Abstract
Background
Despite the high prevalence and morbidity of chronic rhinosinusitis (CRS), little is known about the mechanisms that underlie its pathogenesis. Recent studies have suggested that B cells might play an important role in CRS.
Objective
We sought to thoroughly characterize B lineage cells within sinus tissues of patients with CRS and healthy control subjects and to determine whether levels of EBV-induced protein 2, which is known to play an important role in the development of B-cell responses, were increased in patients with CRS.
Methods
Cells isolated from sinus tissues of patients with CRS and healthy control subjects were characterized by means of flow cytometry and immunohistochemistry. Local production of antibodies was measured in tissue extracts, nasal lavage fluid, and sera by using multiplex bead arrays and ELISA. Quantitative RT-PCR, ELISA, and Western blotting were used to assess gene and protein expression from tissue extracts.
Results
Nasal polyps (NPs) from patients with CRS had increased levels of both B cells and plasma cells compared with uncinate tissue from healthy control subjects (P < .05). NPs also contained significantly increased levels of several antibody isotypes compared with normal uncinate tissue (P < .05), but no differences in circulating antibody levels were found. Interestingly, levels of EBV-induced protein 2 were also increased in NPs (P < .05) and were positively correlated with expression of plasma cell markers (CD138 and B lymphocyte–induced maturation protein) in sinus tissue.
Conclusion
B cells and plasma cells are enriched in NPs, actively produce antibodies locally, and might contribute to chronic inflammation in patients with CRS. Elucidating the mechanisms that underlie this excessive local B-cell response might provide novel insights for the development of improved therapeutic strategies.
Keywords: Chronic rhinosinusitis, B cells, plasma cells, antibodies, EBV-induced protein 2 (EBI2), chronic inflammation
Chronic rhinosinusitis (CRS) is characterized by chronic inflammation of the sinonasal mucosa that persists for at least 12 weeks despite medical therapy and affects up to 10% of the US population.1–3 CRS is often divided into 2 clinically and phenotypically distinct classifications: chronic rhinosinusitis with nasal polyps (CRSwNP) and chronic rhinosinusitis without nasal polyps (CRSsNP). Despite the high incidence of this disease, the pathology of CRS is not well understood, and treatment options are limited, resulting in a significant loss in quality of life and a large financial burden on the health care system. However, it is clear that defects in innate immunity, along with chronic activation of a variety of inflammatory cells, are 2 factors that play roles in the pathogenesis of CRS.4
B cells are a key component of the adaptive immune response and are known to play several important roles in a variety of inflammatory disorders and at mucosal sites.5–7 In addition to their ability to produce antibodies that contribute to disease pathogenesis, B cells can function as antigen-presenting or regulatory cells and produce a variety of cytokines and chemokines that can influence inflammation. Recently, we have demonstrated that levels of B cell–activating factor of the TNF family, a key B-cell survival factor, are highly increased in nasal polyp (NP) tissue from patients with CRSwNP.8 Several reports have demonstrated increased levels of various isotypes of immunoglobulins, including IgG, IgE, and IgA, in sinus tissue from patients with CRS.9–11 In addition, we have recently reported increased levels of autoantigen-specific antibodies in NP tissue.9,12 Together, these studies suggest that polyp tissue might provide a supportive environment for B-cell survival and antibody production, which can play important roles in the pathogenesis of CRSwNP.
EBV-induced protein 2 (EBI2 or GPR183) is known to play a key role in the development of antibody responses in secondary lymphoid organs.13,14 Mice deficient in EBI2 have reduced numbers of plasmablasts and, consequently, diminished ability to mount antibody responses after infection.15,16 Conversely, mice with B cells that overexpress EBI2 generate massive plasmablast responses at the expense of germinal center B-cell development and have enhanced levels of antibody production.15,16 These studies highlight the important role of EBI2 in controlling B-cell responses and antibody production in secondary lymphoid tissues after infection. However, little is known about the role of EBI2 during chronic inflammatory diseases and whether its expression in the periphery might contribute to the generation of pathogenic antibody responses. This study was designed to more fully characterize B-cell responses in the nasal mucosa of patients with CRS and to assess whether EBI2 might play a role in CRS pathogenesis.
METHODS
Patients and tissue sample collection
Healthy control subjects and patients with CRS were recruited from the Allergy-Immunology and Otolaryngology Clinics of the Northwestern Medical Faculty Foundation and the Northwestern Sinus Center at Northwestern Medical Faculty Foundation. Uncinate tissue (UT) and NPs were obtained during routine functional endoscopic sinus surgery from patients with CRS. All patients with CRS met the criteria for CRS, as defined by the American Academy of Otolaryngology–Head and Neck Surgery Chronic Rhinosinusitis Task Force.1 Patients with an aspirin-exacerbated respiratory disease, established immunodeficiency, pregnancy, coagulation disorder, classic allergic fungal sinusitis, or cystic fibrosis were excluded from the study. Tissue samples were obtained from control subjects without a history of sinonasal inflammation during endoscopic skull-base tumor excisions, intranasal procedures for obstructive sleep apnea, and facial fracture repairs. Details of subjects’ characteristics are displayed in Table E1 in this article’s Online Repository at www.jacionline.org. All subjects provided informed consent, and the study was approved by the Institutional Review Board of Northwestern University Feinberg School of Medicine.
Immunohistochemistry
Preparation of nasal tissue for immunohistochemistry (IHC) was performed, as previously described.17–19 For detailed methods, please see the Methods section in this article’s Online Repository at www.jacionline.org.
Cell collection and flow cytometric analysis
Cells were isolated from sinus tissue by using a modified method designed for cutaneous tissue, as previously described.20 Briefly, tissue samples obtained during surgery were weighed and then placed in 5 mL of RPMI-1640 medium (Life Technologies, Grand Island, NY) supplemented with 5% FCS (Atlanta Biologicals, Lawrenceville, Ga), 1 mmol/L sodium pyruvate (Life Technologies), and 60 U/mL IL-2 (C-RPMI; Life Technologies). Tissue was incubated in C-RPMI for 4 days at 378C in a 5% CO2 atmosphere. Cells that emigrated from the tissue into the supernatant were collected, and remaining tissue fragments were removed by straining through a 70-nm nylon mesh (BD Biosciences, San Jose, Calif). Cells were washed with 1 × PBS (without Ca2+ or Mg2+) before counting and staining for flow cytometric analysis. Values were expressed as total numbers of cells obtained per milligram of tissue. For detailed methods, please see the Methods section in this article’s Online Repository. A representative example of the gating strategy can be found in Fig E1 in this article’s Online Repository at www.jacionline.org.
Measurement of antibody isotypes
Antibody levels were measured in tissue and lavage fluid by using the Human Immunoglobulin Isotyping Multiplex kit and the Human IgE Singleplex kit, according to the manufacturer’s instructions (EMD Millipore, Billerica, Mass). Sinus tissue extracts were prepared, as previously described,17 and nasal lavage fluid was collected during surgery. Samples were analyzed for total immunoglobulin isotype levels on a Luminex 200 instrument (Life Technologies). Because the Luminex assay did not discriminate between IgA isotypes, total and secretory IgA1 and IgA2 levels were measured from tissue extracts and lavage fluid by using custom ELISAs with primary and secondary antibodies from GenWay Biotec (San Diego, Calif) or HyTest (Turku, Finland). Standard curves were generated with myeloma IgA1 and IgA2 (EMD Millipore) or secretory IgA1 (sIgA1) and sIgA2 purified from sIgA (MP Biomedicals, Santa Ana, Calif) by using a Jacalin column. Detailed methods are available in this article’s Online Repository.
Quantitative RT-PCR and microarray analysis
RNA was isolated from tissue samples, and cDNA was prepared, as previously described.8 TaqMan real-time PCR was performed with the following primers and probes (all from Life Technologies): EBI2 (Hs00953886_m1), cholesterol 25-hydroxylase (CH25H; Hs02379634_s1), CD138 (Hs00896423_m1), and B lymphocyte–induced maturation protein (Hs00153357_m1). All expression values were normalized to β-glucuronidase and expressed as 2−ΔCt. For polymeric immunoglobulin receptor (pIgR) expression analysis, the following primers and probes (Integrated DNA Technologies, Coralville, Iowa) were used: forward, CTCTCTGGAG-GACCACCGT; reverse, CAGCCGTGACATTCCCTG; probe, 6FAM-AGATCAAGATTATCGAAGGAGAACCAAACCTC - TAMRA. Please see the Methods section in this article’s Online Repository for further details.
A comprehensive microarray analysis was performed, as previously described, and has been deposited with the Gene Expression Omnibus as GSE36830.18,21 For details, please see the Methods section in this article’s Online Repository.
Western blotting
Total lysates of sinus tissue were prepared, as previously described.17 Images were obtained on an Odyssey infrared scanner (LI-COR Biosciences, Lincoln, Neb), and densitometry was performed with Odyssey version 1.2 software. For detailed methods, please see the Methods section in this article’s Online Repository.
Statistical analysis
All data are reported as means ± SEMs, unless otherwise stated, and calculations were done with GraphPad Prism version 5.0b software (GraphPad Software, San Diego, Calif). Differences between groups were analyzed by using the 1-way ANOVA Kruskal-Wallis test, with a Dunn correction for multiple comparisons. Correlations were assessed by using Spearman rank correlation. A P value of less than .05 was considered significant.
RESULTS
Polyp tissue contained increased numbers of inflammatory cells
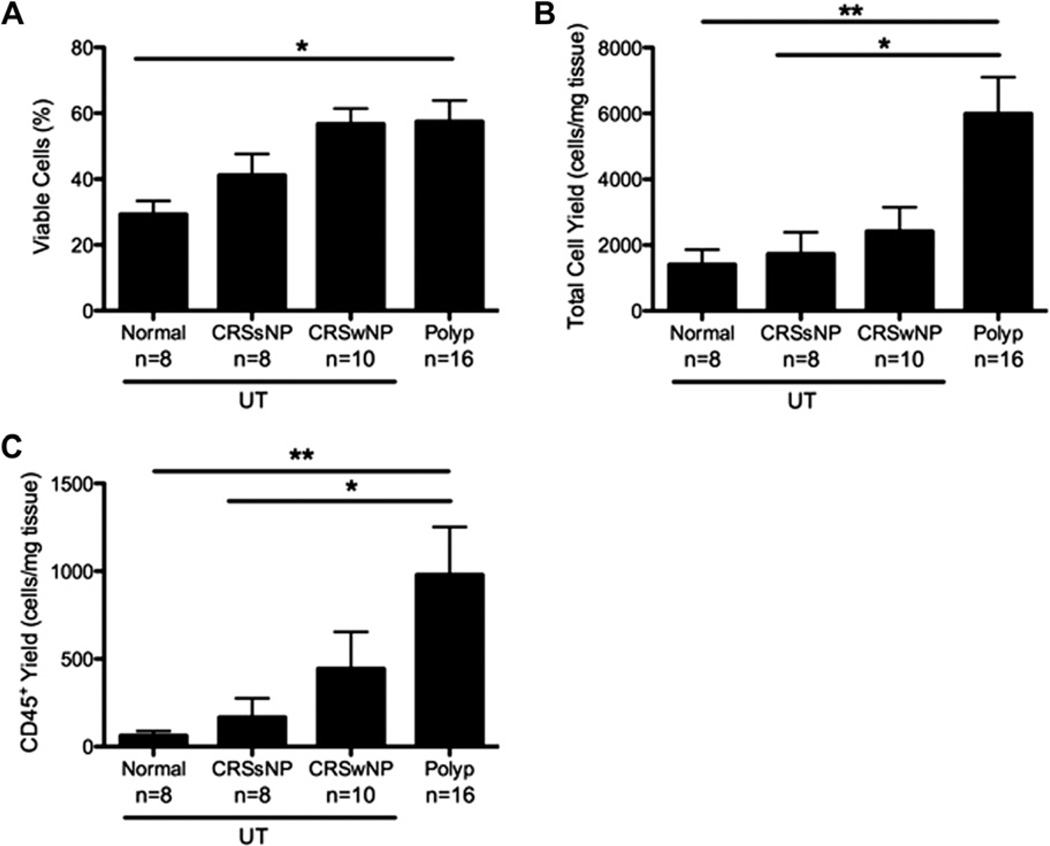
It is well known that CRS is characterized by chronic inflammation of the nasal and paranasal sinuses. We first wanted to assess the quantity and nature of the inflammatory cells found in sinus tissues from patients with CRS and control subjects by using flow cytometry. Overall cell viability, as measured by using of 4′-6-diamidino-2-pnehnylindole dihydrochloride (DAPI) staining, was greater in NPs compared with that seen in control UT (P <.05; Fig 1, A), and there was a significant increase in the total number of cells recovered per milligram of NP compared with that seen in patients with CRSsNP and normal UT (P < .05; Fig 1, B). Furthermore, the total number of CD45+ inflammatory cells recovered from NPs was significantly increased compared with that from patients with CRSsNP and normal UT (P < .01; Fig 1, C). Importantly, the increase in CD45+ cell numbers in NPs was not simply due to the overall increase in total cell numbers because the increase in CD45+ cell numbers was more than 2 times the increase in total cell numbers.
FIG 1.
NPs contain increased numbers of inflammatory cells. Cells were isolated from sinus tissue biopsy specimens after 4 days and analyzed by using flow cytometry. A, Cell viability was assessed by using DAPI exclusion. B, Total cell yield was determined by dividing the number of cells recovered after 4 days by the original weight of the biopsy specimen. C, CD45+ cell numbers were calculated based on the total number of cells recovered and the frequency of CD45+ cells within the DAPI− gate and divided by the original weight of the biopsy specimen. Data represent means ± SEMs. *P < .05 and **P < .01, Kruskal-Wallis test.
B cells and plasma cells accumulated in the nasal tissue of patients with CRS
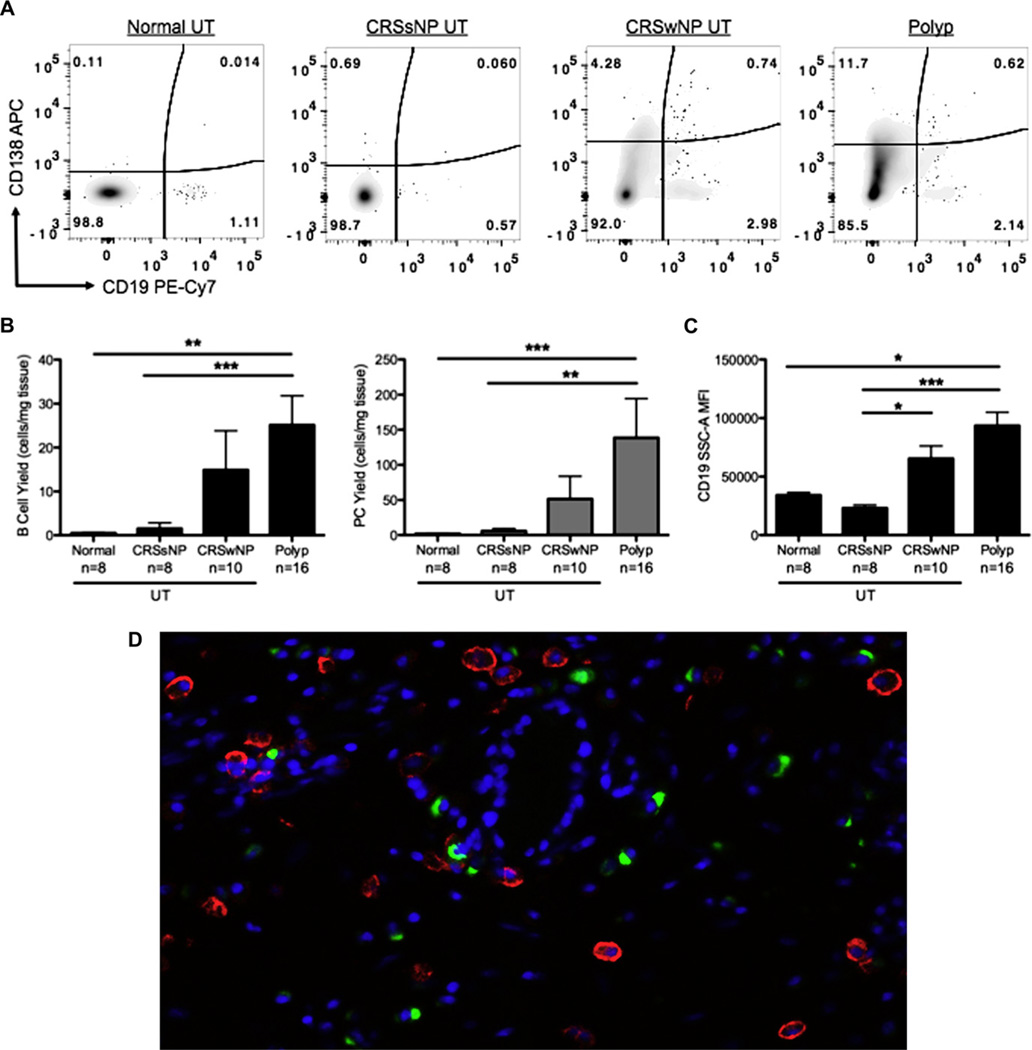
Recent work from our group has suggested that antibody production and B cells might play an important role in the pathogenesis of CRS.8,9 Therefore we used IHC to assess accumulation of CD20+ B cells in sinus tissue from patients with CRS. Similar to what other groups have shown,22 a semiquantitative analysis of the IHC staining revealed no difference in the mean number of CD20+ cells per high-power field (hpf) in any CRS tissue compared with normal control tissue (normal UT, 6.5 cells/hpf; UT from patients with CRSsNP, 13.2 cells/hpf; UT from patients with CRSwNP, 6.9 cells/hpf; NPs, 8.3 cells/ hpf; n = 8 per group). However, it is notoriously difficult to quantitatively assess protein expression by using IHC, and previous work,8 as well as a microarray study from our group, indicated that expression of CD20 was increased in polyp tissues (relative CD20 expression in normal UT, 1392 AU; NPs, 6452 AU; P < .01, Mann-Whitney U test). To more accurately enumerate and characterize the B-cell populations within sinus tissues, we assessed their numbers and phenotype using flow cytometry. We found a significant increase in the number of CD19+ B cells in NPs compared with those seen in the UTof healthy control subjects and patients with CRSsNP (P <.01; Fig 2, A and B). Interestingly, the B cells from NPs also had significantly higher side scatter mean fluorescence intensity, suggesting that they were in an activated state (P <.05; Fig 2, C). There was also a significant increase in the number of CD138+ cells (putative plasma cells) in NPs compared with numbers seen in normal UT (P <.001; Fig 2, A and B). Although it is possible that differential effects of IL-2 on tissue cells from patients and control subjects played some role in these findings, control studies evaluating residual cells in UTand NPs indicated that the majority of CD19+ and CD138+ cells had emigrated out of the tissue by day 4 of culture (data not shown). Together, these data suggest that NPs of patients with CRS contain a local inflammatory environment that is conducive to the accumulation of B cells and plasma cells.
FIG 2.
B cells and plasma cells were more abundant in NPs. Cells were isolated from sinus tissue biopsy specimens after 4 days and analyzed by means of flow cytometry. A, Representative flow cytometric plots. APC, Allophycocyanin; Pe-Cy7, phycoerythrin-cyanin 7. B, Numbers of B cells (CD19+) and plasma cells (PC, CD138+) were calculated as above from within the DAPl− CD45+ population. C, Mean side scatter A (SSC-A) fluorescence intensity of CD19+ B cells. D, Representative ×20 immunofluorescence image of CD19+ (red) and CD138+ (green) cells in NP tissue. Data represent means ± SEMs. *P < .05, **P < .01, and ***P < .001, Kruskal-Wallis test.
Antibody levels were increased in polyps
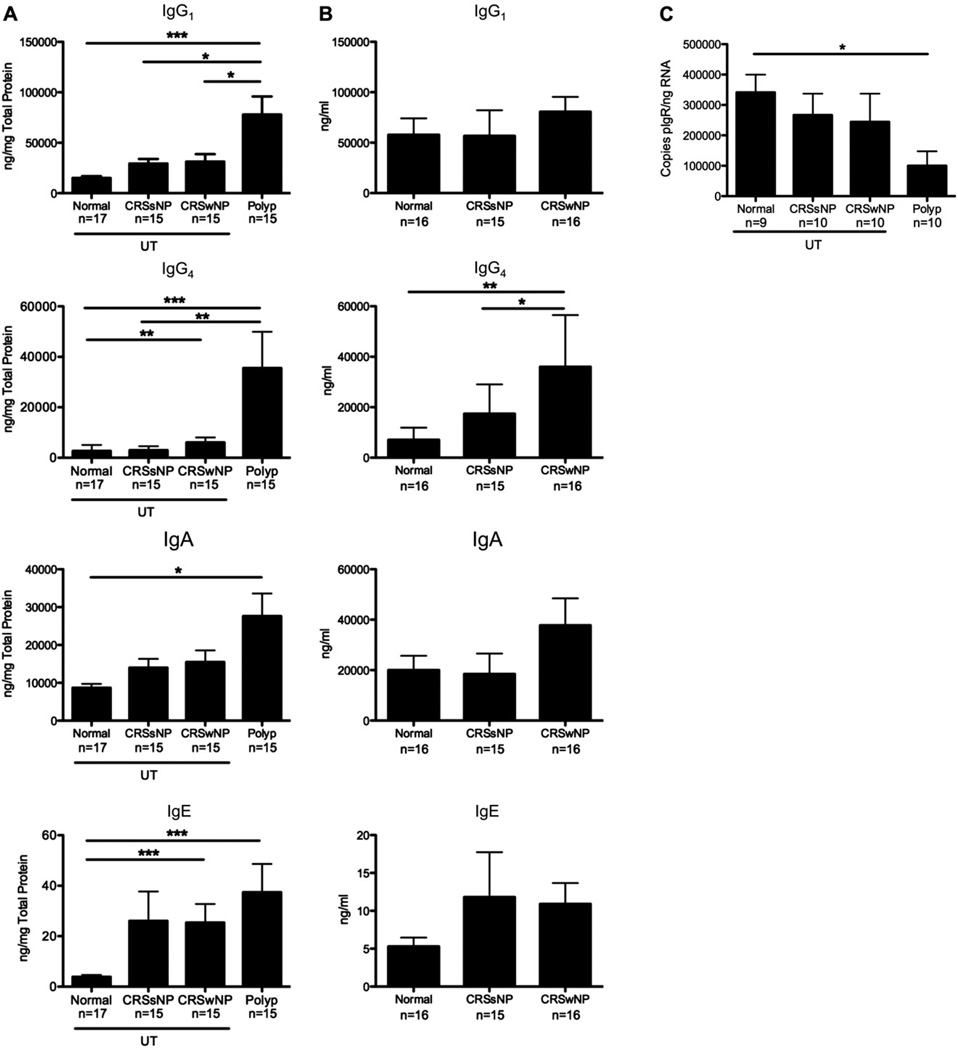
Although B cells can play a variety of roles during ongoing inflammation, an important component of their function is the production and secretion of antibodies.23 We recently reported that levels of a variety of autoantigen-specific antibodies were increased in polyps but not in the systemic circulation of patients with CRSwNP.9 Because the effector functions mediated by secreted antibodies are dependent on their isotype, we set out to assess the levels of different antibody isotypes present in sinus tissue biopsy specimens, as well as in nasal lavage fluid and serum samples. Antibody isotype levels were quantitated in sinus tissue detergent extracts or nasal lavage fluid by using a multiplex bead array kit, and values were normalized to total protein. As expected, NPs contained significantly increased levels of all antibody isotypes (except IgG3) compared with those seen in normal UT (P < .05; Fig 3, A, and see Fig E2 in this article’s Online Repository at www.jacionline.org). Importantly, the only isotype with increased levels in the sera of patients with CRS was IgE (see Fig E3 in this article’s Online Repository at www.jacionline.org); this is likely because of more atopy in this group (see Table E1). However, because antibody concentrations are much higher in serum compared with those in tissue, it is possible that the increased antibody levels in NPs might be due to increased vascular leak. Therefore we normalized the concentration of each isotype in the tissue and serum to albumin to account for this. The data in Fig E4 in this article’s Online Repository at www.jacionline.org demonstrate that only 10% or less of the antibody found in tissue can be attributed to vascular leak. Together, these data suggest that the antibodies found in the tissue were produced locally.
FIG 3.
Antibody levels were increased in NPs. A, Antibody isotype levels in tissue extracts were normalized to total protein. B, Antibody isotype levels measured in nasal lavage fluid. C, Gene expression of pIgR in tissue extracts. Data represent means ± SEMs. *P < .05, **P < .01, and ***P < .001, Kruskal-Wallis test.
Despite the high antibody levels in tissue, there was only a trend toward increased levels in the nasal lavage fluid of patients with CRSwNP compared with those seen in healthy control subjects (Fig 3, B, and see Fig E2). It is well established that the pIgR is responsible for the transport of polymeric IgA and IgM through the epithelium into the lumen of the airways and the gastrointestinal tract.24 Because IgA and IgM levels were increased in tissue from patients with CRS but not in lavage fluid, we examined pIgR levels in sinus tissue. Expression of pIgR was lowest in polyp tissue (P <.05; Fig 3, C). Together, these data suggest that polyp tissue not only contains increased numbers of B cells and plasma cells but that these cells are producing antibodies. Furthermore, in the case of IgA and IgM, inadequate pIgR expression might contribute to an accumulation of antibodies in tissue, with a concomitant reduction in lavage fluid.
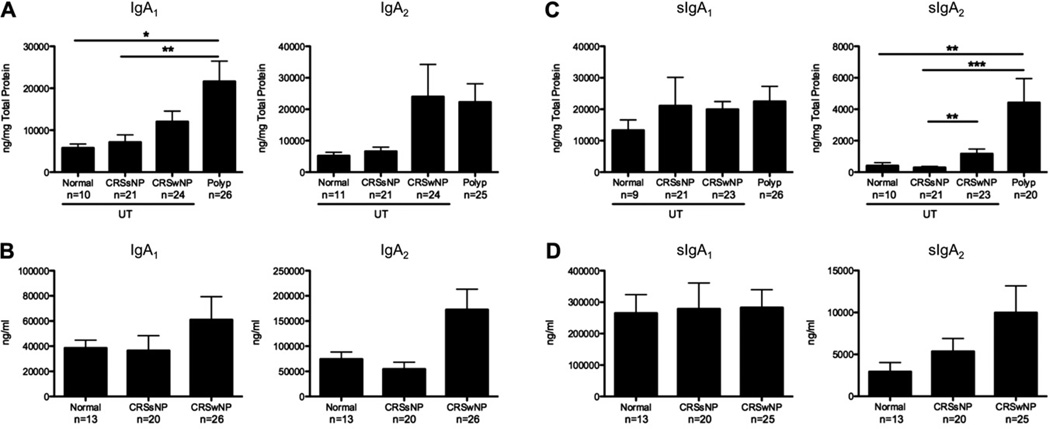
Because human subjects have 2 distinct IgA subclasses, we set out to measure total and secretory IgA1 and IgA2 levels in tissue extracts and nasal lavage samples using ELISA because the Luminex assay did not distinguish between these 2 isotypes. Although IgA1 levels were significantly increased in NPs (P <.05; Fig 4, A), there was only a trend toward increased IgA2 levels. Interestingly, sIgA2 levels were significantly increased in polyp tissue (P <.01; Fig 4, C). Similar to the other immunoglobulin isotypes we measured, neither total nor secretory IgA1 or IgA2 levels were increased in the lavage fluid of patients with CRSwNP (Fig 4, B and D). Although there was a clear trend toward an increase in sIgA2 levels in lavage fluid, these data further support the notion that normal mechanisms for export of antibodies to the lumen might be disrupted in patients with polyps and might contribute to the accumulation of antibodies within the tissue.
FIG 4.
Differential expression of IgA1 and IgA2 in NPs. Total IgA1 and IgA2 levels in tissue extracts (A) and nasal lavage fluid (B) are shown. sIgA1 and sIgA2 levels in tissue extracts (C) and nasal lavage fluid (D) are shown. Data represent means ± SEMs. *P < .05, **P < .01, and ***P < .001, Kruskal-Wallis test.
EBI2 levels were increased in polyps and correlated with plasma cell markers
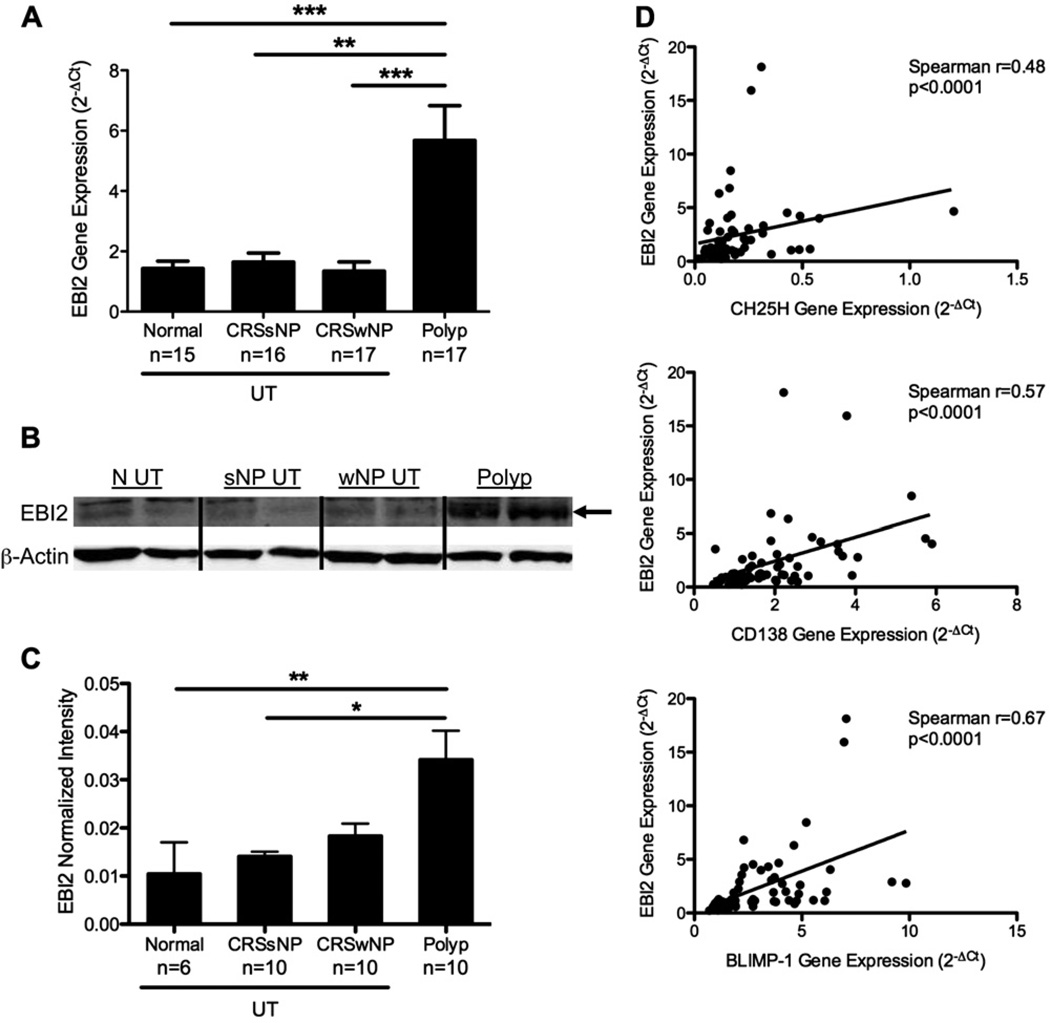
The G protein–coupled receptor EBI2 plays a critical role in the positioning of B cells in secondary lymphoid organs and consequently influences the fate of maturing B cells.13,14 Because we found significant increases in B-cell numbers, plasma cell numbers, and antibody levels in polyp tissues, it was important to determine whether EBI2 levels were increased in the nasal tissue of patients with CRS as well. RNA was extracted from patients’ tissue samples, and EBI2 gene expression was assessed by using TaqMan quantitative RT-PCR. We found a significant increase in the expression of EBI2 in NPs compared with that found in UT from all other groups (P <.001; Fig 5, A). We next wanted to confirm this increase at the protein level. As expected, we found significant increases in EBI2 protein levels using Western blot analysis in NP extracts compared with levels seen in the UT from all other groups (P < .05; Fig 5, B and C).
FIG 5.
EBI2 was highly expressed in NPs. A, Gene expression analysis of EBI2 from sinus tissue biopsy specimens was normalized to β-glucuronidase. B, Representative Western blot showing EBI2 and β-actin. C, Average of EBI2 expression normalized to β-actin in sinus tissue. D, Correlations between normalized EBI2 gene expression and CH25H, CD138, and B lymphocyte–induced maturation protein (BLIMP-1; n = 60 in each plot). Data in Fig 5, A and C, represent means ± SEMs. *P < .05, **P < .01, and ***P < .001, Kruskal-Wallis test.
Recently, 7α-25-dihydroxycholesterol, which can be synthesized by the enzyme CH25H, was identified as a ligand for EBI2.25,26 We assessed gene expression of CH25H in our sinus tissue samples using microarray analysis and quantitative RT-PCR. Interestingly, the expression of EBI2 was significantly positively correlated with expression of CH25H in these tissues, suggesting a relationship between expression of the receptor and local synthesis of the ligand (P < .0001, Spearman r = 0.48; Fig 5, D, upper panel). Moreover, EBI2 expression was significantly positively correlated with expression of the plasma cell markers CD138 and B lymphocyte–induced maturation protein (P < .0001, Spearman r = 0.57 and P < .0001, Spearman r = 0.67, respectively; Fig 5, D, middle and lower panels). These correlations were also validated in a microarray experiment using a distinct set of samples that has been deposited with the Gene Expression Omnibus as GSE36830 (see Fig E5 in this article’s Online Repository at www.jacionline.org). Taken together, these data suggest that EBI2 is associated with the enzyme for local production of its ligand and correlated with the accumulation of B-cell subsets and antibodies in nasal tissues.
DISCUSSION
It is well established that CRSwNP is characterized by TH2 inflammation and eosinophilia.22,27 Yet it is becoming increasingly apparent that B cells might play an important role in the inflammatory response within the sinus tissue of patients with CRSwNP.8,9 The lungs and upper airways represent an important mucosal immune site that is in constant contact with airborne antigens and microbial organisms. Many studies have focused on elucidating the mechanisms involved in the induction and maintenance of B-cell responses within the gut mucosa in animal models and human subjects, but there is a paucity of data regarding these mechanisms in the airway mucosa, especially in human subjects (Kato et al, unpublished data).6 CRS provides us with a unique opportunity to investigate an ongoing inflammatory response in human tissues, in part because the affected tissue is relatively easily accessible, even in healthy control subjects.
In the current study we have expanded on our earlier work and found striking evidence of B-cell inflammation and local antibody production in NPs from patients with CRSwNP.8,9 NP tissue from patients with CRS not only contained increased numbers of inflammatory cells (Fig 1) but also contained significantly more B cells, plasma cells, and antibodies (Fig 2–4) compared with UT. Whether these B cells enter the tissue as naive cells and become activated or if they enter as memory cells primed to respond within the tissue is not yet clear. Elucidating where B cells and plasma cells are activated in this disease can provide valuable insight and potential new avenues of investigation for therapeutic interventions. Because we know that levels of B cell–activating factor of the TNF family are highly increased in polyp tissue and correlate with expression of CD20,8 it is tempting to speculate that B cells that do traffic through the polyp tissue will find a favorable microenvironment for activation and differentiation within the tissue itself. Preliminary experiments from our laboratory indicate that NP tissue supports the survival of plasma cells in vitro (data not shown), but the mechanisms involved are presently under investigation. Further studies are needed to determine whether B cells can be activated within the tissue itself and what mechanisms might be involved.
We have previously shown increased levels of autoantigen-specific antibodies in NPs,9 and others have found increased IgE antibody levels directed against staphylococcal enterotoxins in NPs10 and evidence for local production of IgE,28 strengthening the argument that enhanced local antibody production might be an important component of CRSwNP pathogenesis. We have extended these findings in this study by demonstrating that the levels of many different antibody isotypes were increased in NPs of patients with CRSwNP but not in sera (Figs 3 and 4 and see Fig E3). This broad array of antibodies could have multiple activating effects on cells known to reside in NPs, including mast cells, eosinophils, and neutrophils. Notably, one of the most highly increased isotypes in NPs was IgG4 (Fig 3), which is often thought to play a protective role in allergic disease.29 However, it is also well established that IgE can be generated by a 2-step process that involves an IgG4 intermediate.30 Because levels of both IgE and IgG4 were highly increased in NPs, future studies will investigate whether sequential class-switch recombination through an IgG4 intermediate plays a role in the generation of IgE in NPs.
Interestingly, we found reduced expression of pIgR in NPs (Fig 3), despite the fact that pIgR expression can be induced by local immunoglobulin production.24 Because pIgR is known to be expressed in glandular epithelium, its reduced expression in NPs might be due to the recently documented reduction of glands in NPs.18 Moreover, it is likely that antibody secretion from NPs constitutes only a small fraction of the total antibodies recovered in nasal lavage fluid, and thus it is difficult to say with certainty that this is a good representation of NP-specific secretion. Nevertheless, the reduced pIgR expression fits well with the fact that we did not find significantly increased IgAor IgM levels in the nasal lavage fluid of patients with CRSwNP (Fig 4 and see Fig E3). In fact, the increased IgA and IgM levels in NP extracts might be a reflection of an inability of the pIgR system to upregulate its function and adequately accommodate the increased antibody production within the NP microenvironment. Accumulation of IgA in the tissue might further exacerbate inflammation through degranulation of eosinophils,31 and this could also be an important component of CRS pathogenesis.32 Although the antigen specificity of NP IgA is unknown, our previous studies have shown that at least some of the IgA in NPs is directed against autoantigens.9 Future studies will investigate whether antibodies derived from NPs are capable of activating innate effector cells, as well as assessing their specificities and the extent of clonal expansion within the tissues. In addition, although we have focused on antibody production as a key effector function of B cells in this work, it is possible that B cells and plasma cells within NPs play multiple roles during chronic inflammation (eg, T-cell costimulation, chemokine expression, cytokine expression, and regulatory roles), and further studies are needed to elucidate other mechanisms used by B cells during CRS pathogenesis.
EBI2 is known to play a critical role in the induction of antibody responses in secondary lymphoid tissues.13,14 However, little is known about the expression and function of EBI2 in human peripheral tissues. To our knowledge, this is the first report of increased EBI2 levels in peripheral tissues of a human pathological condition. We have found a significant increase in both EBI2 gene and protein expression in NPs in this study (Fig 5). Given the known role of EBI2 in facilitating plasmablast development and antibody production, and the striking enhancement of local production of antibodies in NPs, it is reasonable to speculate that EBI2 expression could be playing a role in this process in NPs. In addition, this work highlights EBI2 as a potential new therapeutic target that warrants further investigation. We have also shown that EBI2 levels correlate with markers of plasma cells in patients with CRS, lending further support to a potential role in disease pathogenesis, although future studies are needed to confirm whether EBI2 plays a role in activating local plasma cell responses in patients with CRS. The present studies suggest a potential importance of EBI2 expression in peripheral tissues in human subjects and extend our knowledge of the local mechanisms that promote B cells, plasma cells, and immunoglobulins in patients with CRSwNP-induced inflammation.
In summary, we have shown that NPs from patients with CRS had abundant accumulation of inflammatory cells, especially B cells and plasma cells. Not surprisingly, we also found significantly increased levels of several antibody isotypes in NPs. Finally, we have shown that EBI2 was highly expressed in NPs and correlated with expression of plasma cell markers. These findings indicate that B-cell inflammatory responses occur locally in NPs, might play a critical role in the pathogenesis of CRSwNP, and might provide novel insights for the development of improved therapeutic interventions.
METHODS
IHC
Blocked sections were incubated with anti-CD20 antibody (L26, 1:250; Thermo Fisher Scientific, Fremont, Calif) overnight at 4°C. After washing, sections were incubated with biotinylated horse anti-mouse secondary antibody for 1 hour at room temperature (1:500; Vector Laboratories, Burlingame, Calif). Sections were washed and then incubated with avidin-peroxidase complex (Vectastain Elite ABC Kit, Vector Laboratories) for 1 hour at room temperature. Sections were rinsed and incubated with diaminobenzidine chromogen (Life Technologies) for 10 minutes. Counterstaining with Gill Modified hematoxylin (EMD Millipore) was done for 5 to 10 seconds. Sections were then dehydrated, cleared, and mounted.
Semiquantitative analysis was performed with an Olympus IX71 inverted research microscope (Olympus, Center Valley, Pa) and a MicroFire AR digital microscope camera (Optronics, Goleta, Calif). Tissue section photographs were obtained at ×400 magnification by using Picture Frame software (Optronics). The photographs were blinded, and CD20+ cells in 5 random fields were counted in 5 sections from each tissue specimen. The data from each specimen and each observer were averaged.
Flow cytometry
For flow cytometric analysis, cells were first incubated with an Fc Block reagent (Miltenyi Biotec, Auburn, Calif) for 10 minutes at room temperature. All antibodies were obtained from BD Biosciences, unless otherwise stated. The following antibodies and dilutions were used to stain the surface of cells: allophycocyanin-Cy7 CD45 (2D1, 1:20), allophycocyanin CD138 (359103, 1:10; R&D Systems, Minneapolis, Minn), phycoerythrin-Cy7 CD19 (SJ25C1, 1:20), fluorescein isothiocyanate CD20 (2H7, 1:10), and phycoerythrin CD38 (HIT2, 1:20). All samples were stained with DAPI (10 µg/mL, 1:20) as a live/ dead discriminator. Cells were stained for 15 minutes at room temperature in the dark, and washed with PBS/5% BSA/EDTA before acquisition on an LSRII with FACSDiva software (BD Biosciences). A minimum of 10,000 events were collected for each sample. All analysis and compensation was performed with FlowJo software, version 9.4 (TreeStar, Ashland, Ore), and each experiment contained the proper single-stained control beads (BD Biosciences) and fluorescence minus one negative controls. Dead cells were excluded from the analysis.
Quantitative RT-PCR and microarray analysis
Single-strand cDNA was synthesized from 0.5 µg of total RNA with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, Calif) and random primers. Semiquantitative real-time RT-PCR was performed by using the TaqMan method with an Applied Biosystems 7500 Sequence Detection System (Applied Biosystems, Foster City, Calif) in 20-µL reactions (10 µL of 2× TaqMan Master mix [Applied Biosystems], 400 nmol/L of each primer, and 200 nmol/L of TaqMan probe plus 10 ng of cDNA). Primer and probe sets for β-glucuronidase (human β-glucuronidase endogenous control, PN; 4326320E) were purchased from Applied Biosystems. Quantified aliquots of purified PCR fragments of the target genes were serially diluted and used as standards in each experiment to determine the exact copy number of the target genes.
Total RNA from sinus tissue was extracted with QIAzol (Qiagen, Hilden, Germany) and cleaned and treated with DNase I by using RNeasy, according to the manufacturer’s instructions. The quality of total RNA was assessed with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, Calif) by using a RNA 6000 Nano LabChip (Agilent Technologies). NuGen ovation RNA amplification System V2 was used for cDNA amplification and as a biotinlabeling module to prepare cDNA for hybridization. Starting from 100 ng of total RNA, 3.75 µg of labeled cDNA was hybridized on an Affymetrix GeneChip Human Genome U133 Plus 2.0 probe arrays (Affymetrix, Santa Clara, Calif). GeneChips were washed and stained with Affymetrix fluidics station 450. GeneChips were scanned on an Affymetrix 7G GeneChip scanner. Data were analyzed with GeneChip Command Console software, version 3.2.
IgA custom ELISAs
For total IgA1 and IgA2 ELISAs, plates were coated with primary anti-IgA1 (14A/3C10; 1 µg/mL) or IgA2 (14AS; 1 µg/mL; GenWay Biotec, San Diego, Calif) capture antibodies overnight and then washed 3 times with PBS-Tween. For sIgA1 and sIgA2 ELISAs, plates were coated with primary anti-sIgA (5D8; 1 µg/mL; HyTest, Turku, Finland) capture antibody overnight and then washed 3 times with PBS-Tween. For all assays, plates were then blocked with 3% BSA-PBS for 1 hour and washed 3 times. Protein extract or nasal lavage fluid was added and incubated at room temperature for 2 hours. Plates were washed 4 times with PBS-Tween, and horseradish peroxidase–conjugated secondary anti-IgA1 (B3506B4; 1:2000) or IgA2 (A9604D2; 1:5000; GenWay Biotec) was added and incubated for 1 hour in the dark. After 4 washes with PBS-Tween, TMB Peroxidase substrate was added (KPL, Gaithersburg, Md). The reaction was stopped with TMB Stop Solution (KPL) after 30 minutes and read at 450 nm.
Albumin ELISA
Albumin concentrations were measured in tissue lysates and sera with a commercially available ELISA, according to the manufacturers’ instructions (Bethyl Laboratories, Montgomery, Tex).
Western blotting
Samples were combined with 4× LDL sample buffer and 10× sample reducing reagent (Life Technologies) heated to 70°C for 10 minutes before loading on 4–12% bis-tris gels (Life Technologies) and electrophoresed at 120V for 1.5 hours. Protein was transferred onto a polyvinylidene difluoride membrane (EMD Millipore) in cold 20% methanol/TG buffer by using a semidry transfer apparatus (Bio-Rad Laboratories, Hercules, Calif) at 20 V for 45 minutes. Membranes were blocked for 2 hours at room temperature in LI-COR blocking buffer (LI-COR Biosciences) and probed with a primary rabbit anti-human EBI2 antibody (1:1000; Proteintech, Chicago, Ill) and mouse anti-human β-actin (1:10,000; Sigma-Aldrich, St Louis, Mo) overnight at 4°C on a shaker. Blots were washed 3× in Tris-buffered saline–Tween for 5 minutes each and 1× in Tris-buffered saline before adding fluorescent secondary anti-rabbit antibody and anti-mouse antibody (1:10,000, LI-COR Biosciences).
Supplementary Material
Key messages.
B-cell and plasma cell numbers are significantly enhanced in NPs from patients with CRS.
Several antibody isotypes are highly increased in NPs but not systemically; local antibody production can contribute to chronic inflammation.
EBI2 levels are increased in NPs and correlate with expression of plasma cell markers.
The inflammatory environment within NPs can promote the survival, activation, or both of B cells and plasma cells.
Acknowledgments
Supported by National Institutes of Health grants R37 HL068546, R01 HL078860, and R01 AI072570 and the Ernest S. Bazley Trust.
Abbreviations used
- DAPI
4′-6-Diamidino-2-pnehnylindole dihydrochloride
- CH25H
Cholesterol 25-hydroxylase
- CRS
Chronic rhinosinusitis
- CRSsNP
Chronic rhinosinusitis without nasal polyps
- CRSwNP
Chronic rhinosinusitis with nasal polyps
- EBI2
EBV-induced protein 2
- hpf
High-power field
- IHC
Immunohistochemistry
- NP
Nasal polyp
- pIgR
Polymeric immunoglobulin receptor
- UT
Uncinate tissue
Footnotes
Disclosure of potential conflict of interest: J. E. Norton, R. K. Chandra, and A. Kato have received research support from the National Institutes of Health (NIH). A. T. Peters has provided expert witness testimony on drug allergy and has received lecture fees from Baxter. L. C. Grammer III has received research and travel support from the NIH; has received the Bazley Foundation grant; has received consultancy fees from Astellas Pharmaceuticals; is employed by Northwestern University and Northwestern Medical Faculty Foundation; has received research support from the NIH, the Food Allergy Network, and S&C Electric; has received lecture fees from the American Academy of Allergy, Asthma & Immunology (AAAAI); and receives royalties from Lippincott, UpToDate, BMU, and Elsevier. R. P. Schleimer has received research support from the NIH; has received consultancy fees from Intersect ENT, GlaxoSmithKline, and Allakos; and has stock/stock options in Allakos and Avrasense. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, et al. Rhinosinusitis: establishing definitions for clinical research and patient care. J Allergy Clin Immunol. 2004;114:155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kern RC, Conley DB, Walsh W, Chandra R, Kato A, Tripathi-Peters A, et al. Perspectives on the etiology of chronic rhinosinusitis: an immune barrier hypothesis. Am J Rhinol. 2008;22:549–559. doi: 10.2500/ajr.2008.22.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dykewicz MS, Hamilos DL. Rhinitis and sinusitis. J Allergy Clin Immunol. 2010;125(suppl):S103–S115. doi: 10.1016/j.jaci.2009.12.989. [DOI] [PubMed] [Google Scholar]
- 4.Tan BK, Schleimer RP, Kern RC. Perspectives on the etiology of chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg. 2010;18:21–26. doi: 10.1097/MOO.0b013e3283350053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandtzaeg P. Mucosal immunity: induction, dissemination, and effector functions. Scand J Immunol. 2009;70:505–515. doi: 10.1111/j.1365-3083.2009.02319.x. [DOI] [PubMed] [Google Scholar]
- 6.Drolet JP, Frangie H, Guay J, Hajoui O, Hamid Q, Mazer BD. B lymphocytes in inflammatory airway diseases. Clin Exp Allergy. 2010;40:841–849. doi: 10.1111/j.1365-2222.2010.03512.x. [DOI] [PubMed] [Google Scholar]
- 7.Dorner T, Radbruch A, Burmester GR. B-cell-directed therapies for autoimmune disease. Nat Rev Rheumatology. 2009;5:433–441. doi: 10.1038/nrrheum.2009.141. [DOI] [PubMed] [Google Scholar]
- 8.Kato A, Peters A, Suh L, Carter R, Harris KE, Chandra R, et al. Evidence of a role for B cell-activating factor of the TNF family in the pathogenesis of chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2008;121:1385–1392. doi: 10.1016/j.jaci.2008.03.002. e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan BK, Li QZ, Suh L, Kato A, Conley DB, Chandra RK, et al. Evidence for intranasal antinuclear autoantibodies in patients with chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2011;128:1198–1206. doi: 10.1016/j.jaci.2011.08.037. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachert C, Zhang N, Holtappels G, De Lobel L, van Cauwenberge P, Liu S, et al. Presence of IL-5 protein and IgE antibodies to staphylococcal enterotoxins in nasal polyps is associated with comorbid asthma. J Allergy Clin Immunol. 2010;126:962–968. doi: 10.1016/j.jaci.2010.07.007. e1–6. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Segura A, Brieva JA, Rodriguez C. Regulation of immunoglobulin secretion by plasma cells infiltrating nasal polyps. Laryngoscope. 2000;110:1183–1188. doi: 10.1097/00005537-200007000-00022. [DOI] [PubMed] [Google Scholar]
- 12.Jeffe JS, Seshadri S, Hamill KJ, Huang J, Carter R, Suh L, et al. A role for anti-BP180 autoantibodies in chronic rhinosinusitis. Laryngoscope. 2013 doi: 10.1002/lary.24016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira JP, Kelly LM, Cyster JG. Finding the right niche: B-cell migration in the early phases of T-dependent antibody responses. Int Immunol. 2010;22:413–419. doi: 10.1093/intimm/dxq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan TD, Gardam S, Gatto D, Turner VM, Silke J, Brink R. In vivo control of B-cell survival and antigen-specific B-cell responses. Immunol Rev. 2010;237:90–103. doi: 10.1111/j.1600-065X.2010.00942.x. [DOI] [PubMed] [Google Scholar]
- 15.Gatto D, Paus D, Basten A, Mackay CR, Brink R. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 2009;31:259–269. doi: 10.1016/j.immuni.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–1126. doi: 10.1038/nature08226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tieu DD, Peters AT, Carter RG, Suh L, Conley DB, Chandra R, et al. Evidence for diminished levels of epithelial psoriasin and calprotectin in chronic rhinosinusitis. J Allergy Clin Immunol. 2010;125:667–675. doi: 10.1016/j.jaci.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seshadri S, Lin DC, Rosati M, Carter RG, Norton JE, Suh L, et al. Reduced expression of antimicrobial PLUNC proteins in nasal polyp tissues of patients with chronic rhinosinusitis. Allergy. 2012;67:920–928. doi: 10.1111/j.1398-9995.2012.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takabayashi T, Kato A, Peters AT, Suh LA, Carter R, Norton J, et al. Glandular mast cells with distinct phenotype are highly elevated in chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2012;130:410–420. doi: 10.1016/j.jaci.2012.02.046. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato A, Chustz RT, Ogasawara T, Kulka M, Saito H, Schleimer RP, et al. Dexamethasone and FK506 inhibit expression of distinct subsets of chemokines in human mast cells. J Immunol. 2009;182:7233–7243. doi: 10.4049/jimmunol.0801375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61:1280–1289. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 23.Lund FE, Randall TD. Effector and regulatory B cells: modulators of CD4(+) T cell immunity. Nat Rev Immunol. 2010;10:236–247. doi: 10.1038/nri2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4:598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, et al. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Yang XV, Wu J, Kuei C, Mani NS, Zhang L, et al. Oxysterols direct B-cell migration through EBI2. Nature. 2011;475:519–523. doi: 10.1038/nature10226. [DOI] [PubMed] [Google Scholar]
- 27.Bachert C, Van Bruaene N, Toskala E, Zhang N, Olze H, Scadding G, et al. Important research questions in allergy and related diseases: 3-chronic rhinosinusitis and nasal polyposis—a GALEN study. Allergy. 2009;64:520–533. doi: 10.1111/j.1398-9995.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- 28.Gevaert P, Nouri-Aria KT, Wu H, Harper CE, Takhar P, Fear DJ, et al. Local receptor revision and class switching to IgE in chronic rhinosinusitis with nasal polyps. Allergy. 2013;68:55–63. doi: 10.1111/all.12054. [DOI] [PubMed] [Google Scholar]
- 29.Akdis CA. Therapies for allergic inflammation: refining strategies to induce tolerance. Nat Med. 2012;18:736–749. doi: 10.1038/nm.2754. [DOI] [PubMed] [Google Scholar]
- 30.Cameron L, Gounni AS, Frenkiel S, Lavigne F, Vercelli D, Hamid Q. S epsilon S mu and S epsilon S gamma switch circles in human nasal mucosa following ex vivo allergen challenge: evidence for direct as well as sequential class switch recombination. J Immunol. 2003;171:3816–3822. doi: 10.4049/jimmunol.171.7.3816. [DOI] [PubMed] [Google Scholar]
- 31.Abu-Ghazaleh RI, Fujisawa T, Mestecky J, Kyle RA, Gleich GJ. IgA-induced eosinophil degranulation. J Immunol. 1989;142:2393–2400. [PubMed] [Google Scholar]
- 32.Schleimer RP, Kato A, Kern R. Eosinophils and Chronic Rhinosinusitis. In: Lee JJ, Rosenberg HF, editors. Eosinophils in health and disease. 1st ed. San Diego: Elsevier; 2012. pp. 508–518. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.