebrafish at 5 dpf, but not at 30 dpf, which suggests that the dysregulated miRNAs in the zebrafish dystrophic disease progression may occur early in development. (Figure 1a; Supplementary Figures S1A and S1B).
Figure 1.
MiRNAs that are dysregulated in dystrophin-deficient skeletal muscle. (a) Table of dysregulated miRNAs in normal and dystrophic zebrafish at 30 dpf. Induced in expression miRNAs are shown in red, while reduced in expression miRNAs are shown in green. Unchanged or not significantly expressed miRNAs are shown in black. Comparisons in miRNA expression level changes by real-time qPCR are listed between sapje and unaffected zebrafish. Real-time qPCR values were normalized to U6 small nuclear RNA (snRNA) levels, and all experiments were performed in triplicate. (b) Genomic location of the miR-199a-5p transcript that is transcribed in both directions within introns of the dynamin-2 and dynamin-3 genes. Although both miR-199a-1 and miR-199a-2 genomic sequences can result in the formation of mature miR-199a-5p transcript, only miR-199a-2 is transcriptionally linked to miR-214 via its proximity in the genome. Chromosomal locations of the human (H.s.=Homo sapiens) and zebrafish (D.r.=Danio rerio) miR-199a-5p (red arrow) and miR-199a-3p (green arrow) mature miRNA sequences are shown. (c) The left panel shows evolutionary conservation of the human, chimp, mouse, and zebrafish miR-199a-5p mature miRNA sequences in the 5′ to 3′ orientation. Inset box with yellow highlighted area denotes conserved seven-nucleotide seed sequence of the miR-199a-5p mature miRNA. The right panel shows the miRNA stem loop formation of the miR-199a-5p (red lettering) and the miR-199a-3p (green lettering) structure with base-pairings. (d) Real-time qPCR of miR-199a-5p levels in normal and dystrophic muscles isolated from zebrafish (whole fish), mouse, and human biopsies. Normal versus DMD human muscle biopsies, wild-type versus mdx5cv (TA muscles; adult 3–6–month-old males), and unaffected versus sapje (5 dpf, n=10 fish). Real-time qPCR values were normalized to U6 snRNA levels, and all experiments were performed in triplicate. (e) Real-time qPCR of miR-199a-5p expression levels in normal and DMD cell lines during myogenic differentiation of MB into MT. (f) Real-time qPCR of miR-214 expression levels in normal human and DMD myogenic cell lines during myogenic differentiation. Expression normalized to 50% confluent MB and to U6 snRNA loading control. *P-value<0.005; **P-value<0.05. All samples analyzed in triplicate in three separate normal or DMD cell lines. ND, not determined
Using our previous miRNA expression analyses, we compared miRNAs that were dysregulated in both zebrafish (at both 5 and 30 dpf) and human DMD muscle biopsies.12 One of such dysregulated miRNAs was miR-199a-5p, a ubiquitously expressed miRNA that is enriched in muscle during early development12, 20 (Figure 1a; Supplementary Figures S1A and S1B). Mammals have two copies of the mature miR-199a-5p 23 nucleotide sequences embedded within the introns of the dynamin 2 and dynamin 3 loci that can be transcribed as miRNA stem loop structures, referred to as miR-199a-1 and miR-199a-2, respectively (Figure 1b). Another miRNA with a similar sequence homology to miR-199a-5p, miR-199b-5p, is transcribed from the dynamin-1 locus. However, real-time quantitative PCR (qPCR) analysis of the miR-199b miRNA stem loop transcript showed no significant dysregulation of miR-199b levels in human DMD muscle biopsies as compared with normal controls (Supplementary Figure S1E). The miR-199a-5p and antisense miR-199a-3p (previously referred to as miR-199a*) mature miRNA sequences arise from a common stem loop structure that is highly conserved across vertebrate species (Figure 1c). Unlike mammals, which have two miR-199a-5p and a miR-199b-5p mature miRNA sequences, zebrafish have three miR-199a-5p sequences (referred to as miR-199-5p) that lack the one base pair variant that distinguishes miR-199a-5p from miR-199b-5p (Supplementary Figure S1D). The miR-199a-2 stem-loop miRNA precursor sequence can also be co-transcribed on the opposite side of the dynamin-3 locus as a long-non-coding RNA along with miR-214, referred to as DNM3OS (dynamin-3 opposite strand). The trans stem-loop precursor sequence encodes for the opposite strand (or star*, miR-199a-3p) sequence at the dynamin-3 locus (Figures 1b and c). The antisense miR-199a-3p transcript showed a similar increase in expression in DMD MB during myogenic differentiation as compared with normal MB but at a different overall amount than the miR-199a-5p mature miRNA transcript (Supplementary Figure S1C). MiR-199a-5p levels were increased in expression when compared with the normal muscle in three vertebrate species, including dystrophic zebrafish (sapje; 30 dpf), mouse (mdx5cv; 4-months old), and DMD patient biopsies (4–20 years of age) (Figure 1d). During normal myogenic differentiation, miR-199a-5p levels increase but at a higher relative expression level in DMD MB compared with unaffected controls (Figure 1e). To further validate the induction of miR-199a transcript in dystrophic muscle, we performed real-time qPCR on the stem-loop miRNA precursor sequences. The miR-199a-2 stem loop precursor transcript was expressed at greater than twofold higher levels than normal muscle biopsies (Supplementary Figure S1E). The miR-199a-1 and miR-199b stem loop precursor sequences were also induced in DMD muscle biopsies relative to controls but not at significant levels (Supplementary Figure S1E). Additionally, we observed an induction of DNM2 and DNM3 expression in the DMD muscle biopsies, whereas DNM1 showed no induction in expression levels (Supplementary Figure S1F). MiR-214, which is thought to be co-transcribed with the miR-199a-2 stem loop, was also induced during myogenic differentiation but did not differ between unaffected and DMD myogenic cultures (Figure 1f).
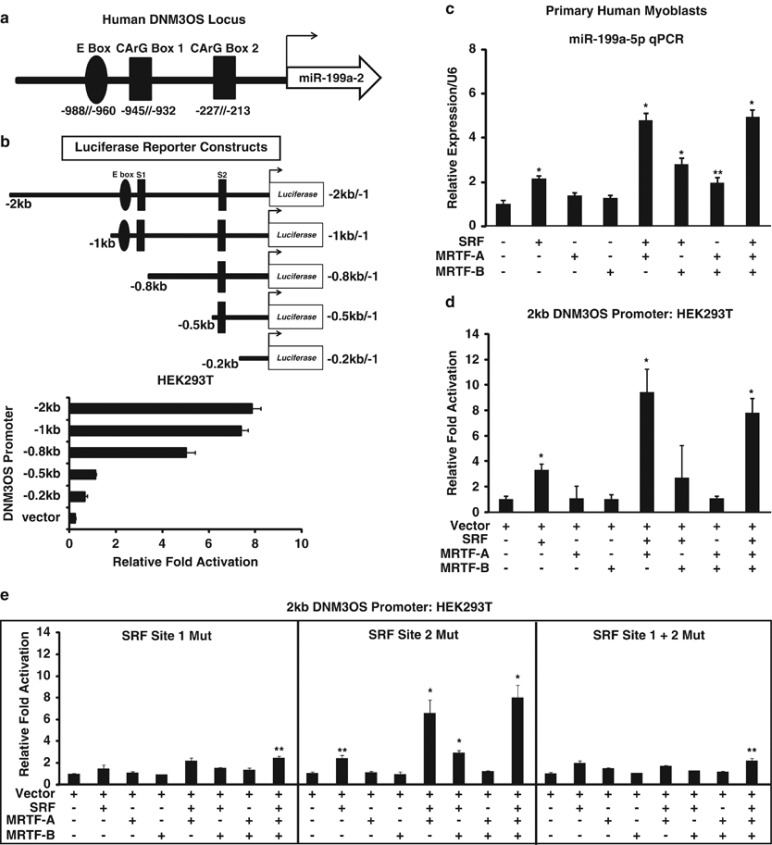
To determine which transcription factors regulate miR-199a-5p expression in muscle, we used the rVISTA 2.0 program and identified two evolutionarily conserved cis serum response factor (SRF) DNA-binding elements (also referred to as SREs or CArG boxes) at the 5′ upstream sequence of the DNM3OS locus. Previous studies demonstrated that miR-199a-5p transcription was activated by Egr1 and Twist1, two transcription factors that are modestly expressed in myogenic progenitors, but are expressed at low levels in differentiated MT.20, 21 However, miR-199a-5p expression levels are the highest in MT, which suggests that a transcriptional activator of miR-199a-5p expression might also increase during myogenic differentiation. In support of this hypothesis, SRF and its transcriptional cofactors MRTF (myocardin-related transcription factor)-A and MRTF-B, have been shown to increase in expression levels during myogenic differentiation.22, 23 SRF and MRTF transcription factors are known to regulate myogenic fusion, differentiation, regeneration, and the dystrophic disease pathology.24, 25, 26 Based on evolutionary conservation, we generated several luciferase reporter constructs (2, 1, 0.8, 0.5, and 0.2 kb) using DNA sequences upstream of the miR-199a-2 locus that contained either the SRF CArG box (labeled S1 or S2 for Site 1 or Site 2, respectively) along with a conserved E-box site to which the myogenic helix-loop-helix factors could potentially bind (Figures 2a and b). Overexpression of SRF and MRTF factors, specifically MRTF-A, was sufficient to strongly induce endogenous miR-199a-5p transcript in human primary MB (Figure 2c). Transfection of the 2-kb DNM3OS-luc reporter revealed low expression in the non-myogenic HEK293T cells and the highest expression of the luciferase reporter was obtained in primary MB (Figure 2d; data not shown). However, when the reporters were co-transfected with a plasmid that overexpressed SRF, a threefold increase of the luciferase reporter activity was observed (Figure 2d). SRF signaling in muscle requires the transcriptional co-factors that belong to the MRTFs MRTF-A and -B (also referred to as MKL1 and MKL2) to fully activate gene transcription. Thus, we overexpressed these factors along with the 2-kb DNM3OS-luc reporter to determine whether these factors were capable of activation of reporter and, in turn, miR-199a-2 transcript levels. Overexpression of SRF and MRTF-A together had the strongest activation of the 2-kb DNM3OS-luc reporter as compared with overexpression of both SRF and MRTF-B individually or together (Figure 2d). To test which CArG box (SRF-binding Site 1 or Site 2) was most critical for activation of the 2-kb hDNM3OS-luc reporter, we mutated three evolutionarily conserved nucleotides essential for SRF protein binding to DNA. Transfection of the mutated CArG boxes (Site 1, Site 2, and the double mutant) along with SRF and MRTF-A/B cofactors revealed a strong preference for SRF activation of the luciferase reporter at Site 1, next to the myogenic E box (Figure 2e). Ablation of the Site 2 CArG box showed little repression of SRF and MRTF-A/B's ability to activate the reporter, suggesting that the CArG box next to the moygenic E box (Site 1) is most essential for activation of the 2-kb hDNM3OS-luc reporter and potentially miR-199a-2 pri-miRNA transcript (Figure 2e). As previous studies have demonstrated that another transcription factor Yin-Yang 1 (YY1) can compete with SRF for binding to CArG boxes, we transfected YY1 along with the 2-kb hDNM3OS-luc reporter in HEK293T cells.27 YY1 overexpression inhibited the 2-kb hDNM3OS-luc reporter, supporting the functional role of YY1 as a repressor of miRNAs that regulate myogenic differentiation28 (Supplementary Figure S3A). Furthermore, overexpression of YY1 in primary human MB and MT showed little effect on overall miR-199a-5p transcript levels, suggesting that other transcription factors might regulate global miR-199-5p expression levels (Supplementary Figures S3B and S3C).
Figure 2.
MiR-199a-2 expression levels are regulated by a SRF/MRTF-dependent pathway. (a) Schematic of the DNM3OS (upstream sequence of hsa-miR-199a-2) 5′ upstream promoter that contains two conserved SRF CArG boxes and a single myogenic E box. Exact genomic locations of each of the transcription factor DNA-binding sites are listed as number of base pairs in the 5′ upstream region from the start of the human miR-199a-2 genomic sequence. (b) Schematic of human DNM3OS promoter deletion constructs fused to a luciferase reporter gene. Site 1 (SRF CArG box 1) is approximately 0.8 kb upstream of the pri-miR-199a-2 genomic sequence. Site 2 (SRF CArG box) is approximately 0.5 kb upstream of the pri-miR-199a-2 genomic sequence. Relative luciferase expression of the DNM3OS promoter deletion constructs in HEK293T cells. Luciferase values are normalized to the empty vector control. (c) Overexpression of the MRTF transcriptional co-factors along with SRF induces an increase in endogenous miR-199a-5p transcript levels in primary human MB. (d) Independent overexpression of SRF and MRTF transcription factors induce activation of the 2-kb DNM3OS luciferase reporter in HEK293T cells. Values are normalized to the vector alone control. (e) The Site 1 SRF CArG box is essential for driving the 2-kb hDNM3OS-luc reporter and miR-199a-2 expression. Transcriptional luciferase reporter assays in HEK293T cells overexpressing SRF, MRTF-A, and/or MRTF-B along with the 2-kb hDNM3OS-luc reporter with or without mutations in the two SRF CArG boxes (Sites 1 and 2). *P-value<0.005; **P-value<0.05. NS, not significant
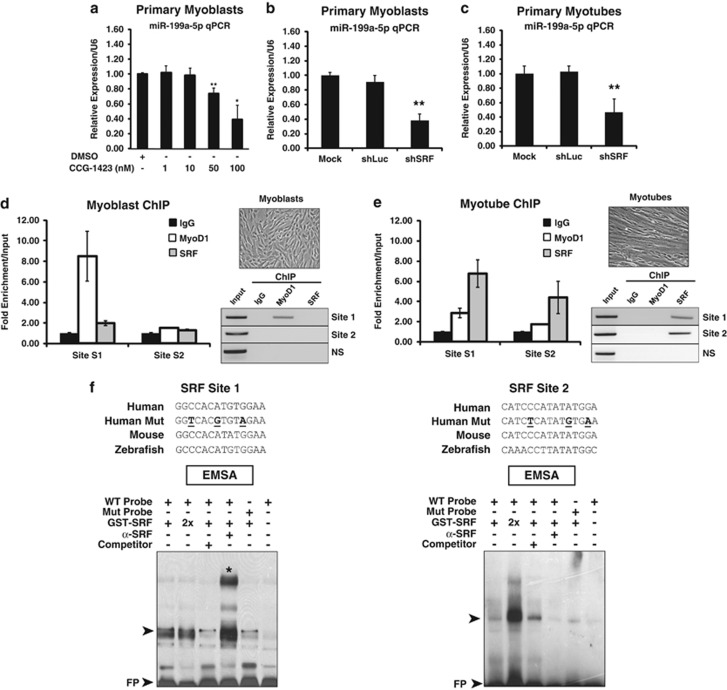
The small molecule CCG-1423 has been previously shown to block SRF/RhoA signaling by reducing nuclear MRTF-A levels and, as a result, preventing SRF from interacting with MRTF-A.29 We tested SRF signaling inhibition using different concentrations of CCG-1423 in MB, and we noticed a strong reduction in overall expression levels of endogenous miR-199a-5p transcripts (Figure 3a). Furthermore, shRNAi inhibition of SRF in both MB and MT resulted in a decrease of miR-199a-5p expression when compared with shLuciferase and mock controls (Figures 3b and c). To further validate these in vitro results, chromatin immunoprecipitation (ChIP) assays for endogenous SRF at the DNM3OS locus were performed on proliferating human primary MB and differentiated MT. Endogenous SRF protein bound to the two CArG box DNA-binding sites (S1 and S2) upstream of the miR-199a-2 sequence with different binding affinities but only in the MT (Figures 3d and e). Further analysis of the two putative SRF-binding sites by electrophoretic mobility shift assay (EMSA) revealed a stronger preference for SRF protein:DNA binding at the Site 1 CArG box as opposed to the Site 2 CArG box (Figure 3f). Thus, it is likely that the miR-199a-2 stem loop precursor miRNA and, subsequently, miR-199a-5p mature miRNA transcripts are induced by SRF-dependent signaling in differentiating human MB.
Figure 3.
SRF expression is essential for inducing miR-199a-5p expression in MB and MT. (a) Pharmacological inhibition of SRF blocks miR-199a-5p transcription levels. Real-time qPCR of miR-199a-5p levels of primary human MB treated with increasing levels of the SRF/Rho small-molecule inhibitor CCG-1423 as compared with a 10-nM DMSO control-treated cells. (b and c) SRF knockdown by shRNAi in MB (b) and MT (c) inhibits levels of miR-199a-5p transcript. A non-targeting (shLuciferase) construct is used as a negative control. *P-value<0.005; **P-value<0.05. (d and e) ChIP analysis of primary MB (d) and MT (e) reveals that SRF binds to the upstream (Sites 1 and 2) CArG boxes in differentiated human MT but not in MB (shown in upper panel). MyoD ChIP is used as a positive control in the myoblast samples. Real-time qPCR samples are normalized to input levels and showed as fold enrichment normalized to 10% input. A non-specific (ns) genomic sequence was also used as a negative control. ChIP PCR products obtained from MB and MT (both phase images shown) were resolved on agarose gels. All real-time qPCR of ChIP products were performed in triplicate. (f) EMSA of SRF protein binding CArG box Sites 1 and 2. Evolutionarily conserved putative CArG box sequence alignments are shown along with the three mutated nucleotides (bold, underlined). A stronger preference for Site 1 is observed. The top arrowhead denotes protein (SRF):DNA complex formation. A non-radiolabeled (competitor) was used to inhibit formation of the protein:DNA complex. The asterisk demarcates the supershift (SS) band. FP, free probe
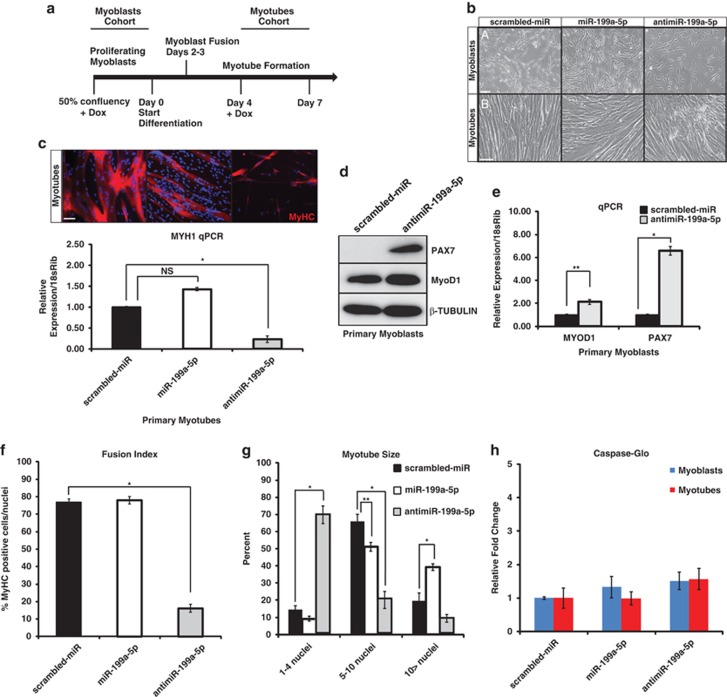
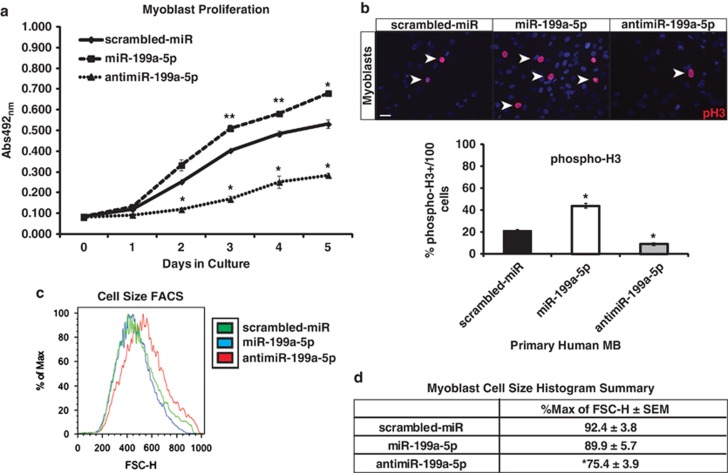
To determine what effects the manipulation of miR-199a-5p expression might have on muscle, normal human primary myoblast cell lines were utilized and miR-199a-5p expression was modulated using constitutively active and doxycycline (Dox)-inducible lentiviral constructs either in proliferating human MB or differentiated human MT (Figure 4a). Overexpression of miR-199a-5p did not affect the ability of normal MB to differentiate into MT, resulting in similar fusion indices as scrambled-miR controls (Figures 4b and c). qPCR of MYH1 (myosin heavy chain 1) showed no differences in expression levels between scrambled-miR and miR-199a-5p overexpressing primary MB (Figure 4c). However, quantification of myotube size by counting myonuclei per myosin heavy chain-positive MT revealed that the miR-199a-5p-overexpressing cultures contained greater numbers of myonuclei within the MT (Figures 4c). Lentiviral inhibition of miR-199a-5p (antimiR-199a-5p) in normal MB resulted in decreased myogenic differentiation as quantified by a myogenic fusion index assay (Figure 4f). This inhibition of miR-199a-5p expression was accompanied by an increase in the protein levels of MyoD1 and Pax7 (Figure 4d). These findings were further validated by qPCR, which showed a strong increase in PAX7 and a slight increase in MyoD1 mRNA expression in the antimiR-199a-5p primary MB as compared with scrambled-miR controls (Figure 4e). We observed no significant increase in the amount of cleaved Caspase-3/7 in MB or MT that were infected with miR-199a-5p-overexpressing or -inhibiting lentivirus, suggesting that the effects on myogenic differentiation by miR-199a-5p expression levels are not due to increased cell death (Figure 4h). Additionally, MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) proliferation assays demonstrated that MB-overexpressing miR-199a-5p displayed increased proliferation as compared with the scrambled-miR (control) MB (Figure 5a). Primary MB-overexpressing miR-199a-5p showed increased numbers of phosphorylated-Histone H3 (Ser10)-positive cells, which confirmed a higher level of cellular mitosis (Figure 5b). Conversely, miR-199a-5p-inhibited MB showed significantly reduced numbers of phospho-Histone H3 positive MB (Figure 5b). Using fluorescence-activated cell sorting (FACS) analysis, we observed a significant increase in myoblast cell size when miR-199a expression was inhibited (Figures 5c and d). These results validate a functional role for miR-199a expression levels as a means of regulation for myoblast proliferation and cell size.
Figure 4.
MiR-199a-5p levels in MB and MT have profound effects on myogenesis and cell proliferation. (a) Schematic for manipulating miR-199a-5p expression in normal human MB and MT. Doxycycline (+Dox) is administered at 50% myoblast confluency for analysis of effects in the proliferative phase. In the other experiment, Dox is given 4 days following the addition of differentiation media for myotube analysis. (b) Phase microscopy (a and b) of Dox-induced MB (a) or MT (b) following infection with scrambled-miR, miR-199a-5p, or antimiR-199a-5p lentivirus. Note the reduced proliferation and myotube destruction in primary human MB infected with antimiR-199a-5p lentivirus. (c) Immunofluorescence of sarcomeric myosin heavy chain (MyHC) in human primary MT. Real-time qPCR of mRNA levels of MYH1 (myosin heavy chain 1) normalized to 18sRib in scrambled-miR-, miR-199a-5p-, or antimiR-199a-5p-infected primary MT. P-value<0.005; **P-value<0.05. NS, non-significant. Bar=50 μm. (d) WB of the myogenic and proliferation markers PAX7 and MyoD1 in MB-overexpressing either scrambled-miR or antimiR-199a-5p. β-Tubulin is used as a loading control. (e) Real-time qPCR of mRNA levels of PAX7, MYOD1 in scrambled-miR, or antimiR-199a-5p-infected primary human MB. Total mRNA levels are normalized to a 18sRib loading control. (f) Fusion index of MyHC-positive MT infected with scrambled-miR, miR-199a-5p, or antimiR-199a-5p. (g) Myotube size as determined by myonuclei counts in MT infected with scrambled-miR, miR-199a-5p, or antimiR-199a-5p in three categories: 1–4 nuclei/fiber; 5–10 nuclei/fiber, and >10 nuclei/fiber. (h) Caspase-glo assay of protein lysates from MB (blue bars) and MT (red bars) that express miR-199a-5p, antimiR-199a-5p, or scrambled-miR control lentivirus. *P-value<0.005; **P-value<0.05
Figure 5.
MiR-199a-5p expression regulates myoblast proliferation and cell size. (a) The proliferation rates of MB infected with scrambled-miR, miR-199a-5p, or antimiR-199a-5p over 5 days post infection and using a MTS cell proliferation assay. (b) Phospho-Histone-H3 (pH3; red) immunofluorescent staining of primary human MB infected with lentivirus-expressing scrambled-miR, miR-199a-5p, or antimiR-199a-5p. Table showing percentage of phospho-Histone H3-postive MB out of 100 DAPI-positive cells. All infections and immunostaining was performed in triplicate. Bar=40 μm. (c) FACS forward scatter height analysis of myoblast cell size. (d) Table of histogram values from FACS forward scatter analysis. *P-value<0.005; **P-value<0.05. All samples analyzed in triplicate in three separate cell lines per lentivirus used
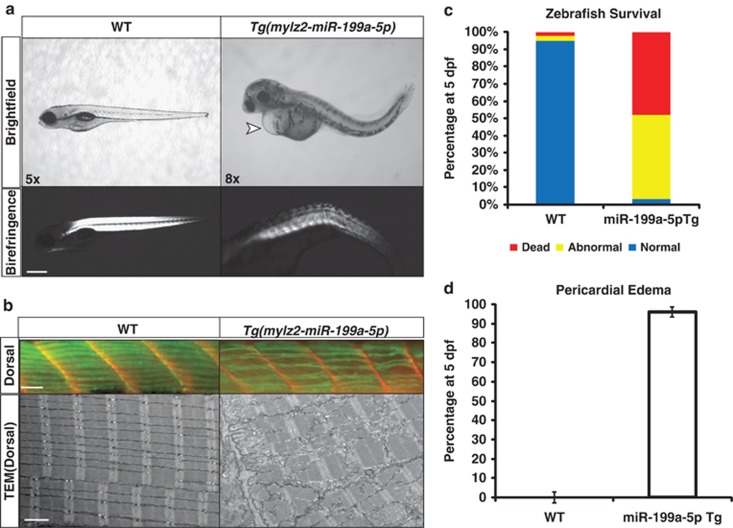
To determine what effects overexpression of miR-199a-5p might have in vivo, a transgenic (Tg) zebrafish line that overexpressed zebrafish miR-199a-5p using the mylz2 (myosin light chain polypeptide 2) promoter (specifically the zebrafish dre-miR-199a-1 miRNA genomic sequence) exclusively in muscle lineages was generated. The mylz2 promoter sequence has been previously shown to drive expression of green fluorescent protein (GFP) reporter constructs specifically in zebrafish skeletal muscle fibers, excluding muscle stem cell progenitors.30 Stable F1 generation miR-199a-5p, henceforth referred to as Tg(mylz2-miR-199a-5p), zebrafish developed enlarged yolk sacs with visible pericardial edema (Figures 6a and d). Immunohistochemical analysis of their skeletal muscles revealed massive fiber disruption, poor muscle birefringence, and detachment of the myofibers from the myosepta (Figure 6b). Additional ultrastructural analysis of the miR-199a-5p Tg zebrafish dorsal muscle revealed massive disorganization of the myofibers and triad regions (Figure 6b). The large amount of myofiber detachment and pericardial edema resulted in high lethality among the Tg(mylz2-miR-199a-5p) by 5 dpf, as compared with zebrafish that did not carry the transgene (Figure 6c). By 5 dpf, the majority (96%) of the Tg(mylz2-miR-199a-5p) had developed noticeable pericardial edema high lethality observed in this Tg line (Figures 6c and d). Given that previous findings have shown a high level of miR-199a-5p expression in the developing mouse and zebrafish mesodermal tissues, it is probable that miR-199a-5p has important myogenic and non-myogenic roles in early embryonic development.20, 31
Figure 6.
Overexpression of miR-199a-5p transcript in vivo results in myofiber disruption and early lethality in zebrafish. (a) Morphological and histological analysis of Tg(mylz2-miR-199a-5p) Tg zebrafish and their wild-type (WT) littermate controls as shown by brightfield microscopy. Patchy birefringence is observed in the miR-199a-5p Tg fish that highlights weakened and disorganized myofibers. Pericardial edema is denoted by the arrowhead (upper panel). (b) Dorsal muscle of Tg(mylz2-miR-199a-5p) Tg zebrafish and their WT littermate controls is immunostained for myosin heavy chain (green) and myosepta are stained with laminin (red). Transmission electron microscopy reveals massive myofiber disruption and disorganization in miR-199a-5p-overexpressing zebrafish. (c) Histogram characterizing the Tg(mylz2-miR-199a-5p) zebrafish survival percentage as compared with WT (Tg negative) littermates. The color codes demarcate zebrafish that are either normal (blue), dead (red), or abnormal (yellow). (d) Percentage of zebrafish with noticeable pericardial edema WT or Tg(mylz2-miR-199a-5p) Tg fish offspring at 5 dpf. Scale bars denote percentage error of the mean, and samples were analyzed in three separate mating cohorts
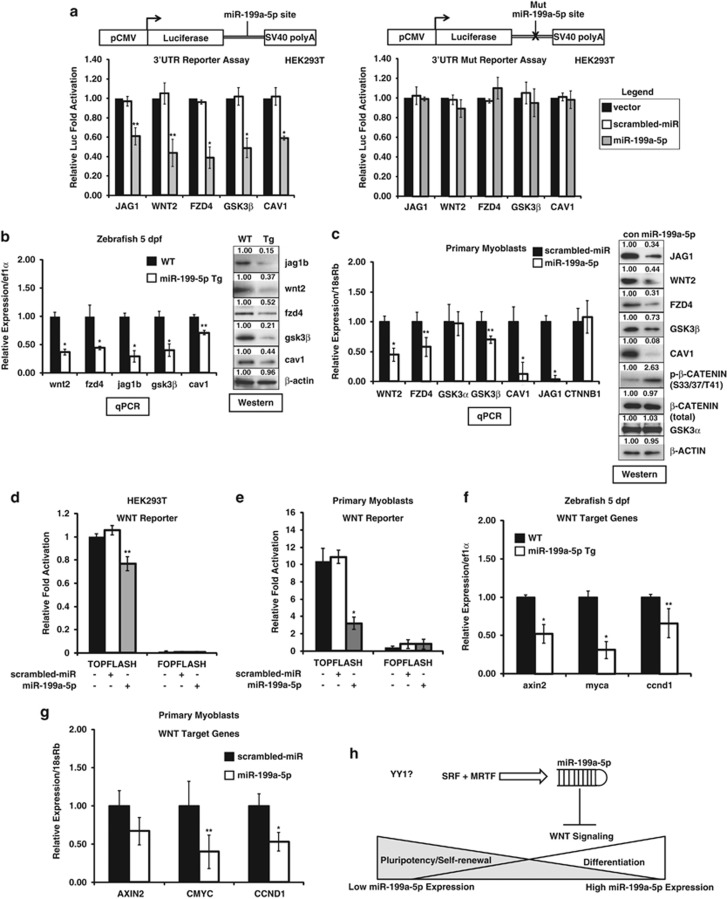
Using miRNA target prediction programs such as TargetScan, MicroCosm, and miRDB, with an emphasis on those targets whose miR-199a-5p-binding sites were evolutionarily conserved and had been defined as having a functional role in muscle, several members of the WNT signaling pathway were identified as predicted downstream targets (Figure 7a and Supplementary Figure S2). Previous work had identified several hypoxia-regulated mRNAs, such as HIF1α and SIRT1, as being direct miR-199a-5p targets in cardiomyocytes.32 Emphasis was placed on the WNT signaling factors WNT2, FZD4, and JAG1, as these three genes have been shown to have an important role in muscle stem cell differentiation and/or proliferation.5, 33, 34, 35, 36 Additionally, these WNT-signaling factors are reduced in expression levels during myogenic differentiation, whereas miR-199a-5p levels reciprocally increase in expression. Most of the WNT-signaling factors have evolutionarily conserved (zebrafish to human) miR-199a-5p seed sites in their 3′ untranslated region (UTR) regions with the exception of CAV1 and GSK3β whose seed sites were conserved only in mammals (Supplementary Figure S2). MiR-199a-5p overexpression in HEK293T cells suppressed the 3′UTR of each predicted WNT signaling target (fused to luciferase open reading frame (ORF)) reporter (Figure 7a). Mutation of the miR-199a-5p-binding site in each of the 3′UTR luciferase reporters was able to ablate the inhibition of the luciferase reporter by miR-199a-5p (Figure 7a). Given that miR-199a-5p overexpression in vivo resulted in profound myofiber structural changes, levels of WNT signaling components were measured in the 199a-5p Tg-overexpressing zebrafish. The Tg(mylz2-miR-199a-5p) zebrafish showed significant reduction of the miR-199a-5p-targeted WNT pathway genes wnt2, fzd4, and jag1b (Figure 7b). Overexpression of miR-199a-5p in normal human MB strongly inhibited endogenous protein levels of each of the miR-199a-5p-predicted WNT pathway genes (Figure 7c). The TOPFLASH (8xTCF/LEF-binding sites) and FOPFLASH (8xTCF/LEF sites mutated) have been used in WNT reporter assays to characterize WNT signaling levels in various cell lines.37, 38 To further validate miR-199a-5p as a regulator of WNT signaling, we overexpressed miR-199a-5p along with the TOPFLASH or the control FOPFLASH luciferase reporters in either HEK293T cells or normal human primary MB. Overexpression of miR-199a-5p inhibited the TOPFLASH (8xTCF/LEF-binding sites) but not the FOPFLASH reporter in both HEK293T cells and human primary MB (Figures 7d and e). These findings indicate that constitutive overexpression of miR-199a-5p in muscle has a strong inhibitory effect on WNT signaling during early vertebrate muscle development. As miR-199a-5p overexpression strongly inhibited several WNT signaling components, we measured β-catenin protein levels by western blot (WB) analysis, which showed an increase in phosphorylated β-catenin but no change in total expression levels (Figure 7c). Additionally, we observed no change in the protein levels of GSK3α in the miR-199a-5p overexpressing MB, which suggests that GSK3 proteins may still phosphorylate β-catenin at critical functional residues (Figure 7c). To further validate miR-199a-5p as a regulator of the WNT signaling pathway, we measured the levels of several well-characterized WNT/β-catenin-downstream target genes axin2, myca (zebrafish c-myc ortholog), and ccnd1 (cyclin D1) in the Tg(mylz2-miR-199a-5p) zebrafish and primary MB overexpressing miR-199a-5p lentivirus.39 The levels of these miR-199a-5p target genes were significantly reduced in miR-199a-5p overexpressing zebrafish compared with wild-type controls (Figures 7f and g). These results suggest that high levels of miR-199a-5p inhibit WNT signaling, which drives myoblast proliferation and differentiation.
Figure 7.
MiR-199a-5p targets several components of the WNT signaling pathway in muscle. (a) The schematic of constructs used for transfection of the 3′UTR luciferase reporter constructs with either the miR-199a-5p-binding site or mutated miR-199a-5p seed site. The reporter was transfected with either scrambled-miR (control; white boxes) or miR-199a-5p overexpression constructs (grey boxes) and normalized to luciferase expression levels of the vector alone (black boxes). The ablation of the miR-199a-5p inhibition is observed when the miR-199a-5p site is mutated in the 3′UTR luciferase reporter constructs. (b) Real-time qPCR of endogenous mRNA expression levels of miR-199a-5p WNT signaling target genes (cav1, wnt2, fzd4, jag1b, and gsk3β) in wild-type (WT) and Tg(mylz2-miR-199a-5p) zebrafish at 5 dpf in three separate Tg or control lines. WB analysis of the same five miR-199a-5p WNT targets along with a β-actin loading control. WB densitometries are shown where each miR-199a-5p Tg zebrafish blot is normalized to the corresponding WT blot. (c) Real-time qPCR and WB analysis of endogenous expression levels of miR-199a-5p WNT targets following overexpression of either control (scrambled-miR) or miR-199a-5p in human primary MB. Additional WBs of total β-catenin, phospho-β-catenin (Ser33/37/Thr41), GSK3α, and a β-actin loading control are also shown. WB densitometries are shown where each miR-199a-5p-overexpressing myoblast blot is normalized to the corresponding control blot. (d and e). Inhibition of the WNT (8xTCF/LEF sites) TOPFLASH and FOPFLASH (8xTCF/LEF sites mutated) luciferase reporter constructs by miR-199a-5p in (d) HEK293T and (e) human primary MB. Luciferase values were normalized to empty vector controls. (f) Real-time qPCR expression of the endogenous levels of the WNT/β-catenin downstream target genes axin2, myca, and ccnd1 in WT (black bars) and Tg(mylz2-miR-199a-5p) (white bars) zebrafish at 5 dpf in three separate Tg or control lines (n=3 replicates per individual sample; all expression values are normalized to an ef1α loading control). (g) Real-time qPCR expression of the endogenous levels of WNT/β-catenin downstream target genes AXIN2, CMYC, and CCND1 in scrambled-miR (white bars) and miR-199a-5p (black bars) overexpressing human primary MB. (n=3 replicates per individual sample; all expression values are normalized to an 18sRb loading control). (h) Schematic of miR-199a-5p regulation of WNT signaling during myogenic differentiation. MiR-199a-5p targets several WNT signaling components that affect myoblast proliferation and differentiation capacity. Low levels of miR-199a-5p exist in proliferating MB, whereas an increase in miR-199a-5p levels that occurs during myogenic differentiation is concordant with miR-199a-5p transcriptional activation by SRF and MRTF transcription factors
Discussion
MiR-199a-5p is a ubiquitously expressed miRNA whose expression is modulated at key time points of development, growth, regeneration, and in various muscle diseases and cancers.12, 21, 40 Another recent study showed that miR-199a-5p expression was upregulated in mice with idiopathic pulmonary fibrosis (IPF), suggesting that miR-199a-5p could regulate tissue fibrosis in different types of fibroproliferative diseases.41 MiR-199a-5p can also regulate cardiomyocyte size during cardiac hypertrophy under ischemic conditions.42 A hypoxic environment, which could trigger the increase in miR-199a-5p expression, has been described in dystrophin-deficient skeletal muscle.43
The results classify miR-199a-5p in the same family of SRF-regulated muscle miRNAs (or myomiRs) miR-1, miR-133a/b, miR-143, miR-145, miR-206, and miR-486.44, 45, 46, 47, 48 Additionally, they suggest a SRF/MRTF-dependent mechanism for the induction of miR-199a-5p transcription and subsequent downregulation of WNT signaling to allow for normal myogenic differentiation to occur (Figure 7h). Another factor capable of binding to the same CArG box DNA-binding site as SRF, Yin-Yang 1, is highly expressed in MB and is displaced from muscle structural gene promoters during myogenic differentiation.27 Our findings suggest that miR-199a-5p transcription, similar to what has been shown for other SRF-regulated miRNAs, is likewise repressed by YY1 expression levels.28 Interestingly, miR-199a-5p expression is also induced in interstitial skeletal muscle side population cells but absent in MSCs.49, 50 This observation may be explained by the fact that SRF and MRTF factors have been detected in the interstitial cells that line the vasculature, which are absent in undifferentiated MSCs before myogenic differentiation.51, 52 The induction of miR-199a-5p expression levels, coinciding with increased SRF expression during myogenic differentiation, further supports the hypothesis that SRF is a regulator of muscle hypertrophic growth while suppressing MSC proliferation.25 Recently, it has been shown that induction of WNT7a can induce muscle hypertrophy by activating the PI3K/AKT pathway in myofibers.53 Given that we demonstrated miR-199a-5p overexpression promotes myoblast proliferation but has a modest effect in MT, one might hypothesize that miR-199a-5p's regulation of WNT signaling may be acting in a similar mechanism. As previous findings have shown a high level of expression of miR-199a-5p in developing zebrafish and mouse mesodermal tissues, it is likely that miR-199a-5p has important non-myogenic roles in early development.20, 31
Genomic removal of both the miR-199a-1 and miR-199a-2 transcripts along with a miR-199a-1/miR-199a-2 compound deletion in mice may elucidate functional roles of each miR-199a-5p transcript. Further characterization of miR-199a-5p under different pathogenic conditions might elucidate its function as a regulator of other non-WNT signaling targets. Additionally, these studies open up the possibility that inhibitors of miR-199a-5p expression might improve dystrophic muscle function and alleviate some of the symptoms associated with these devastating diseases.
Materials and Methods
Zebrafish lines
Wild-type (AB strain) zebrafish were housed in sterile fish tanks. The sapje fish line has been described previously.16, 54 The Tg(mylz2-miR-199a-5p) Tg zebrafish were generated by injecting a modified version pKS-GM2f plasmid that contained the 2 kb mylz2 5′ upstream promoter sequence as described elsewhere.55 A 0.5-kb amplicon containing the zebrafish miR-199-2 genomic sequence was cloned by PCR into the BamH1 and HindIII restriction sites and the final plasmid construct was linearized by Not1 restriction digestion. All zebrafish embryo injections were performed in one-cell AB zebrafish embryos at a DNA concentration of 100 pg/nl. Three Tg(mylz2-miR-199a-5p) F0 founder lines were generated and used for all subsequent experiments. Overexpression of zebrafish miR-199a-5p was confirmed by real-time qPCR for zebrafish miR-199a-5p expression levels. Zebrafish were housed, fed, and maintained as breeding stocks as described elsewhere.56 All zebrafish were housed in the Boston Children's Hospital Aquatics facility (Director Christian Lawrence) under the animal protocol number: 09-10-1534R.
Mouse husbandry and muscle biopsies
Wild-type C57Bl6/J mice and mdx5cv mice were originally obtained from Jackson Laboratories, Bar Harbor, ME, USA. Muscle tissue was homogenized with a FastPrep FP120 bead homogenizer (ThermoScientific, Waltham, MA, USA) and miRNA was extracted using the miRVana miRNA kit (Ambion, Houston, TX, USA). Normal and mdx5cv mice were housed in sterile, pathogen-free conditions in the Boston Children's Hospital ARCH animal facilities under the animal protocol number: 10-12-1852R.
Human biopsies and establishment of primary myoblast cell lines
All normal and DMD skeletal muscle biopsies for the establishment of myoblast cell lines were obtained during either diagnostic skeletal muscle biopsies or other clinical surgeries as described previously.10 All patients gave written and oral consent before surgery and all protocols were approved by the Boston Children's Hospital human subjects internal review board protocol number: 03-12-205R.
Mouse tibialis anterior (TA) muscle extractions
The TA muscles from age-matched normal and mdx5cv male, 4-month-old mice were harvested for miRNA and mRNA following euthanization by CO2 gas. Muscle miRNA and mRNA were extracted using the miRVana (Ambion) kit as previously described.10
MiRNA isolation and real-time qPCR
MiRNA and total mRNA was extracted from cell lines, fish embryos, or mouse/human muscle biopsies using a miRVana Isolation kit (Ambion). Fifty nanograms of total miRNA per sample was copied into complementary DNA (cDNA) using the Multiscribe RT System (Applied Biosystems, Foster City, CA, USA). One microliter of RT reaction was used for each specific miRNA Taqman assay (Applied Biosystems), which was amplified using NoAmp Erase Taqman Polymerase (Applied Biosystems) following the manufacturer's instructions. All human miRNA Taqman precursor assays were obtained commercially (Applied Biosystems). Pri-microRNA Taqman assays were run using a Taqman Universal Master Mix Kit (Applied Biosystems) following the manufacturer's instructions. All samples were analyzed in 96-well optical plates on an ABI 7900HT real-time PCR machine (Applied Biosystems). Cycle times were normalized to U6 (or RNU6-2 for pri-miRNA assay qPCR) small nuclear RNAs as loading controls. Relative fold expression and changes were calculated using the 2−ΔΔCt method as described previously.10
For all SYBR green-based real-time qPCR assays, total RNA from muscle biopsies and cell cultures was extracted using the mirVana miRNA/mRNA extraction kit (Ambion). One microgram of total RNA was copied into cDNA using the First-Strand Synthesis kit (Invitrogen, Carlsbad, CA, USA). Ten-fold serial dilutions of the cDNA were analyzed on 96-well optical plates using the Power SYBR Green Mix system (Applied Biosystems) and analyzed on the ABI 7900HT real-time PCR machine (Applied Biosystems). Primers specific for each gene were designed using NCBI-PrimerBlast, and all cycle time values were normalized to an 18sRb loading control. Real-time qPCR primers sequences for all SYBR green-based assays are listed in Supplementary Figure S4A.
MiRNA microarrays
MiRNA was extracted from unaffected and sapje (3 pools of each zebrafish strain containing approximately 20 fish per pool at 30 dpf, and 2 separate pools of each zebrafish strain at 5 dpf) that were obtained from sapje heterozygous matings. Following miRNA extraction (see miRNA isolation methods above), miRNA samples were flashPAGE fractionated and purified (Ambion) as previously described.12 MiRNA samples were hybridized to zebrafish miRNA microarrays, and all data were gathered, processed, and analyzed by LC Sciences LLC (Houston, TX, USA). Clustal hierarchical analysis was performed using analysis of variance-1 statistical analysis. MiRNA microarray data files were deposited in NCBI GEOmnibus under the accession number GSE40204.
Lentivirus production
Lentivirus vectors that constitutively overexpress a miR-199a-5p precursor, antimiR-199a-5p (miRZip), or scrambled-miR control all three of which also contained bicistronic internal ribosome entry site (IRES)-GFP tags were obtained commercially (System Biosciences Inc., Mountain View, CA, USA) and generated in HEK293T cells using third-generation lentiviral packaging plasmids (MDL/RRE, Rev, and VSV-G) as previously described.10 Dox-inducible lentivirus was generated by cloning a 500 base-pair genomic fragment containing the human miR-199a-2 precursor sequence into the LV-TetO plasmid57 (gift from Dr. Konrad Hochedlindger, Harvard). A shRNAi hairpin-targeting human miR-199a-5p was subcloned by PCR from the miRZip vector into the Dox-inducible LV-TetO plasmid. Cells infected with the LV-miR-199a-5p-overexpressing or -inhibiting (antimiR-199a-5p) virus were also co-infected with a reverse-tetracycline transactivator (rtTA) virus at the same multiplicity of infection (MOI). A YY1-overexpressing lentivirus was generated by cloning a YY1-3xHA ORF into the Xba1 and BamH1 restriction sites of the pCDH-EF1-MCS-IRES-RFP vector (System Biosciences Inc.). Dox hyclate (Sigma-Aldrich, St. Louis, MO, USA) was added at an optimal concentration of 50 ng/ml to induce the expression of miRNA or antimiR-199a-5p hairpin. Most lentiviral infections were done using a MOI ranging from 10 to 100.
Cell cultures
HEK293T cells (GeneHunter Corp., Nashville, TN, USA) were maintained using 10% FBS (Atlanta Biologicals, Lawrenceville, GA, USA)/Dulbecco's Modification of Eagle Medium (Invitrogen) supplemented with antibiotics (Antibiotic-Antimycotic; Invitrogen). Primary human MB from normal and DMD patients were isolated and maintained in cell culture using Human Skeletal Muscle Growth Media (Promocell, Heidelberg, Germany) supplemented with 20% FBS and antibiotics as previously characterized.10
Plasmids
Plasmids overexpressing either a non-targeting (scrambled) miRNA or human miR-199a-2 were obtained commercially (Origene, Rockville, MD, USA). The human SRF ORF was subcloned by PCR into pIRES-2a-hrGFP (Strategene, La Jolla, CA, USA) from a purchased clone MHS1011-9199653 (Open Biosystems, Huntsville, AL, USA). Plasmids overexpressing human MRTF-A or B were obtained from Addgene (Addgene plasmids 19838 and 27175; Cambridge, MA, USA) and were originally generated from the laboratory of Robert Prywes and described elsewhere.23, 58 The TOPFLASH and FOPFLASH WNT-reporter plasmids were a gift from Leonard Zon and have been characterized elsewhere.59 Plasmids overexpressing human MYOD1 and SRF were generated by PCR of the ORFs from commercially obtained ESTs (Open Biosystems) into pIRES-1a-hrGFP or pIRES-2a-hrGFP (Strategene). The human YY1 ORF (NCBI Accession Number: NM_003403.3) was cloned from normal human muscle cDNA into pIRES-2a-hrGFP. The 2, 1, 0.8, 0.5, and 0.2 kb human DNM3OS promoter constructs were cloned into pGL4.20 (Promega, Madison, WI, USA) at the Nhe1/BglII restriction sites. Mutagenesis of the CArG box DNA sequences from (Site 1) 5′-GGCCACATGTGGAA-3′ to 5′-GGTCACGTGTAGAA-3′ and (Site 2) 5′-CATCCCATATATGGA-3′ to 5′-CATCTCATATGTGAA-3′ was performed using the Quik Change II kit (Stratagene). All luciferase-based promoter assays were performed using the Dual-Glo Assay system (Promega).
Antisera
A table of primary antibodies and their concentrations in different applications can be found in Supplementary Figure S5. Secondary antibodies used in this study for WB analysis include anti-mouse and anti-rabbit horse radish peroxidase (HRP)-conjugated (Cell Signaling Technology, Beverly, MA, USA; 1 : 2000 WB dilution), along with anti-goat HRP-conjugated (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1 : 1000 WB dilution). Secondary antibodies (Invitrogen) in mouse and rabbit Alexa-Fluor fluorochromes that were conjugated to fluorochromes to be excited at either 488 or 568 nm were used for immunofluorescent detection of primary antisera.
FACS analysis
Muscle MB were sorted using FACS for detection and were stained with propidium iodide (2 μg/ml; BD Biosciences, San Diego, CA, USA) to exclude dead cells. All cells were analyzed on a FACSVantage flow cytometer (BD Biosciences), and all FACS plots were analyzed using FloJo software (Version 6.4.7; Tree Star Inc., Ashland, OR, USA).
Western Blots
All protein preparations were quantified using a bicinchoninic acid kit (ThermoScientific). Thirty micrograms of whole-cell protein lysate was resolved on 4–20% gradient Novex tris-glycine gels (Invitrogen). Following a transfer of proteins onto PVDF membranes (Invitrogen), the membranes were then blocked with 5% non-fat milk/1 × tris-buffered saline (TBS)-Tween 20 (Sigma-Aldrich), before being incubated overnight at 4 °C with primary antisera diluted in 5% milk/TBS-Tween 20 (Boston BioProducts Inc., Ashland, MA, USA). The membranes were washed in a series of three TBS-Tween 20 washes. The secondary HRP-conjugated antibodies were diluted in 5%milk/TBS-Tween 20 before incubation with the PVDF membranes for 1 h at room temperature. Following additional washes before application of chemoluminscent substrate (Millipore, Billerica, MA, USA), the membranes were exposed on blue film (Genesee Scientific, San Diego, CA, USA) using varying exposure times. Uncropped image scans of WBs are located in Supplementary Figure S6.
Immunohistochemistry/fluorescence
Each group of cells was fixed at each of the time points with 4% paraformaldehyde for 10 min at 4 °C with gentile agitation. Following fixation, the cells were washed twice in 1 × phosphate-buffered saline (PBS), three times in 0.1% Triton-X/1 × PBS, and then incubated in blocking buffer (10% horse serum, Invitrogen; 0.1% Triton-X, Sigma-Aldrich; mixed in 1 × PBS, Mediatech, Inc, Manassas, VA, USA) for 1 h at room temperature with light shaking. Following blocking, the cells were incubated with primary antisera overnight at 4 °C with gentle shaking. Cells were incubated in secondary antisera of either AlexaFluor-488 or AlexaFluor-568 (Invitrogen) conjugated to either mouse or rabbit immunoglobulin G (IgG). Cells were then washed three times with 0.1% Triton-X/1x PBS before being mounted with Vectashield containing DAPI (4′,6-diamidino-2-phenylindole; Vector Labs, Burlingame, CA, USA). Cells that had been stained were photographed with a Nikon II fluorescent microscope (Nikon Instruments Inc., Melville, NY, USA) using SPOT software (Spot Imaging Solutions, Sterling Heights, MI, USA).
Non-Tg and Tg(mylz2-miR-199a-5p) Tg 5 dpf zebrafish were fixed in 4% paraformaldehyde (EM Sciences, Gibbstown, NJ, USA) overnight with gentle rocking at 4 °C. The samples were then rehydrated with a series of methanol washes (75–50%) and blocked in a buffer consisting of 2% casein, 1% bovine serum albumin fraction V, and 0.05% Tween-20 to reduce non-specific binding. Following overnight incubation at 4 °C in primary antisera, zebrafish samples were incubated in secondary antisera of either AlexaFluor-488 or AlexaFluor-568 (Invitrogen) conjugated to either mouse or rabbit IgG. The samples were then imaged with a Nikon II fluorescent microscope using OpenLab software Version 3.1.5 (Improvision/Perkin Elmer, Waltham, MA, USA).
SRF inhibition
Primary MB were treated with the SRF signaling small-molecule inhibitor CCG-1423 (Cayman Chemical, Ann Arbor, MI, USA), diluted in DMSO (Sigma-Aldrich), at concentrations of 1, 10, 50, and 100 nM, and incubated for 48 h. To accomplish SRF shRNAi knockdown, a double-strained small hairpin DNA oligo was ligated into the BamH1 and EcoR1 restriction sites of the pGreenPuro shRNAi lentivector (Catalog number SI506A-1; System Biosciences Inc.). The cloning primers used to design the SRF shRNAi lentivector included: sense: 5′-AGAGAATGAGTGCCACTGG-3′ and antisense 5′-CCAGTGGCACTCATTCTCT-3′ 19-nt sequences targeting the human SRF ORF. Following infection with shRNAi targeting either SRF or a shLuciferase (System Biosciences Inc.), the cells were incubated with puromycin (RPI Corp., Mt. Prospect, IL, USA) at a concentration of 1 μg/ml before selection 48 h post infection.
Caspase-glo assays
A Caspase-glo 3/7 assay (Promega) kit was used to measure the amounts of cleaved-Caspase 3/7 following the manufacturer's protocol. Normal human primary MB were seeded at 5 × 104 cells in six-well plates, infected with lentivirus, and assayed 48 h following infection in a protocol previously described.10
GST expression and protein purification
Recombinant GST-tagged SRF and control GST protein were generated by first cloning the full-length ORF of human SRF into the EcoR1 and Xho1 restriction sites of pGEX6P-1 (GE Healthcare Life Sciences, Piscataway, NJ, USA). Recombinant GST-tagged proteins were purified from BL21Gold (Agilent Technologies, Santa Clara, CA, USA) Escherichia coli following a 6-h IPTG (isopropyl β-D-1-thiogalactopyranoside; Sigma-Aldrich) induction using the GST GraviTrap kit in accordance with the manufacturer's protocol (GE Healthcare Life Sciences).
EMSA
EMSA assays were performed using recombinant GST-tagged SRF or control GST protein and oligos corresponding to the CArG DNA-binding elements located 5′ upstream of the human DNM3OS promoter (Supplementary Figure S4C). Oligos containing ⩾2 5′ guanine overhangs were resuspended in sterile water to a concentration of 100 μM. Oligos were then annealed to each other by first boiling them in 1X annealing buffer (100 mM Tris-HCl, pH=8.0; 150 mM NaCl; and 10 mM MgCl2) at 100 °C for 5 min, followed by a 1 h cool-down to room temperature. The annealed primer pairs were radiolabeled using ATP-γ-32P (Perkin Elmer) and T4 polynucleotide kinase (Promega) for 30 min at 37 °C before purification on nucleic acid-binding columns (Roche Applied Sciences; Basel, Switzerland). One microliter of radiolabeled oligos (5 × 104 counts per minute) was incubated at 37 °C for 30 min, with 1 μl 20 mM DTT, 1 μl of poly dI-dC (1 μg/μl), 4 μl of EMSA buffer (0.1 M HEPES, 200 mM KCl, 5 mM EDTA, 20 mg/ml BSA, 15% glycerol), and 5 or 10 μg (2 × ) of recombinant protein extract, brought to a final volume of 20 μl with sterile water. For the supershift of the DNA:protein complex, the sample was incubated with 3 μg of SRF antibody (Santa Cruz Biotechnology) for 2 h at 4 °C before incubation with the radiolabeled oligo. The samples were then resolved on 5% TBE/polyacrylamide gels for 2 h, before being dried, and exposed overnight at −80 °C on blue film (Genesee Scientific). Some gels were exposed on phospho-imaging cassettes, and images were acquired using a Typhoon Variable Mode Imager (Amersham Pharmacia, Amersham, UK).
MiRNA reporter assays
Following prediction of the transcripts that were thought to be targets of miR-199a-5p, the 3′UTR regions of these genes were amplified by PCR from a human total mRNA library (Ambion). The 3′ UTR amplicons were then cloned into the SpeI and NotI restriction sites of a modified version of the pGL2Basic vector containing a novel multi-cloning site from the pCR4-TOPO vector as described previously.10 To start the miRNA 3′UTR-luciferase reporter assays, 20 000 HEK293T cells per well were seeded in 48-well plates. The following day, the cells were transfected (Lipofectamine 2000; Invitrogen) with 30 ng of 3′UTR miRNA luciferase reporter constructs and 100 ng of miR-486 over-expression plasmid (Origene; pCMVmiR-IRES-GFP vector) or scrambled miRNA controls (Origene). Forty-eight hours post transfection, cells were lysed in RIPA buffer (ThermoScientific), and 20 μl of whole cell lysate assayed with 50 μl of luciferase substrate using the Dual Reporter Assay (Promega). Luciferase levels were measured on a single-tube luminometer (Berthold Technologies, Bad Wildbad, Germany). The seed sites in the 3′UTR luciferase reporters were mutated from 5′-GTACAGG-3′ to 5′-GgAaAtG-3′ using the Quik Change II kit (Stratagene).
ChIP
The myoblast fraction and primary human MT were harvested for ChIP using the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology) following the manufacturer's instructions. Briefly, MB were seeded at 4 × 105 cells/15 cm dish in 10 dishes per cohort. The myoblast fraction consisted of normal human MB grown to approximately 70% confluency. For the myotube fraction (five 15 cm dishes), after reaching 90% confluency, MB were differentiated into MT by serum withdrawal (2% FBS) for 4 days. Cells were harvested following three washes in ice-cold 1 × DPBS, were crosslinked with 1% formaldehyde (Sigma-Aldrich) for 1 h at RT, and then chromatin was sheared using by incubating the DNA pellets with micrococcal nuclease for 20 min at 37 °C. Ten percent of chromatin was removed for input controls, while the approximately 200 μg of total chromatin was used to purify DNA fragments following immunoprecipitation with 5 μg of either anti-MyoD1 (Santa Cruz Biotechnology; sc-760) or anti-SRF (Santa Cruz Biotechnology; sc-335) antibodies overnight at 4 °C under gentle rotation. An IgG (5 μg total; BD Pharmingen, San Diego, CA, USA) was used as an internal control. The following morning, the samples were incubated with 30 μl of ChIP Grade Protein G magnetic beads (Cell Signaling Technology) for 2 h at 4 °C under gentle rotation. Following several stringent washes, the chromatin was eluted off the columns in 1 mM Tris-HCL pH 8.0 (Sigma-Aldrich) and purified on DNA-binding columns before a final elution in sterile water. Following quantification on the NanoDrop spectrophotometer (ThermoScientific), the chromatin was amplified using primers specific for the SRF CArG box-binding site(s) or a non-specific DNA sequence (ChIP primer sequences are listed in Supplementary Figure S4B).
Transmission electron microscopy
Zebrafish were fixed in a mixture of glutaraldeyhyde-formaldeyhyde-picric acid mixture (EM Sciences) overnight at 4 °C. Samples then underwent osmication and uranyl acetate staining before undergoing a series of ethanol washes. The samples were then embedded in TAAB embedding resin (Marivac Ltd., Halifax, Nova Scotia, Canada) and one-micron sections were cut on a Leica ultracut microtome (Leica Microsystems; Wetzlar, Germany). The sections were then placed on 100 μm copper grids and stained with 0.2% lead citrate. The sections were viewed on a Philips Tecnai BioTwin Spirit Electron Microscope (Philips, Amsterdam, The Netherlands) and imaged by the Harvard Medical School Electron Microscopy Core.
MTS proliferation assays
MB were seeded at 1000 cells per 96-well gelatin-coated (0.1% porcine; Sigma-Aldrich) dish. MTS assay was performed using the Cell Titer 96 Non-Radioactive Cell Proliferation Assay (Promega) in accordance with the manufacturer's instructions. Absorbance values were measured using a Synergy 2 (BioTek, Winooski, VT, USA) plate reader and calculated using Gen5 1.08 software (BioTek). All absorbance readings were normalized to either blank, non-infected, or scrambled-miR internal control values.
Statistical analysis
Unless otherwise stated, all statistical analyses were performed using a Student's t-test (two-tailed).
URLs
GEO Database: http://www.ncbi.nlm.nih.gov/geo/
Sanger miRBase: http://mirbase.org/
MicroCosm (v5): http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/
miRDB: http://mirdb.org/miRDB/
Primer-BLAST (NCBI): http://www.ncbi.nlm.nih.gov/tools/primer-blast/
rVista (v2.0): http://rvista.dcode.org/
TargetScan (v6.2): www.targetscan.org
Acknowledgments
Funding for this work was generously provided by the Bernard F. and Alva B. Gimbel Foundation (LMK) and through a NIH grant (P50 NS040828-10). MSA was funded by a traineeship awarded by the NIH through the Harvard Stem Cell Institute and supported by a Muscular Dystrophy Association (MDA) Development Grant MDA255059. PBK is supported by the Muscular Dystrophy Association (MDA186796 and MDA 114353) and the Genise Goldenson Fund. ATK is supported by an NIH grant HL091124. The F59 (developed by F.E. Stockdale), MF20 (developed by D.A. Fischman), and Pax7 (developed by A. Kawakami) antibodies were obtained from the Developmental Studies Hybridoma Bank (DSHB) developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA, USA. We thank Louise Trakimas for assistance with the electron microscopy and Christoph Eicken at LC Sciences LLC for assistance with the microarray data analysis and processing. Flow cytometry was performed in the IDDRC Stem Cell Core Facility at Boston Children's Hospital that is supported by National Institutes of Health award NIH-P30-HD18655. We thank Emanuela Gussoni for a critical reading of the manuscriptOur thanks to Teresa Bowman for sharing zebrafish WNT signaling primer sequences and Bryan MacDonald for helpful discussions about the WNT signaling pathway. We thank Dr. Morris White and Dr. Kyle Copps for our usage of imaging equipment. We also thank all the families and patients who donated their biopsies and time for these experiments.
Author contributions
MSA designed and performed the experiments and analyzed the data. JCC, NM, JAM, MJG, SM, and GK performed the experiments and helped in the analysis of the data. IE performed the experiments and contributed new reagents. ATK analyzed the microRNA microarray data. EAE collected patient samples, family histories, and reagents. FS isolated muscle biopsies from normal and DMD patients. PBK contributed new reagents and edited the manuscript. MSA and L.M.K. analyzed all of the data and wrote the manuscript. All authors have read and approved of the manuscript before submission.
Glossary
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- DMD
Duchenne muscular dystrophy
- DNM3OS
dynamin-3 opposite strand
- Dox
doxycycline
- EMSA
electrophoretic mobility shift assay
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- HRP
horse radish peroxidase
- IgG
immunoglobulin G
- IRES
internal ribosome entry site
- MB
myoblasts
- miRNA
microRNA
- MOI
multiplicity of infection
- MRTF
myocardin-related transcription factor
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- MSC
muscle satellite cell
- MT
myotubes
- mylz2
myosin light chain polypeptide 2
- ORF
open reading frame
- PBS
phosphate-buffered saline
- qPCR
quantitative PCR
- snRNA
small nuclear RNA
- SRF
serum response factor
- TA
tibialis anterior
- TBS
tris-buffered saline
- Tg
transgenic
- YY1
Yin-Yang 1
- WB
western blot
- WNT
wingless-type MMTV integration site family
- UTR
untranslated region
LMK is a consultant for Pfizer Inc. for muscle disease drug therapies. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Bakay M, Zhao P, Chen J, Hoffman EP.A web-accessible complete transcriptome of normal human and DMD muscle Neuromuscul Disord 200212: SupplementS125–S141. [DOI] [PubMed] [Google Scholar]
- Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, et al. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci USA. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Webster C, Pavlath GK. Defective myoblasts identified in Duchenne muscular dystrophy. Proc Natl Acad Sci. 1983;80:4856–4860. doi: 10.1073/pnas.80.15.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121:2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- Le Grand F, Jones AE, Seale V, Scimè A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- Pescatori M, Broccolini A, Minetti C, Bertini E, Bruno C, D'amico A, et al. Gene expression profiling in the early phases of DMD: a constant molecular signature characterizes DMD muscle from early postnatal life throughout disease progression. FASEB J. 2007;21:1210–1226. doi: 10.1096/fj.06-7285com. [DOI] [PubMed] [Google Scholar]
- Alexander M, Casar JC, Motohashi N, Myers JA, Eisenberg I, Gonzalez RT, et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skeletal Muscle. 2011;1:27. doi: 10.1186/2044-5040-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M, et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne Muscular Dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad f Sci. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D, Currie PD. Identification of a zebrafish model of muscular dystrophy. Clin Exp Pharmacol Physiol. 2004;31:537–540. doi: 10.1111/j.1440-1681.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- Kawahara G, Karpf JA, Myers JA, Alexander MS, Guyon JR, Kunkel LM. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc Natl Acad Sci. 2011;108:5331–5336. doi: 10.1073/pnas.1102116108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslett J, Kang P, Han M, Kho AT, Sanoudou D, Volinski JM, et al. The influence of muscle type and dystrophin deficiency on murine expression profiles. Mamm Genome. 2005;16:739–748. doi: 10.1007/s00335-005-0053-8. [DOI] [PubMed] [Google Scholar]
- Roberts TC, Blomberg KEM, McClorey G, Andaloussi SE, Godfrey C, Betts C, et al. Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol Ther Nucleic Acids. 2012;1:e39. doi: 10.1038/mtna.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-B, Bantounas I, Lee D-Y, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Furukawa C, Haraguchi T, Inada K, Shiogama K, Tagawa T, et al. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res. 2011;71:1680–1689. doi: 10.1158/0008-5472.CAN-10-2345. [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C, Vandromme M, Tuil D, Lautredou N, Morris M, Soulez M, et al. Expression and activity of serum response factor is required for expression of the muscle-determining factor MyoD in both dividing and differentiating mouse C2C12 myoblasts. Mol Biol Cell. 1996;7:719–729. doi: 10.1091/mbc.7.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj A, Prywes R. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J Biol Chem. 2003;278:41977–41987. doi: 10.1074/jbc.M305679200. [DOI] [PubMed] [Google Scholar]
- Soulez M, Rouviere CG, Chafey P, Hentzen D, Vandromme M, Lautredou N, et al. Growth and differentiation of C2 myogenic cells are dependent on serum response factor. Mol Cell Biol. 1996;16:6065–6074. doi: 10.1128/mcb.16.11.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerci A, Lahoute C, Hébrard S, Collard L, Graindorge D, Favier M, et al. Srf-dependent paracrine signals produced by myofibers control satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2012;15:25–37. doi: 10.1016/j.cmet.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Mokalled MH, Johnson AN, Creemers EE, Olson EN. MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Dev. 2012;26:190–202. doi: 10.1101/gad.179663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvagni F, Lestingi M, Cartocci E, Oliviero S. Serum response factor and protein-mediated DNA bending contribute to transcription of the dystrophin muscle-specific promoter. Mol Cell Biol. 1997;17:1731–1743. doi: 10.1128/mcb.17.3.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, et al. A novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS ONE. 2012;7:e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Goldfine AB, Boes T, Henry RR, Ciaraldi TP, Kim EY, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Clin Invest. 2011;121:918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B, Chong SW, He J, Wang X, Xu Y, Wan H, et al. Recapitulation of fast skeletal muscle development in zebrafish by transgenic expression of GFP under the mylz2 promoter. Dev Dyn. 2003;227:14–26. doi: 10.1002/dvdy.10273. [DOI] [PubMed] [Google Scholar]
- He X, Yan Y-L, DeLaurier A, Postlethwait JH. Observation of miRNA gene expression in zebrafish embryos by in situ hybridization to microRNA primary transcripts. Zebrafish. 2011;8:1–8. doi: 10.1089/zeb.2010.0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1α and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozopas KM, Nusse R. Direct flight muscles in Drosophila develop from cells with characteristics of founders and depend on DWnt-2 for their correct patterning. Dev Biol. 2002;243:312–325. doi: 10.1006/dbio.2002.0572. [DOI] [PubMed] [Google Scholar]
- Wang H, Gilner JB, Bautch VL, Wang DZ, Wainwright BJ, Kirby SL, et al. Wnt2 coordinates the commitment of mesoderm to hematopoietic, endothelial, and cardiac lineages in embryoid bodies. J Biol Chem. 2007;282:782–791. doi: 10.1074/jbc.M606610200. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Gnocchi VF, White RB, Ono Y, Ellis JA, Zammit PS. Further characterisation of the molecular signature of quiescent and activated mouse muscle satellite cells. PLoS ONE. 2009;4:e5205. doi: 10.1371/journal.pone.0005205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, et al. XTcf-3 transcription factor mediates β-catenin-induced axis formation in xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman DA. β-catenin/XTcf-3 complex binds to thesiamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman Teresa V, Lawton Lee N, Fan ZP, Wu DC, DiBiase A, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HH, Davis AJ, Lee TL, Pang AL, Nagrani S, Rennert OM, et al. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene. 2011;30:3404–3415. doi: 10.1038/onc.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, et al. miR-199a-5p is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 2013;9:e1003291. doi: 10.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X-W, Li Q, Lin L, Wang XC, Li DF, Wang GK, et al. MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol. 2010;225:437–443. doi: 10.1002/jcp.22217. [DOI] [PubMed] [Google Scholar]
- Sander M, Chavoshan B, Harris SA, Iannaccone ST, Stull JT, Thomas GD, et al. Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc Natl Acad Sci. 2000;97:13818–13823. doi: 10.1073/pnas.250379497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Azhar G, Helms S, Wei J. Regulation of cardiac microRNAs by serum response factor. J Biomed Sci. 2011;18:15. doi: 10.1186/1423-0127-18-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N, Alexander MS, Casar JC, Kunkel LM. Identification of a novel microRNA that regulates the proliferation and differentiation in muscle side population cells. Stem Cells Dev. 2012;21:3031–3043. doi: 10.1089/scd.2011.0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goupille O, Pallafacchina G, Relaix F, Conway SJ, Cumano A, Robert B, et al. Characterization of Pax3-expressing cells from adult blood vessels. J Cell Sci. 2011;124:3980–3988. doi: 10.1242/jcs.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Honore A, Lamb NJ, Vandromme M, Turowski P, Carnac G, Fernandez A. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol Biol Cell. 2003;14:2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J, Bentzinger CF, Rudnicki MA. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat Cell Biol. 14:186–191. doi: 10.1038/ncb2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon JR, Goswami J, Jun SJ, Thorne M, Howell M, Pusack T, et al. Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Human Mol Genet. 2009;18:202–211. doi: 10.1093/hmg/ddn337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C, Best J, James A, Maloney K. The effects of feeding frequency on growth and reproduction in zebrafish (Danio rerio) Aquaculture. 2012;368-369:103–108. [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlich S, Wang R, Lee S-M, Lewis TC, Dai C, Prywes R. Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol Cell Biol. 2008;28:6302–6313. doi: 10.1128/MCB.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RTA. Transgenic Lef1/β-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.