Abstract
Deregulation of transcriptional networks contributes to haematopoietic malignancies. The transcription factor Spi-1/PU.1 is a master regulator of haematopoiesis and its alteration leads to leukaemia. Spi-1 overexpression inhibits differentiation and promotes resistance to apoptosis in erythroleukaemia. Here, we show that Spi-1 inhibits mitochondrial apoptosis in vitro and in vivo through the transcriptional repression of Bim, a proapoptotic factor. BIM interacts with MCL-1 that behaves as a major player in the survival of the preleukaemic cells. The repression of BIM expression reduces the amount of BIM-MCL-1 complexes, thus increasing the fraction of potentially active antiapoptotic MCL-1. We then demonstrate that Spi-1 represses Bim transcription by binding to the Bim promoter and by promoting the trimethylation of histone 3 on lysine 27 (H3K27me3, a repressive histone mark) on the Bim promoter. The PRC2 repressive complex of Polycomb is directly responsible for the deposit of H3K27me3 mark at the Bim promoter. SUZ12 and the histone methyltransferase EZH2, two PRC2 subunits bind to the Bim promoter at the same location than H3K27me3, distinct of the Spi-1 DNA binding site. As Spi-1 interacts with SUZ12 and EZH2, these results indicate that Spi-1 modulates the activity of PRC2 without directly recruiting the complex to the site of its activity on the chromatin. Our results identify a new mechanism whereby Spi-1 represses transcription and provide mechanistic insights on the antiapoptotic function of a transcription factor mediated by the epigenetic control of gene expression.
Keywords: apoptosis, epigenetic regulation, leukaemia, Spi-1/PU.1 oncogene, transcription factor
Acute myeloid leukaemia (AML) develops through a multistep process driven by the progressive accumulation of mutations, leading to deregulation of cell proliferation and differentiation. Most frequently, abnormalities in cell differentiation are the result of loss-of-function mutations in lineage-specific transcription factors, whereas alterations in cell proliferation are associated with gain-of-function mutations in signalling pathways.1 Mutations that target components of the spliceosomal machinery2 and epigenetic regulators3 have been identified as well, although their functional consequences in leukaemogenesis are not clear. To date, AML treatments are largely determined by the nature of the identified mutated proteins and a major challenge is to design molecules that can target each molecular alteration contributing to transformation. For that, understanding the molecular mechanisms controlled by an oncoprotein is one essential step. We have previously described a mouse model of erythroleukaemia (Spi-1 transgenic mice) that recapitulates the multistep development of human AML.4 Spi-1 is a master transcription factor of haematopoiesis that is also involved in pre-mRNA splicing regulation.5 During the preleukaemic stage of the disease, the differentiation blockage of the erythroid progenitors is the first oncogenic mark associated with Spi-1 activity.6 Spi-1 is also responsible for resistance to apoptosis, acceleration of replication and increased genetic instability.7, 8 The leukaemic stage of the disease is characterized by the emergence of malignant cells that have acquired Kit mutations promoting constitutive activation of signalling pathways.4, 9
Apoptosis is a major defence in response to the loss of growth control, and many cancer cells develop antiapoptotic activities.10 BCL-2 family members are major regulators of the commitment to programmed cell death and have pro- or antiapoptotic activities.11, 12 The balance between pro- and antiapoptotic molecules is disrupted in many leukaemic cells, conferring resistance to apoptosis. Consequently, proteins of the BCL-2 family appear to be suitable targets for therapies in cancer.13 To this end, it is crucial to identify the molecules that regulate the apoptotic machinery as well as their downstream effectors.
Here, we explored how Spi-1 induces resistance to apoptosis in the preleukaemic erythroid progenitors of the Spi-1 transgenic mice. We found that Spi-1 inhibits mitochondrial apoptosis both in vitro and in vivo through transcriptional repression of Bim. We then demonstrate that interaction of Spi-1 with the polycomb repressive complex 2 (PRC2) promotes epigenetic modifications that have a major role in repressing Bim transcription. Our results argue for a new mechanism of transcriptional repression by Spi-1 that involves the modification of PRC2 activity to create a repressive chromatin context in the preleukaemic cells.
Results
Spi-1 inhibits mitochondrial apoptosis
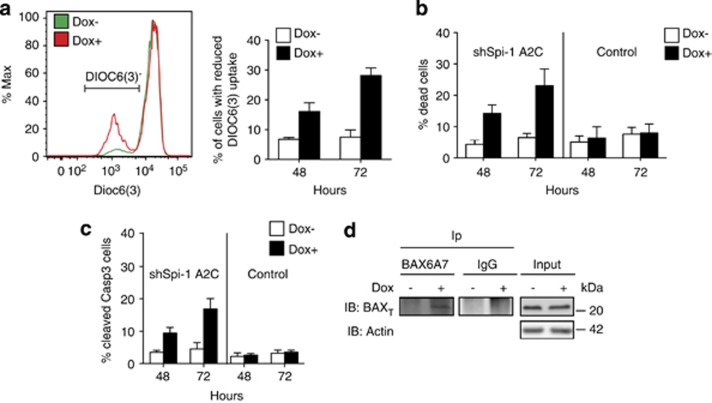
Here, we investigated whether Spi-1 protects preleukaemic cells from apoptosis by influencing the apoptotic mitochondrial pathway. We used preleukaemic Spi-1 transgenic cell lines previously established with a doxycycline (dox)-inducible shRNA against Spi-1 (shSpi-1-A2B and shSpi-1-A2C cells).8 The percentage of cells with reduced mitochondrial membrane potential (ΔΦ) (Figure 1a), of dead cells (Figure 1b) and of cells that expressed cleaved-caspase-3 (the active form) (Figure 1c), was increased when Spi-1 was silenced. Upon activation, BAX undergoes a conformational change detectable using the anti-BAX monoclonal antibody 6A7.14, 15 Immunoprecipitation (IP) experiments showed that Spi-1 downregulation by dox treatment induced the exposure of the 6A7 epitope (Figure 1d). Altogether, these results indicate that Spi-1 inhibits the mitochondrial apoptotic pathway.
Figure 1.
Spi-1 protects erythroleukaemic cells against apoptosis through the mitochondrial pathway. (a–c) shSpi-1-A2C cells and control cells were seeded at 5 × 103 cells per ml and cultured with or without dox for the indicated periods of time (h). (a) Dissipation of the mitochondrial membrane potential (ΔΦ) was measured by using the DiOC6(3) dye. The percentage of cells with reduced membrane potential (DiOC6(3)−) was determined by flow cytometry. The curve of a representative experiment is shown (left panel) and the histogram (right panel) represents the mean±S.D. of three independent experiments. (b) Dead cells were counted by using the Trypan blue exclusion test. Data are the mean±S.D. of at least six independent experiments. (c) Apoptosis was detected by flow cytometry using an anti-cleaved-caspase-3 antibody. Data are the mean±S.D. of at least three independent experiments. (d) Extracts from shSpi-1-A2C cells grown with or without dox for 3 days were immunoprecipitated (Ip) with the anti-BAX 6A7 monoclonal antibody or immunoglobulin G (IgG) as indicated and immunoblotted with an anti-BAX polyclonal antibody (BAXT). Input (10% of the lysate) was immunoblotted with the anti-BAX and anti-Actin (loading control) antibodies. The western blot is representative of three independent experiments
Spi-1 decreases BIM expression in preleukaemic cells
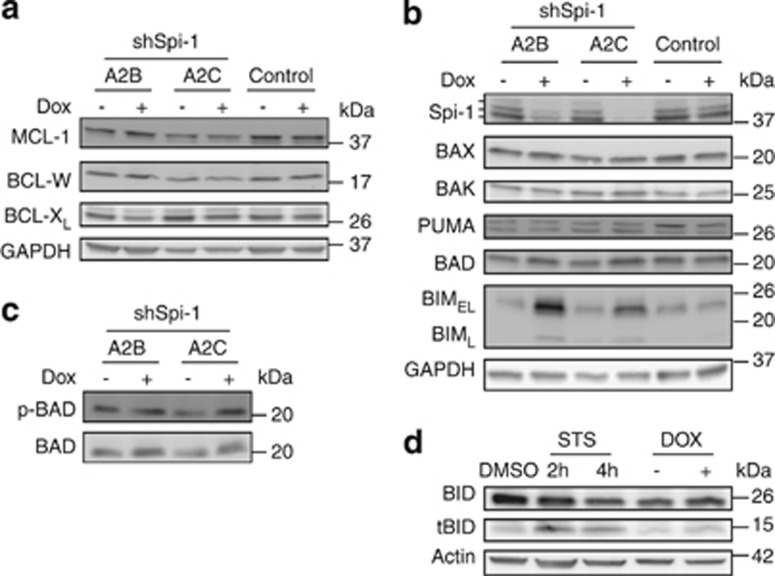
Mitochondrial apoptosis is regulated through the balance between pro- and antiapoptotic BCL-2 family members. Transcriptomic data we previously obtained16 indicated that the proapoptotic Bmf, Hrk and Noxa and the antiapoptotic Bcl2 and A1 genes were not expressed in shSpi cells cultured with or without dox (see Supplementary Figure 1A and Ridinger-Saison et al.16). Therefore, we evaluated the protein level of the other BCL-2 family members in two independent shSpi cell lines (shSpi-1-A2B and shSpi-1-A2C) and control cells cultured with or without dox for 2 days. Among the antiapoptotic factors, BCL-XL expression was slightly decreased in cells treated with dox (x0.75), whereas the protein level of MCL-1 and BCL-W was not changed (Figure 2a). Among the proapoptotic factors, the expression of PUMA, BAX and BAK was not modified by dox treatment (Figure 2b). The BAD protein level and phosphorylation (Figures 2b and c) were slightly higher in cells grown with dox (x1.5) than in cells cultured without dox. As phosphorylated BAD is inactive,17 the increase in its phosphorylation should not be related to the apoptosis induced by Spi-1 knockdown. BID and its activated form tBID were slightly increased in cells treated with dox compared with untreated cells. This increase appears poorly relevant when compared with that induced by staurosporine (STS), which was used as an apoptosis inducer (Figure 2d). The most prominent effect associated with Spi-1 silencing in preleukaemic cells concerned BIM expression. BIM is a proapoptotic factor with three major isoforms (short, BIMS; long, BIML; and extra-long, BIMEL).18 In preleukaemic cells, BIMEL was the most abundant isoform, whereas BIML expression was modest and BIMS was barely detectable. The addition of dox strongly increased BIMEL and BIML protein levels in the two shSpi cell lines (3–4 times) (Figure 2b), suggesting that the antiapoptotic effect of Spi-1 may result from its ability to maintain BIM expression low.
Figure 2.
BCL-2 family protein expression as a function of Spi-1 level of expression. Expression of antiapoptotic (a) and proapoptotic (b–d) BCL-2 family members was determined by immunoblotting using lysates from shSpi-1-A2B, shSpi-1-A2C and control cells cultured with or without dox for 2 days. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or Actin were used as a loading control. The apparent molecular weights (kDa) are indicated on the right of the panels. When indicated, cells were treated with STS
Spi-1 exerts its antiapoptotic function through repression of BIM expression
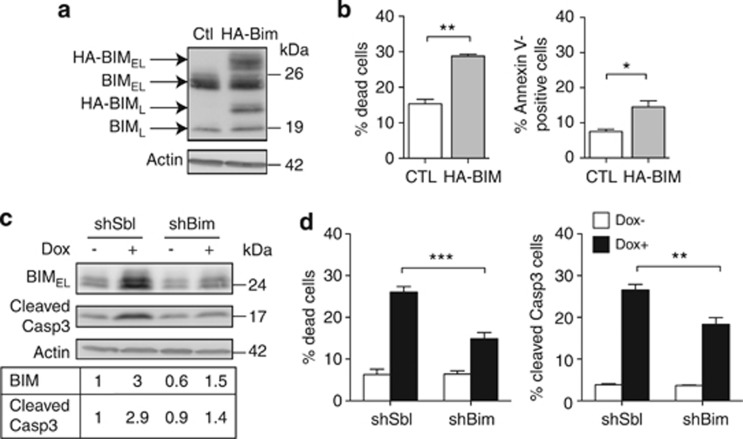
To assess whether low BIM expression is required for Spi-1 antiapoptotic function, we transfected a HA-tagged Bim expression vector in the preleukaemic cells (Figure 3a). An increase (twofold) in the percentage of dead and apoptotic cells was detected (Figure 3b), indicating that BIM overexpression is sufficient to trigger apoptosis in preleukaemic cells. To determine whether increased BIM expression is required for apoptosis induced by Spi-1 knockdown, we introduced shRNA against Bim (shBim) or scrambled shRNA (shSbl) in the preleukaemic cells and cultured them with or without dox. The shBim expression reduced by 50% the upregulation of Bim associated with Spi-1 knock-down (Figure 3c). This results in a decrease in the percentage of dead cells (Figure 3d) and of cells that expressed active caspase-3 (Figures 3c and d).
Figure 3.
Spi-1 inhibits apoptosis by reducing BIM expression. (a and b) Overexpression of BIM in shSpi-1-A2B cells. Cells were transfected with the pcDNA-HA-Bim (HA-Bim) construct or pcDNA vector (CTL) as a control. (a) Twenty-four hours after transfection, cells lysates were immunoblotted with the anti-BIM antibody and the anti-Actin antibody as loading control. (b) Dead cells and apoptotic cells were measured by flow cytometry after DAPI staining (left panel) or Annexin V/DAPI staining (right panel), respectively. Bars represent the mean±S.D. of three independent experiments. (c and d) Bim knockdown in shSpi-1-A2B cells. Cells were infected with lentiviral vectors that encoded shBim or control shSbl. Cells were plated at 5 × 103 cells per ml and cultured with or without dox for 72 h. (c) Whole-cell extracts were analysed by immunoblotting with anti-BIM and anti-cleaved-caspase-3 antibodies and the anti-Actin antibody as loading control (upper panels). The lower panel shows the quantification of the intensity of the protein bands after normalization to Actin and relative to shSbl Dox−. (d) The percentage of dead cells and of apoptotic cells was evaluated by flow cytometry after DAPI staining (left panel) or after incubation with the anti-cleaved-caspase-3 antibody (right panel), respectively. Bars represent the mean±S.D. of five independent experiments. Statistical differences were assessed with the Student's t-test: ***P<0.001, **P<0.01, *P<0.05
Thus, Bim silencing significantly overcame apoptosis induced by Spi-1 knockdown. These results demonstrate that the antiapoptotic function of Spi-1 in preleukaemic cells requires low BIM expression.
Spi-1 inhibits Bim expression and protects preleukaemic cells from apoptosis also in vivo
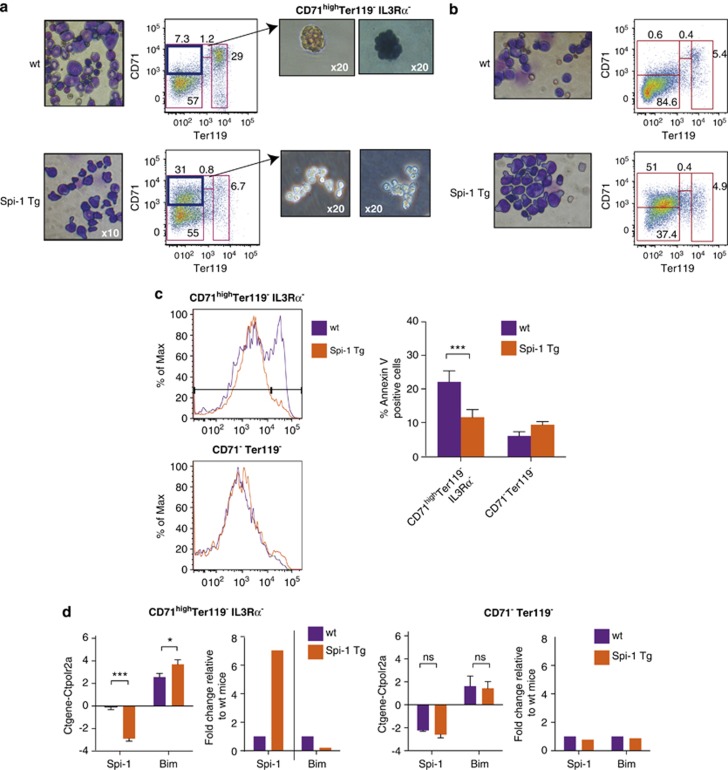
Then, we investigated whether resistance to apoptosis and low BIM expression were also observed in primary preleukaemic cells isolated from Spi-1 transgenic mice.6 As Spi-1 induces resistance to apoptosis in erythroid progenitors, we used flow cytometry to purify such cells. Specifically, the earliest erythropoietin (EPO)-dependent progenitors (the colony-forming unit-erythroid, CFU-E) are CD71highTer119−IL3Rα−, proerythroblasts are CD71highter119med and terminally differentiated erythroblasts are Ter119high.19, 20, 21 Accordingly, CD71highTer119−IL3Rα− cells purified from the bone marrow of wild-type (wt) mice and grown in methylcellulose supplemented with EPO gave rise to CFU-E, as indicated by their clonogenicity, morphology, EPO dependence and haemoglobinization (blue staining with benzidine) (Figure 4a). Compared with wt mice, the bone marrow (Figure 4a) and spleen (Figure 4b) of Spi-1 transgenic leukaemic mice contained a higher fraction of CD71highTer119− cells (around 30 and 55 versus 10% and 1%, respectively) that were mainly IL3Rα− (around 80%). Only CD71highTer119−IL3Rα− cells required EPO to expand in methylcellulose, leading to consider them as preleukaemic blasts.
Figure 4.
Spi-1 decreases Bim expression and prevents apoptotic death in vivo. (a and b) Cytospin preparations of cells from bone marrow (a) and spleen (b) of wt and Spi-1 transgenic (Spi-1 Tg) mice with erythroleukaemia were stained with May–Grünwald–Giemsa (left panels). Images were acquired with an × 10 objective lens (Nikon Eclipse TE300 microscope, Champigny-Sur-Marne, France). Representative fluorescence profiles of cells incubated with antibodies against Ter119 and CD71 are shown. Dead cells were excluded by DAPI positivity. Sorted CD71highTer119−IL3Rα− cells were plated on methylcellulose supplemented with FBS and EPO and CFU-Es from wt bone marrow were examined at day 3. Haemoglobinized cells were detected by staining with 0.2% benzidine (right panels). (c) Representative flow cytometric profiles of living CD71highTer119− IL3Rα− and CD71−Ter119− cells (DAPI−) that were Annexin V-positive. The histograms present the mean of eight independent experiments of Annexin V staining in each population. (d) Bim and Spi-1 mRNA levels in CD71highTer119− IL3Rα− (left) and CD71−Ter119− (right) cells from wt and Spi-1 Tg mice were quantified by real-time qPCR and normalized to the Polr2a mRNA level (Ctgene-Ctpolr2a). Bars correspond to the mean ΔCt±S.D. of at least three independent experiments. Statistical analysis of the ΔCt values was carried out by using the Student's t-test; ***P<0.001 and *P<0.05. For easier interpretation, we added on the right the histograms representing the fold change relative to wt mice, calculated from the 2−ΔΔCt. NS, nonsignificant
We compared apoptosis in CD71highTer119−IL3Rα− cells from wt and Spi-1 transgenic leukaemic mice. The percentage of Annexin V-positive apoptotic cells from preleukaemic mice (11%) was lower than in wt mice (22%) (Figure 4c). No significant difference in the number of Annexin V-positive apoptotic CD71−Ter119− non-erythroid cells was observed in wt and preleukaemic mice. In parallel, we found that Spi-1 expression was higher (sevenfold, P<0.001) in CD71highTer119−IL3Rα− cells from Spi-1 transgenic preleukaemic mice than in wt animals (Figure 4d). Conversely, Bim mRNA expression was reduced (2.2-fold, P<0.05) in CD71highTer119−IL3Rα− cells from Spi-1 transgenic preleukaemic mice compared with wt animals. We did not detect any significant difference of Bim and Spi-1 mRNA expression in CD71−Ter119− non-erythroid cells of wt and preleukaemic mice. These results validate the relationship between the high expression of Spi-1, the weak expression of BIM and the resistance to apoptosis in primary preleukaemic cells.
MCL-1 contributes to the survival of preleukaemic cells
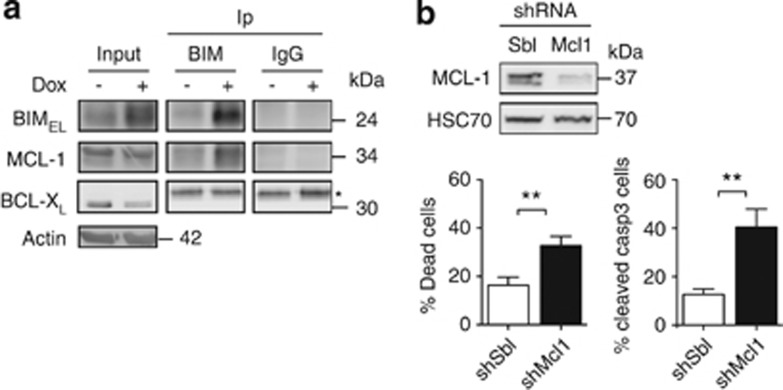
BIM could carry out its proapoptotic function by interacting with the antiapoptotic factors BCL-XL or MCL-1 that are expressed in the preleukaemic cells, thereby acting as an antagonist of these proteins.22, 23, 24, 25, 26 We investigated whether MCL-1 and BCL-XL interacted with BIM by co-IP. MCL-1, but not BCL- XL, was detected in BIM immunoprecipitates (Figure 5a). Moreover, in dox-treated cells (Spi-1 knocked down) the amount of MCL-1 complexed with BIM was clearly increased. As MCL-1 expression was not influenced by Spi-1 knockdown (Figure 2), the increased amount of BIM-MCL-1 complexes strongly suggests that the pool of free, active MCL-1 is reduced upon Spi-1 silencing. Consequently, BIM proapoptotic effect may, at least in part, occur through neutralization of the antiapoptotic MCL-1. These results prompted us to quantify the effect of MCL-1 silencing on survival of preleukaemic cells using Mcl-1-shRNA. The reduction of MCL-1 expression (80% of decrease) increased the number of dead cells (twofold) and of cells that expressed cleaved-caspase 3 (threefold) in comparison with shSbl cells (Figure 5b). Thus, MCL-1 contributes decisively to the survival of preleukaemic cells.
Figure 5.
Spi-1 inhibits apoptosis by increasing the pool of free MCL-1. (a) Protein lysates from shSpi-1-A2C cells cultured with or without dox for 72 h were immunoprecipitated (Ip) with anti-BIM antibody and immunoblotted with anti-BIM, anti-MCL-1 and anti-BCL-XL antibodies. Input corresponds to 10% of the lysates and was analysed by immunoblotting with the same antibodies and the anti-Actin antibody as loading control. Data are representative of three independent experiments. *Aspecific band. (b) shSpi-1-A2C cells were infected with lentiviral vectors encoding an shRNA against Mcl-1 (shMcl1) or control shSbl. Cells were plated at 5 × 103 cells per ml and cultured for 24 h. Whole-cell extracts were analysed by immunoblotting with the anti-MCL-1 and the anti-heat-shock cognate 70 (HSC70) (loading control) antibodies (upper panel). The percentage of dead cells and of apoptotic cells was evaluated by flow cytometry after DAPI staining (bottom left panel) or after incubation with the anti-cleaved-caspase-3 antibody (bottom right panel), respectively. Bars represent the mean±S.D. of five independent experiments. Statistical differences were assessed with the Student's t-test: **P<0.01
Altogether, our data suggest that Spi-1 promotes resistance to apoptosis in preleukaemic cells by inhibiting the expression of BIM. This in turn leads to an increase of the free pool of MCL-1 that can protect cells from apoptosis.
Downregulation of Bim in preleukaemic cells is due to transcriptional repression by Spi-1
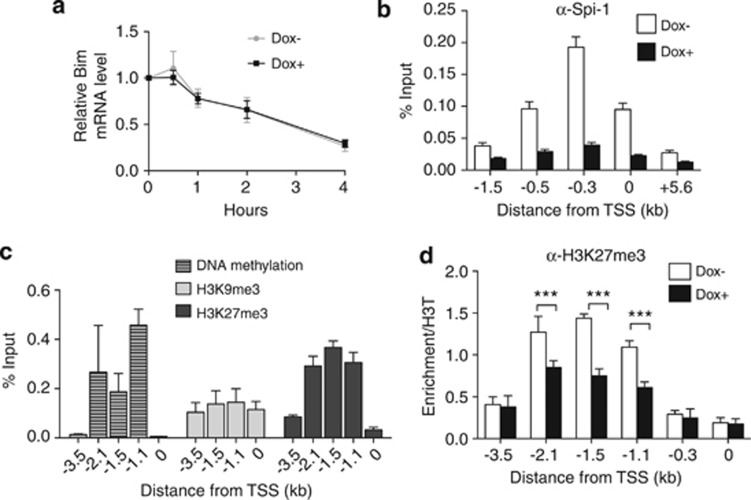
We aimed to determine the mechanism of BIM downregulation associated with Spi-1 expression. Bim transcript and protein expression was induced as soon as 26 h after dox treatment (Supplementary Figures 2A and B), showing that the regulation of Bim expression occurs first at the transcriptional level. As Bim mRNA can be stabilized through binding of the heat-shock cognate 70 to the Bim 3′-UTR,27 we examined whether Spi-1 reduces the stability of Bim mRNA. Dox-treated or untreated shSpi-1-A2B cells were cultured in the presence of the transcription inhibitor actinomycin D. Quantification of Bim mRNA in actinomycin D-treated cells revealed a comparable decrease of Bim expression with or without dox, indicating that Spi-1 did not influence Bim RNA degradation (Figure 6a). Thus, the inhibition of Bim expression by Spi-1 occurs at the level of the initiation of transcription.
Figure 6.
Spi-1 represses Bim by controlling transcription initiation. (a) ShSpi-1-A2B cells were cultured with or without dox for 24 h and then treated with 1 μg/ml of actinomycin D for the indicated times. Bim mRNA expression was measured by real-time qPCR and normalized to the Gapdh mRNA level. The levels of Bim mRNA in actinomycin D-treated cells are relative to the levels in untreated cells. The graph corresponds to the mean±S.D. of at least three independent experiments. (b) Chromatin from shSpi-1-A2C and shSpi-1-A2B cells cultured with or without dox for 72 h was immunoprecipitated using an anti-Spi-1 antibody. Immunopurified DNA was quantitated by real-time qPCR using primers that amplify the regions of the Bim promoter located at the indicated distance from the TSS. Bars represent the mean±S.E.M. of at least five independent experiments. (c) Chromatin and DNA isolated from shSpi-1-A2C and shSpi-1-A2B cells were sonicated and methylated DNA or DNA associated with H3K27me3 or H3K9me3 was immunoprecipitated. The same primer pairs were used to amplify the Bim promoter regions to assess H3K27me3, H3K9me3 and DNA methylation. The histogram bars represent the enrichment relative to the Input (%Input) determined by real-time qPCR. The mean±S.D. of at least three independent experiments are shown. (d) Chromatin from shSpi-1-A2C cells cultured with or without dox for 72 h was immunoprecipitated using an anti-H3K27me3 antibody. The %Input was normalized to the enrichment obtained with the anti-histone 3 antibody. Immunopurified DNA was quantitated by real-time qPCR using primers that amplify the regions of the Bim promoter located at the indicated distance from the TSS. The bars represent the mean±S.E.M. of at least four independent experiments. ***P<0.001 by analysis of variance (ANOVA) test followed by post hoc test (pairwise t-test adjusted for multiple tests)
Spi-1 binds to the Bim promoter and promotes H3K27 trimethylation, a repressive histone mark
To further investigate how Spi-1 restrains the Bim transcription, we screened transcription factor binding sites on the Bim promoter using the Genomatix-MatInspector module. We found one putative Spi-1 binding site located 0.3 kb upstream of the Bim transcriptional start site (TSS). To determine the biological relevance of this Spi-1 binding site, we performed chromatin IP (ChIP) experiments using an anti-Spi-1 antibody in shSpi-1-A2C and shSpi-1-A2B cells. The Bim promoter region was immunoprecipitated with the highest enrichment at 0.3 kb upstream of the TSS (Figure 6b). This enrichment was strongly reduced in dox-treated cell extracts, consistent with Spi-1 knockdown. These data indicate that Spi-1 directly binds to the Bim promoter.
Bim transcription is sensitive to epigenetic modifications of chromatin, particularly trimethylation of histone 3 on lysine 27 (H3K27me3) or DNA methylation.28, 29, 30, 31, 32 We first evaluated DNA methylation at the Bim promoter in shSpi-1-A2C and shSpi-1-A2B cells by IP of methylated DNA (MeDIP) using an antibody against 5-methylcytidine. We detected DNA methylation at the Bim promoter between 2.1 and 1.1 kb upstream of the TSS (Figure 6c). Then, we used bisulphite sequencing to determine accurately cytosine methylation at the Bim promoter, which includes 45 CpG dinucleotides. Comparison of the methylation profiles after the addition or not of dox showed that Spi-1 did not modify the frequency or the profile of cytosine methylation (Supplementary Figure 3). Thus, the modulation of Bim transcription by Spi-1 was not due to a change in DNA methylation.
Next, we evaluated the trimethylation on lysine 9 (H3K9me3) or H3K27me3 by ChIP using appropriate antibodies. We did not detect any peak of H3K9me3 enrichment at the Bim promoter (Figure 6c). Conversely, we observed H3K27me3 along the Bim promoter, with the highest enrichment 1.5 kb upstream of the TSS (Figure 6c). Furthermore, H3K27me3 was significantly reduced in dox-treated cells compared with untreated cells, particularly in the region from 2.1 to 1.1 kb upstream of the Bim TSS (Figure 6d). These data indicate that Spi-1 promotes the deposit of the H3K27me3 repressive mark further upstream on the Bim promoter.
Spi-1 represses Bim transcription through functional interference with the PRC2
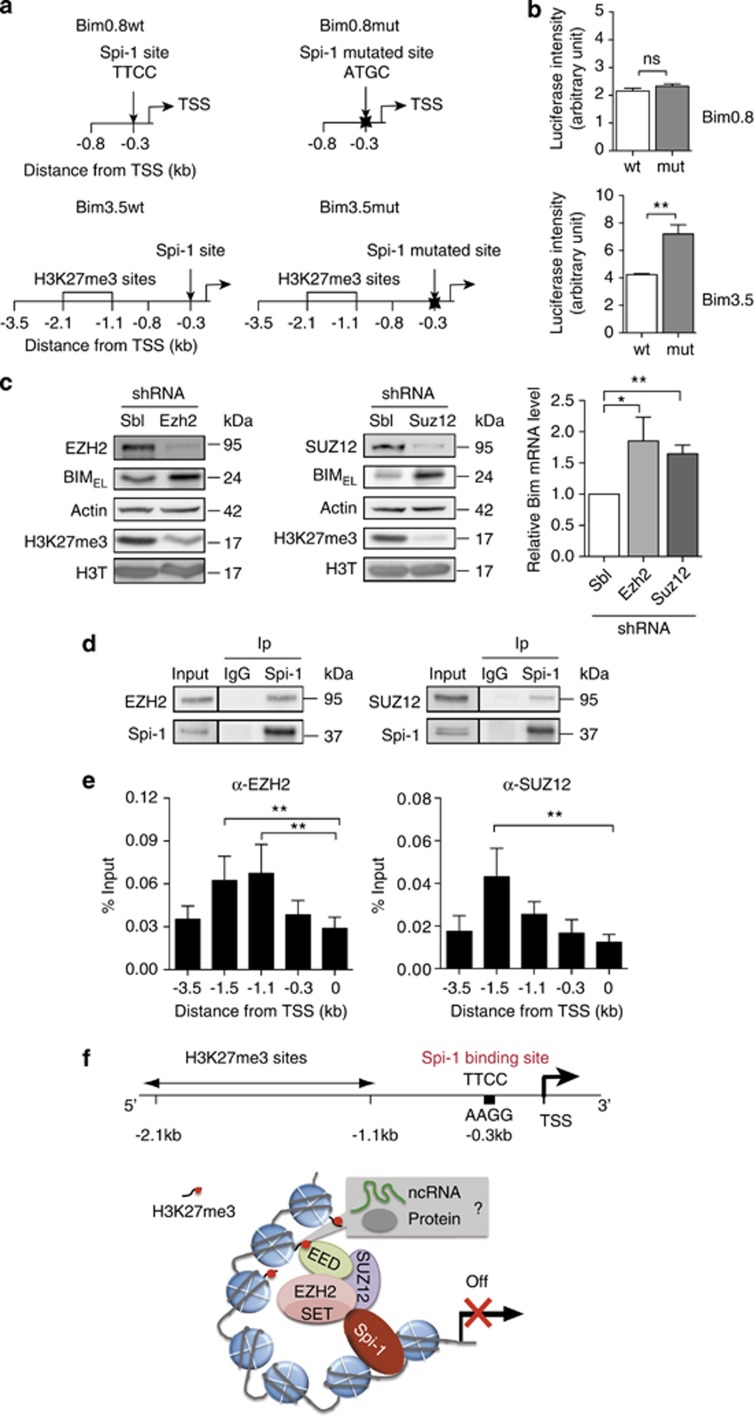
Then, we speculated that Spi-1 might repress Bim expression by contributing to the deposition of repressive histone marks on the Bim promoter. To check this hypothesis, we generated constructs in which a luciferase cDNA was inserted downstream of the 0.8 kb Bim promoter region that included the Spi-1 DNA binding motif (Bim0.8wt), or the 3.5 kb region that further includes the H3K27me3 sites (Bim3.5wt) (Figure 7a). We also produced two constructs in which the Spi-1 DNA binding site was mutated (Bim0.8mut and Bim3.5mut). The mutation abolished Spi-1 binding to DNA in the EMSA assay (Supplementary Figure 4), confirming that Spi-1 binds directly to this DNA motif. We stably transfected shSpi-A2B cells with the luciferase constructs and pools of cells were selected to analyse the promoter activity in a chromatin context. The luciferase expression was driven similarly by Bim0.8wt and Bim0.8mut promoters (Figure 7b). In contrast, when luciferase transcription was driven by Bim3.5 promoter, mutation of the Spi-1 binding site increased luciferase expression (twofold).
Figure 7.
Both Spi-1 binding to the Bim promoter and H3K27 trimethylation by PRC2 are required for the repression of Bim transcription. (a) Schematic representation of the Bim promoter–luciferase reporter constructs. Two fragments (0.8 and 3.5 kb) of the mouse Bim promoter were cloned in the pGL4 reporter vector to generate, respectively, the Bim0.8wt and Bim3.5wt vectors. The Spi-1 binding site (underlined) in the two vectors was then mutated (from 5′-AGTTCCGC-3′ to 5′-AGATGGGC-3′) to generate the Bim0.8mut and Bim3.5mut vectors. The TSS, the Spi-1 DNA binding site and the H3K27me3 sites are indicated. (b) shSpi-A2B cells were stably transfected with the different vectors (puromycin resistance) and luciferase assays were performed using exponentially growing cells 24 h after seeding. Data are reported as the relative firefly luciferase activity normalized to the number of cells and they represent the mean±S.D. of three experiments. Statistical differences were assessed by using the Student's t-test: **P<0.01. (c) shSpi-1-A2C cells were stably infected using lentiviral vectors encoding shRNAs against Ezh2 (shEZH2), Suz12 (shSUZ12) or control shSbl. The expression of EZH2, SUZ12, BIM, H3K27me3 and Actin was assessed by western blot (left) and the expression of Bim mRNA by real-time qPCR (right) in drug-selected cells. Bim mRNA level was normalized to Polr2α mRNA and to the level in shSbl. The bars represent the mean±S.E.M. of at least three independent experiments. Statistical analysis of the ΔCt values was carried out with the Student's t-test; **P<0.01 and *P<0.05. (d) Lysates from shSpi-1-A2B and shSpi-1-A2C cells were immunoprecipitated (Ip) with an anti-Spi-1 antibody or immunoglobulin G (IgG) as indicated. Immunoprecipitates were analysed by immunoblotting with anti-EZH2, anti-SUZ12 and anti-Spi-1 antibodies. (e) Chromatin isolated from shSpi-1-A2C or shSpi-1-A2B cells was IP using anti-EZH2 or anti-SUZ12 antibodies. The histogram bars represent the enrichment relative to the Input (%Input) determined by real-time qPCR using the indicated Bim promoter primers. The bars represent the mean±S.E.M. of three independent experiments. **P<0.01 by one-way analysis of variance (ANOVA) model followed by the post hoc test (pairwise t-test adjusted for multiple tests). (f) Working hypothesis for repression of Bim transcription by Spi-1. Spi-1 and PRC2 activities are necessary to favour trimethylation of H3K27 and to repress Bim transcription. The binding of Spi-1 and PRC2 to DNA occurs at distinct locations into the Bim promoter. These data indicate that Spi-1 promotes PRC2 activity without directly recruiting the complex to the site of its activity on the chromatin. Non-coding RNAs (ncRNAs) or other proteins may serve as intermediates in the PRC2 targeting at the methylation site and Spi-1 may act on these intermediates through stabilizing their interactions with PRC2
These results demonstrate that Spi-1 binding to the Bim promoter is necessary but not sufficient to repress Bim transcription in preleukaemic cells. The transcriptional repression of Bim requires also the repressive H3K27me3 marks.
This prompted us to investigate whether H3K27me3 contributes to Bim repression in preleukaemic cells. As PRC2 is a major regulator of H3K27me3, we asked whether inhibition of PRC2 activity affected Bim transcription. To this aim, we reduced the expression of SUZ12 or EZH2,33 two PRC2 subunits, using shRNA against Ezh2 or Suz12 or shSbl in shSpi-1-A2C cells. Suz12 or Ezh2 silencing reduced H3K27me3 (Figure 7c). Moreover, ChIP experiments using anti-H3K27me3 antibodies indicated that H3K27me3 at the Bim promoter was significantly reduced when SUZ12 or EZH2 was depleted (Supplementary Figure 5). In parallel, the levels of Bim transcript and BIM protein were increased in cells in which Suz12 or Ezh2 was silenced (Figure 7c). These data demonstrate that PRC2 contributes to the deposition of the H3K27me3 mark on the Bim promoter, causing the repression of Bim transcription in preleukaemic cells.
We then asked whether Spi-1 facilitates the enrichment of H3K27me3 at the Bim promoter by recruiting and/or modulating the activity of PRC2 at the Bim promoter. Co-IP assays using shSpi-1-A2B and shSpi-1-A2C cell lysates and the anti-Spi-1 antibody indicated that EZH2 and SUZ12 interact with Spi-1 (Figure 7d). We then examined whether EZH2 and SUZ12 bound to the Bim promoter by ChIP using appropriate antibodies. We detected EZH2 and SUZ12 at the Bim promoter at the same location as the H3K27me3 marks, with the highest enrichment at −1.5 kb from the TSS (Figure 7e). Conversely, we never found any EZH2 or SUZ12 bound to the Spi-1 binding site (0.3 kb downstream of the TSS). Thus, PRC2 and Spi-1 bind to distinct regions of the Bim promoter. These results suggest that Spi-1 promotes the PRC2 activity on histone 3 located at a genomic location distinct from the Spi-1 binding sites.
Discussion
An imbalance in the interplay between pro- and antiapoptotic proteins of the BCL-2 family34, 35 can favour cell survival, contributing to AML development and conferring resistance to therapy.36 In this study, we established that Spi-1 inhibits the apoptotic mitochondrial pathway through transcriptional repression of the proapoptotic factor Bim in preleukaemic cells cultured in vitro. Indeed, BIM overexpression is sufficient to kill cells and Bim knockdown protects cells from apoptosis induced by Spi-1 silencing. Similarly, we found a significant reduction of the number of apoptotic cells among primary preleukaemic cells isolated from preleukaemic mice in comparison with wt mice, which was associated with higher Spi-1 expression and lower BIM expression. Altogether, these results demonstrate the essential role of Bim downregulation in mediating Spi-1 antiapoptotic effect.
It is worth noting that the protection of apoptosis using Bim shRNA in preleukaemic cells was partial. This may be due to the residual BIM expression. However, other pro- or antiapoptotic factors might be affected by Spi-1. In particular, the expression of BCL-XL was slightly reduced, whereas the expression of tBID was slighly increased with depletion of Spi-1, two modifications that might promote cellular death.
We found that the antiapoptotic protein MCL-1 interacts with BIM in preleukaemic cells and that upon Spi-1 silencing, the more quantity of BIM neutralizes MCL-1 by physical interaction. Moreover, the increased apoptosis associated with MCL-1 depletion using shRNA indicates that MCL-1 is the main player in the survival of preleukaemic cells. Thus, the BIM-MCL-1 interaction appears critical in controlling the pool of free MCL-1 that will determine whether a preleukaemic cell will survive or die. The free pool of MCL-1 in a low BIM context might inhibit apoptosis by neutralizing other proapoptotic factors such as BAX/BAK, tBID and PUMA that are expressed in preleukaemic cells and are capable to interact with MCL-1.37
BIM expression is regulated at the transcriptional and translational level in many cell types. The EPO signalling pathway through ERK-mediated phosphorylation of BIM induces its proteasomal degradation in erythroid cells.38, 39 However, we previously established that EPO signalling was not involved in the antiapoptotic function of Spi-1 in preleukaemic cells. Moreover, BIM phosphorylation and stability were not changed following Spi-1 silencing in preleukaemic cells (data not shown) that is inconsistent with a role of Spi-1 in BIM degradation. Moreover, Bim is a target of the miR-17-92 and miR106b-25 clusters40 and Spi-1 was reported to modify the expression of the pri-miR-17-92.41, 42 We found that the expression of the pri-miR-17-92 and pri-miR-106b-25 in preleukaemic cells was not affected by Spi-1 (data not shown), leading to exclude a role of these miRNAs in Bim repression by Spi-1 in preleukaemic cells.
We show here that transcriptional repression of Bim is the main mechanism mediated by Spi-1 to maintain BIM expression to a low level in preleukaemic cells. We demonstrate that Bim is a direct Spi-1 transcriptional target through DNA binding and transactivation assays. The occupancy of the Bim promoter by Spi-1 in macrophages was previously reported but not functionally examined.43 It would be interesting to know whether Bim is a Spi-1 target gene in haematopoietic lineages wherein Spi-1/PU.1 behaves as a master differentiation regulator.
Studying epigenetic components of transcriptional regulation provides important clues for understanding Spi-1 function in repressing Bim transcription. Spi-1 silencing has no impact on the repressive DNA methylation of the Bim promoter, but provokes an enrichment in the repressive H3K27me3 mark at the Bim promoter. Moreover, the transcriptional repression of Bim requires Spi-1 binding to the Bim promoter and also the region containing the repressive H3K27me3 marks further upstream of the Spi-1 binding site. Finally, we identify PRC2 as a regulator of Bim expression by Spi-1 through promoting the H3K27me3 transcriptional repressive mark.
Other studies reported that Spi-1 represses gene transcription by epigenetic mechanisms involving the formation of repressive H3K9 trimethylation44 and the recruitment of histone deacetylases.45 Recent data also identified Spi-1 binding to enhancers as a primary event that facilitates H3K4me1 deposition.43 Our work reveals a new mechanism whereby Spi-1 represses gene transcription, by promoting H3K27me3 deposition at chromatin.
It is still not well understood how PRCs are targeted to specific genes in mammals. As PRCs do not contain inherent specific DNA-binding activity, additional factors must mediate their chromatin recruitment, such as JARID2, non-coding RNAs and transcriptional regulators.29, 33, 46, 47 Recruitment of PRC2 by GATA-1 has been reported during normal erythroid differentiation.37 Abnormal recruitment of PRCs by the oncogenic transcription factors PML-RARα48 and PLZF-RARα49 participates in leukaemic transformation. PRC2 might also associate with CG-rich sequences in genes devoid of DNA motifs for transcriptional activators in ES cells.50 We found that SUZ12 and EZH2 interact with Spi-1 and bind to the Bim promoter at the H3K27me3 sites that are 0.9 kb distant from the Spi-1 binding motif. As both Spi-1 and PRC2 are required to favour H3K27m3 at the Bim promoter, at least, two scenarios might fit with these findings (Figure 7f). In the first one, Spi-1 is not responsible for PRC2 recruitment. Spi-1 interaction with PRC2 would strengthen PRC2 activity or maintain PRC2 at the Bim promoter. The second scenario is based on the hypothesis that Spi-1 recruits PRC2 to the chromatin and that this indirect binding of PRC2 would not be detectable because of the structure of the chromatin or to the weakness of the indirect PRC2 binding to Spi-1 binding site. In both hypothesis, the PRC2 activity on histone 3 may involve intermediates, such as proteins or non-coding RNAs that may target PRC2 to the −2.1 to −1.1 kb region on DNA. As non-coding RNAs interact with PRC2 at repressed genes51, 52 and because Spi-1 binds to RNA,53 it is tempting to speculate that Spi-1 may affect PRC2 repressive activity or maintain PRC2 at the Bim promoter through its RNA-binding activity. In conclusion, our results provide evidence that a transcription factor can modulate the activity of PRC2 and epigenetic regulation without recruiting PRC2 at the site of its activity on the chromatin. Further studies are needed to determine the precise mechanism of the functional cooperation between PRC2 and Spi-1.
Genetic alterations of members of the polycomb gene (PcG) complex that confer increased or reduced activity can promote leukaemic transformation.54, 55 We describe here the repressive consequence of PRC2 activation at the Bim promoter and associate this molecular deregulation with a functional impact on apoptosis resistance in preleukaemic erythroid progenitor cells. This illustrates how deregulation of the epigenetic activity of PcG by an oncogenic transcription factor can lead to aberrant gene silencing. It will be essential in the future to define whether this Spi-1 activity extends to other target genes that may influence Spi-1 oncogenic function as well.
Materials and Methods
Cell lines, clonogenic assays, pharmacological inhibitors and antibodies
Spi-1 transgenic cells were derived from Spi-1 transgenic mice with erythroleukaemia as described previously.6 Cells were grown in α-MEM supplemented with 5% foetal bovine serum (FBS) and 1 U/ml EPO. Cells producing anti-Spi-1 shRNA (ShSpi-1-A2B and ShSpi-1-A2C) in the presence of 100 ng/ml dox and control cells expressing shRNA with scrambled sequence have been described previously.8
For colony assays with purified cells from bone marrow or spleen, 105 cells per ml were plated in MethoCult M3334 (StemCell Technologies, Glasgow, UK) and analysed at day 3 for CFU-E.
Cells were treated with STS (10 μM) for 2 or 4 h (Calbiochem, Nottingham, UK). Actinomycin D (Sigma-Aldrich, Saint Quentin, France) were added in the culture medium at the indicated concentrations. Antibodies used in this study are listed in Supplementary Table 1.
Cell viability, apoptosis assay and flow cytometry
For measurement of viability and apoptosis, cells were seeded at 5 × 104 cells per ml in liquid medium with or without dox. Cell viability was measured by using the Trypan blue exclusion test with a Vi-Cell analyser (Beckman Coulter, Roissy, France) or by flow cytometry analysis after staining with DAPI (4′,6′-diamidino-2-phenylindole) (Invitrogen, Saint Aubin, France).
Apoptosis was monitored with three different methods based on flow cytometry. Cells were stained with Annexin V-APC (BD Biosciences, Le Pont de Claye, France) and DAPI according to the manufacturer's instructions. Annexin V-positive cells were defined as apoptotic among the living cells (DAPI-negative). Detection of the active form of caspase-3 (cleaved-caspase-3) was performed as described previously.9 To determine the loss of mitochondrial transmembrane potential, cells were incubated with 20 nM of 3,3′-Dihexyloxacarbocyanine iodide (DiOC6(3); Invitrogen) at 37 °C for 30 min.
Flow cytometry analyses were conducted on an LSR II flow cytometer (BD Biosciences) with the DIVA software (Beckton Dickinson). Data were analysed using the the FlowJo 9.5 software (Tree Star, Ashland, OR, USA).
In vivo staining
Bone marrow cells were flushed from femurs and washed in cold phosphate-buffered saline with 0.5% bovine serum albumin (PBS/0.5% BSA). Freshly isolated bone marrow or spleen cells were immunostained at 4 °C in PBS/0.5% BSA with APC-conjugated anti-Ter119 (1:100; eBioscience, Paris, France), FITC-conjugated anti-CD71 (1:100; BD Biosciences) and biotin-conjugated anti-IL3R (1:200; BD Biosciences) antibodies for 20 min, followed by a 15-min incubation with PE-conjugated streptavidin (1:1000; BD Biosciences). DAPI was used to exclude dead cells. For apoptosis measurement, stained cells using the fluorescent antibodies were incubated for additional 15 min with APC-conjugated Annexin V and DAPI according to the manufacturer's instructions (BD Biosciences).
Cell sorting was performed on a FACSAria instrument (BD Biosciences) using the medium speed sorting settings with a 85 μm nozzle. Analysis of Ter119, CD71 and Annexin V labelling was carried out on an LSR II flow cytometer (BD Biosciences) with the DIVA software (Beckton Dickinson). Data were analysed using the FlowJo 9.5 software (Tree Star).
Transfection and lentiviral transduction
Cells were transfected using Amaxa Nucleofector (Lonza, Levallois Perret, France) as described previously.8 pLKO.1-Puro vectors transducing shRNA against Bim, Mcl-1, Suz12 or Ezh2 or control shSbl were selected from the MISSION shRNA lentivirus library (Sigma-Aldrich). Lentiviral particles were generated as described earlier.9 Preleukaemic cells were transduced with freshly prepared lentiviral particles and 30 h after infection 1 mg/ml puromycin (CAYLA, Toulouse, France) was added to the cells for selection. Selected cells were used for experiments 2.5 days after puromycin addition.
Plasmid construction and luciferase assay
The Bim promoter regions (0.8 or 3.5 kb) were PCR amplified from the mouse Bim promoter plasmid (kindly provided by Dr. Bouillet18) and subcloned into the pGL4basic luciferase vector (Promega, Madison, WI, USA) to obtain the Bim0.8 kb and Bim3.5 kb vectors. The Spi-1 binding site (underlined) in the two vectors was then mutated (from 5′-AGTTCCGC-3′ to 5′-AGATGGGC-3′) using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA, USA), according to the manufacturer's recommendations. The following mutagenic oligonucleotides were used for PCR: primer forward, 5′-CTGAGCGCCCCCTAAGATGGGCTCTGCGAGGTGGCC and reverse, 5′-GGCCACCTCGCAGAGCCCATCTTAGGGGGCGCTCAG. shSpi-A2B cells were then stably transfected with the luciferase plasmids and selected with puromycin. Luciferase assays were performed using exponentially growing cells 24 h after seeding and the Luciferase Assay System (Promega), according to the manufacturer's protocol. Luminescence was recorded with a Fluostar Optima microplate reader (BMG Labtech, Champigny sur Marne, France).
IP and immunobloting
For IP of BIM, MCL-1, BCL-XL and activated BAX, 2 × 107 cells were lysed in CHAPS lysis buffer (10 mM HEPES pH 7.4, 150 mM NaCl and 1% CHAPS) supplemented with protease inhibitors (complete Mini EDTA-free; Roche, Mannheim Germany). For IP of Spi-1, EZH2 and SUZ12, 4 × 107cells were lysed in the following buffer: 20 mM HEPES, 2 mM MgCl2, 0.5% NP-40, 100 mM NaCl and 0.1% Triton. Supernatants were incubated with 2 μg of antibody and 10 μl of magnetic beads (Dynabeads; Invitrogen). Whole-cell extracts or immunoprecipitates were fractionated by SDS-PAGE and immunoblotted as described previously.4 Images of blots were acquired using a Fujifilm LAS4000 digital imager and quantified with the Fujifilm Multi-Gauge Imaging software (Fujifilm, Tokyo, Japan).
RNA extraction and quantification by real-time quantitative PCR
Total RNA from cells was extracted using the RNeasy Mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. One microgram of total RNA was reverse transcribed using the iScript Reverse Transcription Kit (Bio-Rad, Hercules, CA, USA). Real-time quantitative PCR (qPCR) was performed using TaqMan Expression Assays and TaqMan Master Mix (Applied Biosystems, Alameda, CA, USA) on a Chromo4 System (Bio-Rad). Data were analysed using an Opticon Monitor (Bio-Rad) and normalized to Gapdh mRNA level or to Polr2a mRNA level. Fold changes were calculated using the 2−ΔΔCt method.
ChIP and MeDIP assays
ChIP was performed as described previously.16 Precleared chromatin from 8 × 106 cells and 10 μg of antibody were used for each ChIP assay.
MeDIP experiments were performed using the MeDIP kit (Diagenode, Liege, Belgium) following the manufacturer's protocol. Briefly, genomic DNA from 3 × 106 cells was extracted by shaking cells in digestion buffer containing 0.1 μg/μl Proteinase K for 12–18 h at 50 °C and then purified by phenol–chloroform extraction. DNA was sonicated using the Bioruptor apparatus (Diagenode), denatured at 95 °C for 10 min and quickly chilled on ice. Two micrograms of sonicated DNA was incubated with 2 μg of mouse monoclonal anti-5-methylcytidine antibody and 20 μl of beads at 4 °C with rotation. After washing, DNA was eluted, purified by phenol/chloroform extraction, precipitated with ethanol and resuspended in 50 μl water.
Enrichment of immunoprecipitated DNA (from ChIP or MedIP) was determined by real-time qPCR using Power SYBR Green Master Mix (Applied Biosystems) on a Chromo4 System apparatus (Bio-Rad). The enrichment of specific genomic regions was assessed relative to the input DNA as follows: 2^(Ctinput−CtChIP/MedIP), where Ct is the threshold cycle; Ct input is the Ct for input sample; and Ct ChIP/MedIP is the Ct for ChIP or MedIP sample. Primer sequences are listed in Supplementary Table 2.
Statistical analysis
The results are expressed as mean±standard deviation and differences between values of two groups were determined by using the paired Student's t-test. The statistical significance of Spi-1 knockdown in the H3K27me3 ChIP experiment was tested using a two-way ANOVA model followed by the post hoc test (pairwise t-test adjusted for multiple tests). The statistical significance of enrichments at the specific location on the Bim promoter in the SUZ12 and EZH2 ChIP experiments was tested using a one-way ANOVA model followed by the post hoc test (pairwise t-test adjusted for multiple tests). For all tests, a P-value <0.05 was considered statistically significant.
Acknowledgments
We thank N Brandon, N Denis and Z Maciorowski for technical assistance; R Duffie and D Bourc'his for help with bisulphite sequencing. We also thank H Harada for kindly providing the HA-Bim expression vector.
Glossary
- bp
base pairs
- AML
acute myeloid leukaemia
- Dox
doxycycline
- CFU-E
colony-forming unit-erythroid
- wt
wild-type
- ChIP
chromatin immunoprecipitation
- MeDIP
immunoprecipitation of methylated DNA
- shSbl
scrambled shRNA
- STS
staurosporine
- IP
immunoprecipitation
- TSS
transcription start site
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by P Bouillet
Supplementary Material
References
- Renneville A, Roumier C, Biggio V, Nibourel O, Boissel N, Fenaux P, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22:915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- Visconte V, Makishima H, Maciejewski JP, Tiu RV. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia. 2012;26:2447–2454. doi: 10.1038/leu.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Kosmider O, Denis N, Lacout C, Vainchenker W, Dubreuil P, Moreau-Gachelin F. Kit-activating mutations cooperate with Spi-1/PU.1 overexpression to promote tumorigenic progression during erythroleukemia in mice. Cancer Cell. 2005;8:467–478. doi: 10.1016/j.ccr.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Guillouf C, Gallais I, Moreau-Gachelin F. Spi-1/PU.1 oncoprotein affects splicing decisions in a promoter binding-dependent manner. J Biol Chem. 2006;281:19145–19155. doi: 10.1074/jbc.M512049200. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmele P, Komatsu J, Hupe P, Roulin C, Barillot E, Dutreix M, et al. Spi-1/PU.1 oncogene accelerates DNA replication fork elongation and promotes genetic instability in the absence of DNA breakage. Cancer Res. 2010;70:6757–6766. doi: 10.1158/0008-5472.CAN-09-4691. [DOI] [PubMed] [Google Scholar]
- Rimmele P, Kosmider O, Mayeux P, Moreau-Gachelin F, Guillouf C. Spi-1/PU.1 participates in erythroleukemogenesis by inhibiting apoptosis in cooperation with Epo signaling and by blocking erythroid differentiation. Blood. 2007;109:3007–3014. doi: 10.1182/blood-2006-03-006718. [DOI] [PubMed] [Google Scholar]
- Buet D, Gallais I, Lauret E, Denis N, Lombard B, Guillonneau F, et al. Cotargeting signaling pathways driving survival and cell cycle circumvents resistance to Kit inhibitors in leukemia. Blood. 2012;119:4228–4241. doi: 10.1182/blood-2011-07-368316. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zivny J, Klener P, Jr., Pytlik R, Andera L. The role of apoptosis in cancer development and treatment: focusing on the development and treatment of hematologic malignancies. Curr Pharm Des. 2010;16:11–33. doi: 10.2174/138161210789941883. [DOI] [PubMed] [Google Scholar]
- Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Wang HG. The protein kinase PKB/Akt regulates cell survival and apoptosis by inhibiting Bax conformational change. Oncogene. 2001;20:7779–7786. doi: 10.1038/sj.onc.1204984. [DOI] [PubMed] [Google Scholar]
- Ridinger-Saison M, Boeva V, Rimmele P, Kulakovskiy I, Gallais I, Levavasseur B, et al. Spi-1/PU.1 activates transcription through clustered DNA occupancy in erythroleukemia. Nucleic Acids Res. 2012;40:8927–8941. doi: 10.1093/nar/gks659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, et al. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome. 2001;12:163–168. doi: 10.1007/s003350010242. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pop R, Sadegh C, Brugnara C, Haase VH, Socolovsky M. Suppression of Fas–FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood. 2006;108:123–133. doi: 10.1182/blood-2005-11-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Terszowski G, Waskow C, Conradt P, Lenze D, Koenigsmann J, Carstanjen D, et al. Prospective isolation and global gene expression analysis of the erythrocyte colony-forming unit (CFU-E) Blood. 2005;105:1937–1945. doi: 10.1182/blood-2004-09-3459. [DOI] [PubMed] [Google Scholar]
- Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Matsui H, Asou H, Inaba T. Cytokines direct the regulation of Bim mRNA stability by heat-shock cognate protein 70. Mol Cell. 2007;25:99–112. doi: 10.1016/j.molcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein–Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathogen. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Jose-Eneriz E, Agirre X, Jimenez-Velasco A, Cordeu L, Martin V, Arqueros V, et al. Epigenetic down-regulation of BIM expression is associated with reduced optimal responses to imatinib treatment in chronic myeloid leukaemia. Eur J Cancer. 2009;45:1877–1889. doi: 10.1016/j.ejca.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Zheng SS, Li ZM, Qiao YY, Aau MY, Yu Q. Polycomb protein EZH2 regulates E2F1-dependent apoptosis through epigenetically modulating Bim expression. Cell Death Differ. 2010;17:801–810. doi: 10.1038/cdd.2009.162. [DOI] [PubMed] [Google Scholar]
- Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abutin RM, Chen J, Lung TK, Lloyd JA, Sawyer ST, Harada H. Erythropoietin-induced phosphorylation/degradation of BIM contributes to survival of erythroid cells. Exp Hematol. 2009;37:151–158. doi: 10.1016/j.exphem.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Agami R. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev. 2010;24:1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayali S, Giraud G, Morle F, Spi-1 GuyotB. Fli-1 and Fli-3 (miR-17-92) oncogenes contribute to a single oncogenic network controlling cell proliferation in friend erythroleukemia. PLoS One. 2012;7:e46799. doi: 10.1371/journal.pone.0046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisil V, Vargova K, Kokavec J, Rybarova J, Savvulidi F, Jonasova A, et al. Epigenetic silencing of the oncogenic miR-17-92 cluster during PU.1-directed macrophage differentiation. EMBO J. 2011;30:4450–4464. doi: 10.1038/emboj.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M. Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU.1-mediated transcriptional repression. Oncogene. 2003;22:8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Cory S, Zhang LC, Strasser A, Adams JM. Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Dev Cell. 2001;1:645–653. doi: 10.1016/s1534-5807(01)00083-1. [DOI] [PubMed] [Google Scholar]
- Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Boukarabila H, Saurin AJ, Batsche E, Mossadegh N, van Lohuizen M, Otte AP, et al. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23:1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallier M, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb. J Biol Chem. 1996;271:11177–11181. doi: 10.1074/jbc.271.19.11177. [DOI] [PubMed] [Google Scholar]
- Martin-Perez D, Piris MA, Sanchez-Beato M. Polycomb proteins in hematologic malignancies. Blood. 2010;116:5465–5475. doi: 10.1182/blood-2010-05-267096. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.