Abstract
During meiosis, DNA double-strand breaks (DSBs) are physiologically induced to start the recombination process and promote the formation of interhomologue crossovers (COs), which are required to ensure faithful chromosome segregation into the gametes. The timely repair of DSBs is an essential part of the meiotic programme, as accumulation of unprocessed DSBs during the pachytene stage of meiotic prophase triggers a DNA damage checkpoint response that induces apoptosis of damaged cells. We show that CO-promoting factors MSH-4, MSH-5, and ZHP-3, but not COSA-1, are required for the apoptotic response of the meiotic DNA damage checkpoint. Lack of MSH-4 or MSH-5 suppresses the apoptotic response observed in some DNA repair-defective mutants such as fcd-2 and brc-1 (orthologues of FANCD2 and BRCA1), irrespectively of the amount of DSBs present in pachytene nuclei. Although ionizing radiation fails to induce apoptosis in msh-4/5-mutant backgrounds, it induces transcriptional activation of the apoptosis-activator egl-1, which is controlled by the Caenorhabditis elegans p53 orthologue CEP-1. This finding suggests that MSH-4/5 involvement in the apoptotic response occurs downstream or independently of damage sensing and checkpoint activation. This study establishes a role for pro-CO factors MSH-4/5 and ZHP-3 in the execution of apoptosis at late meiotic prophase following the accumulation of exogenous or endogenous DNA damage.
Keywords: apoptosis, crossover, DNA damage, genome stability, meiosis
Eukaryotes execute meiosis to ensure the proper partition of chromosomes into the gametes. Crossing-overs (COs) between homologous chromosomes are essential, along with sister chromatid cohesion, to ensure proper chromosome segregation at meiosis I. All studied organisms exhibit an excess of double-strand breaks (DSBs), generated by type II topoisomerase-like SPO-11, with just a few being ultimately repaired as interhomologue COs. Once DSBs arise, they undergo resection to produce single-stranded DNA, a substrate for the loading of the recombinase RAD-51. This protein, homologous to the Escherichia coli recombinase RecA, promotes strand exchange and invasion of the homologous DNA template, allowing homologous DNA repair to take place.1 Faithful repair of DSBs during meiosis is crucial to maintain genomic integrity and to allow formation of functional gametes, as unrepaired DSBs trigger activation of the DNA damage checkpoint, which results in a block to cell cycle progression or removal of damaged cells by apoptosis.2
The Caenorhabditis elegans germ line exhibits a complete time course of meiotic prophase in which nuclei at the different stages of oogenesis can be easily identified based on their position and appearance, and it has been shown that DSB repair is differently modulated along the germ line. For example, loading of RAD-51, the only RecA-like protein responsible for the strand exchange step in C. elegans meiosis,3, 4 is RAD-50 independent in the premeiotic region of the germ line and in late-pachytene nuclei,5 but it is RAD-50 dependent during early meiotic prophase (leptotene to mid-pachytene). During this period, there is a clear bias towards repair using the homologous chromosome as a template, thus promoting CO formation.6 Later, residual DSBs can also be repaired using the sister chromatid.5 During meiotic prophase, a number of proteins cooperate to ensure CO formation: some are involved in promoting pairing and synapsis of homologous chromosomes,7 whereas others, known as pro-CO factors, seem to be involved in stabilizing recombination intermediates between homologous chromosomes.8, 9
MSH-4 and MSH-5 are known to form heterodimers in all eukaryotes and bind as clamps to stabilize recombination intermediates. The interdependence relationships of four different pro-CO factors (MSH-4, MSH-5, ZHP-3, and COSA-1) have been recently described in C. elegans.10 High numbers of MSH-5 foci start to appear in mid-pachytene, probably labelling most recombination intermediates, but by late pachytene, MSH-5 foci diminish in number to one focus per homologue pair, forming a complex that also contains COSA-1 and ZHP-3. These foci are thought to mark the single CO that is formed in each homologue pair during C. elegans meiosis. Before becoming limited to the ‘pro-CO focus', ZHP-3 is first observed forming stretches along chromosomes during pachytene,11, 12 whereas COSA-1 appears directly at the transition from mid-pachytene to late pachytene forming a single focus per chromosome pair. The function of COSA-1 is required for recruiting MSH-5 and ZHP-3 to designated CO sites and, correspondingly, COSA-1 foci are absent in msh-5 mutants.
By the end of pachytene, about half of the oocytes in the C. elegans germ line undergo physiological cell death.13 In addition, two different checkpoints enhance apoptosis during pachytene in response to either DNA damage or defects in synaptonemal complex (SC) assembly.14, 15 The DNA damage checkpoint induces cell cycle arrest at the G2 phase when DNA damage occurs in the premeiotic proliferative region of the germ line.16 The DNA damage checkpoint requires the 9-1-1 complex, a DNA damage–sensor complex that contains the conserved proteins RAD-9, RAD-1, and HUS-1.17 Following 9-1-1 loading, activation of the p53 orthologue CEP-1 is required to elicit apoptosis.18 It is important to note that apoptosis occurs in cells only in the late pachytene stage of meiosis, suggesting that there must be specific signals that make late-pachytene nuclei competent for apoptosis and that prevent apoptosis at earlier meiotic stages. Restricting DNA damage-induced apoptosis to late pachytene stage could prevent the inappropriate loss of nuclei undergoing the earlier stages of meiotic recombination, during which DSBs are present.3, 19 It has been reported that GLD-1 represses the translation of CEP-1 in the early stages of meiosis,20 and that MPK-1 is required for CEP-1 activation.21 Once CEP-1 is activated, it triggers EGL-1 expression, which disrupts the interaction between CED-4, the nematode orthologue of mammalian Apaf1, and the Bcl2 family member CED-9, causing a relocalization of CED-4 to the nuclear periphery and activation of the caspase protein CED-3, which leads to apoptosis execution.22, 23, 24 Finally, SIR-2.1, a member of the sirtuin family, besides being implicated in aging and stress response, is also required for the execution of apoptosis in response to DNA damage.25, 26
Mutants of DNA repair genes, such as rad-51, brc-1 (orthologue of human BRCA1), or fcd-2 (orthologue of human FANCD2), show elevated levels of apoptosis that are spo-11 and cep-1 dependent.14, 27, 28 Similarly, mutants deficient in structural components of the SC, such as syp-2,19 show an increase in germ-nuclei apoptosis that is in part dependent on the DNA damage checkpoint and in part on the synapsis checkpoint.15 Worms carrying mutations in CO-promoting factors, such as him-14 (the C. elegans orthologue of Msh4; called msh-4 hereafter), msh-5, and zhp-3, show an abnormal accumulation of RAD-51 foci that are interpreted as unresolved recombination intermediates.11, 19 Surprisingly, despite the accumulation of recombination intermediates, msh-5 mutants do not exhibit increased apoptosis above the basal level observed in wild-type worms8 (also shown in this study). Similarly, mice lacking Mlh1, a protein that is also required for CO formation and localizes to CO sites in late pachytene, do not exhibit apoptosis induction during meiotic prophase.29 These observations provide an indication that pro-CO factors may be necessary for inducing apoptosis in response to recombination defects. In this study, we investigate the crosstalk and interdependence between pro-CO factors and the DNA damage-dependent apoptosis pathway, and the roles of MSH-4/5 and ZHP-3 in germ-line apoptosis when exogenous DSBs are introduced.
Results
MSH-4 and MSH-5 are required to induce germ-nuclei apoptosis after exogenously induced DNA damage
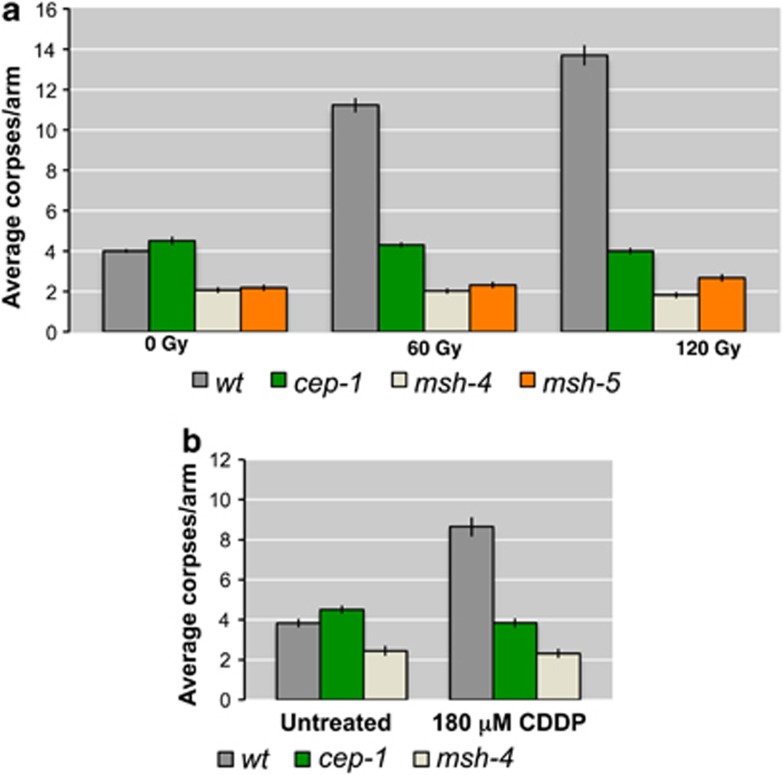
As previously mentioned, mutants lacking CO-promoting factors MSH-4 or MSH-5 show an increase in RAD-51 foci,19 demonstrating a defect in the timely processing of recombination intermediates. We quantified the number of apoptotic corpses in germ lines of msh-4 and msh-5 mutants and confirmed, as previously described for msh-5,8 that neither mutant shows an increase in the average number of apoptotic corpses compared with wild-type worms (Figure 1a). To assess whether exogenously induced DNA damage was able to enhance apoptosis in these mutants, we induced DSBs by exposing the worms to increasing doses of ionizing radiation (IR) and quantified the number of apoptotic corpses 24 h post irradiation. Although the levels of apoptosis were significantly increased in wild-type oocytes after 60 and 120 Gy, no enhancement was observed in irradiated msh-4 or msh-5 mutants (Figure 1a and Supplementary Table S1). A very similar result was obtained treating msh-4-mutant nematodes for 48 h with 180 μM cis-diamminedichloroplatinum(II) (CDDP) that induces interstrand crosslinking (ICL, Figure 1b). We conclude that both MSH-4 and MSH-5 proteins are required for eliciting apoptosis in pachytene cells after genotoxic insults.
Figure 1.
(a) Average apoptosis levels scored before and 24 h after irradiation with IR at 60 and 120 Gy. The y axis shows average number of SYTO-12-labelled nuclei per gonad arm. Genotypes are indicated in the colour legend at the bottom of the chart. Unlike what occurs in wild type, gamma irradiation does not induce germ-line apoptosis in pachytene nuclei of cep-1, msh-4, and msh-5 mutants. Number of observed gonads: wt 0 Gy: 185, cep-1 0 Gy: 155, msh-4 0 Gy: 160, msh-5 0 Gy: 183, wt 60 Gy: 156, cep-1 60 Gy: 185, msh-4 60 Gy: 167, msh-5 60 Gy: 184, wt 120 Gy: 153, cep-1 120 Gy: 198, msh-4 120 Gy: 180, msh-5 120 Gy: 183. (b) Average apoptosis levels scored in untreated nematodes and 48 h after treatment with 180 μM CDDP. Unlike what occurs in wild type, CDDP, an ICL agent, does not induce germ-line apoptosis in pachytene nuclei of the cep-1 and msh-4 mutants. Number of observed gonads: wt untreated: 92, cep-1 untreated: 60, msh-4 untreated: 74, wt 180 μM CDDP: 54, cep-1 180 μM CDDP: 67, msh-4 180 μM CDDP: 59. Error bars correspond to standard error of the means (S.E.M.) calculated from at least three independent experiments. Student's t-tests are shown in Supplementary Table S1
Ionizing radiation triggers activation of the DNA damage checkpoint in the mitotic and meiotic region of msh-4- and msh-5-mutant germ lines
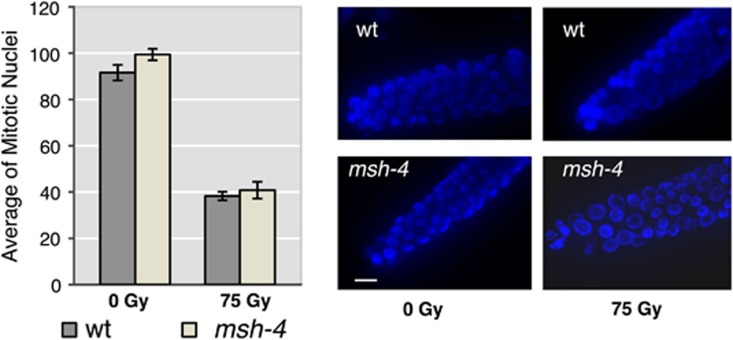
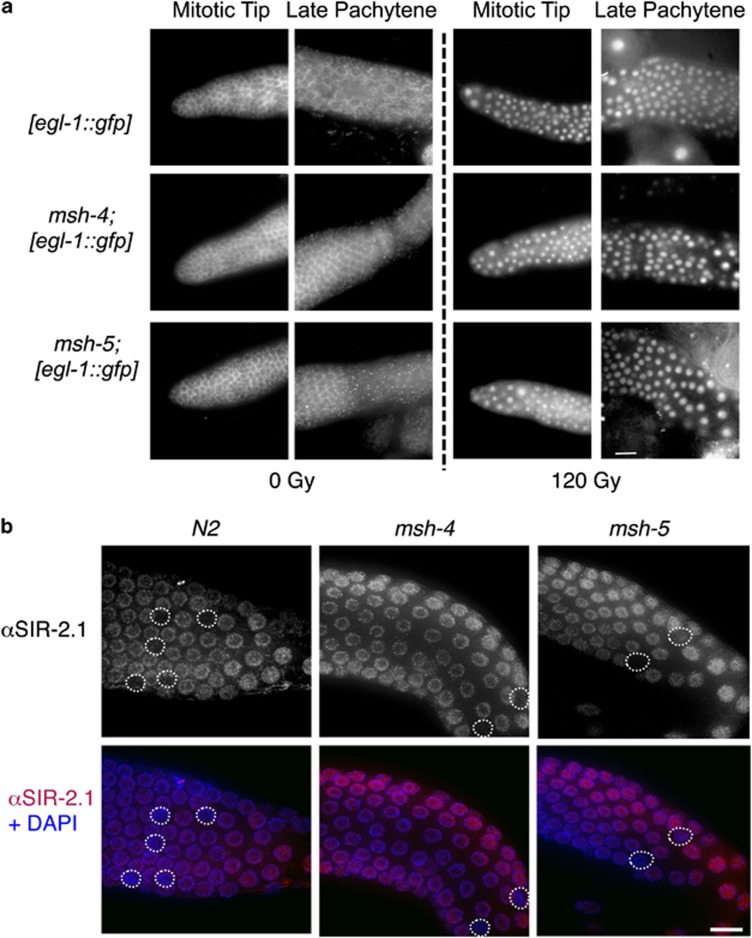
Although the DNA damage checkpoint exerts its action in pachytene by removing cells carrying DNA damage through apoptosis execution, nuclei that are engaged in mitotic proliferation, such as those residing in the tip of the C. elegans germ line, respond to DNA damage triggering a temporary block in cell cycle progression to allow DNA repair. As a consequence of the cell cycle arrest, nuclei in the mitotic region of the germ line of irradiated worms appear enlarged and their overall number greatly reduced. To assess whether MSH-4 was also involved in the damage response in the proliferative region of the germ line, we irradiated wild-type and msh-4-mutant worms and then quantified the number of nuclei in the mitotic tip of the germ line. As shown in Figure 2, msh-4 mutants are proficient in activating the DNA damage response following IR, with the average number of mitotic nuclei dropping to the same levels as those seen in wild-type controls. Given the functionality of the DNA damage response in the proliferative region of msh-4 mutants, we next investigated if the DNA damage pathway, that leads to CEP-1 activation and EGL-1 expression to induce apoptosis in late pachytene, was active in msh-4 and msh-5 mutants. To this end, we used a transgene carrying a transcriptional reporter that drives green fluorescent protein (GFP) expression under the control of the egl-1 promoter and shows nuclear accumulation of GFP in mitotic and late-pachytene nuclei of wild-type worms following IR.17 As expected, GFP expression was never detected in the mitotic region of wild-type control worms and only rarely (and weakly) in late-pachytene nuclei; however, irradiation of these worms induced robust GFP accumulation in the nuclei of germ cells in the mitotic tip and late pachytene regions of most observed germ lines (Figure 3a and Supplementary Table S2). Similar to wild-type controls, GFP expression was not detected in the mitotic region of msh-4 or msh-5 mutants and only rarely in late-pachytene nuclei, suggesting that the recombination intermediates that accumulate in these mutants fail to elicit a response leading to the transcriptional activation of egl-1 (Figure 3a and Supplementary Table S2). However, irradiation of msh-4 and msh-5 mutants induced clear GFP expression in both mitotic and late-pachytene nuclei, demonstrating that these mutants are competent in the damage-dependent transcriptional activation of egl-1 in response to surplus exogenously induced DSBs (Figure 3a and Supplementary Table S2).
Figure 2.
Left: average number of mitotic nuclei before and after treatment with IR (75 Gy). Error bars correspond to standard error of the means (S.E.M.) calculated from at least three independent experiments. Student's t-tests: P (wt (0 Gy)=msh-4 (0 Gy))=0.08; P (wt (75 Gy)=msh-4 (75 Gy))=0.54; P (wt (0 Gy)=wt (75 Gy))<0.0001; P (msh-4 (0 Gy)=msh-4 (75 Gy))<0.0001. Right: representative images of DAPI-stained nuclei in the distal portion of the germ line (mitotic area) before and 15 h after irradiation with IR (75 Gy), scale bar 10 μm. As in wild type, IR induces cell cycle arrest in the premeiotic proliferating area of the gonad of msh-4 mutants
Figure 3.
(a) Analysis of the expression of the Pegl-1::GFP reporter in the indicated genotypes, under physiological conditions and upon exposure to IR (120 Gy). Both msh-4 and msh-5 mutants are competent for the induction of the Pegl-1::GFP reporter after IR in the premeiotic tip and in late pachytene. Scale bar 15 μm. (b) Immunolocalization of the SIR-2.1 protein in pachytene nuclei of the indicated genotypes under physiological conditions. Scale bar 10 μm
SIR-2.1 is a member of the sirtuin family implicated in aging, stress response, and execution of apoptosis in response to DNA damage. In wild-type nematodes, SIR-2.1 accumulates in germ-line nuclei, and, in the few nuclei undergoing physiological apoptosis, it translocates from the nucleus to the cytoplasm.25 As displayed in Figure 3b, SIR-2.1 shows a robust signal in the nucleoplasm of cells in both msh-4 and msh-5 mutants, suggesting that the localization of this protein is not MSH-4/5 dependent. Furthermore, in the few nuclei undergoing physiological apoptosis in msh-4 and msh-5 mutants, as well as in wild-type worms, the staining for SIR-2.1 is absent, demonstrating normal nucleus to cytoplasm translocation of SIR-2.1 in dying cells (Figure 3b).
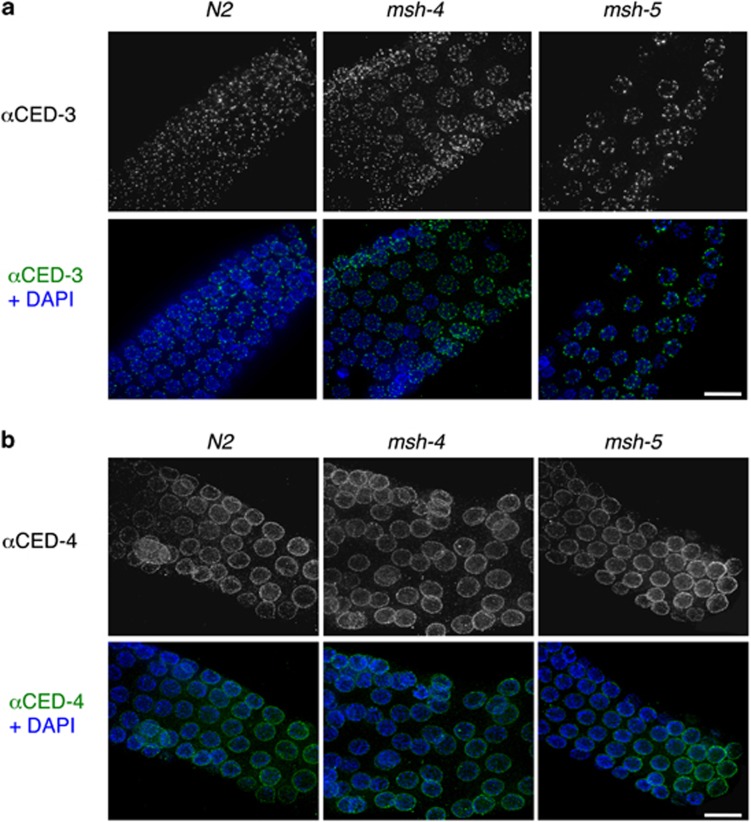
As the DNA damage checkpoint seems to be activated in irradiated msh-4 and msh-5 nematodes, we wondered whether failure in apoptosis execution might depend on defective expression/localization of the apoptosis effectors placed downstream of EGL-1. We therefore performed immunolocalization experiments with antisera against the CED-3 and CED-4 proteins (Figure 4). Both CED-3 and CED-4 proteins properly localize in the pachytene region of msh-4- and msh-5-mutant germ lines, as well as in wild-type controls, suggesting that neither the expression nor the localization of CED-3 or CED-4 requires MSH-4/5.
Figure 4.
Immunolocalization of (a) the CED-3 protein and (b) the CED-4 protein in pachytene nuclei of the indicated genotypes under physiological conditions. Scale bar 10 μm
Lack of msh-4 or msh-5 suppresses activation of damage-dependent apoptosis in some but not all DNA repair-defective mutants
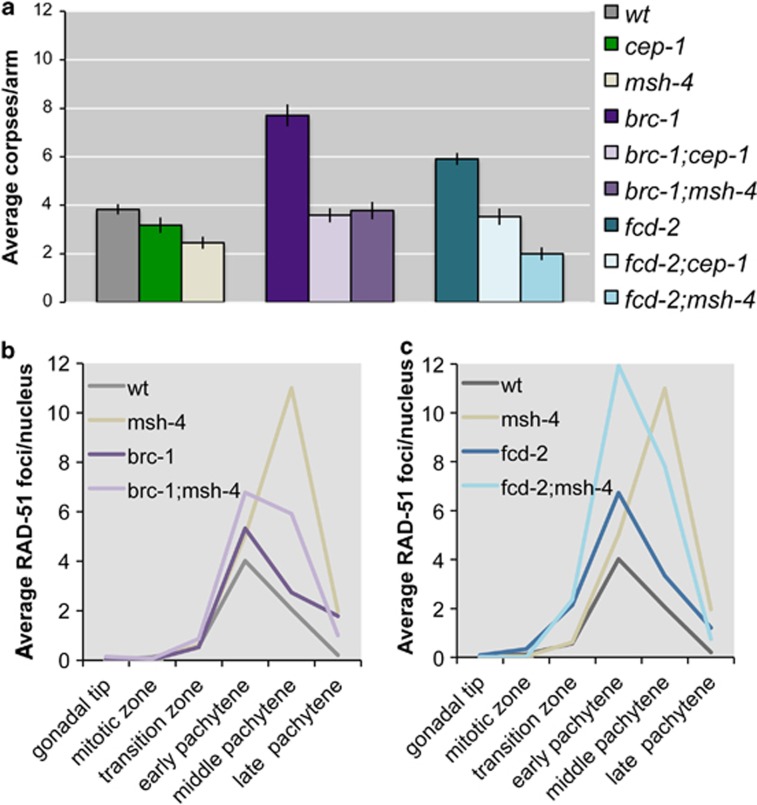
Mutants defective in DNA repair show high levels of germ-nuclei apoptosis due to the activation of the pachytene DNA damage checkpoint. For example, brc-1 and fcd-2 mutants are competent in CO formation but are defective in the non-crossover (NCO) repair pathway that normally deals with the excess DSBs generated during meiotic recombination. As a consequence, mutants of either gene have a significant increase of germ-nuclei apoptosis that is cep-1 and spo-11 dependent.27, 28 We investigated whether apoptosis in these mutants also required MSH-4. Both brc-1;msh-4 and fcd-2;msh-4 double mutants display apoptotic levels lower than the brc-1 and fcd-2 single mutants (Figure 5a). RAD-51 foci levels are higher in both brc-1;msh-4 and fcd-2;msh-4 double mutants compared with single brc-1 and fcd-2 mutants (although accumulation is not additive) (Figures 5b and c). Therefore, MSH-4 appears to be required for inducing apoptosis in response to the DNA damage that accumulates in fcd-2 and brc-1 mutants.
Figure 5.
(a) Average apoptosis levels scored in various genetic combinations. The y axis shows average number of SYTO-12-labelled nuclei per gonad arm. Genotypes are indicated in the colour legend next to the chart. brc-1 or fcd-2 mutants show high levels of damage-induced (cep-1/p53 dependent) apoptosis. In the double mutant brc-1;msh-4 or fcd-2;msh-4, apoptosis decreases. Number of observed gonads: wt: 92, cep-1: 89, msh-4: 74, brc-1: 73, brc-1;cep-1: 94, brc-1;msh-4: 73, fcd-2: 109, fcd-2;cep-1: 76, fcd-2;msh-4: 82. Error bars correspond to standard error of the means (S.E.M.) calculated from at least three independent experiments. Student's t-tests are shown in Supplementary Table S1. (b and c) Quantitative analysis of RAD-51 foci. The y axis refers to the average number of RAD-51 foci/nucleus in the corresponding region of the C. elegans gonad. Genotypes are indicated in the colour legend on the top left of each chart. An average of 100 nuclei for each gonad zone was scored for each genotype. Note that although in the double mutant brc-1;msh-4 there is a relative decrease of RAD-51 foci compared with that in the single msh-4 mutant, removing MSH-4 from brc-1 or fcd-2 single mutants induces an increase in the levels of RAD-51 foci compared with either single mutant, demonstrating that removal of MSH-4 induces an extra accumulation of DNA damage in brc-1 and fcd-2 mutants. Moreover, although the single msh-4 mutant shows diakinesis oocytes with an asset of 12 properly repaired univalents,9 the brc-1;msh-4 double mutant shows high levels of unrepaired chromosome fragments in diakinesis oocytes, further demonstrating that brc-1;msh-4 mutants accumulate DNA damage28
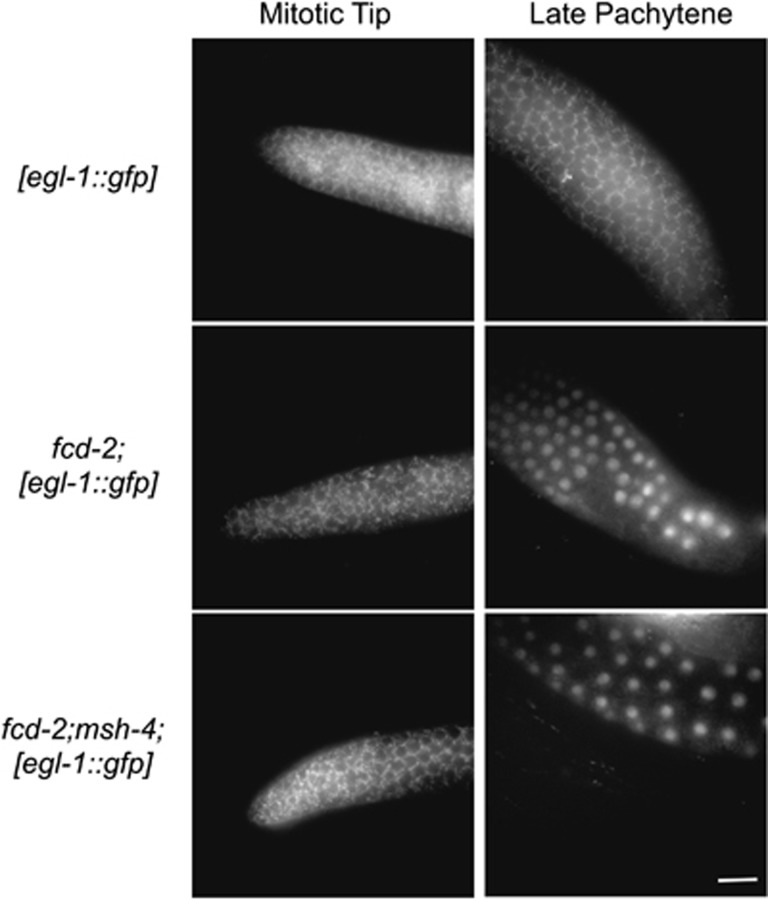
We have shown that the SPO-11-dependent recombination intermediates that accumulate in msh-4 and msh-5 mutants fail to trigger egl-1 expression (Figure 3). These data suggest the possibility that MSH-4/5 functions upstream of checkpoint activation for endogenous DSBs. We therefore tested whether expression of egl-1 was triggered in the double mutant fcd-2;msh-4. Both fcd-2 single and fcd-2;msh-4 double mutants showed expression of the egl-1 transcriptional reporter in late-pachytene nuclei (Figure 6 and Supplementary Table S1). Thus, the DNA damage checkpoint is activated in an fcd-2 background, even in the absence of the MSH-4 protein, but despite this, the apoptotic response in fcd-2 mutants is MSH-4 dependent.
Figure 6.
Analysis of the expression of the Pegl-1::GFP reporter in fcd-2- and fcd-2;msh-4-mutant backgrounds under physiological conditions. The fcd-2 mutants express the Pegl-1::GFP reporter in late-pachytene nuclei irrespectively of the presence of the MSH-4 protein. Scale bar 15 μm
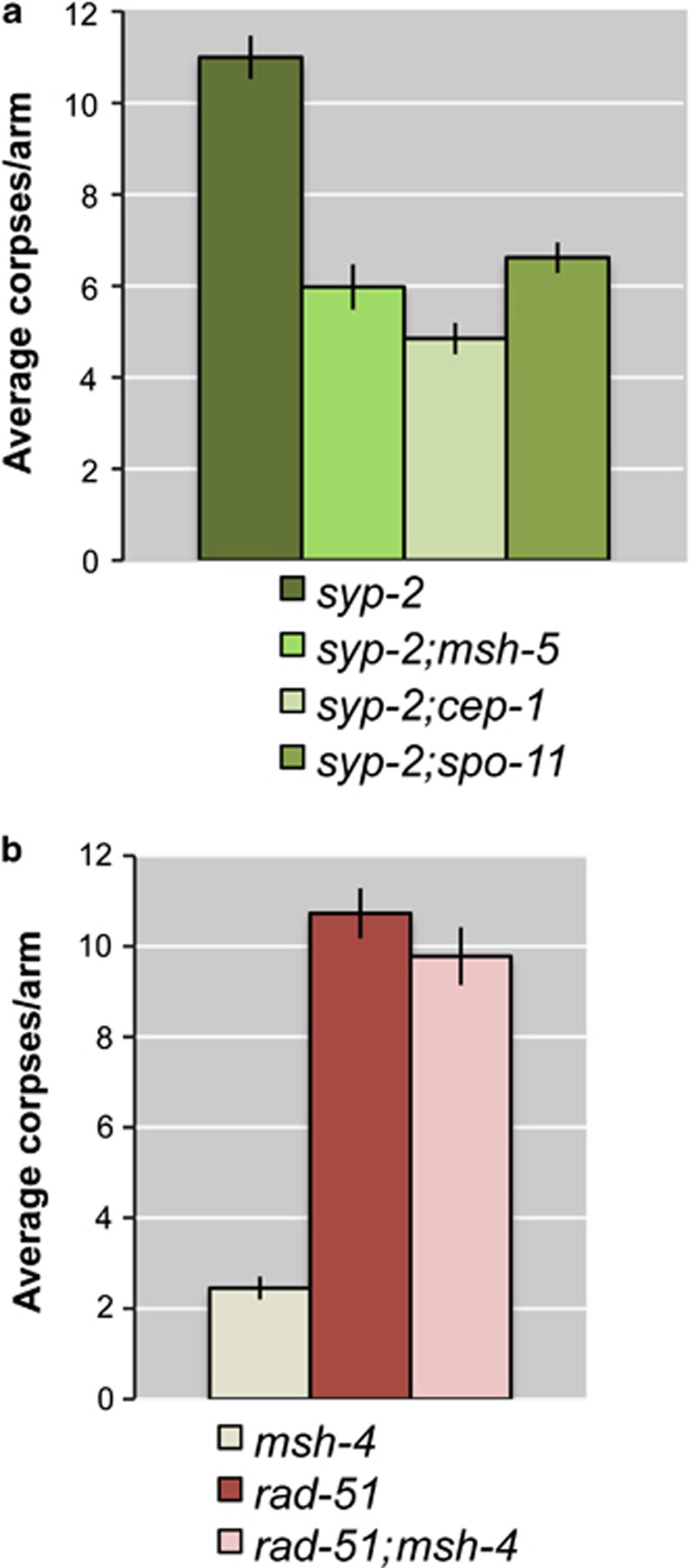
The SYP-2 protein is an essential component of the central region of the SC, and therefore syp-2-mutant nematodes lack synapsis between homologous chromosomes, impairing DSB repair and inducing an accumulation of RAD-51 foci until late pachytene, when DSBs are likely to be repaired using the sister chromatid as a template.19 In this genetic background, two different checkpoints are activated: the synapsis checkpoint15 and the pachytene DNA damage checkpoint,14 causing a dramatic enhancement of apoptosis. We measured the number of apoptotic corpses in syp-2;msh-5 double mutants and found that the level of apoptosis in these worms was significantly reduced compared with that in syp-2 single mutants (Figure 7a). Furthermore, the level of apoptosis detected in syp-2;msh-5 double mutants was similar to that seen in syp-2;cep-1 and syp-2;spo-11 double mutants, in which just the synapsis checkpoint is activated (Figure 7a). These data suggest that DNA damage-induced apoptosis, but not apoptosis due to synapsis defects, is msh-5 dependent in syp-2 mutants.
Figure 7.
Average apoptosis levels scored in various genetic combinations. (a) In the double mutants syp-2;msh-5, apoptosis is reduced at the same level of syp-2, spo-11, and syp-2;cep-1 double mutants, suggesting that DNA damage-induced apoptosis, but not apoptosis due to synapsis defects, is msh-5 dependent. Number of observed gonads: syp-2: 91, syp-2;msh-5: 59, syp-2;cep-1: 53, syp-2;spo-11: 58. (b) In the double mutants rad-51;msh-4, apoptosis remains at very high levels, as observed in rad-51 single mutants. Number of observed gonads: msh-4: 74, rad-51: 41, rad-51;msh-4: 42. Error bars correspond to standard error of the means (S.E.M.) calculated from at least three independent experiments. Student's t-tests are shown in Supplementary Table S1
Taken together, the experiments described above show that the apoptotic response of the DNA damage checkpoint observed in mutants that accumulate recombination intermediates during meiosis depends on the presence of the pro-CO factors MSH-4/5.
We next investigated whether MSH-4 and MSH-5 are also required for the apoptotic response seen in mutants lacking the RAD-51 recombinase, which are unable to undergo any form of homologous recombination/repair. As a consequence, oocytes that are not eliminated by apoptosis in rad-51 mutants display chromatin diffusion and fragmented DNA in diakinesis.3, 30 Surprisingly, rad-51;msh-4 double mutants maintain the same high level of germ-nuclei apoptosis seen in the rad-51 single mutant (Figure 7b), suggesting that the MSH-4/5 dependency of the apoptotic response requires the presence of RAD-51-dependent recombination intermediates.
Crossover-promoting factor ZHP-3, but not COSA-1, is required for DNA damage-dependent apoptosis in pachytene cells
Given the requirement of MSH-4/5 in the apoptotic response of the pachytene DNA damage checkpoint, we asked whether other pro-COs factors that colocalize with them at late pachytene, such as ZHP-3 and COSA-1, might also be required for an efficient apoptotic response.
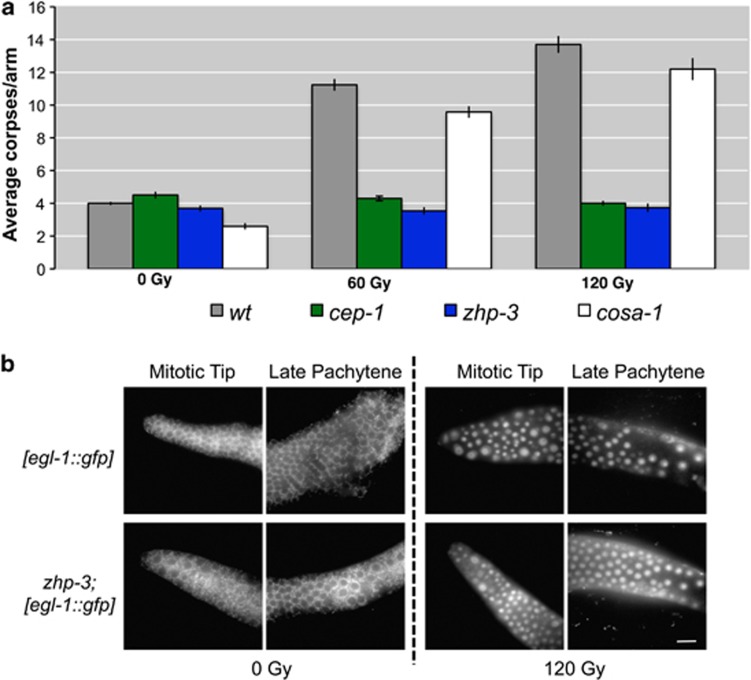
zhp-3 mutants show delayed removal of RAD-51 foci from chromosomes and an impairment in chiasma formation, as shown by the presence of 12 univalents in diakinesis oocytes, suggesting that this protein is crucial for CO formation.12 We exposed zhp-3 mutants to increasing doses of IR and, as shown in Figure 8a, no increase in apoptosis was detectable, indicating that, as MSH-4 and MSH-5, ZHP-3 is also required for the apoptotic response to DNA damage. We next investigated the transcriptional activation of egl-1 in zhp-3 mutants to determine if, similar to msh-4 and msh-5 mutants, they displayed a differential activation of the DNA damage checkpoint depending on whether SPO-11-dependent or IR-induced damage was present. Non-irradiated zhp-3 mutants rarely displayed accumulation of the GFP reporter in any germ-line region, showing that SPO-11-dependent damage failed to efficiently induce egl-1 expression (Figure 8b). However, 24 h after irradiation, GFP accumulated in mitotic and late-pachytene nuclei of zhp-3-mutant germ lines (Figure 8b and Supplementary Table S2). Although we and others17, 31 have observed that IR does not induce robust GFP expression in all irradiated germ lines, we noticed that the number on GFP-negative germ lines following IR was higher in zhp-3 mutants than in other genetic backgrounds (Supplementary Table S2). This could be due to differences in the number of copies of the reporter transgene or partial silencing of the transgene in the population. In either case, the fact that we can only see robust GFP expression of the egl-1 transcriptional reporter following IR treatment is consistent with MSH-4/5 and ZHP-3 being required for activation of the DNA damage checkpoint in response to accumulated SPO-11-dependent intermediates.
Figure 8.
(a) Average apoptosis levels scored in mutants of the pro-CO factors zhp-3 and cosa-1 before and 24 h after irradiation with IR at 60 and 120 Gy. zhp-3 mutants do not respond to genotoxic insult as the apoptosis levels remain unperturbed at different doses of IR, suggesting that the ZHP-3 protein is required for apoptosis execution along with MSH-4/5. COSA-1, on the other hand, is not required for execution of apoptosis in pachytene cells after DNA damage. Number of observed gonads: wt 0 Gy:185, cep-1 0 Gy: 155, zhp-3 0 Gy: 149, cosa-1 0 Gy: 74, wt 60 Gy: 156, cep-1 60 Gy: 185, zhp-3 60 Gy: 139, cosa-1 60 Gy: 48, wt 120 Gy: 153, cep-1 120 Gy: 198, zhp-3 120 Gy: 110, cosa-1 120 Gy: 48. Error bars correspond to standard error of the means (S.E.M.) calculated from at least three independent experiments. Student's t-tests are shown in Supplementary Table S1. (b) Analysis of the expression of the Pegl-1::GFP reporter in a zhp-3-mutant background under physiological conditions and upon exposure to IRs. The zhp-3 mutants are competent for the induction of the Pegl-1::GFP reporter after IR in the premeiotic tip and in late pachytene cells. Scale bar 15 μm
COSA-1 has been recently shown to be required for CO formation. COSA-1 is loaded onto chromosomes at late pachytene forming six foci where it colocalizes with MSH-5 and ZHP-3.10 Therefore, we assayed the apoptotic response of cosa-1 mutants following IR. Unlike msh-4/5 and zhp-3 mutants, the apoptotic response of cosa-1 mutants followed the same trend observed in wild-type worms (Figure 8a), suggesting that COSA-1 is not required for apoptosis execution after DNA damage.
Discussion
We have shown that the pro-CO factors MSH-4, MSH-5, and ZHP-3 are required for executing apoptosis of germ-line nuclei in response to the presence of DNA damage originated by defective meiotic recombination/repair, IR, and CDDP treatment. These three proteins are required at a late step of the recombination process to promote the formation of interhomologue COs, and their absence results in the accumulation of SPO-11-dependent recombination intermediates. Despite this accumulation of DNA damage, single mutants lacking any of the three proteins show impaired activation of the late pachytene DNA damage checkpoint, as evidenced by their failure to induce transcriptional activation of the BH3-only gene egl-1. Although this defect could in principle explain the absence of apoptotic corpses in msh-4, msh-5, or zhp-3 mutants, irradiation of these mutants, or removal of FCD-2 in a msh-4-mutant background, restores the expression of egl-1 without inducing apoptosis. Therefore, the apoptotic failure of msh-4, msh-5, or zhp-3 mutants cannot be simply attributed to deficient checkpoint activation. Furthermore, CEP-1-dependent apoptosis is triggered when silencing by RNAi or cosuppression is activated in the germ line,32 and we have observed that MSH-4 is also required for this kind of selective culling of germ-line nuclei (data not shown). We propose that pro-CO factors are required for apoptosis execution in a pathway that must act downstream or independently of checkpoint activation.
The apoptotic culling of meiotic nuclei carrying DNA damage is executed by a well-conserved pathway that includes the damage sensors ATM, ATR, and the 9-1-1 complex; the kinases CHK-1 and CHK-2; the p53 homologue CEP-1; and the core apoptotic machinery composed of EGL-1, CED-9, CED-4, and the caspase CED-3. Although all the components of this pathway are required for the apoptotic response of the pachytene DNA damage checkpoint, recent investigations have revealed the existence of other pathways that appear to act in parallel or downstream of the CEP-1/EGL-1 pathway to promote apoptosis execution. These pathways are defined by the following proteins: the sirtuin SIR-2.1, which is exported from the nucleus to the cytoplasm in apoptotic cells;25 HYL-1 and LAGR-1,33 which are part of ceramide biogenesis and may be involved in reorganization of the mitochondrial outer membrane in apoptotic cells; the KRI-1 protein, which regulates apoptosis in a cell-nonautonomous manner;34 a worm homologue of the Holliday junction resolvase Gen1;35 and the EEL-1 ubiquitin ligase.36 Irradiation of mutants lacking the activity of any of these proteins induces increased egl-1 transcription, demonstrating activation of the canonical CEP-1-dependent DNA damage pathway, but fails to induce apoptosis. Our investigations have revealed that irradiation of msh-4/5 and zhp-3 mutants results in a similar phenotype, egl-1 expression without apoptosis, revealing that pro-CO factors must also act in parallel or downstream of checkpoint activation to promote apoptosis. As CED-3, CED-4, and SIR-2.1 are properly localized in the germ lines of msh-4/5 mutants, we rule out a direct effect of MSH-4/5 on the expression or localization of these proteins. Future studies should address if MSH-4/5 and ZHP-3 act in a common pathway with any of the apoptosis-licensing proteins listed above or if they define an independent proapoptotic pathway.
A fourth pro-CO factor, COSA-1, is normally recruited onto late pachytene chromosomes where it colocalizes with MSH-4/5 and ZHP-3, probably labelling the sites where COs have occurred. Interestingly, the absence of COSA-1 does not perturb the execution of the apoptotic programme. The apoptotic response of cosa-1 mutants followed the same trend observed in wild type after irradiation. However, in wild-type untreated nematodes, COSA-1 is normally loaded on chromosomes at a later stage than MSH-4/5 and ZHP-3. Furthermore, msh-5 and zhp-3 mutants do not show any COSA-1 loading, whereas both MSH-5 and ZHP-3 are present on the pachytene chromosomes of cosa-1 mutants.10 These observations suggest that the requirement for MSH-4/5 and ZHP-3 in the apoptotic response to DNA damage dwells in a spatial/temporal interval that precedes COSA-1 licensing/loading, implying that MSH-4/5 and ZHP-3 may play an important role in fine-tuning the timing of apoptosis licensing.
Controlling the timing of apoptosis licensing is essential during meiotic prophase, where premature licensing could lead to the elimination of most meiocytes undergoing the early steps of meiotic recombination. Interestingly, we have observed that the absence of RAD-51 bypasses the requirement of MSH-4/5 in the execution of apoptosis, suggesting that MSH-4 and MSH-5 are involved in licensing apoptosis only when recombination intermediates downstream of RAD-51-mediated strand invasion are present. When too many unrepaired recombination events persist in the presence of MSH-4/5-labelled recombination intermediates at late pachytene, the nucleus is doomed to die.
How may MSH-4/5 participate in apoptosis licensing during meiosis? In human cells, hMSH5 has been described to induce apoptosis via a complex interaction with the cAbl protein that results in autophosphorylation of cAbl and the cAbl-dependent phosphorylation of hMSH5 in response to IR, which in turn triggers p73 accumulation and phosphorylation.37 However, in contrast to what we have observed in C. elegans meiosis, hMSH5 has a protective effect to cisplatin treatment in human cells.38 Therefore, further studies will be required in C. elegans to identify the specific partners and steps of the apoptotic response during meiosis at which MSH-4/5 and ZHP-3 are required. Elucidating the proapoptotic roles of MSH-4/-5 and ZHP-3 will increase our understanding of how these repair proteins contribute to preserve genome integrity during gametogenesis.
Materials and Methods
C. elegans strain maintenance
Maintenance and cultivation of worms were based on a standard protocol.39, 40 The following strains were used in this study:
N2: C.elegans wild isolate (Bristol)
AV308: him-14(it21)/mnC1 dpy-10(e128) unc-52(e444) II
AV115: msh-5(me23) IV/nT1[unc-?(n754) let-?] (IV;V)
UV1: zhp-3(jf61::unc-119+)/+ I; unc-119(ed3) III
AV596: cosa-1(tm3298)/qC1[qIs26] III
AV106: spo-11(ok79) IV/nT1[unc-?(n754) let-?](IV;V)
TG9: dpy-13(e184) rad-51(lg8701) IV/nT1[let-?(m435)] (IV;V)
DW102: brc-1(tm1145) III
NB105: fcd-2(tm1298) IV
AV276: syp-2(ok307) V/nT1[unc-?(n754) let-?(m435)] (IV;V)
VC172: cep-1(gk138) I
WS1973: egl-1 V; unc-119(ed3) III; opIs56
Quantitative analysis of DAPI-stained germ lines
Adult hermaphrodites were suspended in 15 μl of M9 solution on glass slides, fixed in absolute ethanol, and stained in 4′, 6′-diamidino-2-phenylindole hydrochloride (DAPI) (2 ng/μl) diluted in M9. Mitotic nuclei were scored 15 h after IR. The quantitative analysis was performed on z series of images acquired using a Leica DM6000 fluorescence microscope and Leica DC 350 FX camera under the control of Leica LAS AF 6000 software. Optical sections were collected at 0.50 μm increments.
Apoptosis quantification
Adult worms were collected into M9 solution containing 33 μM SYTO-12 (Molecular Probes, Eugene, OR, USA) for 90 min in the dark. Worms were transferred to seeded plates for 30 min and then mounted on 2% agarose pads in 2 mM levamisole. The quantitative analysis was performed using a Leica DM6000 fluorescence microscope and Leica DC 350 FX camera under the control of Leica LAS AF 6000 software.
Immunolocalization
The α-RAD-51 antibody was described in a previous study.19 Immunostaining was performed using whole-mount preparations of dissected gonads as described by Colaiacovo et al.19 Quantitative analysis of RAD-51 foci was performed on z series of images acquired using a Leica DM5000 fluorescence microscope and a Leica DC 350 FX camera under the control of Leica LAS AF 6000 software. Optical sections were collected at 0.25 μm increments. Goat anti-SIR-2.1 was used at a dilution of 1 : 50, and the immunostaining was performed as described by Greiss et al.25 Goat anti-CED-4 and goat anti-CED-3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; catalogue no. sc-9193 and sc-9192, respectively) were both used at a dilution of 1 : 100, and the immunostaining was performed as described by Colaiacovo et al.19
Treatment with ICL-inducing agent
Young adult worms were treated with 180 μM CDDP (Sigma-Aldrich, St Louis, MO, USA) and were scored for germ-line apoptosis after 48 h.
Expression of the Pegl-1::GFP transcriptional reporter
Hermaphrodites were exposed to ionizing irradiation from a 137Cs source, and GFP localization was observed 24 h postirradiation in dissected germ lines.
Acknowledgments
We thank Aurora Storlazzi, Stefania Crispi, and John Pulitzer for useful discussions and critical reading of the paper and Sarah Testori for helping with irradiation experiments. Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank Anton Gartner, Verena Jantsch, and Anne Villeneuve for strains and reagents and the IGB-ABT for the use of the integrated microscopy facility. This study was started as part of the doctoral thesis of N Silva, who was the recipient of a predoctoral fellowship from MIUR (PhD, School of Molecular Genetics and Medicine, Naples). A La Volpe's laboratory is funded by CNR Progetto Merit RBNE08YFN3, Associazione Italiana Ricerca sul Cancro Progetto IG11422, and Telethon Grant GGP11076, and E Martinez-Perez's laboratory is funded by an MRC core-funded grant.
Glossary
- CO
crossover
- DSBs
double-strand breaks
- SC
synaptonemal complex
- IR
ionizing radiation
- CDDP
cis-diamminedichloroplatinum(II)
- GFP
green fluorescent protein
- NCO
non-crossover
- DAPI
4′, 6′-diamidino-2-phenylindole hydrochloride
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- La Volpe A, Barchi M. Meiotic double strand breaks repair in sexually reproducing eukaryotes: we are not all equal. Exp Cell Res. 2012;318:1333–1339. doi: 10.1016/j.yexcr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol. 2005;25:7203–7215. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–479. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C, Ederle S, Rocco V, La Volpe A. The Caenorhabditis elegans RAD51 homolog is transcribed into two alternative mRNAs potentially encoding proteins of different sizes. Mol Gen Genet. 1998;260:289–294. doi: 10.1007/s004380050897. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S, Libuda DE, Villeneuve AM. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science. 2011;334:1286–1289. doi: 10.1126/science.1212424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP. The many facets of SC function during C. elegans meiosis. Chromosoma. 2006;115:195–211. doi: 10.1007/s00412-006-0061-9. [DOI] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, et al. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- Moser SC, von Elsner S, Bussing I, Alpi A, Schnabel R, et al. Functional dissection of Caenorhabditis elegans CLK-2/TEL2 cell cycle defects during embryogenesis and germline development. PLoS Genet. 2009;5:e1000451. doi: 10.1371/journal.pgen.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, et al. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Rutkowski R, Dickinson R, Stewart G, Craig A, Schimpl M, et al. Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS Genet. 2011;7:e1002238. doi: 10.1371/journal.pgen.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Yan N, Gu L, Kokel D, Chai J, Li W, et al. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Greiss S, Hall J, Ahmed S, Gartner A. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 2008;22:2831–2842. doi: 10.1101/gad.482608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK. Germline survival and apoptosis. WormBook. 2008;4:1–20. doi: 10.1895/wormbook.1.145.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Adamo A, Montemauri P, Silva N, Ward JD, Boulton SJ, et al. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008;9:287–292. doi: 10.1038/sj.embor.7401167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaker S, Cobb J, Pyle A, Handel MA. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: insights into regulation of spermatogenic progress. Dev Biol. 2002;249:85–95. doi: 10.1006/dbio.2002.0708. [DOI] [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Woglar A, Silva N, Penkner A, Jantsch V, et al. Transgene-mediated cosuppression and RNA interference enhance germ-line apoptosis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:3440–3445. doi: 10.1073/pnas.1107390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Yin X, Allan R, Lu DD, Maurer CW, et al. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science. 2008;322:110–115. doi: 10.1126/science.1158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Greiss S, Gartner A, Derry WB. Cell-nonautonomous regulation of C. elegans germ cell death by kri-1. Curr Biol. 2010;20:333–338. doi: 10.1016/j.cub.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly AP, Freeman A, Hall J, Declais AC, Alpi A, et al. The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS Genet. 2010;6:e1001025. doi: 10.1371/journal.pgen.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AJ, Li M, Yu B, Gao MX, Derry WB. The EEL-1 ubiquitin ligase promotes DNA damage-induced germ cell apoptosis in C. elegans. Cell Death Differ. 2011;18:1140–1149. doi: 10.1038/cdd.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins JD, Wu X, Chu YL, Her C. Evidence for a direct involvement of hMSH5 in promoting ionizing radiation induced apoptosis. Exp Cell Res. 2009;315:2420–2432. doi: 10.1016/j.yexcr.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins JD, Wu X, Her C. MutS homologue hMSH5: role in cisplatin-induced DNA damage response. Mol Cancer. 2012;11:10. doi: 10.1186/1476-4598-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin J.MethodsIn: Wood WB, (ed.)The Nematode Caenorhabditis elegans Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA; 1988588–589. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.