Abstract
We report here the development of 65 novel microsatellite loci and construction of a composite genetic linkage map for Culex pipiens complex mosquitoes. Microsatellites were identified by in silico screening of the Culex quinquefasciatus genome assembly. Cross-species utility of 73 microsatellites for population studies in C. pipiens sensu stricto and C. quinquefasciatus was evaluated by genotyping a subset of samples collected in Indiana, United States, and Point Fortin, Trinidad. Allele frequencies of 67 microsatellites were within Hardy–Weinberg expectations in both population subsets. A composite linkage map was constructed based on restriction fragment length polymorphism and microsatellite polymorphisms in 12 independent F1 intercross mapping populations. The composite map consists of 61 marker loci totaling 183.9 cM distributed across the 3 linkage groups. These loci cover 29.5, 88.8, and 65.6 cM on chromosomes I–III, respectively, and allow for assignment of 10.4% of the genome assembly and 13.5% of the protein coding genes to chromosome position. Our results suggest that these microsatellites will be useful for mapping and population studies of 2 pervasive species in the C. pipiens complex. Moreover, the composite map presented here will serve as a basis for the construction of high-resolution genetic and physical maps, as well as detection of quantitative trait loci to aid in the investigation of complex genetic traits influencing phenotypes of interest.
Key words: composite linkage map, Culex quinquefasciatus, genome assembly, house mosquito, SSR, supercontigs
The Culex pipiens complex of mosquitoes includes some of the most geographically widespread and medically important vectors of human disease. They are key vectors of West Nile and St. Louis encephalitis viruses, as well as Wuchereria bancofti, the parasite responsible for most periodic lymphatic filariasis (Day 2001; Turell et al. 2001; Fonseca et al. 2004; Michalski et al. 2010). Members of this species complex can be found on every continent except Antarctica (Vinogradova 2000), making them among the most pervasive mosquitoes known. Culex pipiens sensu stricto (L.), the northern house mosquito, inhabits temperate regions where females are able to survive the adverse conditions associated with winter by entering a programmed developmental arrest (i.e., reproductive diapause) (Vinogradova 2000). In contrast, Culex quinquefasciatus (Say), the southern house mosquito, inhabits most tropical and subtropical regions of the world and is genetically unable to enter diapause. Most members of this species complex live above ground where females must take a blood meal, usually from birds, prior to vitellogenesis and oviposition (anautogenous). However, the “molestus” form of C. pipiens lives predominantly underground and does not require a blood meal to lay an initial batch of eggs (autogenous). When they do blood feed, they tend to feed more readily on mammals (Byrne and Nichols 1999). These physiological and behavioral traits exemplify the ability of the C. pipiens complex to adapt to diverse environments. A greater understanding of the molecular underpinnings of these and other complex traits associated with their vectorial capacity could provide additional tools to help prevent the transmission of pathogens by these important disease vectors.
Mori et al. (1999) were the first to report a detailed genetic linkage map of C. pipiens. They utilized restriction fragment length polymorphism (RFLP) markers using random complementary DNAs (cDNAs) from the yellow fever mosquito Aedes aegypti to construct a genetic linkage map. Of the 22 cDNA clones used, 21 were mapped to scaffolds (supercontigs) in the C. quinquefasciatus genome assembly (Arensburger et al. 2010). This not only provided genetic landmarks, it also helped illustrate the congruence among chromosome arms between the C. pipiens and A. aegypti genomes. Consequently, we were able to utilize data from previous mapping studies in C. pipiens (Mori et al. 1999, 2007) and a composite linkage map for A. aegypti (Severson et al. 2002) to develop microsatellites broadly distributed across all 3 chromosomes in C. pipiens sensu latu. We used these microsatellites to analyze 6 independent F1 intercross mapping populations, and we then used the JoinMap program to integrate these data with 6 independent F1 intercross mapping populations analyzed using RFLP markers to construct a composite linkage map for C. pipiens complex mosquitoes. This also allowed for assignment of associated genome assembly supercontigs to chromosome positions.
Materials and Methods
Microsatellite Identification and Characterization
Microsatellites were identified as previously described in Hickner et al. (2010). Briefly, loci were identified by screening the C. quinquefasciatus genome assembly (CpipJ1) for di- and trinucleotide repeats using Tandem Repeats Finder (Benson 1999) or by pasting a supercontig nucleotide sequence into a Word document and using the “find” function to search for specific motifs. To help eliminate multiple-copy genome sequences, a 400–600 nucleotide region containing a target microsatellite sequence was subjected to BLASTn analysis against the CpipJ1 assembly. Primers were designed to produce polymerase chain reaction (PCR) amplicons ranging from 110 to 350bp using Primer3 (Rozen and Skaletsky 2000), Primer3 plus (Untergasser et al. 2007), or OligoCalc (Kibbe 2007). To detect potential nontarget amplification and to determine if a given microsatellite was within a protein coding region, primer sequences were subjected to BLASTn analysis against the CpipJ1assembly and the CpipJ1.2 gene build at VectorBase (Lawson et al. 2009).
Most of the supercontigs screened for microsatellites were chosen because their chromosome position had been previously determined based on RFLP mapping in C. pipiens or A. aegypti (Mori et al. 1999, 2007; Severson et al. 2002). However, we also chose several supercontigs based on their size and their having genes of interest. Supercontigs 3.1, 3.2, and 3.4 were screened because they are among the largest genomic scaffolds in the assembly. Supercontigs 3.119, 3.163, and 3.1500 were chosen in association with ongoing independent studies because they contained the genes shaggy (CPIJ006114), period (CPIJ007193), and cryptochrome 2 (CPIJ018859), respectively.
Primers were then tested in PCRs using total genomic DNA from 1 individual from each of 7 laboratory colonies: Boane, Johannesburg, and Trinidad (C. quinquefasciatus); Shasta and South Bend (C. pipiens); Gose (Culex pipiens pallens); and Shinkura (C. pipiens form molestus) (see Supplementary Table S1 online). DNA was extracted using a simple alkaline extraction method adapted from Rudbeck and Dissing (1998). Mosquitoes were homogenized in 80 µL 0.2M NaOH and incubated at 75°C for 10min. Thereafter, 28.8 µL 1M Tris, pH 8.0 and 91.2 µL sterile double-distilled water (ddH2O) were added to the mixture bringing the volume to 200 µL. Sterile ddH2O was added to produce a final volume of 800 µL for adult females and 500 µL for adult males. One microliter of the DNA preparation was used in each PCR. PCR amplification was performed in 25 µL reactions in 96-well PCR plates (Dot Scientific Inc. Burton, MI, USA). Each reaction contained 1 × Taq buffer (50mM KCl, 10mM Tris pH 9.0, 0.1% Triton X), 1.5mM MgCl2, 200mM dNTPs, 5 pmoles of each primer, 1 unit of Taq DNA polymerase, and 1 µL of genomic DNA preparation (~20ng). Thermal cycling was performed using Mastercycler® thermocyclers (Eppendorf AG, Hamburg, Germany) under the following conditions: initial denaturation for 5min at 94°C followed by 30 cycles of denaturation for 1min at 94°C, annealing for 1min at 60°C, extension for 2min at 72°C, and a final extension at 72°C for 10min. Fluorophore-labeled (6-FAM®, HEX®, NED®) forward primers were prepared for microsatellites that amplified well in individuals from all 7 colonies based on UV visualization on ethidium bromide–stained 2% agarose gels.
We tested individual microsatellite loci for Mendelian inheritance in one or more families, when possible. Alleles were scored using the ABI PRISM 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and ROX 400HD size standard. GeneMapper v.4.0 software (Applied Biosystems, Foster City, CA, USA) was used to call the alleles, with subsequent visual verification of each sample. Different primers flanking the microsatellite were designed when null alleles were evident in a family. Each microsatellite was named based on the supercontig assembly where it was found, the repeat motif, and the primer version (C134AC1b: C = C. quinquefasciatus genome, 134 = supercontig 3.134, AC = AC repeat, 1 = first microsatellite developed with this motif from this supercontig, b = alternate primer sequence of a previously published microsatellite). To ensure that the microsatellites were truly novel, BLASTn analysis of previously published C. pipiens and C. quinquefasciatus microsatellite sequences was performed.
Cross-species utility of these microsatellites for population studies was evaluated by genotyping ~30 individual C. pipiens s.s. collected from Indianapolis, Indiana, USA, and C. quinquefasciatus collected from Point Fortin, Trinidad. The Indianapolis (IN) samples were a subset of samples collected in 2008 and previously determined to be in Hardy–Weinberg equilibrium (HWE) based on allele frequencies of 12 microsatellite loci (Hickner et al. 2010). The Point Fortin (PF) samples were a subset from samples collected during 2010 using the same method described for the IN samples. This population had not been previously characterized. An exact test for HWE was performed following Guo and Thompson (1992) with 100 000 dememorization steps followed by 1000000 iterations using the program Arlequin (Excoffier et al. 2005). The program CONVERT was used to format the input files for Arlequin (Glaubitz 2004).
Genetic Crosses
The laboratory strains used for generation of the mapping populations were isolated from field collections of C. quinquefasciatus (Boane, Johannesburg, Trinidad, and Vero Beach), C. pipiens s.s. (Shasta and South Bend), C. pipiens pallens (Gose), and C. pipiens form molestus (Chicago and Shinkura) (see Supplementary Table S1 online). Pairwise matings of the F0 and F1 generations were carried out using the following method: pupae were separated and placed in individual 500mL cups containing ~250mL of water until emergence, at which time ~5 females and 1 male were placed in a cheesecloth cage (30×20×20cm) and allowed to mate. Approximately 7 days later the females were blood fed on rats, and each female was placed in a vial (22×46mm) containing 1–2cm of water until the eggs were laid. The number of F1 females used to produce each mapping population varied from 1 to 3. The F0 strains and the number of markers used for each mapping family are listed in Table 1.
Table 1.
Summary of F1 intercross mapping populations used for construction of the composite linkage map
| F0 laboratory strains | No. of F2 scored | No. of markers | Marker type | Citation | |
|---|---|---|---|---|---|
| ♀ | ♂ | ||||
| South Benda | Johannesburgb | 94 | 34 | Microsatellite | — |
| Shinkurac | Boaneb | 98 | 20 | Microsatellite | — |
| Chicagoc | Boane | 95 | 21 | Microsatellite | — |
| Gosed | Johannesburg | 95 | 20 | Microsatellite | — |
| Trinidadb | Shastaa | 95 | 29 | Microsatellite | — |
| Shinkura | Chicago | 95 | 28 | Microsatellite | — |
| Shinkura | Vero Beachb | 92 | 24 | RFLP | — |
| Nile Deltac | Iowaa | 191 | 13 | RFLP | Mori et al. (1999) |
| Akohc | Iowa | 123 | 6 | RFLP | Mori et al. (1999) |
| Akoh | Iowa | 192 | 10 | RFLP | Mori et al. (1999) |
| South Bend | Vero Beach | 96 | 8 | RFLP | Mori et al. (2007) |
| Gose | Vero Beach | 192 | 9 | RFLP | Mori et al. (2007) |
See Supplementary Table 1 online for additional information on the parental (F0) strains.
a Culex pipiens.
b Culex quinquefasciatus.
c Culex pipiens form molestus.
d Culex pipiens pallens.
Linkage Mapping
We constructed individual linkage maps and combined them using the Kosambi function (Kosambi 1943) in JoinMap 3.0 (Van Ooijen and Voorrips 2001). Microsatellites were anchored to RFLP or other microsatellite markers when they shared the same supercontig and they mapped to a common locus. The fixed order function in JoinMap 3.0 was used to maintain the order of the markers established in the individual maps. Because the mapping families were generated to investigate the genetic basis of female traits (diapause, autogeny, blood feeding), only F2 females were genotyped and used for calculation of the composite map. Chromosomes were designated following Mori et al. (1999).
Results
Microsatellite Identification and Characterization
Sixty-five novel microsatellites were identified and charac terized in this present study (see Supplementary Table S2 online). Mendelian inheritance of 5 microsatellites (C32TC1b, C32TGC1, C66TGT1, C111CAG1, and C550TGC1) was not tested in our laboratory colonies due to the absence of informative polymorphisms in the families. BLASTn analysis against the CpipJ1.2 gene build revealed that 14 microsatellites (21.5%) map to protein coding genes, with 13 of those being within exons (Table 2). Two microsatellites (C660CTC1 and C660GTG1) were within different exons of a single gene (CPIJ014662). Analysis of the proteins comprising these microsatellites revealed that 10 of the 14 (71%) repeats represented runs of the amino acid glutamine. Nine of the 11 codons are CAG, with only 2 being the alternative codon CAA. One microsatellite (C111CAG1) is in the 5′ untranslated region of the protein CPIJ005853.
Table 2.
Microsatellites within protein coding regions
| Microsatellite | Repeat motif | Amino acid | Gene ID | Gene name/description |
|---|---|---|---|---|
| C36GTT1 | CAA | Glutamine | CPIJ002765 | conserved hypothetical protein |
| C38GCT1 | CAG | Glutamine | CPIJ003062 | cornetto |
| C65CAG1 | CAG | Glutamine | CPIJ004312 | conserved hypothetical protein |
| C66TGT1 | AAC | Asparagine | CPIJ004404 | conserved hypothetical protein |
| C95GCA1 | CAG | Glutamine | CPIJ005607 | conserved hypothetical protein |
| C108CAG1 | CAG | Glutamine | CPIJ005224 | conserved hypothetical protein |
| C143TGC1 | CAG | Glutamine | CPIJ007159 | forkhead box protein |
| C186TGT1 | CAA | Glutamine | CPIJ007969 | conserved hypothetical protein |
| C309TGC1 | CAG | Glutamine | CPIJ010840 | mitochondrial inner membrane translocase |
| C550TGC1 | CAG | Glutamine | CPIJ013972 | trithorax |
| C660CTC1 | GAG | Glutamic acid | CPIJ014662 | ubiquitin specific protease |
| C660GTG1 | CAC | Histidine | CPIJ014662 | ubiquitin specific protease |
| C1500CAG1 | CAG | Glutamine | CPIJ018859 | cryptochrome 2 |
A total of 73 microsatellites were characterized in population subsets from IN and PF (see Supplementary Table S3 online). These included 65 microsatellites described here and 8 microsatellites described previously (Hickner et al. 2010). Of these, 2 were monomorphic in IN and 5 were monomorphic in PF. None were monomorphic in both populations. Six microsatellites had allele frequencies that deviated (P < 0.05) from Hardy–Weinberg expectations—or amplified poorly—in one of the population subsets tested. Five of these (C32TC1b, C38AC1, C66GT1, C309TGC1, and C446TC1) deviated in the IN population, whereas only 1 (C205GAC1) deviated in the PF population (see Supplementary Table S3 online). Each of these had heterozygote deficiencies suggesting the presence of null alleles at these loci. The total number of alleles identified for all markers was 364 in the IN population and 263 in the PF population. The number of alleles per locus ranged from 1 to 13 (mean = 5.1) in IN and from 1 to 11 (mean = 3.6) in PF.
Linkage Mapping and Genome Assignment
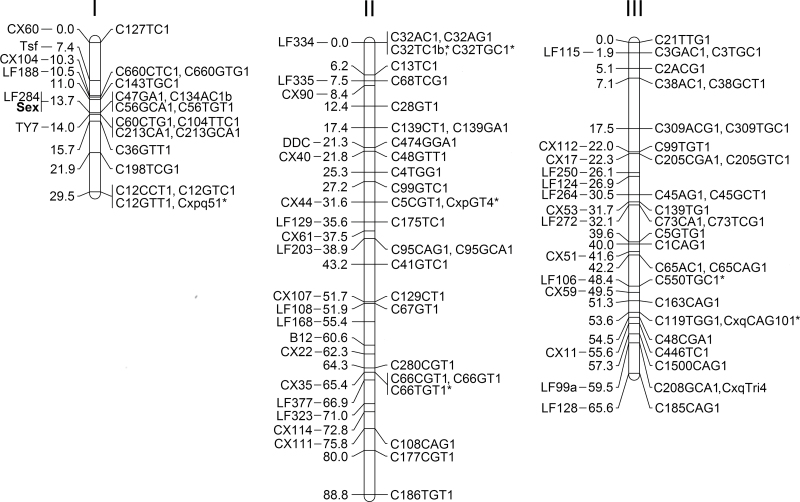
We were able to map 69 microsatellites and 39 cDNA clones to 63 genetic loci distributed across 3 linkage groups with a combined length of 183.9 cM (Figure 1). The lengths of chromosomes I–III are 29.5, 88.8, and 65.6 cM, respectively. We assigned the map locations of 8 microsatellites (C32TC1b, C32TGC1, C66TGT1, C550TGC1, CxpGT4, Cxpq51, CxqCAG101, and CxqTri4) based on the position of other markers within the same supercontig assembly. The microsatellites CxpGT4 (Keyghobadi et al. 2004), Cxpq51 (Abreu et al. 2012), CxqCAG101 (Edillo et al. 2007), and CxqTri4 (Smith et al. 2005) were described previously and found within supercontigs mapped in our study. Because we did not genotype males in the F2 generations, the sex-determining locus was placed on the composite map based on its association with marker LF284 (Mori et al. 1999).
Figure 1.
Composite RFLP and microsatellite linkage map for Culex pipiens sensu lato based on 12 independent F1 intercross mapping populations. The cDNA clones used for RFLP mapping and the map distances (Kosambi centiMorgans, cM) are listed on the left side of the chromosomes—microsatellites are listed on the right. Markers with positions predicted based on mapping of other markers in the supercontig are indicated by an asterisk.
Linkage mapping suggested the presence of misassemblies in 9 supercontigs (Table 3). Misassemblies were evident when different markers from 1 supercontig mapped to disparate positions on the linkage map. Markers in 7 supercontigs (3.5, 3.28, 3.48, 3.65, 3.99, 3.134, and 3.139) mapped to locations on different chromosomes, whereas markers in 3.177 and 3.309 mapped to different positions on the same chromosome. BLASTn analysis of the cDNAs and microsatellites against the contigs suggested that the errors were made while assembling the contigs into supercontigs rather than assembling the reads into contigs. The map reported here assigns chromosome position to 10.4% (56.4 of 540.0Mb) of the C. quinquefasciatus genome assembly and 13.5% (2541 of 18 883) of the protein coding genes (Lawson et al. 2009; Arensburger et al. 2010).
Table 3.
Putative misassembled supercontigs identified by genetic mapping of multiple markers within a supercontig assembly
| Supercontig | Marker | Map position | Marker | Map position |
|---|---|---|---|---|
| 3.5 | CX44, C5CGT1 | 2–31.6 | C5GTG1 | 3–39.6 |
| 3.28 | C28GT1 | 2–12.4 | CX51 | 3–41.6 |
| 3.48 | CX40, C48GTT1 | 2–21.8 | C48CGA1 | 3–54.5 |
| 3.65 | CX90 | 2–8.4 | C65AC1, C65CAG1 | 3–42.2 |
| 3.99 | C99GTC1 | 2–27.2 | CX112, C99TGT1 | 3–22.0 |
| 3.134 | C134AC1b | 1–13.7 | CX61 | 2–37.5 |
| 3.139 | C139CT1, C139GA1 | 2–17.4 | CX53, C139TG1 | 3–31.7 |
| 3.177 | CX114 | 2–72.8 | C177CGT1 | 2–80.0 |
| 3.309 | LF250 | 3–26.1 | C309ACG1, C309TCG1 | 3–17.5 |
Supercontigs with their corresponding markers and map positions are listed in each row.
Discussion
The number of loci on the C. pipiens linkage map has been increased from 38 to 63, therefore increasing marker density from one marker every 5.1 cM to one marker every 3.1 cM (Arensburger et al. 2010). The combined length increased marginally from 177.6 to 183.9 cM. The length of chromosome I increased from 12.5 to 29.5 cM. This was a modest gain considering that we added microsatellites from 8 supercontigs, and only 3 supercontigs (3.36, 3.198, 3.12) spanned beyond the markers previously mapped. Microsatellites on 9 supercontigs mapped to a 5.4 cM region on chromosome I that includes the sex-determining locus, confirming earlier results suggesting there is considerable reduction in recombination on chromosome I compared with chromosomes II and III, and chromosome I in A. aegypti (Mori et al.1999). The length of chromosome II increased slightly from 85.9 to 88.8. This was due to the addition of 2 loci (C177CGT1 and C186TGT1) beyond CX111/C108CAG1 on the current map. The length of chromosome III decreased from 79.2 to 65.6 cM despite an increase in the number of loci from 15 to 25. This was due to differences largely in the 2-point recombination estimates between the same markers among the different mapping populations. The linear order of the markers reported in Arensburger et al. (2010) was maintained in the current map with the exception of LF99a/C208GCA1, which was initially placed above CX11/C446TC1 in the composite map. The order reported here (CX11/C446TC1—LF99a/C208GCA1—LF128/C185CAG1) represents the only data for these markers where the 3-point estimates were derived from a single mapping family.
Despite the increased marker density in the current map, substantial gaps remain that preclude high-resolution genome analysis. Filling these gaps using the strategy employed here is limited by the fragmented state of the current genome assembly. A combination of high-resolution linkage mapping and physical mapping is necessary to substantially enhance the current assembly. Genotyping large mapping families or advanced intercross lines using methods such as restriction-site associated DNA tag sequencing (Baird et al. 2008) could potentially assign a large number of supercontigs to their respective chromosome positions. Physical mapping of A. aegypti metaphase chromosomes via in situ hybridization of BAC clones carrying genetic markers facilitated the construction of an integrated linkage, chromosome, and genome map (Timoshevskiy et al. 2013). A BAC library has been constructed that would make this a feasible approach for improving the C. quinquefasciatus genome assembly (Hickner et al. 2011).
As expected, most of the microsatellites are not within protein coding regions of the genome. However, 26% of the microsatellites with trinucleotide repeats were mapped to exons, with most (11 of 14) representing repeats of glutamine. Katti et al. (2001) compared the frequencies and types of amino acid repeats in Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cerevisiae and found that glutamine repeats make up the greatest proportion of amino acid repeats (≥7) in all 3 organisms. Moreover, D. melanogaster has a comparatively high number and proportion, with 1555 glutamine repeats out of a total of 2993 (Katti et al. 2001). Hyperexpansions of glutamine have been linked to some human diseases called polyglutamine diseases. These are predominantly neurodegenerative diseases and include Huntington’s disease and some spinocerebellar ataxias (Zoghbi and Orr 2000).
This present study provides the first composite linkage map for C. pipiens complex mosquitoes, which includes microsatellites and their corresponding genome scaffolds. This effort represents an ~41% increase in genome assignment to chromosome positions, for example, from 5.7% to 10.5% of the existing genome assembly (Arensburger et al. 2010). It also brings the total number of microsatellites developed for this species complex to over 100 (Fonseca et al. 1998; Keyghobadi et al. 2004; Edillo et al. 2007; Hickner et al. 2010; Smith et al. 2005; Abreu et al. 2012). In summary, we utilized data from 12 F1 intercross mapping families ranging in size from 94 to 192 to construct a composite linkage map for C. pipiens complex mosquitoes. The integration of microsatellites—a practical alternative to RFLP—should help facilitate additional mapping and population studies, as well as provide landmarks for enhancing the genome assembly in this important disease vector.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Institutes of Health, National Institute of Allergy and Infectious Diseases, grant no. RO1-AI079125-A1 .
Supplementary Material
Acknowledgments
We thank the field crew in Trinidad: Stephen “Billy” Deonarine, Doolam Lalla and Dexter Maraj for their assistance in sample collections from Point Fortin, Trinidad, and Jim Erwin and Shawn Moore for their assistance in sample collections in Indianapolis, IN, USA. We also thank Ned Walker for providing us with the Chicago strain and Anton Cornel for providing us with the Johannesburg, Shasta and Boane strains.
References
- Abreu AA, Albaina A, Alpermann TJ, Apkenas VE, Bankhead-Dronnet S, Bergek S, Berumen ML, Cho C-H, Clobert J, Coulon A, et al. 2012. Permanent genetic resources added to molecular ecology resources database 1 October 2011–30 November 2011. Mol Ecol Res. 12:374–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, et al. 2010. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 330:86–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 3:e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K, Nichols RA. 1999. Culex pipiens in London Underground tunnels: differentiation between surface and subterranean populations. Heredity. 82:7–15 [DOI] [PubMed] [Google Scholar]
- Day JF. 2001. Predicting St. Louis encephalitis virus epidemics: lessons from recent, and not so recent, outbreaks. Annu Rev Entomol. 46:111–138 [DOI] [PubMed] [Google Scholar]
- Edillo FE, Tripet F, McAbee RD, Foppa IM, Lanzaro GC, Cornel AJ, Spielman A. 2007. A set of broadly applicable microsatellite markers for analyzing the structure of Culex pipiens (Diptera: Culicidae) populations. J Med Entomol. 44:145–149 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 1:47–50 [PMC free article] [PubMed] [Google Scholar]
- Fonseca DM, Atkinson CT, Fleischer RC. 1998. Microsatellite primers for Culex pipiens quinquefasciatus, the vector of avian malaria in Hawaii. Mol Ecol. 7:1617–1619 [PubMed] [Google Scholar]
- Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Mogi M, Fleischer RC, Wilkerson RC. 2004. Emerging vectors in the Culex pipiens complex. Science. 303:1535–1538 [DOI] [PubMed] [Google Scholar]
- Glaubitz JC. 2004. CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes. 4:309–310 [Google Scholar]
- Guo SW, Thompson EA. 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 48:361–372 [PubMed] [Google Scholar]
- Hickner PV, Debruyn B, Lovin DD, Mori A, Behura SK, Pinger R, Severson DW. 2010. Genome-based microsatellite development in the Culex pipiens complex and comparative microsatellite frequency with Aedes aegypti and Anopheles gambiae . PLoS One. 5:e13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickner PV, Debruyn B, Lovin DD, Mori A, Saski CA, Severson DW. 2011. Enhancing genome investigations in the mosquito Culex quinquefasciatus via BAC library construction and characterization. BMC Res Notes. 4:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katti MV, Ranjekar PK, Gupta VS. 2001. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 18:1161–1167 [DOI] [PubMed] [Google Scholar]
- Keyghobadi N, Matrone MA, Ebel GD, Kramer LD, Fonseca DM. 2004. Microsatellite loci from the northern house mosquito (Culex pipiens), a principal vector of West Nile virus in North America. Mol Ecol Notes. 4:20–22 [Google Scholar]
- Kibbe WA. 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35:W43–W46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. 1943. The estimation of map distances from recombination values. Ann Eugen. 12:172–175 [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. 2009. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 37:D583–D587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski ML, Erickson SM, Bartholomay LC, Christensen BM. 2010. Midgut barrier imparts selective resistance to filarial worm infection in Culex pipiens pipiens. PLoS Negl Trop Dis. 4:e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Romero-Severson J, Severson DW. 2007. Genetic basis for reproductive diapause is correlated with life history traits within the Culex pipiens complex. Insect Mol Biol. 16:515–524 [DOI] [PubMed] [Google Scholar]
- Mori A, Severson DW, Christensen BM. 1999. Comparative linkage maps for the mosquitoes (Culex pipiens and Aedes aegypti) based on common RFLP loci. J Hered. 90:160–164 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa (NJ): Humana; p. 365–386 [DOI] [PubMed] [Google Scholar]
- Rudbeck L, Dissing J. 1998. Rapid, simple alkaline extraction of human genomic DNA from whole blood, buccal epithelial cells, semen and forensic stains for PCR. Biotechniques. 25:588–90, 592 [DOI] [PubMed] [Google Scholar]
- Severson DW, Meece JK, Lovin DD, Saha G, Morlais I. 2002. Linkage map organization of expressed sequence tags and sequence tagged sites in the mosquito, Aedes aegypti. Insect Mol Biol. 11:371–378 [DOI] [PubMed] [Google Scholar]
- Smith JL, Keyghobadi N, Matrone MA, Escher RL, Fonseca DM. 2005. Cross-species comparison of microsatellite loci in the Culex pipiens complex and beyond. Mol Ecol Notes. 5:697–700 [Google Scholar]
- Timoshevskiy VA, Severson DW, Debruyn BS, Black WC, Sharakhov IV, Sharakhova MV. 2013. An integrated linkage, chromosome, and genome map for the yellow fever mosquito Aedes aegypti. PLoS Negl Trop Dis. 7:e2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turell MJ, O’Guinn ML, Dohm DJ, Jones JW. 2001. Vector competence of North American mosquitoes (Diptera: Culicidae) for West Nile virus. J Med Entomol. 38:130–134 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorrips RE. 2001. JoinMap 3.0, Software for the calculation of genetic linkage maps. Wageningen, the Netherlands: Plant Research International [Google Scholar]
- Vinogradova EB. 2000. Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control. Moscow (Russia): Pensoft; p. 250 [Google Scholar]
- Zoghbi HY, Orr HT. 2000. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 23:217–247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.