Abstract
It is recognized now that intrinsically disordered proteins (IDPs), which do not have unique 3D structures as a whole or in noticeable parts, constitute a significant fraction of any given proteome. IDPs are characterized by an astonishing structural and functional diversity that defines their ability to be universal regulators of various cellular pathways. Programmed cell death (PCD) is one of the most intricate cellular processes where the cell uses specialized cellular machinery and intracellular programs to kill itself. This cell-suicide mechanism enables metazoans to control cell numbers and to eliminate cells that threaten the animal's survival. PCD includes several specific modules, such as apoptosis, autophagy, and programmed necrosis (necroptosis). These modules are not only tightly regulated but also intimately interconnected and are jointly controlled via a complex set of protein–protein interactions. To understand the role of the intrinsic disorder in controlling and regulating the PCD, several large sets of PCD-related proteins across 28 species were analyzed using a wide array of modern bioinformatics tools. This study indicates that the intrinsic disorder phenomenon has to be taken into consideration to generate a complete picture of the interconnected processes, pathways, and modules that determine the essence of the PCD. We demonstrate that proteins involved in regulation and execution of PCD possess substantial amount of intrinsic disorder. We annotate functional roles of disorder across and within apoptosis, autophagy, and necroptosis processes. Disordered regions are shown to be implemented in a number of crucial functions, such as protein–protein interactions, interactions with other partners including nucleic acids and other ligands, are enriched in post-translational modification sites, and are characterized by specific evolutionary patterns. We mapped the disorder into an integrated network of PCD pathways and into the interactomes of selected proteins that are involved in the p53-mediated apoptotic signaling pathway.
Keywords: intrinsically disordered proteins, programmed cell death, protein–protein interaction, interaction network, protein structure, protein function, molecular recognition
In a multicellular organism, exposure of a cell to a set of environmental factors may start specific intracellular programs that trigger a chain of biochemical events that could lead to the characteristic changes in cellular morphology and ultimately to the cell death. These cell-killing intracellular events constitute programmed cell death (PCD) phenomenon, which includes at least three different mechanisms, apoptosis, autophagy, and programmed necrosis (necroptosis).1, 2, 3 These mechanisms are regulated/executed by different signaling pathways and are easily distinguished by their morphological differences and specific biochemical changes in dying cells.1, 4 Since apoptosis and necroptosis invariably contribute to the cell death, and since autophagy can have either pro-survival or pro-death roles, these three forms of PCD jointly decide the fate of cells, being responsible for a fine balance between the cell death and survival of normal cells.3
Morphologically, apoptosis, or type I PCD, is characterized by chromatin condensation and fragmentation, chromosomal DNA fragmentation, nuclear fragmentation, cell shrinkage, and disintegration of the cell into apoptotic bodies.3 Autophagy, or type II PCD, is an evolutionarily conserved catabolic process beginning with formation of autophagosomes, double- or multimembrane-bound structures surrounding cytoplasmic macromolecules and organelles destined for recycling.3, 5, 6, 7, 8, 9 Finally, necroptosis or type III PCD is a programmed necrosis, which is a genetically controlled event that involves cell swelling, organelle dysfunction, and cell lysis.10
At the molecular level, apoptosis is a tightly controlled and ordered cellular suicide program that is critical for the development, immune regulation, and homeostasis of a multicellular organism. Although several different pathways and signals can lead to apoptosis, the major mechanism that actually causes the cell to die is associated with the organized degradation of cellular organelles by activated members of the caspase family of cysteine proteases.11 A wide range of cell signals of either extracellular or intracellular origin can positively (i.e., trigger) or negatively (i.e., repress, inhibit, or dampen) affect apoptosis, leading to its initiation or repression, respectively. Each of the two major apoptotic pathways (extrinsic and intrinsic) can be regulated at multiple levels.
Autophagy mediates the turnover of long-lived proteins, the elimination of damaged organelles and misfolded proteins, and the recycling of cellular building blocks following nutrient deprivation. Under certain circumstances it can be causative for the programmed cellular suicide.1, 12 Furthermore, autophagy is required during periods of starvation or stress due to growth factor deprivation and therefore it has a crucial pro-survival role in cell homeostasis.12, 13 This defines the Janus role of autophagy and its ability to control a wide range of physiological processes, such as starvation, cell differentiation, cell survival, and death.14
Finally, necroptosis is characterized by the relatively smallest number of regulators. Typically, necroptosis is modulated by c-Jun N-terminal kinase (JNK), apoptosis inducing factor (AIF), death-associated protein kinase (DAPK), and reactive oxygen species,15, 16, 17, 18 whereas in the apoptosis-incompetent cells, necroptosis is activated by death receptors and involves the RIP1 kinase.19
Among the various roles of PCD in maintaining the homeostasis of living organisms is its involvement in the advancement of immune responses.20, 21, 22 In fact, the development of adaptive and innate immune responses depends on strict regulation of PCD signaling pathways, which are crucial for shaping and maintaining the immune system and ensuring functional immune responses.20, 21, 22 For example, PCD is involved in clonal deletion of lymphocytes during an infection, whereas the misregulation of PCD is known to lead to the defective clearance of autoreactive T cells and autoimmune disease.23
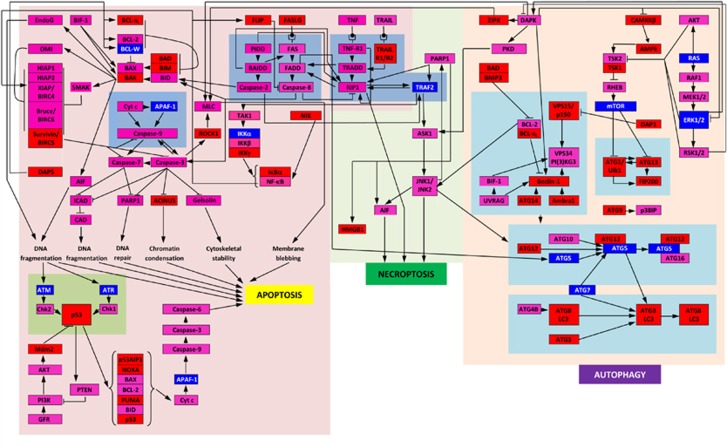
Figure 1 represents some major events taking place within a cell undergoing apoptosis, or autophagy, or necroptosis and clearly shows that these three types of PCD are heavily interlinked.1 In fact, three PCD modules are integrated into a common PCD network, where pathways of these functional modules are interconnected and where many death regulatory proteins are common to more than one module.1 This tight control of the various PCD processes and strong connectivity of the involved proteins suggest that PCD-related proteins should possess specific (and potentially common) structural characteristics. These characteristics would allow them to be uniquely and effectively modulated via multiple specific interactions with various partners and to control the regulation and execution of different PCD modules. In a search for these specific structural characteristics, we analyzed the peculiarities of intrinsic disorder distribution in proteins involved in various pathways related to the PCD.
Figure 1.
Schematic representation of three PCD modules. Diagram shows a map of the regulators and molecular components of the apoptosis, autophagy, and necroptosis death pathways, constituting the three known modules of the PCD network. The proteins are color coded according to their intrinsic disorder content evaluated by PONDR-FIT, with highly ordered ((IDP score)<10%), moderately disordered (30%>(IDP score)>10%), and highly disordered proteins ((IDP score)>30%) being shown as blue, pink, and red bars, respectively. This diagram is based on the PCD maps published in1, 56, 57
It is recognized now that the well-being of any living cell relies on the functionality of intrinsically disordered proteins (IDPs) and IDP regions (IDPRs). IDPs/IDPRs do not have a unique 3D structure as a whole or in part and exist as dynamic ensembles characterized by different degree and depth of disorder.24 IDPs are abundant in all proteomes25, 26 and possess a wide spectrum of biological functions that are typically related to regulation, signaling, and control pathways, promote the assembly of supra-molecular complexes, and complement the functions of ordered proteins.24 IDPs/IDPRs possess complex ‘anatomy' (they contain multiple, relatively short functional elements), which contributes to their unique ‘physiology' (an ability to be involved in interaction with, regulation of and control by multiple structurally unrelated partners). Functions of IDPs are further controlled by alternative splicing, which generates a set of protein isoforms with a highly diverse set of regulatory elements.27 The complexity of the disorder-based interactomes is further increased due to the ability of a single IDPR to bind to multiple partners gaining potentially very different structures in the bound state.28 Because of their critically important roles in regulation, signaling, and control pathways, misbehavior of IDPs is commonly associated with the pathogenesis of various diseases.29
Since IDPs are ‘control freaks', they commonly act as important regulators of protein–protein interaction networks. In fact, intrinsic disorder is intimately associated with ‘hubness' of proteins; that is, their ability to be involved in multiple interactions with unrelated partners via one-to-many and many-to-one binding mechanisms.30 This general commonness of intrinsic disorder in proteins involved in control and regulation suggests that the PCD could belong to the crucial biological processes that are controlled and regulated by IDPs/IDPRs. To test this hypothesis, we applied a broad spectrum of modern computational techniques to analyze abundance and functional roles of intrinsic disorder in proteins related to the different types of PCD, with the major focus on proteins involved in apoptosis, autophagy, and necroptosis.
Results
Characterization of the intrinsic disorder in human proteins associated with the PCD
Analysis of the compositional biases in human PCD-related proteins
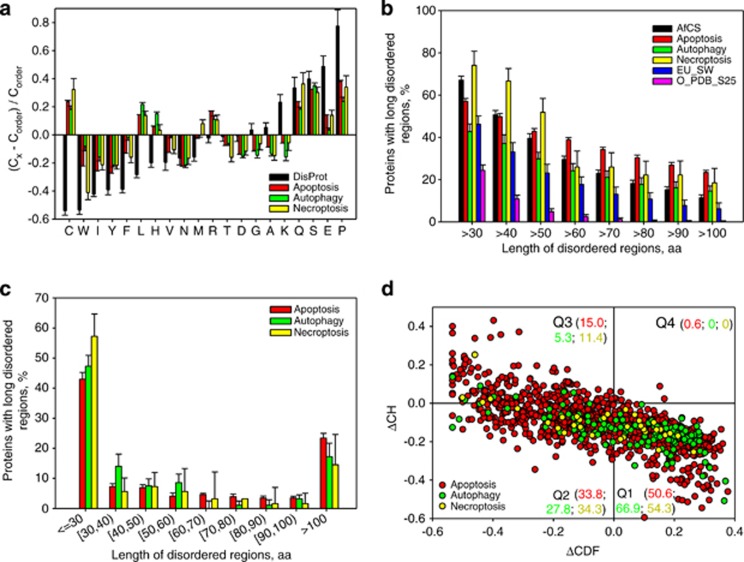
At the amino-acid composition level, IDPs/IDPRs are significantly depleted in order-promoting amino acids, C, W, I, Y, F, L, H, V, and N, and substantially enriched in disorder-promoting residues, A, G, R, T, S, K, Q, E, and P.31 Figure 2a shows that the PCD-related human proteins are depleted in some major order-promoting residues and enriched in some major disorder-promoting residues, suggesting that these proteins might contain multiple signatures characteristic for the IDPs.
Figure 2.
Peculiarities of intrinsic disorder distribution in human PCD proteins. (a) Fractional difference in the amino-acid composition between the different PCD-related proteins (apoptosis, red bars; autophagy, green bars; and necroptosis, yellow bars) and a set of ordered/structured proteins calculated for each amino-acid residue (compositional profiles). The fractional difference was evaluated as (Cx−Corder)/Corder, where Cx is the content of a given amino acid in a query set, and Corder is the corresponding content in the data set of fully ordered proteins. Composition profile of typical IDPs from the DisProt database is shown for comparison (black bars). Positive bars correspond to residues found more abundantly in PCD-related proteins, whereas negative bars show residues, in which PCD-related proteins are depleted. Amino-acid types are ranked according to their increasing disorder-promoting potential.31 (b) Abundance of predicted long disordered regions in human PCD proteins (apoptosis, red bars; autophagy, green bars; and necroptosis, yellow bars) in comparison with long disordered regions in 2329 proteins involved in cellular signaling (AfCS, black bars), 53 630 eukaryotic proteins from SWISS-PROT (EU_SW, blue bars), and 1138 sequences corresponding to ordered parts of proteins from PDB Select 25 (O_PDB_S25, pink bars). (c) Distribution of the length of the disordered segments in human PCD-related proteins (apoptosis, red bars; autophagy, green bars; and necroptosis, yellow bars). (d) CH-CDF analysis of the human PCD proteins (apoptosis, red circles; autophagy, green circles; and necroptosis, yellow circles). Here, the coordinates of each point were calculated as a distance of the corresponding protein in the CH plot from the boundary (Y-coordinate) and an average distance of the respective CDF curve from the CDF boundary (X-coordinate). The four quadrants correspond to the following predictions: Q1, proteins predicted to be disordered by CH plots, but ordered by CDFs; Q2, ordered proteins; Q3, proteins predicted to be disordered by CDFs, but compact by CH plots (i.e., putative molten globules or mixed proteins); Q4, proteins predicted to be disordered by both methods (i.e., proteins with extended disorder). The values shown next to the quadrant name denote the corresponding color coded fractions of chains from each of the three types of PCD-related proteins
Abundance of long disordered regions in human PCD-related proteins
Previous study revealed that 66% of signaling proteins contain predicted regions of disorder of 30 residues or longer.32 Figure 2b illustrates that intrinsic disorder is prevalent in the PCD-related proteins too, being comparable to the prevalence observed for signaling and eukaryotic proteins. In fact, the fraction of human PCD-related proteins with long regions of predicted disorder is 3- to 6-fold higher than that of non-homologous ordered proteins from PDB,32 being also a bit higher than the corresponding fraction of eukaryotic proteins. Figure 2c further illustrates the peculiarities of the disorder content distribution in the three PCD-related data sets and shows that although ∼50% of human PCD proteins contain disordered regions shorter than 30 consecutive residues, sizable fractions of these data sets (in a range of 15–20%) correspond to proteins with very long disordered regions (longer than 100 consecutive residues).
Disorder propensity of human PCD-related proteins by the binary disorder predictors
Sequences of the 1138, 137, and 35 human proteins associated with apoptosis, autophagy, and necroptosis, respectively, were used to predict whether they are likely to be mostly disordered using two binary predictors of intrinsic disorder: charge-hydropathy plot (CH plot)33, 34 and cumulative distribution function (CDF) analysis, both of which perform binary classification of whole proteins as either mostly disordered or mostly ordered.33 Figure 2d represents the results of the combined CH-CDF analysis of the human PCD-related proteins. Here, the coordinates of each spot are calculated as a distance of the corresponding protein in the CH plot from the boundary (Y coordinate) and an average distance of the respective CDF curve from the boundary (X coordinate).35 Figure 2d shows that ∼50, ∼33, and ∼45% of proteins associated with apoptosis, autophagy, and necroptosis, respectively, are expected to behave as native coils or native pre-molten globules or native molten globules or mixed proteins in their unbound states (see Supplementary Materials for further clarifications).
Overall, this analysis revealed that human PCD proteins possess relatively large amount of disorder, with apoptosis- and necroptosis-related proteins being noticeably more disordered (on average) than proteins involved in autophagy.
Overall characteristics of the intrinsic disorder in Deathbase proteins
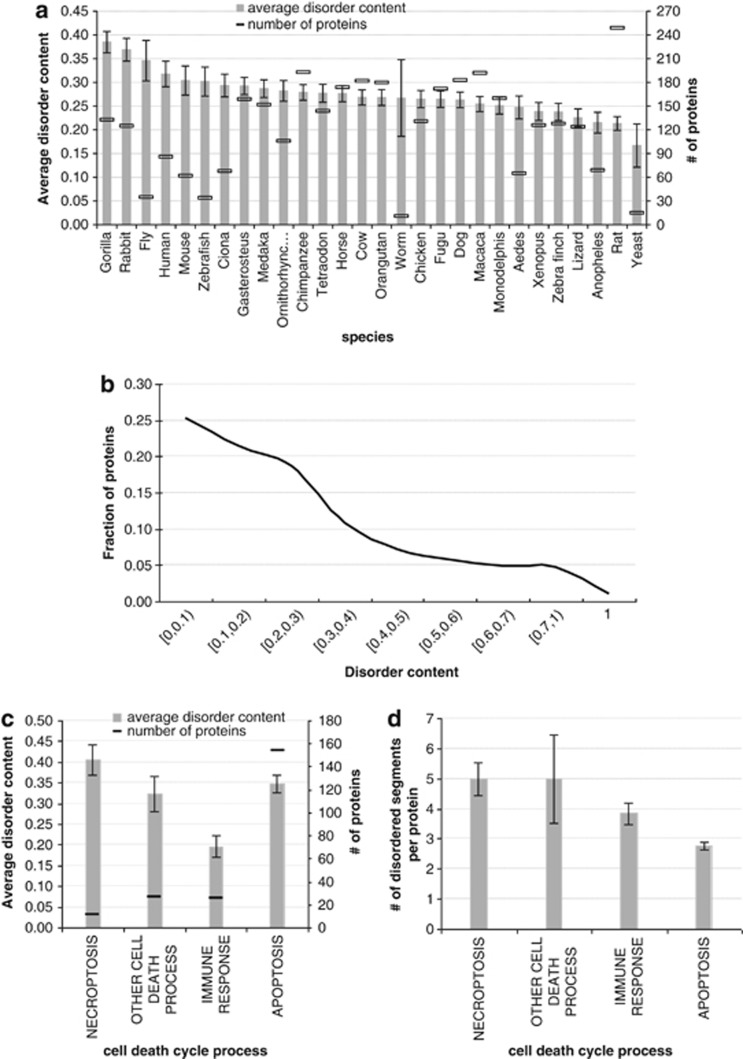
At the next stage, we performed a comprehensive analysis of the manually curated and well-annotated PCD-related proteins from the Deathbase. The average disorder content (i.e., fraction of disordered residues in a protein chain) in these proteins ranged between 0.17 and 0.38 across the considered 28 species; see Figure 3a. The average content in these proteins across the 28 species was 0.27, which was substantially larger than the overall disorder content in eukaryotic species that was estimated to be 0.19.25 Side-by-side comparison of disorder content for individual species between the entire proteome and the cell death proteins showed consistent enrichment of the disorder in the latter protein sets. Specifically, in human, the overall disorder content was reported at 0.2225 while in human cell death proteins the content was 0.32, in worm they were 0.16 versus 0.27, and in fly 0.22 versus 0.35.
Figure 3.
Evaluation of abundance of intrinsic disorder in death-related proteins from the Deathbase. (a) The average disorder content (grey bars) with standard errors (error bars) and the number of proteins for the 28 considered species shown on the x axis. (b) Distribution of disorder content for the cell death proteins for the 28 considered species. (c) The average disorder content (grey bars) with standard errors (error bars) and the number of proteins for different cell death processes. (d) The average number of disordered segments per protein (grey bars) with standard errors (error bars) for different cell death processes
The distribution of the disorder content is shown in Figure 3b. We observed that close to half cell death proteins contained up to 20% disordered residues. On the other hand, 17% of these proteins had at least half of their amino acids disordered and about 1% (39 proteins) was fully disordered.
Next, we investigated the enrichment of disorder across various cell death processes. Figure 3c reveals that proteins involved in necroptosis and apoptosis have larger amounts of disorder at about 0.41 and 0.35, respectively, compared with proteins involved in immune responses, for which the average disorder content equals to 0.2.
Figure 3d compares the average number of disordered segments per proteins and demonstrates that the apoptosis-related proteins have fewer numbers of (longer) disordered segments, about 2.8 per chain, compared with proteins implicated in necroptosis and immune responses that have on average about 5 and 3.8 disordered segments per chain, respectively. Overall, there were about 3.3 disordered segments per chain when considering all cell death proteins. Our analysis indicated that disorder is important across all cell death processes.
Functional analysis of disordered segments
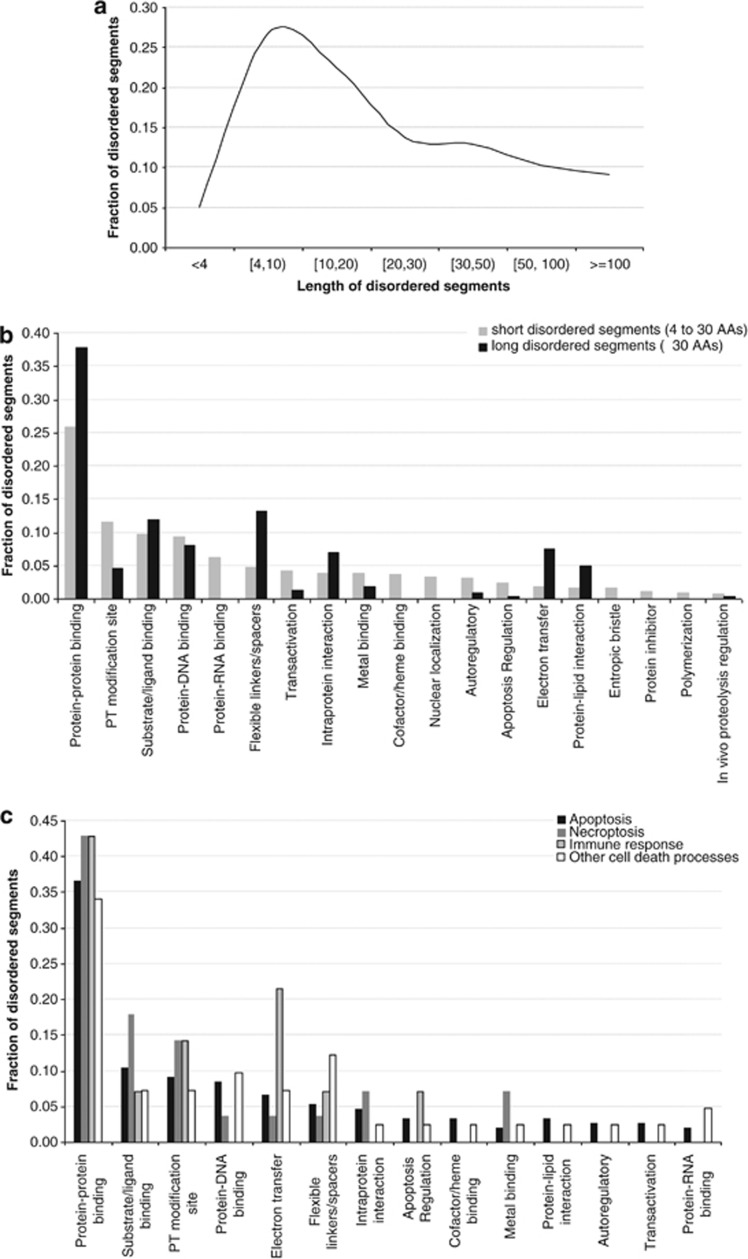
Distribution of the sizes of the disordered segments in cell death proteins across the considered 28 species is given in Figure 4a. Interestingly, we observed that the segment sizes followed a bimodal distribution with a relatively large number of short disordered segments, between 4 and 25 consecutive amino acids, and with a second peak for longer sizes, at over 30 amino acids. Thus, we analyzed the functional repertoire of the disordered segments separately for short (<30 consecutive amino acids) and long (at least 30 consecutive amino acids) segments.
Figure 4.
Correlation between intrinsic disorder in cell death proteins and their functions. (a) Distribution of the length of the disordered segments in cell death proteins. (b) Fraction of short, 4–30 consecutive amino acids (AAs), and long, ≥30 consecutive AAs, disordered segments for a given function; x axis shows 19 considered functions sorted in descending order by the number of the short segments. (c) Fraction of disordered segments for a given function for the cell death proteins in the curated species that are annotated with cell death processes; x axis shows 14 considered functions sorted in descending order by the overall number of the disordered segments
To this end, we considered 26 functions associated with disorder, which were annotated based on the data in the DisProt database,36 that are summarized in Supplementary Table S2. We excluded functions with <20 annotations for both short and long disordered segments, as they do not offer enough data to draw statistically sound conclusions. Figure 4b compares the annotations of the 19 remaining functions for the short and long disordered segments and reveals that disorder has a diverse set of roles in cell death proteins, from facilitating interactions with other proteins, DNA, RNA, and other ligands, to involvement in post-translational modification (PTM) sites, intraprotein interactions, and implementation of linker regions. Both long and short disorder segments were implicated with a similar frequency in several functions including interactions with DNA and ligands and regulation of proteolysis. The short disordered segments were more prevalent in a larger number of functions compared with the long segments, including protein–RNA and cofactor/heme binding, nuclear localization, protein inhibition, polymerization, trans-activation, regulation of apoptosis, and in auto-regulatory functions. They were also implicated in entropic bristle activities and were twice more often associated with the PTM sites. On the other hand, long disordered segments more often served as linkers, were involved in electron transfer, and had a strong role in protein–protein, protein–lipid, and intraprotein interaction.
Furthermore, we investigated the functional roles of disordered segments for the proteins associated with specific cell death processes. Since there were substantially fewer (only 128) disordered segments with annotated function for the five curated species that included cell death process annotations (compared with results in Figure 4b), we excluded the functions with <5 annotations; the remaining 14 functions are summarized in Figure 4c. Disordered segments found in all cell death processes were shown to be responsible for five functions: protein–protein and protein–ligand binding, electron transfer, linker regions, and were also prevalent in PTM sites. The remaining functions were specific to selected subsets of cell death processes. Disorder implemented all considered 14 functions in proteins involved in apoptosis. In contrast, the disordered regions in necroptosis-related proteins were responsible for fewer functions, such as protein–protein, protein–ligand, and protein–DNA binding, intraprotein interactions, and metal binding. Finally, immune response triggered cell death utilized disorder to facilitate electron transfer and protein–protein and protein–ligand interactions. To sum up, disorder is utilized to facilitate a different set of cellular functions for different cell death processes.
Molecular recognition feature regions
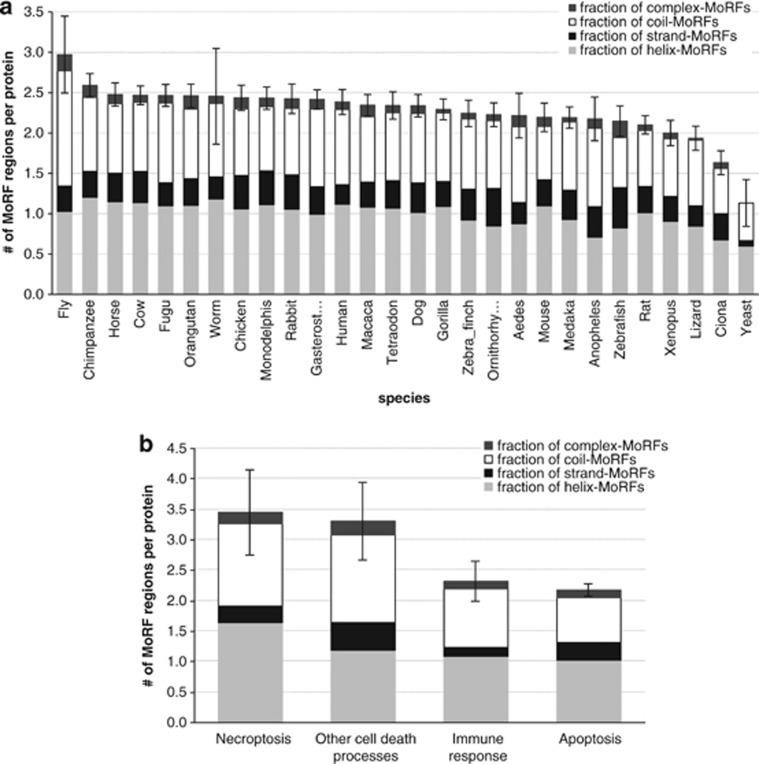
The most prevalent function of disorder in cell death proteins is facilitation of protein–protein interactions; Figure 4b shows that about 30% of the functionally annotated disordered segments were implicated in these binding events. This motivated our analysis of molecular recognition feature (MoRF) regions24, 37 that undergo coupled binding and folding upon interaction with protein partners. Figure 5a demonstrates that there are on average between 1.1 and 3 MoRF regions per protein across the considered 28 species. The average number of MoRFs per chain over all cell death proteins is at 2.3, which means that about two-thirds of all disordered segments, which there are on average 3.3 per chain, include MoRFs. This means that protein–protein interactions dominate the function of the disorder in these proteins. Furthermore, Figure 5a suggests that MoRFs primarily fold into helical or irregular conformations upon binding; this is consistent across all considered species.
Figure 5.
Intrinsic disorder in cell death proteins and MoRFs. (a) Average number of MoRF regions per protein, shown using bars, with standard errors (error bars) across the considered 28 species shown on the x axis. The bars are subdivided to represent different MoRF types. (b) Average number of MoRF regions per protein, shown using bars, with standard errors (error bars) across the four types of cell death processes. The bars are subdivided to represent different MoRF types
Figure 5b shows that the proteins that control apoptotic pathways have the smallest number of MoRFs, at about 2.2 per chain. On the other hand, proteins involved in the necroptosis-driven cell death have the largest number MoRFs per sequence, which equals 3.5. These counts correlate with the overall number of the disordered segments, see Figure 3d, resulting in a similar ratio between the numbers of disordered and MoRF segments.
Evolutionary conservation of disorder
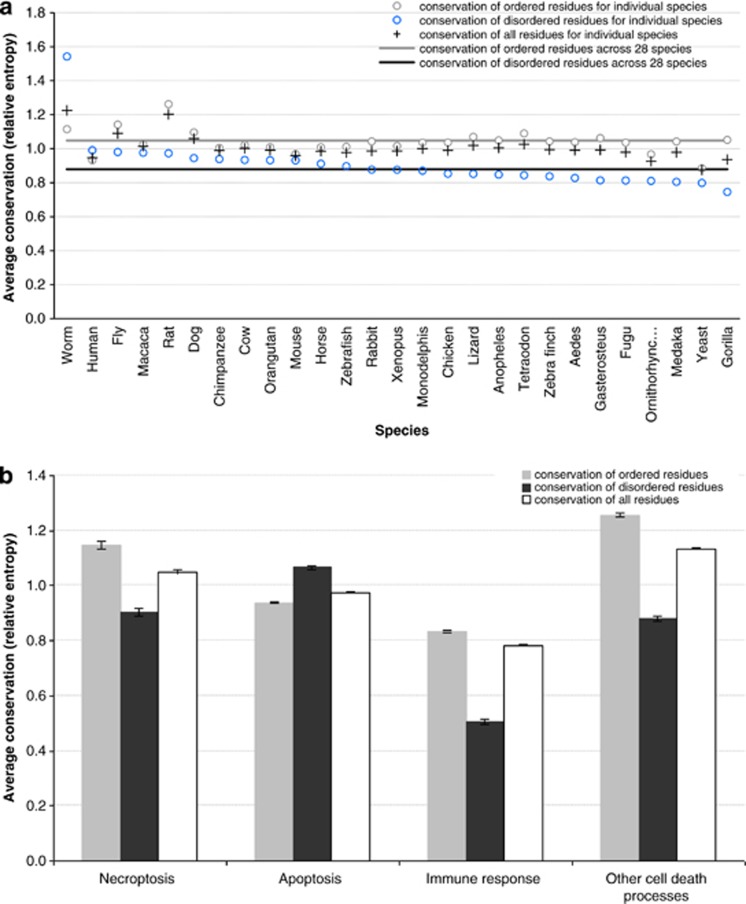
Finally, we investigated evolutionary conservation of the disorder in cell death proteins across various species, Figure 6a, and across various processes, Figure 6b. The conservation was quantified with the relative entropy computed from the WOP profiles generated by PSI-BLAST. Higher values of the relative entropy indicate a higher degree of evolutionary conservation. Figure 6a shows that for significant majority of species, except for worm and human, disordered residues were characterized by a consistently lower level of conservation compared with the structured amino acids. This suggested that disordered regions are under higher evolutionary pressure compared with structured regions. Figure 6b demonstrates that the above trend holds for all cell death processes, except for the apoptosis where disordered amino acids have higher conservation. Immune response-related proteins were characterized by the highest difference in conservation where the disordered residues had conservation lower by about 40% compared with the ordered residues. As immune-related processes require dynamic adjustments in response to relatively rapid evolution of pathogens, we speculate that these dynamics are facilitated through the use of disorder.
Figure 6.
Intrinsic disorder and evolution of cell death proteins. (a) The average evolutionary conservation, quantified with relative entropy, for ordered (hollow circles), disordered (solid circles), and all (crosses) residues across the considered 28 species that are shown on the x axis. Solid horizontal lines show average conservation for ordered (gray line) and disordered (black line) residues. (b) Average relative entropy, which quantifies evolutionary conservation for ordered, disorder and all residues for each cell death process, shown using bars, with standard errors (error bars)
Discussion
Figure 1 schematically represents major events happening in a cell undergoing PCD via one of the three modules, apoptosis, necroptosis, and autophagy, and clearly shows that many proteins involved in these three modules contain significant amount of intrinsic disorder. Here, the proteins are color coded according to their intrinsic disorder content evaluated by PONDR-FIT, where two arbitrary cutoffs for the levels of intrinsic disorder were used to classify proteins as highly ordered ((IDP score)<10%, blue bars), moderately disordered (30%>(IDP score)>10%, pink bars), and highly disordered ((IDP score)>30%, red bars).38 According to this classification, only 11 human proteins related to the controlled cell death pathways are characterized by low disorder scores, whereas the absolute majority of the PCD-related proteins is moderately or highly disordered. As it follows from the functional analysis of the proteins from Deathbase, the most prevalent function of IDPRs in the PCD-related proteins is protein–protein interaction. In agreement with this general observation, Figure 1 shows that the highly connected PCD-related proteins are typically more disordered than proteins with lesser number of interaction partners.
It is important to emphasize here that intrinsic disorder is very common in human PCD-related proteins despite the fact that many of these proteins (some of which are shown in Figure 1) are enzymes (kinases, ribonucleases, deoxyribonuclease, proteases, protein and ubiquitin ligases, polymerases, oxidoreductase, GTPases and so on).
To delve deeper into the roles of intrinsic disorder in functions of PCD-related protein, we consider in more detail a part of the PCD protein–protein interaction network, namely the p53-mediated intrinsic apoptotic signaling pathway shown at the bottom left corner of Figure 1. Detailed discussion of the major regulators of the p53-mediated intrinsic apoptotic signaling pathway and roles of intrinsic disorder in their functions can be found elsewhere (manuscript in preparation).
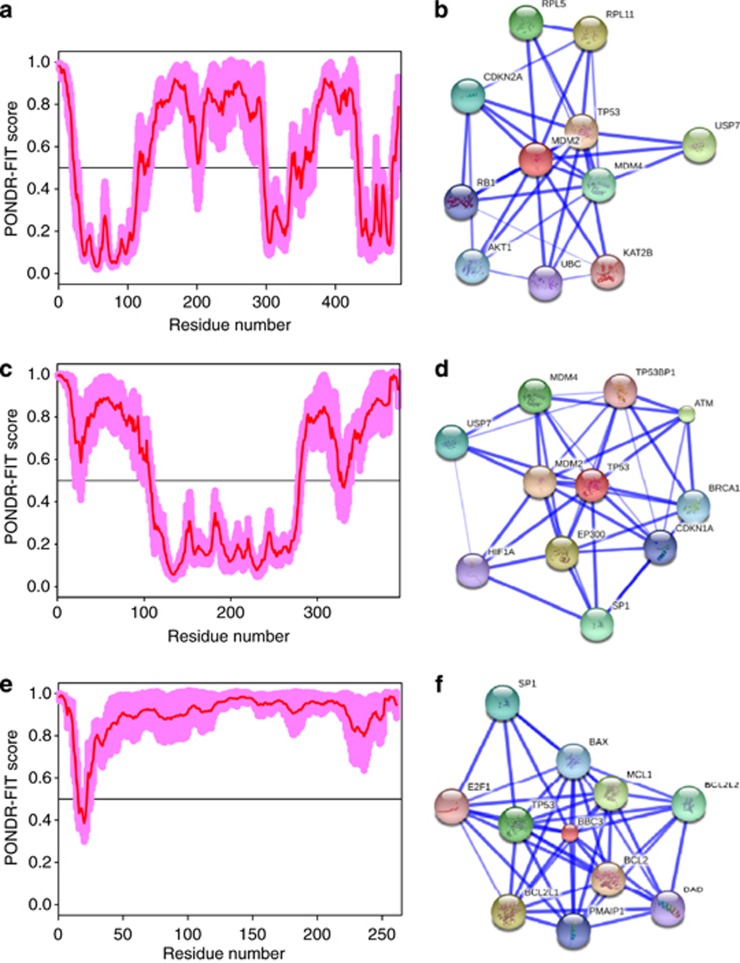
Figure 7 represents the PONDR-FIT disorder profiles of three human proteins known to be involved in p53-mediated apoptotic signaling pathway, and also shows the results of the functional analysis of these proteins using the STRING database.39 This database acts as a ‘one-stop shop' for all information on functional links between proteins, and version 9.0 of STRING (accessible at http://string-db.org) covers >1100 completely sequenced organisms, including Homo sapiens. Analogous data for the remaining 20 proteins from the p53-mediated apoptotic signaling pathway are shown in Supplementary Figure S1. Figure 7 and Supplementary Figure S1 show that all the members of the p53-controlled apoptosis signaling pathway are involved in multiple interactions and therefore can be considered as hub proteins. Since many of these 23 proteins contain significant amount of predicted disorder and since almost all of them interacts with other apoptosis-related proteins, which are often predicted to be disordered, Figure 7 and Supplementary Figure S1 suggest that the hubness of the major players of the p53-modulated apoptosis is related to their intrinsically disordered nature and/or to the intrinsic disorder of their partners.
Figure 7.
Intrinsic disorder (plots a, c, and e) and STRING analysis (plots b, d, and f) of the interactomes of some proteins involved in the p53-mediated apoptotic signaling pathway. (a and b) Mdm2 (UniProt ID: Q00987); (c and d) p53 (UniProt ID: P04637); (e and f) PUMA (UniProt ID: Q96PG8). Intrinsic disorder propensity was evaluated by PONDR FIT (red curves). Shadow around PONDR FIT curves represents distribution of statistical errors. STRING database is the online database resource Search Tool for the Retrieval of Interacting Genes, which provides both experimental and predicted interaction information.39 For each protein, STRING produces the network of predicted associations for a particular group of proteins. The network nodes are proteins. The edges represent the functional associations evaluated based on the experiments, search of databases and text mining. The thickness of edges is proportional to the confidence level39
Furthermore, many of the proteins involved in the p53 signaling (as well as other proteins related to all the PCD modules) are regulated via various PTMs and many of them have several alternatively spliced isoforms. Both of these features are typical for IDPs/IDPRs.24, 27 These observations provide further support to the general conclusion of this study that intrinsic disorder is crucial for the regulation and control of all the PCD types. Here, proteins regulating necroptosis seem to be more disordered than protein involved in other types of the controlled cell death.
Finally, it is important to emphasize here that there are many examples in the literature where PCD-related proteins were experimentally found to be disordered or possess long IDPRs. Since the literature on this topic is vast, only several most characteristic cases of the experimentally validated disorder in PCD-regulating proteins are briefly discussed below. A well-documented example is the transcription factor p53, which possesses functionally important IDPRs.40, 41 Here, ∼70% of the interactions of p53 are mediated by IDPRs containing 86, 90, and 100% of observed acetylation, phosphorylation, and protein conjugation sites of this protein, respectfully.42 Early on, X-ray and NMR structural analysis revealed that an important inhibitor of PCD, BCL-xL, contains a 60-residue IDPR connecting helices αl and α2.43 Other members of the BCL-2 family, BH3-only proteins (such as BIM, BAD, and BMF) that serve as key initiators of PCD are largely disordered in solution.44 Interaction of the disordered BIM, BAD, BMF, BAK, and tBID with several pro-survival BCL-2 family members leads to the formation of an α-helical segment that anchors these BH3-only proteins to the hydrophobic groove of their binding partners.45, 46 In addition to BH3-only proteins shown in Figure 1, pro-survival BCL-2 family members include MCL-1, BFl-1, and BCB-B, all of which are expected to contain long functional IDPRs.45 Overall, structural and sequence analyses revealed that many pro-apoptotic and pro-survival proteins of the BCL-2 family (which, in their turn, are tightly regulated by various PTMs among other means) are either IDPs or contain functionally important IDPRs.46 Furthermore, conformational plasticity and ability to fold upon binding were shown to have a crucial role in multifarious interactions of the BCL-2 family members with their numerous partners.47
NMR spectroscopy in conjunction with circular dichroism spectroscopy and limited proteolysis revealed that a large central region (∼1500 residue) of the BRCA1 tumor suppressor protein is a long IDPR that lacks any pre-existing independently folded globular domains and serves as ‘an intrinsically disordered scaffold for multiple protein–protein and protein–DNA interactions'.48 NMR analysis revealed that an important regulator of proliferation and apoptosis, cAMP-responsive (CRE)-binding (CREB) protein (CBP), contains an intrinsically disordered ACTR-binding domain (residues 2059–2117) that completely folds upon binding.49 Combined experimental and computational analysis revealed that N- and C-terminal regions of the human Nogo proteins50 and C-terminal domain of a mitochondrial pro-apoptotic protein ARTS51 are typical IDPRs, and that the prostate apoptosis response factor-4 (PAR-4),52 prostate-associated gene 4 (PAGE4) protein,53 and important oncoprotein c-Myc54 are mostly disordered apoptosis-related proteins.
Materials and methods
Data sets
In this work, several data sets of proteins related to the PCD were analyzed. These include 1138, 137, and 35 human proteins associated with apoptosis, autophagy, and necroptosis, as well as 3458 proteins from Deathbase.55 Peculiarities of data set assembly are described in Supplementary Materials.
Computational characterization of disorder
Computational tools and approaches used for the characterization of the amino-acid compositions, peculiarities of intrinsic disorder distribution, functions, PTMs, and evolutionary patterns of PCD-related proteins are described in Supplementary Materials.
Glossary
- AIF
apoptosis inducing factor
- DAPK
death-associated protein kinase
- CDF
cumulative distribution function
- CH plot
charge-hydropathy plot
- CRE
cAMP responsive
- CREB
CRE binding
- CBP
CRE-binding protein
- IDP
intrinsically disordered protein
- IDPR
intrinsically disordered protein region
- JNK
c-Jun N-terminal kinase
- MoRF
molecular recognition feature
- PAGE4
prostate-associated gene 4
- PAR-4
prostate apoptosis response factor-4
- PCD
programmed cell death
- PTM
post-translational modification
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Bialik S, Zalckvar E, Ber Y, Rubinstein AD, Kimchi A. Systems biology analysis of programmed cell death. Trends Biochem Sci. 2010;35:556–564. doi: 10.1016/j.tibs.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vanden Berghe T, Vanlangenakker N, Buettner S, Eisenberg T, Vandenabeele P, et al. Programmed necrosis from molecules to health and disease. Int Rev Cell Mol Biol. 2011;289:1–35. doi: 10.1016/B978-0-12-386039-2.00001-8. [DOI] [PubMed] [Google Scholar]
- Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ML, Ooi JP, Ismail N, Moad AI, Muhammad TS. Programmed cell death pathways and current antitumor targets. Pharm Res. 2009;26:1547–1560. doi: 10.1007/s11095-009-9895-1. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Huett A, Goel G, Xavier RJ. A systems biology viewpoint on autophagy in health and disease. Curr Opin Gastroenterol. 2010;26:302–309. doi: 10.1097/MOG.0b013e32833ae2ed. [DOI] [PubMed] [Google Scholar]
- Chen S, Rehman SK, Zhang W, Wen A, Yao L, Zhang J. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010;1806:220–229. doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Li ZY, Yang Y, Ming M, Liu B, Mitochondrial ROS. generation for regulation of autophagic pathways in cancer. Biochem Biophys Res Commun. 2011;414:5–8. doi: 10.1016/j.bbrc.2011.09.046. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Lin M, Yu JY, Liu B, Bao JK. Targeting apoptotic and autophagic pathways for cancer therapeutics. Cancer Lett. 2011;300:105–114. doi: 10.1016/j.canlet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Henriquez M, Armisen R, Stutzin A, Quest AF. Cell death by necrosis, a regulated way to go. Curr Mol Med. 2008;8:187–206. doi: 10.2174/156652408784221289. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326 (Pt 1:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Cheng Y, Liu Q, Bao JK, Yang JM. Autophagic pathways as new targets for cancer drug development. Acta Pharmacol Sin. 2010;31:1154–1164. doi: 10.1038/aps.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Vandenabeele P. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim Biophys Acta. 2006;1757:1371–1387. doi: 10.1016/j.bbabio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Boujrad H, Gubkina O, Robert N, Krantic S, Susin SA. AIF-mediated programmed necrosis: a highly regulated way to die. Cell Cycle. 2007;6:2612–2619. doi: 10.4161/cc.6.21.4842. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Kimchi A. DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ. 2007;14:1908–1915. doi: 10.1038/sj.cdd.4402212. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Kim YS, Liu ZG. TNFalpha and reactive oxygen species in necrotic cell death. Cell Res. 2008;18:343–349. doi: 10.1038/cr.2008.31. [DOI] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Cohen JJ, Duke RC, Fadok VA, Sellins KS. Apoptosis and programmed cell death in immunity. Annu Rev Immunol. 1992;10:267–293. doi: 10.1146/annurev.iy.10.040192.001411. [DOI] [PubMed] [Google Scholar]
- Hedrick SM, Ch'en IL, Alves BN. Intertwined pathways of programmed cell death in immunity. Immunol Rev. 2010;236:41–53. doi: 10.1111/j.1600-065X.2010.00918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JV, Walsh CM. Programmed necrosis and autophagy in immune function. Immunol Rev. 2012;249:205–217. doi: 10.1111/j.1600-065X.2012.01147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzka M, Walsh CM. Apoptotic signal transduction and T cell tolerance. Autoimmunity. 2007;40:442–452. doi: 10.1080/08916930701464962. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. BBA Proteins Proteom. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WL, Oldfield C, Meng J, Huang F, Xue B, Uversky VN, et al. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput. 2012. pp. 116–127. [PubMed]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are ‘natively unfolded' proteins unstructured under physiologic conditions. Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Huang F, Oldfield C, Meng J, Hsu WL, Xue B, Uversky VN, et al. Subclassifying disordered proteins by the CH-CDF plot method. Pac Symp Biocomput. 2012. pp. 128–139. [PubMed]
- Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, et al. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35 (Database issue:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem. 2011;112:3256–3267. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39 (Database issue:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R, Muller L, Dehner A, Klein C, Kessler H, Buchner J. The N-terminal domain of p53 is natively unfolded. J Mol Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, et al. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9 (Suppl 1:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- Rautureau GJ, Day CL, Hinds MG. Intrinsically disordered proteins in bcl-2 regulated apoptosis. Int J Mol Sci. 2010;11:1808–1824. doi: 10.3390/ijms11041808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds MG, Day CL. Regulation of apoptosis: uncovering the binding determinants. Curr Opin Struct Biol. 2005;15:690–699. doi: 10.1016/j.sbi.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B, et al. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions. J Mol Biol. 2005;345:275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- Li M, Song J. The N- and C-termini of the human Nogo molecules are intrinsically unstructured: bioinformatics, CD, NMR characterization, and functional implications. Proteins. 2007;68:100–108. doi: 10.1002/prot.21385. [DOI] [PubMed] [Google Scholar]
- Reingewertz TH, Shalev DE, Sukenik S, Blatt O, Rotem-Bamberger S, Lebendiker M, et al. Mechanism of the interaction between the intrinsically disordered C-terminus of the pro-apoptotic ARTS protein and the Bir3 domain of XIAP. PLoS ONE. 2011;6:e24655. doi: 10.1371/journal.pone.0024655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libich DS, Schwalbe M, Kate S, Venugopal H, Claridge JK, Edwards PJ, et al. Intrinsic disorder and coiled-coil formation in prostate apoptosis response factor 4. FEBS J. 2009;276:3710–3728. doi: 10.1111/j.1742-4658.2009.07087.x. [DOI] [PubMed] [Google Scholar]
- Zeng Y, He Y, Yang F, Mooney SM, Getzenberg RH, Orban J, et al. The cancer/testis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J Biol Chem. 2011;286:13985–13994. doi: 10.1074/jbc.M110.210765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen C, Helander S, Lemak A, Fares C, Csizmok V, Carlsson J, et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012;40:6353–6366. doi: 10.1093/nar/gks263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez J, Walter D, Munoz-Pinedo C, Gabaldon T. DeathBase: a database on structure, evolution and function of proteins involved in apoptosis and other forms of cell death. Cell Death Differ. 2010;17:735–736. doi: 10.1038/cdd.2009.215. [DOI] [PubMed] [Google Scholar]
- Van Herreweghe F, Festjens N, Declercq W, Vandenabeele P. Tumor necrosis factor-mediated cell death: to break or to burst, that's the question. Cell Mol Life Sci. 2010;67:1567–1579. doi: 10.1007/s00018-010-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Jochemsen AG. p53: a guide to apoptosis. Curr Cancer Drug Targets. 2008;8:87–97. doi: 10.2174/156800908783769337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.