Abstract
Proteoglycans represent a structurally heterogeneous family of proteins that typically undergo extensive posttranslational modification with sulfated sugar chains. Although historically believed to affect signaling pathways exclusively as growth factor coreceptors, proteoglycans are now understood to initiate and modulate signal transduction cascades independently of other receptors.
From within the extracellular matrix, proteoglycans are able to shield protein growth factors from circulating proteases and establish gradients that guide cell migration. Extracellular proteoglycans are also critical in the maintenance of growth factor stores and are thus instrumental in modulating paracrine signaling.
At the cell membrane, proteoglycans stabilize ligand–receptor interactions, creating potentiated ternary signaling complexes that regulate cell proliferation, endocytosis, migration, growth factor sensitivity, and matrix adhesion. In some cases, proteoglycans are able to independently activate various signaling cascades, attenuate the signaling of growth factors, or orchestrate multimeric intracellular signaling complexes. Signaling between cells is also modulated by proteoglycan activity at the cell membrane, as exemplified by the proteoglycan requirement for effective synaptogenesis between neurons.
Finally, proteoglycans are able to regulate signaling from intracellular compartments, particularly in the context of storage granule formation and maintenance. These proteoglycans are also major determinants of exocytic vesicle fate and other vesicular trafficking pathways.
In contrast to the mechanisms underlying classical ligand-receptor signaling, proteoglycan signaling is frequently characterized by ligand promiscuity and low-affinity binding; likewise, these events commonly do not exhibit the same degree of reliance on intermolecular structure or charge configurations as other ligand-receptor interactions. Such unique features often defy conventional mechanisms of signal transduction, and present unique challenges to the study of their indispensable roles within cell signaling networks.
1. Overview of Proteoglycan Structure, Nomenclature, and Function
The myriad contributions made by proteoglycans to physiological processes such as cellular proliferation, migration, immunity, and matrix assembly are largely owed to the structural diversity within this molecular superfamily. Originally characterized by their ability to facilitate ligand–receptor interactions and confer structural stability upon tissues, proteoglycans have more recently been shown to hold profound influence over cell physiology by independently engaging in the initiation and modulation of numerous cell signaling networks (Schaefer and Schaefer, 2010).
Proteoglycans consist of a protein core to which linear, negatively charged polysaccharide chains known as glycosarninoglycans (GAGs) are covalently linked. GAGs are comprised of disaccharide repeats and vary with respect to their sulfation patterns. GAGs are categorized by the structure of their disaccharide chain sequences; among the GAG subtypes are heparan sulfates, chondroitin sulfates, keratin sulfates, and dermatan sulfates. These structurally distinct chains confer different binding properties and signaling capabilities upon the nascent core proteins to which they become attached (Kjellén and Lindahl, 1991).
Some protein cores, such as those of neuropilins and betaglycan/transforming growth factor beta receptor III, may be expressed without the addition of GAG chains (Mythreye and Blobe, 2009), whereas those of others, such as the syndecans, are not found on the cell surface without GAG modifications (Tkachenko et al., 2005). Furthermore, certain proteoglycans are capable of undergoing modification by multiple GAG types, while other proteoglycans are limited in the type of GAG that may become attached.
Tissue distribution of proteoglycans also varies significantly among types. While some (including syndecan 4) are ubiquitously expressed, others are only found within particular tissues, as in the cases of neurocan, exclusively expressed in the nervous system (Rauch et al., 2001). The numerous cellular localization patterns of proteoglycans likewise reflect their functional versatility; these molecules may perform their various roles in signaling, vesicular trafficking and structural support from the cell membrane, intracellular compartments, or the extracellular matrix.
2. Growth Factor Signaling
The range of disaccharide modifications, protein core structures, and expression patterns across different tissues results in a high degree of structural variability and functional versatility within the proteoglycan family. Negative charges carried by proteoglycan GAG chains also facilitate ionic interactions with growth factors, a diverse family of soluble proteins that induce cellular responses such as migration, proliferation, and endocytosis once they bind membrane receptors and induce their dimerization (Bernfield et al., 1999; Murakami et al., 2008; Ornitz et al., 1992). This process of receptor activation results in subsequent receptor phosphorylation on intracellular residues, thus communicating the external stimulatory impulse toward the cytosol, where protein components are mobilized to initiate various parallel signaling cascades.
Proteoglycans influence growth factor signal transduction in several capacities. On account of their ability to directly bind growth factors such as vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), and epidermal growth factor (EGF), proteoglycans in the extracellular matrix are able to maintain high-concentration stores of growth factors that would otherwise be diluted by passive diffusion. These proteoglycans also facilitate the formation of growth factor gradients and prevent their premature degradation (Grünert et al., 2008; Saksela et al., 1988).
At the cell membrane, proteoglycans are able to stabilize interactions between growth factor ligands and their receptors, thus potentiating the resultant intracellular signaling events. In certain contexts, proteoglycans are required for effective signaling to occur, as with syndecans and the fibroblast growth factor receptors (FGFRs) (Yayon et al., 1991); in others they serve to inhibit signaling, as shown with betaglycan's ability to act as a coreceptor with type II activin receptor for the binding of inhibin (Lewis et al., 2000).
In contrast to the high degree of specificity generally required for receptor–ligand interactions, proteoglycan coreceptors typically exhibit a lower affinity for growth factors (Nugent and Iozzo, 2000). The specificity of these proteoglycan–growth factor interactions has comprised the basis for intense research, with variable results reported for different systems (Ashikari-Hada et al., 2004; Guimond and Turnbull, 1999; Kreuger et al., 2006). Although the structural features and charge conformations required to create proteoglycan–protein specificity remain undefined, it appears likely that overall GAG charge distribution and steric structure, rather than particular disaccharide sequences, determine specificity for protein binding (Dreyfuss et al., 2009).
3. Integrin Interactions
In parallel with their influence over growth factor signaling, proteoglycans also affect multiple intracellular signaling networks via the integrins. Named for their ability to integrate outside signals toward the cytoplasm, integrins represent a dichotomous family of alpha and beta glycoprotein subunits that sense and bind to specific extracellular matrix components. At least 18 alpha and 8 beta subunits have been identified to date, and integrins are found as a noncovalent pairing of one alpha and one beta subunit (van der Flier and Sonnenberg, 2001). Each paired combination, of which 24 have been identified, is able to bind with high affinity to a specific matrix protein, such as fibronectin, laminin, von Willebrand factor, and collagen, although there is some degree of overlap among the pairing specificities (Hood and Cheresh, 2002).
Integrins are capable of relaying extracellular signals toward intracellular compartments, and are conversely able to transform intracellular signaling into alterations of extracellular attachment (Hynes, 2002). Similarly, proteoglycans and integrins often bind identical extracellular matrix components, and influence the same signaling networks. Such cross talk between ligand binding and signal transduction has been reported for years, although only recently have the notions of mutual dependence and synergy been ascribed to the signaling properties of these two protein families. Assembling focal contacts between cells and the extracellular matrix, establishing cell polarity, and the processes of wound healing and angiogenesis, for example, are all dependent upon the cooperative activity of both integrins and proteoglycans in various cell types (Morgan et al., 2007).
4. Autonomous Signaling
Beyond the coreceptor functions served by proteoglycans in growth factor and integrin signaling are numerous circumstances under which proteoglycans also participate in signal transduction independently (Kirn-Safran et al., 2009; Tkachenko et al., 2005). Because proteoglycans frequently initiate signaling events within multimeric complexes comprised of other receptors, the isolation of independent proteoglycan capabilities represents a nontrivial experimental challenge.
One method to overcome this issue is to induce proteoglycan activation by way of ligands that do not concurrently initiate signaling via other receptors. In the study of syndecan 4, a heparan sulfate proteoglycan known to initiate intracellular signaling cascades, this has been accomplished by addition of a particular fibronectin fragment (Bass et al., 2007) or antibodies against syndecan 4 (Tkachenko et al., 2006). Such antibodies (directed against the extracellular proteoglycan domain) do not induce simultaneous activation of other receptors, and their multivalency facilitates proteoglycan oligomerization. The extracellular epitopes of numerous proteoglycans, however, have proven notoriously difficult to immunologically target with great efficacy. This is likely due to the variable steric effects of their GAG chains, and has perhaps resulted in the underestimation of proteoglycan signaling potency when induced solely by means of antibody clustering.
For this reason, models of chimeric proteoglycan activation have been developed for the study of their downstream signaling effects (Tkachenko and Simons, 2002). These chimeras contain native proteoglycan transmembrane and intracellular domains, thus preserving their intracellular signaling capabilities, and have a substituted GAG-free extracellular receptor domain. In the study of syndecans, the extracellular domain has been successfully replaced with the human Fc receptor (FcR), a molecule exclusively found in cells of the immune system. By introducing an exogenous, immunogenic target to proteoglycan core domains, it is possible to study their interactions, localization, and signaling capabilities with a high level of specificity.
As detailed in Section 8, this model of proteoglycan oligomerization first involves expression of the chimeric construct and confirmation of its appropriate localization at the cell membrane. Nonimmune IgG is used to occupy the chimeric FcR receptor sites, and oligomerization is subsequently induced by multivalent anti-IgG antibodies or antibody fragments, such as F(ab)2 subunits. The use of fluorescently labeled antibodies in such experiments also enables the visualization of both spatial distributions and kinetics of oligomerization at the cell surface.
In the case of syndecan 4, clustering of a chimeric receptor construct as described above leads to an enhanced rate of cell migration (Tkachenko et al., 2006). This is due to the unique signaling capabilities afforded by this proteoglycan's cytoplasmic domains, involving the downstream activation of protein kinase Ca (PKCα) and two Rho family GTPases: RhoG and Rac1 (Elfenbein et al., 2009; Tkachenko et al., 2005). The active form of Rac1 is typically found at the leading edge of migrating cells, where it coordinates the actin polymerization machinery required for membrane protrusion and directional migration (Vicente-Manzanares et al., 2005).
Syndecan 4 assembles multimeric protein complexes that suppress the activity of such GTPases until activation during cell migration; inactive pools of Rho GTPases are thus sequestered until syndecan 4 both releases and activates them at specifically localized regions cell regions (Elfenbein et al., 2009). Chimeric syndecan 4 constructs have been used to demonstrate that protein core mutations lead to an inability to form GTPase-inhibitory complexes, resulting in GTPase dysregulation and a cell migration defect (Elfenbein et al., 2009). In this way, the autonomous signaling potential of proteoglycan receptors has been shown to supersede that of simple ligand-dependent activation, and additionally encompasses the ability to orchestrate multimeric, localized signaling complexes (Naccache et al., 2006; Zimmermann et al., 2005). Assembly of these multiprotein complexes at proteoglycan cytoplasm domains, in concert with integrin activation, likely underlie the mechanism by which GTPases are precisely activated and regulated in response to extracellular signaling cues (Morgan et al., 2007).
Despite effectively excluding other receptors from antibody-mediated clustering, the proteoglycan chimera model of signal transduction bears several limitations. First, the stereochemistry of antibody-mediated chimera oligomerization represents an approximation of physiological activation by ligands such as growth factors or extracellular matrix components. Whether this corresponds to comparable molecular proximities and conformational interactions with native ligand binding is unknown; both factors are critical for effective activation and deactivation in other receptor systems, and are likely to impact proteoglycan signaling similarly. Second, the spatial distribution of proteoglycan clustering is not controlled by antibody treatment in the same way as it commonly is, for example, at the leading edge of a migrating cell. Instead, the chimera becomes clustered on all apical and lateral surfaces, partially undermining the proteoglycan function of spatial guidance and the establishment of cell polarity for directional migration (Elfenbein et al., 2009; Pankov et al., 2005). Third, it is unclear whether the immunological isolation of proteoglycans during oligomerization also functionally excludes the activation of other receptors; this is potentially a confounding issue that occurs by mass effect of multiprotein oligomerization, thus diminishing the specificity of antibody-directed proteoglycan clustering. Finally, as with other model systems involving receptor overexpression, chimeric proteoglycan oligomerization is fraught with issues of the stoichiometric imbalance between the receptor and downstream signaling molecules. This stoichiometric inconsistency may further be complicated by a high degree of intercellular variability, depending on whether cells used for assay exhibit stable and similar levels of chimeric construct expression.
For these reasons, while chimeric oligomerization models provide insight into the signaling potential of cell surface proteoglycans, characterization of their respective signaling networks is best achieved when supported by complementary methodologies. In the case of syndecans, the generation of syndecan 4 genetic knockout models have validated the role of this proteoglycan and its associated signaling proteins in cell migration, and demonstrated their importance in physiological processes such as wound healing, neurodevelopment, and maintenance of vascular integrity (Chittenden et al., 2006; Gorovoy et al., 2007; Matthews et al., 2008; Partovian et al., 2008).
In such animal models of targeted genetic deletion, the redundancy of other expressed proteoglycan receptors often serves to compensate for defects generated by a particular receptor's absence. It follows that alternative approaches may be used in vitro to better define proteoglyans’ signaling roles. These include RNA interference (RNAi)-mediated genetic knockdown of the studied proteoglycan(s), the addition of antibodies that preclude ligand binding, introduction of dominant-negative constructs, and the use of specific proteases and/or pharmacological inhibitors. Collectively, such approaches enhance the ability to identify the requirement and define the specific involvement of proteoglycans within signaling networks.
5. Proteoglycan Downregulation: Endocytosis and Ectodomain Shedding
Following membrane-bound receptor activation, resultant signaling complexes are generally subjected to endocytosis in order to downregulate cell surface expression. Once internalized, these protein complexes may be recycled back to the membrane, directed toward lysosomal vesicles for degradation, or sorted toward numerous other vesicle types and cell compartments. In recent years, the role of proteoglycans as key regulators of endocytic processes has become more apparent (Kobialka et al., 2009; Tkachenko et al., 2004; Valdembri et al., 2009).
Endocytosis is studied by cell surface labeling of the protein or membrane region of interest with a fluorescent dye, fluorophore, or immunologic tag. Because the internalization of labeled protein increases over time, endocytosis assays should be performed within consistently and strictly timed intervals. Synchronization for the start of endocytosis is therefore performed by bringing cells to a temperature of 0 °C, at which point all endocytic machinery is arrested. After incubation with the labeling agent, cells are returned to 37 °C to permit endocytosis to occur for a set period of time.
Several techniques exist for microscopic observation of endocytic processes or the quantification of internalized protein (Fig. 1.2), and are detailed in Section 8. Microscopic approaches are taken when the goal is to dissect the particular endocytic pathway taken by internalized proteoglycans, or to ascertain whether certain other labeled proteins are found within close proximity to labeled endocytic vesicles. More quantitative approaches rely on measurements of rate and amount of internalized receptor.
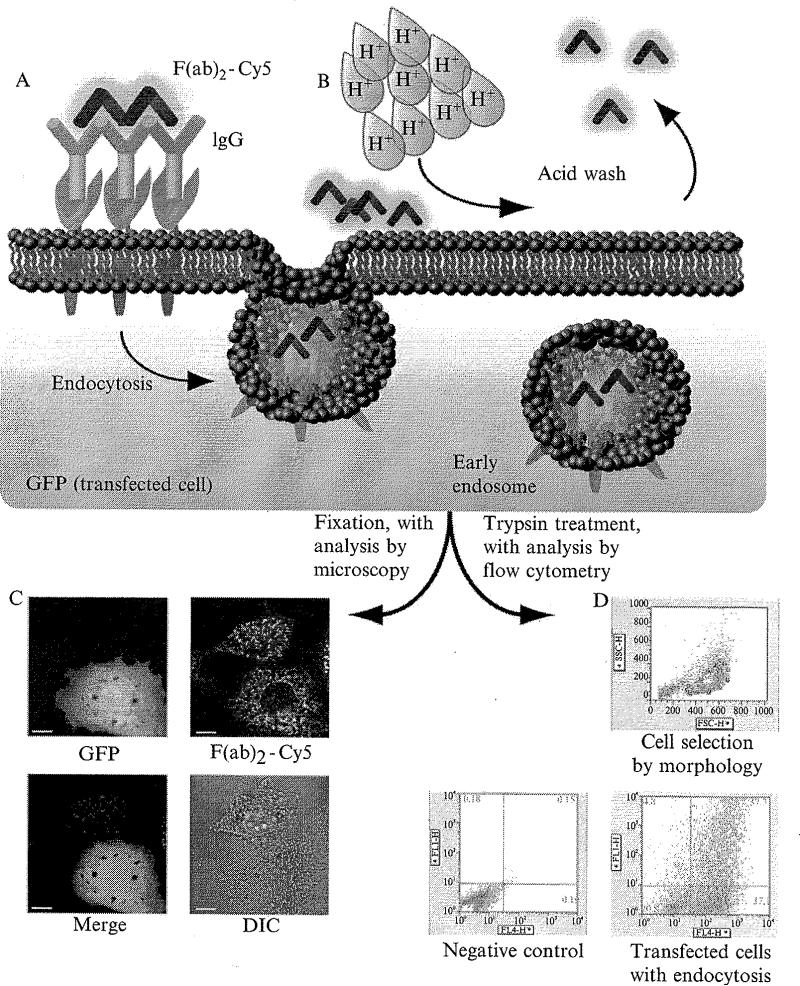
Figure 1.2.
Quantification of proteoglycan endocytosis. Internalization of proteoglycans following oligomerization is quantified by first inducing receptor oligomerization, as described in Fig. 1.1. For these experiments, fluorescently labeled antibodies (either IgG or F(ab)2 fragements) are used to label endocytosed proteoglycans. In this figure, the fluorescent dye Cy5 is conjugated to F(ab)2 fragments, and cells used for assay express both the S4-FcR chimera and green fluorescent protein (GFP) (A). Incubations are performed on ice, and endocytosis is permitted to proceed at 37 °C for specified periods of time. At the conclusion of an endocytosis time point, cells are brought to ice again to arrest all endocytic processes, and washed with PBS that has been titrated with HCl to pH = 2.5 (B). This disrupts antibody–antigen binding, and permits the effective washing of remaining surface-bound fluorescent antibodies. One approach for characterizing endocytosed vesicles is fixation and analysis by microscopy (C). These panels show two cells, both of which express S4-FcR, and have undergone clustering-mediated endocytosis for 15 min (scale bar = 10 μm). Vesicles are apparent throughout the cytosol, and are not found in nuclear compartments. One of the two cells also coexpresses GFP, and this technique of construct coexpression permits the investigation of other endocytosis-specific plasmids, such as clathrin, dynamin or Rab mutants, or other endocytic markers. All the visualized fluorescent vesicles are known to originate from the cell surface in this technique, as the source of vesicular fluorescence is derived from the dye-conjugated clustering antibodies added to the cells prior to fixation. Note that the cell membrane contains minimal fluorescent signal, on account of the acid wash step described above; this acid wash effectively excludes signals generated by endocytic vesicles from that of the cell membrane. A second approach involves treatment of the cells by trypsin to remove them from their culture dishes, with subsequent analysis performed by flow cytometry (D). The first panel demonstrates morphologic selection of cells for analysis using side-scattered (SSC) and forward-scattered (FSC) light. The region selected represents ~ 64% of the total cell population in this example, excluding debris and cells that have been inadvertently lysed. Of this subpopulation, fluorescence is measured in the bottom two panels with GFP represented on the y-axis (FL1-H) and Cy5 on the x-axis (FL4-H). The left panel shows a negative control condition of untransfected cells that were not exposed to fluorescent antibodies, used to establish a baseline of autofluorescence for gating of experimental samples. The right panel demonstrates a cell population that was transfected with a GFP construct, and underwent clustering-mediated endocytosis of the S4-FcR chimera for 15 min. Variability in GFP expression and fluorescent antibody internalization is readily apparent, and median or mean values are typically used to characterize fluorescence values in each condition. GFP in this experiment may also be replaced with endocytosis-related fluorescence constructs to specifically quantifY the endocytic rates of cells expressing such constructs.
Cell surface receptors, including proteoglycans, may be internalized by several endocytic pathways (Mosesson et al., 2008). The kinetics, accompanying proteins, impact on signal duration and eventual receptor fate differ significantly from pathway to pathway. It is for this reason that a receptor's preference for one route of internalization will often dictate its signaling properties.
Proteoglycans primarily influence endocytosis of other cell surface receptors through the recruitment of proteins involved with directing vesicular traffic. Perhaps the best characterized of these proteins are the Rabs, GTPases that coordinate the translocation of nascent vesicles to specific cellular compartments. A reported connection between Rab5 and syndecan 1 ectodomain cleavage also implies the presence of an additional mechanism by which a proteoglycan receptor's recruitment of endocytic machinery results in subsequent receptor downregulation by nonendocytic means (Hayashida et al., 2008).
Ectodomain shedding in transmembrane proteoglycans has been previously described as a constitutive process that is accelerated during conditions such as tissue inflammation (Alexopoulou et al., 2007). Cleavage of extracellular domains, which is catalyzed by a variety of metalloproteases and other proteases, results in the extracellular accumulation of GAG chains that retain their protein-binding properties. The physiological significance of ectodomain shedding ranges from the formation of growth factor gradients to the attenuation of escalating inflammatory responses (Alexopoulou et al., 2007).
In contrast to proteoglycan endocytosis, methods to study extracellular domain cleavage generally rely on biochemical techniques of quantifying both GAG production and the enzymatic activation of relevant proteases. Among the remaining elusive issues in this field is how proteoglycan removal from the membrane is regulated (via either endocytosis or ectodomain shedding) and how balanced proteoglycan turnover is consistently maintained.
6. Cell Adhesion
Matrix attachment and subsequent migration are among the cellular processes largely mediated by proteoglycan signaling. Because of the significant overlap in signaling mediators and temporal characteristics of these discrete processes, they represent a functional continuum that is often experimentally challenging to dissect. Migration and matrix attachment take place simultaneously when cells are exposed to novel surfaces, thus temporally obscuring the distinction between both events. Likewise, the characterization of cells with a decreased ability to migrate is frequently confounded by the presence of a concurrent deficit in matrix adhesion. The differentiation between adhesion and migration is even further encumbered by a large degree of overlap between the growth factors, receptors, and activation pathways that mediate each event.
Three predominant mechanisms underlie the proteoglycan contribution to cell adhesion, and each is exemplified by the syndecans. First, these proteoglycans are able to directly bind extracellular matrix components, as demonstrated in the cases of fibronectin, laminin, and vitronectin engagement (Couchman, 2003). Second, the syndecans function synergistically with integrins during cell adhesion; this cooperation involves downstream signaling cross talk, the formation of vinculin-rich focal adhesions, and the activation of Rac1 (Morgan et al., 2007). In some systems, the function of syndecans and integrins is even more directly related, as evidenced by the requirement for syndecan 1 in αvβ3 integrin activation (Beauvais et al., 2004). Finally, growth factor signaling in the context of adhesion is partially mediated by syndecans (Gopal et al., 2010), particularly as it applies to the described Rho GTPases and their maintenance of balance between adhesion and motility.
The above mechanisms afford a wide range of experimental approaches for studying cellular adhesion to a novel matrix. Among them are the assay of various adhesion substrates, incubation with proteoglycan-binding growth factors, treatment with pharmacologic modulators of proteoglycan signaling, and antibodies that either oligomerize or functionally block the matrix-binding properties of proteoglycans and/ or integrins.
Cell adhesion studies in the context of proteoglycan signaling typically involve timed measurements of attachment strength. In such experiments, cells in suspension are plated above a substrate coated with a selected matrix. At specified time points, nonattached cells are washed away with an isotonic medium, leaving the adherent cells for quantification. Although the fixation, staining, and counting of individual cells is a commonly employed approach to measure the percentage of remaining adherent cells, fluorescent membrane dyes may also be used to provide a more sensitive readout of this value; these dyes are generally used as part of a preincubation solution prior to the time of assay. Other variations of this technique involve fluorogenic enzymatic reactions that correlate linearly with adherent cell number (Tolosa and Shaw, 1996). On account of the inherent variability in the force of washing and between plated samples, statistically meaningful results may only be realized upon assay of multiple duplicated experimental and control conditions.
Related to the described whole-cell assay are several microscopic approaches to studying cell–matrix adhesion quantitatively. One such method serves to characterize the extent of cell spreading (total area of interface contact) upon a two-dimensional matrix such as fibronectin-coated glass. The degree and morphology of spreading on a particular matrix differs significantly among cell types, thus necessitating appropriate cell type-matched controls. Experiments involve fixation of cells after a time period during which matrix adhesion occurs, with subsequent microscopic imaging and quantification of the two-dimensional region of contact between the cell membrane and matrix. It is important to note that although increased adhesive surface areas imply a greater degree of surface interaction between the cell and extracellular matrix, such values are strictly morpho-metric and do not necessarily reflect adhesion strength.
Live-cell microscopic imaging approaches may also be used to study cell attachment, and these techniques yield particular insight into the morphologic and temporal characteristics of adhesion. The use of total internal reflection fluorescence microscopy (TIRFM) represents one frequently employed method for studying cell signaling events, morphological changes, and cell adhesion processes occurring exclusively at the membrane–matrix interface. TIRFM imaging relies on light refraction properties to illuminate a distance above a reflecting glass surface, typically less than 200 nm in depth (Axelrod, 2003). This is accomplished by controlling a laser beam's angle of incidence, which is related to light refraction according to Snell's law. When directed at angles of total internal reflection, an “evanescent wave” is formed, which illuminates basal membrane surfaces. Evanescent wave intensity decays exponentially with distance above the reflective surface, thus enabling the effective exclusion of all structures not within immediate proximity to the cell's basal membrane. For TIRFM studies, cells must be plated upon thin surfaces with amenable indices of refraction, typically glass coverslips. TIRFM also requires objective lenses with wide numerical apertures (NAs) that permit the manipulation of laser light toward oblique angles of incidence.
When coupled with fluorescent probes for proteoglycans that localize to adhesion sites and mediate adhesion signaling events, TIRFM represents a valuable tool for studying the intersection between signaling and localized generation of adhesive force. Perhaps the best-studied nexus between these processes is the focal adhesion, a structure that serves to anchor cells within their surrounding matrices. Focal adhesions are dynamic multiprotein complexes that establish points of structural contact between the cytoskeleton and the extracellular matrix, and are required for the generation of tensile strength required for cell movement. These multimeric complexes also represent sites of numerous localized signaling processes, and generally contain enriched pools of proteins such as vinculin, talin, paxillin, integrins, in addition to proteoglycans, which may be used experimentally as focal adhesion markers (Albiges-Rizo et al., 2009). In this way, early studies of proteoglycan involvement revealed that chondroitin sulfate and hyaluronic acid are not associated with focal adhesions, while heparan sulfate is intimately associated with these cell structures (Turley, 1984).
Heparan sulfate proteoglycans are now appreciated as requisite components of focal adhesion assembly, where their action is mutually dependent upon (and synergistic with) the activity of integrins (Couchman, 2003). Syndecan 4 overexpression causes increased focal adhesion formation with a subsequent decrease in cell motility (Longley et al., 1999); this proteoglycan is also hypothesized to directly interface between the extracellular matrix and the actin cytoskeleton at focal adhesions on account of its ability to bind both fibronectin and the actin bundling protein, α-actinin (Greene et al., 2003). The quantification of focal adhesion formation after cell fixation and focal adhesion immunostaining has been used to characterize cell-matrix adherence (Partridge and Marcantonio, 2006). TIRFM's applicability to live-cell imaging holds great potential to expand upon these quantitative studies by enabling real-time study of proteoglycan-mediated fluxes of focal adhesion generation and disassembly.
The forces exerted by regions of cells during adhesion may also be discretely measured in studies of cell–matrix interactions to precisely quantify the vectors of tensile strength. This is accomplished either by measurement of the ensuing matrix deformity (passive) or by the direct probing of cellular resistive forces (active). The former principle generally entails the creation of an observable and malleable extracellular array, such as sheets constructed from silicone or similarly pliable materials, or beads embedded within a matrix. Microdeformities or shifts in bead arrays resulting from cellular adhesion are subsequently interpolated to their respective forces and mapped to their corresponding cellular sites (Harris et al., 1980; Munevar et al., 2001). Direct probing of cell resistance and adhesive strength, on the other hand, involves a direct challenges to the integrity of a cell's adherence, and is exemplified by the techniques of shear flow generation, atomic force microscopy, magnetic tweezers, and optical tweezers. Such techniques have permitted the localization and quantification of cellular forces to piconewton levels of resolution, and have been extensively reviewed elsewhere (Addae-Mensah and Wikswo, 2008).
Although the technical considerations of subcellular adhesion force measurement is beyond the scope of this chapter, the application of force mapping to proteoglycan signaling is significant for two reasons. First, as a collectively predominant component of the extracellular matrix, proteoglycans are largely responsible for both the cell-to-matrix and intercellular adherent forces that sustain tissue growth. The structural integrity of most tissues is therefore strongly influenced by proteoglycan support and signaling, the mechanisms of which are likely to be more clearly delineated by future studies of subcellular force generation. Second, emerging evidence in recent years has led to the implication of proteoglycans in mechanotransduction within various signaling roles. In endothelial cells, for example, the detection of shear flow is also likely to be partly accomplished by heparan sulfate proteoglycans (Moon et al., 2005). It has furthermore been postulated that the bulky mass of these proteoglycans’ heparan chains serve as sail-like sensors for shear stress, leading to flow-dependent oligomerization. Such mechanisms currently remain without definitive demonstration, and will likely rely upon the intersection of live-cell imaging modalities and force reactivity assays for complete characterization (Florian et al., 2003; Mochizuki et al., 2003).
7. Migration
Cell adhesion is a requisite stage of migration, although the fonner represents only one element of the coordinated process needed to propel a cell through a given matrix. In order for effective and directional migration to occur, a cell must first establish opposing leading and retracting edges. As desclibed above, this cell polarity is largely mediated by proteoglycans and their effects on Rho family GTPase signaling (Burridge and Wennerberg, 2004).
Studies of cell polarization may be performed by either morphologic characterization (as with directionality of cell protrusions or migration) (Pankov et al., 2005) or on a molecular level by spatial mapping of enzymatic activity over time. In recent years, the mechanistic insight yielded by the latter approach has been significantly expanded by the development of fluorescent biosensors; such engineered constructs allow investigators to observe spatial and temporal features of signaling downstream of proteoglycans within living, migrating cells.
Among these are probes that exploit the potential transfer of energy that occurs when two molecules come within close proximity to each other (Nakamura et al., 2006). This physical property, known as Förster resonance energy transfer (FRET), has permitted the engineering of constructs containing both a donor and acceptor fluorophore, with the spatial proximity of both dictated by construct conformation. Intramolecular FRET -based probes are designed to shift conformation in a reversible manner that is dependent upon ligand binding or enzymatic activity. Cells expressing such probes are subjected to light wavelengths that exclusively excite the donor fluorophores, and the resultant emission from both the donor and acceptors are subsequently measured. Regions of acceptor emission indicate that energy transfer has occurred. Thus, this technique allows the mapping of donor–acceptor proximity throughout the living cell (directly correlating with probe conformation), and so far have successfully been used to charactelize cell polality with respect to phosphoinositide concentrations, Rho family GTPase activity, kinase activity, cell surface receptor activation, among others (Aoki and Matsuda, 2009).
For effective migration to occur, cells must also form new points of matrix adhesion for leverage, dismantle existing adhesion points at the trailing edge, disengage the cell–cell connections that maintain tissue structural integrity, and perform these events in synchrony with neighboling cells. The mechanisms underlying the mobilization and orchestration of all involved signaling networks remain incompletely understood, although the contributions made by proteoglycans are indispensable. In cells lacking syndecan 4, for example, Rac1 activity is not appropriately suppressed at regions of the cell uninvolved with leading edge formation, resulting in noncoordinated membrane protrusion and a subsequent defect in cell migration ability (Bass et al., 2007; Elfenbein et al., 2009; Matthews et al., 2008; Pankov et al., 2005).
Several approaches may be employed for the characterization and quantification of cell migration. Perhaps the most basic characterization of cell migration involves the tracking of single-cell movement in two dimensions by low-magnification live-cell microscopy over time. Cellular displacement over time is recorded by sequential image acquisition, with a readily identifiable cell structure such as the nucleus generally used for tracking motion. From these images, characteristics of migration such as velocity, distance, and directionality may be calculated for comparison among various experimental conditions.
This technique requires the plating of living cells on surfaces such as glass, or plastics that are engineered to exhibit similar refractive indices. Because migration occurs over the course of hours, microscope configurations amenable to migration studies necessitate thermal control and the maintenance of a stable pH in the cell culture medium. Thermal regulation is best achieved with an insulated enclosure and thermostat-heater circuitry, while control of pH may be accomplished by several methods. Culturing chambers may be hermetically sealed at the initiation of assay to prevent changes in CO2 partial pressure within the medium over time. Alternatively, a layer of mineral oil may be added above the culturing medium, providing a barrier to CO2-dependent acidification. Cells used for assay may also be grown in media containing buffers such as 100 mM HEPES, which render the cultures more resistant to pH fluxes than most conventional culturing medium formulations. The latter approach requires controls to exclude the possibility that buffering agents alone influence the cell signaling events under investigation. Finally, environmental O2 and CO2 partial pressures may be kept constant by control of their supply by direct gas perfusion. When combined with a programmable motorized stage, the migration of multiple adherent cells in different microscopic fields may be tracked over time, providing a well-controlled and statistically meaningful comparison of cells’ migration behaviors.
Tracking the migration of individual cells enables the study of proteoglycan roles in whole-cell displacement over time. It largely in this manner that the ability of proteoglycans to direct cell migration via Rho family GTPases has been characterized (Morgan et al., 2007; Pankov et al., 2005; Tkachenko et al., 2006). Despite the wide range of experimental flexibility and described future potential offered by single-cell migration assays, however, several inherent limitations also exist with these techniques. The first is related to characterizing a cell's direction of migration: migration is a purposeful and energy-intensive process that typically occurs in response to a spatially directed signal, often established as a protein gradient. Although single-cell tracking techniques using conventional cell culture chambers (dishes and/or wells) often serve to identify migration deficits or enhanced proclivities, such experimental setups do not facilitate the formation of gradients that guide migration under physiological conditions. To that end, a variety of cell culture devices have been designed for the purpose of establishing a linear gradient of chemoattractant or migration-inhibiting factors (Boyden, 1962; Zicha et al., 1991; Zigmond, 1988). In the study of proteoglycan signaling, these gradients may involve extracellular matrix components, growth factors, and/or antibodies that induce oligomerization at the cell membrane.
Commonly used devices for this purpose are Boyden and Dunn chambers, and their respectively modified variants. These methods of culturing cells involve two interfacing chambers, one in which cultured cells are placed and the one for the chemoattractant(s) being assayed. In Dunn chambers, cells are adherent to the chamber surface at the time of assay and migrate horizontally in two dimensions toward the established gradient. In contrast, the Boyden chamber model of migration has suspended cells in an upper chamber, with a porous filter separating it from a lower chamber in which chemoattractant is typically placed. Cellular migration through the porous filter is generally quantified in this assay by counting cells that traverse the membrane (see Section 8). Alternatively, cells may be preincubated with a fluorescent membrane dye, with subsequent lower chamber fluorescence quantification of trans-filter migration. It is important to note that in measuring migration, the Boyden model actually serves to quantify the entire continuum of adhesion and migration, as cells must first successfully attach to the interchamber filter before migrating to the lower chamber. For this reason, such experiments do not clearly discriminate between cellular adhesion and migration. Given this constraint of interpretation, the ability to modifY which extracellular signaling ligands are (1) immobilized upon the filter and (2) soluble within the lower chamber medium nevertheless confers enormous experimental versatility to this method.
The second limitation of single-cell migration assays relates to the fundamental behavioral difference between cells that migrate in isolation and those that move as part of a multicellular front. The migration of numerous cells is by far more commonly observed within physiologically intact tissues than is lone cell migration, and the former is exquisitely sensitive to factors such as intercellular contact inhibition and paracrine signaling. Multicellular migration may be studied by quantifying en masse movement of cells in confluent two-dimensional monolayers. In such experiments, an acellular discontinuity, or “wound” is created within a monolayer, and the rate of cellular reinfiltration of the void is subsequently quantified. This is most simply accomplished by subjecting the monolayer to a uniform linear disruption or scratch, and imaging the resultant migration into this region at defined time points.
Results of multicellular migration studies are particularly sensitive to artifacts of proliferation; a multiplication of total cell number during the course of an assay can result in falsely accelerated level of migration. It is for this reason that cell cycles should be synchronized by adequate serum-starvation prior to the time of assay. Additionally, the degree of cellular confluence profoundly impacts migration rate; cultured cells that have been permitted to overgrow (subsequent divisions upon reaching complete confluence) exhibit severe migration defects, even after several subsequent cycles of cell passaging. It is for this reason that cells to be used for multicellular migration studies are recommended to invariably undergo passaging as they approach 75–80% confluence, thus minimizing such intercellular contact inhibition artifacts. These and other considerations for tissue culture of monolayer cells are addressed in Section 8.
An alternative approach to the study of cell migration involves the creation of a three-dimensional matrix for the quantification of cell movement. Such techniques were historically used to assay tumor cell invasive potential, and initially involved basement membrane preparations from the amnion (Liotta et al., 1980). Development of reconstituted extracellular matrices, such as Matrigel, and refinement of three-dimensional imaging techniques have since facilitated the ability to readily characterize the velocity, directionality, and morphology of cells as they negotiate defined extracellular matrices in three dimensions.
Migration assays involving Matrigel and related reconstituted matrices permit the selection of numerous experimental conditions, including growth factors, structural proteins, concentration gradients, and matrix viscosity. These create an environment that ideally mimics physiological migration better than two-dimensional cultures. These three-dimensional matrices may be used in conjunction with techniques such as Boyden chamber assays, providing a three-dimensional medium above the filter that separates upper from lower chambers (Albini et al., 2004). Reconstituted matrices may furthermore be used as implants in animal models of migration, particularly as they apply to nerve regeneration or angiogenesis (Malinda, 2009). Finally, such three-dimensional models of migration are indispensable for the assay of morphological features not observed in two-dimensional cultures, such as endothelial tube, which is also partially mediated by proteoglycan signaling (Ferrari do Outeiro-Bernstein et al., 2002; Ponce, 2009).
8. Experimental Procedures
8.1. Immunoblotting for heparan sulfate proteoglycans
On account of their variable GAG chain modifications, proteoglycans typically exhibit patters of poorly defined streaks when subjected to gel electrophoresis. This prevents their effective separation and visualization by Western blot analysis, and necessitates several modifications to classical immunoblot techniques.
GAG chain cleavage is typically performed prior to electrophoresis in order to resolve molecular weights and quantities of proteoglycan naked protein cores. Samples to be analyzed by immunoblotting are incubated with heparitinase (otherwise known as heparanase III) or chondroitinase ABC, depending on the proteoglycan under investigation. The former cleaves heparan sulfate chains, leaving dermatan, keratan, and chondroitin sulfate modifications intact. On the other hand, incubation with chondroitinases generally results in the exclusive cleavage of dermatan and chondroitin sulfates.
Cell lysates are incubated in one of two buffers, depending upon whether the enzyme of choice is a heparitinase or chondroitinase. In the case of heparitinase, the final buffer concentration is: 0.1 mM calcium acetate and 0.1 M sodium acetate with pH adjusted to 7.0. Depending upon whether heparitinase is isolated from a microbial source, the addition of a protease inhibitor is frequently recommended (Couchman and Tapanadechopone, 2001). If chondroitinase is to be used, the following final buffer concentrations are preferred: 50 mM Tris–HCl, 20 mM ethylenediaminetetraacetic acid (EDTA), 30 mM sodium acetate, 10 mM N-ethylmaleimide (NEM), and 0.2 mM phenylmethylsulfonyl fluoride (PMSF). Concentrations of enzyme should be adjusted in accordance with estimated proteoglycan concentration. Heparitinase is added in approximate quantities of 1 mU for up to 1-5 μg of proteoglycan, whereas chondroitinase ABC is added at of 1 mU for every 2-10 μg of proteoglycan. An incubation time of 3 h at 37 °C is recommended for complete digestion. Experimental conditions of optimal enzymatic reaction times and concentrations may be conducted with known quantities of purified proteoglycan.
Samples are added to 2× SDS-PAGE sample buffer in a 1:1 ratio (final concentrations: 10% (v/v) glycerol, 62.5 mM Tris–HCl (pH = 6.8), 2% sodium dodecyl sulfate (SDS), 0.01 mg/ml bromophenol blue, 5% (v/v) β-mercaptoethanol (BME)). All samples are heated to 95 °C for 5 min, and either frozen at –80 °C for future electrophoresis or run as follows.
Samples are loaded onto a poly-acrylamide gel (concentration should be optimized for the proteoglycan of interest), subjected to electrophoresis and then transfer onto a polyvinylidene fluoride (PVDF) membrane.
Nonspecific membrane binding sites are blocked by either 5% nonfat milk in Tris-buffered saline (TBS: 50 mM Tris/HCl (pH = 7.4), 150 mM NaCl) or 5% bovine serum albumin (BSA), also dissolved in TBS. Primary antibodies directed against proteoglycan epitopes are added at dilutions according to their specifications (in either 1% nonfat milk/TBS or 1% BSA/TBS), and incubated for 1 h at room temperature or overnight at 4 °C.
Following three sequential, 10-min washes with TBS containing 0.1% Tween-20 (TBST), secondary antibodies (conjugated with either horseradish peroxidase or a specified fluorescent dye) are added in either 1% nonfat milk/TBS or 1% BSA/TBS at concentrations determined by their specifications.
The membranes are washed three times for 10 min with TBST, and once for 10 min with TBS. They are then imaged by standard luminance-based or fluorescence-based methods, as determined by the secondaty antibody conjugates.
8.2. Proteoglycan chimera oligomerization
In order to isolate the signaling effects of proteoglycan clustering, a chimeric proteoglycan model of antibody-induced oligomerization may be employed (Fig. 1.1). To begin, DNA expression vectors should be designed to conserve both transmembrane and intracellular domains, and the proteoglycan extracellular domain should be substituted with a nonendogenously expressed antibody receptor.
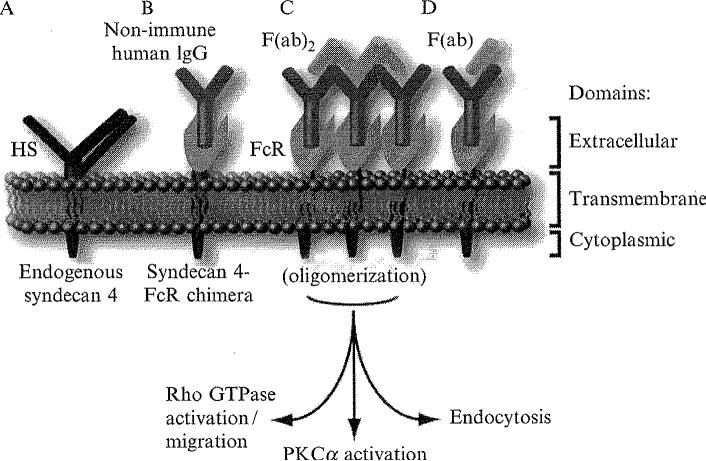
Figure 1.1.
Clustering of proteoglycan chimeras. Endogenous syndecan 4 is membrane bound, and contains heparan sulfate chain modifications on its extracellular domain (A). The syndecan 4-FcR chimera consists of the human Fc receptor extracellular domains substituted for the native (heparan sulfate (HS)-bearing) extracellular domain of syndecan 4 (B). Clustering is initiated by the addition of nonimmune human IgG. Excess antibody is removed by washing, and F(ab)2 fragments are added to induce oligomerization (C). This leads to multiple downstream signaling events, as described in the figure. As a negative control, univalent F(ab) fragments may be added, which do not induce proteoglycan clustering (D).
For stoichiometric consistency among cells undergoing antibody clustering and between experiments, we recommend the generation of clonal populations of cells exhibiting stable expression of the chimera construct. Alternatively, pooled cell populations with comparable stable chimera expression, generated by fluorescence-assisted cell sorting (FACS), may be used for these studies. The following experimental procedures apply to a model chimera receptor system: rat fat pad endothelial cells (RFPECs) with stable expression of a syndecan 4/human Fc receptor chimera (Tkachenko and Simons, 2002).
Prior to assay, cells are plated after sequentially passaging, with confluence not exceeding 75–80%. Because of the described cross talk between proteoglycans and integrins, an extracellular matrix component, such as fibronectin, should be chosen to control for integrin activation. Plates may be coated with fibronectin by covering plating surfaces with a solution of 10 μg/ml fibronectin dissolved in sterile phosphate-buffered saline (PBS) for 30 min at 37 °C. In our experience, failure to select an extracellular matrix (and thus, reliance upon ionically treated plastic substrates for adhesion) results in unacceptable levels of interexperimental variability on account of uncontrolled integrin engagement.
Cells are serum-starved in a culturing medium, such as Dulbecco's modified Eagle medium (DMEM) containing 0.5% fetal bovine serum (FBS) and unchanged antibiotic concentrations from those of typical culture conditions (100 U/ml penicillin, 100 μg/ml streptomycin).
Human nonimmune IgG (Jackson ImmunoResearch Laboratories) is added to cells at a final concentration of 1 μg/ml. Cells are incubated with these antibodies for 10 min at 37 °C.
Excess IgG is washed away three times with serum-free culture medium that has been prewarmed to 37 °C avoid thermal disruption.
The cells are left to incubate in serum-free medium for 30 min at 37 °C to allow shear-mediated signaling to return to baseline. In our experience, this does not result in significant downregulation of chimera-bound IgG at the cell membrane, and is critical for establishing an unstimulated baseline for various signaling pathways.
Clustering is initiated by addition of F(ab)2 fragments at a final concentration of 2-5 μg/ml while keeping cells at 37 °C. As a negative control, monovalent F(ab) fragments may be added at 4–10 μg/ml in place of an F(ab)2 incubation.
At determined time points, cells are placed on ice and lysed with lysis buffers appropriate for subsequent assay(s). For assay of Rho family GTPase activity (as described below), a time course of 15–20 min after clustering is recommended to detect the primary wave of activation (Elfenbein et al., 2009). Upon lysis, samples may be flash frozen in liquid nitrogen and stored at –80 °C for future assay, or run immediately.
8.3. Qualitative and quantitative endocytosis assays
Studies of proteoglycan internalization involve characterizing both the rate of endocytosis and pathway involved in transport. Several common principles apply to studies of both, and the following experimental methods detail such qualitative and quantitative approaches. RFPECs with stable expression of the described syndecan 4 Fc receptor chimera will again be used as a model system for these studies (Fig. 1.2).
For purposes of microscopy, cells are plated on glass-bottom dishes or glass coverslips (in either case precoated with an extracellular matrix component, such as fibronectin (10 μg/ml in PBS, coated at 37 °C for 30 min)). For cells to be analyzed by flow cytometry, matrix-coated 12-well or 24-well dishes may be used for plating. A confluence of 75–80% should not be exceeded, and cells are serum-starved in 0.5% FBS/DMEM with antibiotics (100 U/ml penicillin, 100 μml streptomycin) for 24 h prior to assay.
Chimera clustering is performed as described above, with two modifications. First, the 10 min incubation with prima1y antibody (IgG) and subsequent washes must be performed on ice in order to halt all endocytic processes. Addition of F(ab)2 fragments (chimera oligomerization and initiation of endocytosis) is performed at 37 °C. Second, either the primary antibody (IgG) or the clustering antibody fragment F(ab)2 must be conjugated to a fluorescent dye compatible with available microscope filters.
Endocytosis is allowed to proceed after chimera oligomerization for set time points, with optimal experiments containing several samples in a time course. Typical endocytic time points might span anywhere from 30 s to 30 min, depending upon the route of internalization. Following the completion of a time point, the sample is again placed on ice to arrest endocytosis.
In order to remove noninternalized antibody from the cell surface, a solution of ice-cold PBS titrated with HCl to a pH of 2.5 is added to the cells as a first wash. The cells are subsequently washed three times more with ice-cold standard PBS (pH = 7.4).
For samples to be used in microscopy analyses, a solution of 4% paraformaldehyde diluted in PBS is added to cells for fixation. The cells are then brought to room temperature and permitted to incubate for 10 min. At that time, the paraformaldehyde solution is replaced by PBS. Cells may then be mounted for future analysis, or visualized immediately by fluorescent microscopy. Endocytic vesicles may be quantified by imaging sampled cells, and their subcellular localization is most effectively determined by this technique. Vesicular characterization (with designations such as early endosome, recycling endosome, macropinosomes, etc.) may be accomplished with costaining using additional fluorescent dyes or conjugated antibodies. See notes below for preparation of samples for staining with additional markers.
For samples to be analyzed by flow cytomet1y, cells are removed from their dishes by addition of trypsin–EDTA (0.25%) in a volume of approximately 20–25 μl. Trypsin activity is slowed by transfer of cells to ice and addition of 150 μl of 1% BSA. The activity of trypsin may also optionally be quenched by the addition of trypsin soybean inhibitor. Cells are then either fixed with 4% parafonnaldehyde/PBS for 10 min, followed by centrifugation, resuspension in PBS, and storage at 4 °C until assay, or immediately analyzed for internalized fluorescence by flow cytometry. Appropriate negative controls for autofluorescence, including chimera clustering in the absence of fluorescent antibodies, is critical to establish baseline fluorescence.
Notes
Because of cellular sensitivity to excessive periods of exposure to 4 °C ambient temperatures and endocytic differences at various temperature ranges, experimental timing of each step should be strictly observed for the generation of reliably reproducible data.
In microscopic analysis and flow cytometry experiments, sample fixation with paraformaldehyde may in some cases diminish the fluorescence of dyes and fluorophores used for vesicular identification.
In the case of endogenous proteoglycan endocytosis, antibodies generated against extracellular epitopes may be used as above to visualize and/or characterize their internalization.
In order to stain cytoplasmic protein markers of endosomal stage or function, cells must first be permeabilized to allow diffusion of antibodies across the membranes toward their targets. This is performed after fixation with 4% paraformaldehyde, and after the subsequent PBS washes. Triton X-100 (0.1%) dissolved in PBS is added to samples for 10 min at room temperature, after which three successive washes with PBS are performed. Nonspecific sites are blocked with 5% BSA in PBS for 30 min, after which three more washes with PBS are again performed. Incubation with primary antibody ensues, with antibody dilution being dependent upon affinity specifications. This is performed in 1% BSA/PBS for 1 h, after which the primary antibody solution is washed three times with PBS. Secondary antibody incubation follows (also in 1% BSA/PBS), with three washes of PBS concluding the 1-h incubation. The cells are then visualized by microscopy.
Live-cell microscopy may be performed as above in a temperature-controlled stage environment to visualize proteoglycan endocytosis with greater temporal resolution than that provided with fixed samples. In these experiments, the addition of acidic PBS (pH = 2.5) is not advised. Instead, endocytic vesicles are differentiated from residual membrane-associated antibody fluorescence by morphologic features rather than by acidic removal of the latter.
8.4. Rac1 pull-down assay
Rho family GTPases function as molecular switches, alternating between GDP-bound inactivity and the active, GTP-bound form. Determination of Racl activity will be considered here to exemplifY the GTPase pull-down technique.
Rac1 activity is typically quantified as a ratio of the cellular fraction of active Rac1 to total expressed Rac1. Active Rac1 is isolated by exploiting the enhanced affinity it has for its effector, p21 activated kinase (PAK). Immobilized, PAK-binding domain (PBD) peptides are therefore used as bait to bind active (GTP-bound) Rac1, and to separate this protein pool from all other cellular proteins. The immobilized Rac1 PBD is purified as a GST-fusion construct (GST-PBD), and agarose glutathione beads are subsequently used for pull-down assay (del Pozo et al., 2000).
As described in the experimental procedures for syndecan 4 FcR chimera oligomerization, cells are grown to approximately 75–80% confluence. For the purpose of Rac1 activation assays, these are plated on 10 cm or larger plastic dishes coated with a given extracellular matrix component, such as fibronectin.
The cells are serum-starved in 0.5% FBS/DMEM with unchanged antibiotic concentrations for a minimum of 24 h. Although longer serum-starvation periods (and those of lower serum concentrations) result in a furthennore diminished baseline of Rac1 activity, the ability to tolerate such conditions is cell-type dependent and should be determined prior to assay. After stimulation (as with chimera clustering or growth factor addition), lysis is performed on ice using cell lifters/scrapers and the following lysis buffer: 50 mM Tris (pH = 7.4), 150 mM NaCl, 1 mM EDTA, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS). A substitution of 0.5–1% NP-40 for CHAPS may be made, although stronger detergents such as SDS are not recommended, as they are more likely to disrupt GTPase–bait binding interactions. The lysis buffer is also supplemented with a protease inhibitor cocktail, or addition of 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mM PMSF. The lysis buffer may also be supplemented with 1 U/ml benzonase nuclease (Novagen) to prevent DNA agglutination. A relatively large lysis volume is recommended (750–1000 ml) for each sample. Ten microliters of aliquots are then taken for protein concentration measurement prior to flash freezing of the sample for future assay. Protein concentration measurement may be performed by standard methods, such as Bradford, bicinchoninic acid (BCA), or Low1y protein assays. (3) Agarose glutathione beads with conjugated GST-PAK are prepared prior to thawing samples for assay. The frozen samples are thawed on ice, with corrections for concentration variabilities made with lysis buffer. Once the samples have thawed, they may be precleared with addition of agarose glutathione beads alone (50 μl slurry) and centrifugation for 2 min at 16,000 rpm in a microcentrifuge that is precooled to 4 °C. The supernatant is transferred to new sample tubes, also at 4 °C. At this point, a small, equal volume of each supernatant is removed for immunoblotting of total Rac1 expression (20 μl is sufficient for most cell types).
50 μl of glutathione agarose bead slurry and 50 μg of recombinant GST-PAK is added to each sample, and the tubes are subjected to gentle agitation or rotation for 20–30 min at 4 °C.
The beads are then centrifuged using a precooled (4 °C) microcentrifuge at 16,000 rpm for 2 min, and washed with ice-cold lysis buffer. This sequence of washing and centrifugation is performed sequentially three times. An equal volume of 2× sample buffer (final concentrations: 10% (v/v) glycerol, 62.5 mM Tris–HCl (pH = 6.8), 2% SDS, 0.01 mg/ml bromophenol blue, 5% (v /v) BME) is added to the beads and to the total protein samples taken in step 3. All samples are heated to 95 °C for 5 min, and either frozen at –80 °C for future electrophoresis or run on an acrylamide gel that is optimized for resolution of this 24 kDa protein (e.g., 12% acrylamide). Immunoblotting for Rac1 is performed as described in the above Western blotting protocol, with anti-Rac1 primary antibodies and horseradish peroxidase- or fluorescent dye-conjugated secondary antibodies used for detection of active and total Rac1 bands.
The ratios between active and total Rac1 for each sample are quantified using densitometry.
Notes
The intrinsic GTPase activity of Rac1 renders this assay particularly sensitive to time and temperature; upon thawing flash-frozen samples, all steps until the addition of sample buffer should be performed at 4 °C or on ice, and without excessive delays.
Alternative protocols based upon the enzyme-linked immunosorbent assay (ELISA) method have been developed, and may similarly be used to quantifY the activity of Rho family GTPases (Elfenbein et al., 2009; Zhou et al., 2007). These protocols also rely upon bait peptides to isolate pools of active (GTP-bound) enzyme within each sample, and have a lower requirement for total protein input.
Because pull-down methods measure total cellular pools of active GTPases, they are unable to aid in characterizing the spatial localization of these signaling events. For example, the activation of Rac1 exclusively at the leading edge of migrating cells, although comprising a small proportion of total Rac1 activity, is critical for effective directional migration. It is therefore advisable to conduct experiments complementary to pull-downs in the study of Rho GTPase signaling, including those involving live-cell imaging with GTPase-specific biosensors. The development, technical considerations, and applications of such biosensors have been extensively reviewed elsewhere (Ibraheem and Campbell, 2010).
8.5. Multicellular migration assay
This technique measures the distance that a migrating front of cells in a monolayer traverse in two dimensions over time.
Cells are plated in 12-well format and grown until reaching ~ 50% confluence, at which time they are serum-starved for a minimum of 24 h with 0.5% FBS in DMEM, supplemented with penicillin and 100 μg/ml streptomycin. Because high-power microscopy is not required for this technique, plastic dishes are acceptable for this assay. Dishes should be premarked with a grid or pattern of demarcated lines on the microscope objective side; this enables the accurate imaging of identical regions over time. Proteoglycan and integrin engagement of extracellular matrix components is a critical factor in migrations. For this reason, dishes should invariably be precoated with a known matrix substrate, such as fibronectin, collagen, or vitronectin.
Assay is optimally begun as cells approach 90–95% confluence, as overconfluent cells frequently exhibit diminished migrational capabilities. At the time of assay, a uniform linear disruption is created in the monolayer. This may be accomplished by using a sterile plastic pipette tip and scratching the dish surface in a linear fashion.
Upon formation of the monolayer discontinuity, the dish is imaged at each site marked by the underside grid. Attempts should be made to maximize the number of imaged fields, as this enhances the statistical robustness of migration measurements.
Cell stimulation or inhibition is performed, as with the addition of growth factor, pharmacologic agent, proteoglycan-oligomerizing antibody, or proteoglycan-blocking antibody. Cells are then returned to 37 °C. Reagents targeting integrins may also be used for these experiments in combination with those affecting proteoglycan function.
At determined time points, identical fields marked by the dish markings are imaged under low power. These regions should ideally register perfectly with those imaged in step 3. Optimal migration time points are cell type dependent. Ranges of 12–36 h typically yield reproducible results.
The distance migrated is quantified by either determining the linear distance traversed (difference between division edges) or the approximately columnar area invaded by migrating cell fronts. Either method is appropriate if identical regions are measured consistently, with a statistically meaningful number of data points.
8.6. Two-chambered migration assay (adhesion and migration)
In contrast to multicellular migration assays, the use of a two-chambered assay enables the measurement of single-cell adhesion and invasion through a given matrix. These devices, such as the Boyden chamber and its variants, contain an upper chamber into which cells are placed, and a lower chamber that often contains a chemoattractant or stimulatory molecule such as a clustering antibody. Between the two chambers is a porous membrane that permits cell migration, or a three-dimensional matrix such as Matrigel. The ability of cells to adhere to the membrane/matrix, or migrate from the top chamber to the bottom is quantified in this assay.
Cells are grown to approximately 75–80% confluence on plastic dishes precoated with specified matrix components, removed from their plates by treatment with trypsin–EDTA (0.25%), and their concentration is then determined by a cell counter.
An equal number of cells is added to the top chamber of the device described above, and cells are stimulated by addition of growth factor, pharmacological agent, or receptor-clustering antibody to the lower chamber. The effect of function-blocking antibodies (preventing ligand–proteoglycan interactions) may also be assayed as an experimental condition.
For adhesion studies, the supernatant is aspirated after a specified period of time, and the top chambers are washed three times sequentially with equal force, using PBS or culture medium. Cells adherent to the porous filter or matrix separating both chambers are then fixed by treatment with 4% paraformaldehyde in PBS for 10 min. They are stained with a dye such as crystal violet and quantified by low-power microscopy. Typically, a time course of 20–60 min is used for adhesion studies, although this is highly dependent upon cell type and matrix composition. For studies of cell migration, cells are transferred to the top well and stimulated as described above. At the specified migration endpoint, the upper chamber is aspirated and cells adhering to the upper surface of the membrane/matrix are removed with a cotton swab. Cells in the lower chamber are quantified by a cell counter. Alternatively, if migration has not occurred for long enough for complete invasion into the lower chamber, cells that have migrated through the filter/matrix and remain adherent to the lower aspect of this barrier may be stained with a dye (as above) and quantified by low-power microscopy.
Quantification is typically expressed as percent adherent or migrated, as compared to total cell number per sample.
Note
An alternative method of quantification involves the preincubation of cells with a fluorescent membrane dye. Upon conclusion of the migration assay, the fluorescence of the bottom chamber is measured with a fluorimeter. This value likewise correlates linearly with number of successfully migrated cells.
ACKNOWLEDGMENTS
This work was supported by the National Heart, Lung, and Blood Institute (grant HL62289 to M. Simons). The authors thank the members of the Simons laboratory and the Matsuda laboratory (Kyoto University, Japan) for their experimental guidance and expertise.
REFERENCES
- Addae-Mensah KA, Wikswo JP. Measurement techniques for cellular biomechanics in vitro. Exp. Biol. Med. (Maywood) 2008;233:792–809. doi: 10.3181/0710-MR-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiges-Rizo C, et al. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 2009;122:3037–3049. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini A, et al. The “chemoinvasion assay”: A tool to study tumor and endothelial cell invasion of basement membranes. Int. J. Dev. Biol. 2004;48:563–571. doi: 10.1387/ijdb.041822aa. [DOI] [PubMed] [Google Scholar]
- Alexopoulou AN, et al. Syndecans in wound healing, inflammation and vascular biology. Int. J. Biochem. Cell Biol. 2007;39:505–528. doi: 10.1016/j.biocel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Aoki K, Matsuda M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat. Protoc. 2009;4:1623–1631. doi: 10.1038/nprot.2009.175. [DOI] [PubMed] [Google Scholar]
- Ashikari-Hada S, et al. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Methods Enzymol. 2003;361:1–33. doi: 10.1016/s0076-6879(03)61003-7. [DOI] [PubMed] [Google Scholar]
- Bass MD, et al. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 2007;177:527–538. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais DM, et al. The syndecan-1 ectodomain regulates alphavbeta3 integrin activity in human mammary carcinoma cells. J. Cell Biol. 2004;167:171–181. doi: 10.1083/jcb.200404171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M, et al. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Chittenden TW, et al. Selective regulation of arterial branching morphogenesis by synectin. Dev. Cell. 2006;10:783–795. doi: 10.1016/j.devcel.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Couchman JR. Syndecans: Proteoglycan regulators of cell-surface microdomains? Nat. Rev. Mol. Cell Biol. 2003;4:926–937. doi: 10.1038/nrm1257. [DOI] [PubMed] [Google Scholar]
- Couchman JR, Tapanadechopone P. Detection of proteoglycan core proteins with glycosaminoglycan lyases and antibodies. Methods Mol. Biol. 2001;171:329–333. doi: 10.1385/1-59259-209-0:329. [DOI] [PubMed] [Google Scholar]
- del Pozo MA, et al. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 2000;19:2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss JL, et al. Heparan sulfate proteoglycans: Structure, protein interactions and cell signaling. An. Acad. Bras. Cienc. 2009;81:409–429. doi: 10.1590/s0001-37652009000300007. [DOI] [PubMed] [Google Scholar]
- Elfenbein A, et al. Suppression of RhoG activity is mediated by a syndecan 4-synectin-RhoGDI1 complex and is reversed by PKC{alpha} in a Rac1 activation pathway. J. Cell Biol. 2009;186:75–83. doi: 10.1083/jcb.200810179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari do Outeiro-Bernstein MA, et al. A recombinant NH(2)-terminal heparin-binding domain of the adhesive glycoprotein, thrombospondin-1, promotes endothelial tube formation and cell survival: A possible role for syndecan-4 proteoglycan. Matrix Biol. 2002;21:311–324. doi: 10.1016/s0945-053x(02)00010-0. [DOI] [PubMed] [Google Scholar]
- Florian JA, et al. Heparan sulfate proteoglycan is a mechanosensoron endothelial cells. Circ. Res. 2003;93:e136–e142. doi: 10.1161/01.RES.0000101744.47866.D5. [DOI] [PubMed] [Google Scholar]
- Gopal S, et al. Heparan sulfate chain valency controls syndecan-4 function in cell adhesion. J. Biol. Chem. 2010;285:14247–14258. doi: 10.1074/jbc.M109.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovoy M, et al. RhoGDI-1 modulation of the activity of monomeric RhoGTPase RhoA regulates endothelial barrier function in mouse lungs. Circ. Res. 2007;101:50–58. doi: 10.1161/CIRCRESAHA.106.145847. [DOI] [PubMed] [Google Scholar]
- Greene DK, et al. Syndecan-4 associates with alpha-actinin. J. Biol. Chem. 2003;278:7617–7623. doi: 10.1074/jbc.M207123200. [DOI] [PubMed] [Google Scholar]
- Grünert M, et al. Stem cell fate decisions: The role of heparan sulfate in the control of autocrine and paracrine signals. Curr. Stem Cell Res. Ther. 2008;3:1–8. doi: 10.2174/157488808783489417. [DOI] [PubMed] [Google Scholar]
- Guimond SE, Turnbull JE. Fibroblast growth factor receptor signalling is dictated by specific heparan sulphate saccharides. Curr. Biol. 1999;9:1343–1346. doi: 10.1016/s0960-9822(00)80060-3. [DOI] [PubMed] [Google Scholar]
- Harris AK, et al. Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- Hayashida K, et al. Syndecan-1 ectodomain shedding is regulated by the small GTPase Rab5. J. Biol. Chem. 2008;283:35435–35444. doi: 10.1074/jbc.M804172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JD, Cheresh DA. Role ofin:tegrins in cell invasion and migration. Nat. Rev. Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Ibraheem A, Campbell RE. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010;14:30–36. doi: 10.1016/j.cbpa.2009.09.033. [DOI] [PubMed] [Google Scholar]
- Kirn-Safran C, et al. Multifunctionality of extracellular and cell surface heparan sulfate proteoglycans. Cell. Mol. Life Sci. 2009;66:3421–3434. doi: 10.1007/s00018-009-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellén L, Lindahl U. Proteoglycans: Structures and interactions. Annu. Rev. Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- Kobialka S, et al. Glycosaminoglycan chains affect exocytic and endocytic protein traffic. Traffic. 2009;10:1845–1855. doi: 10.1111/j.1600-0854.2009.00987.x. [DOI] [PubMed] [Google Scholar]
- Kreuger J, et al. Interactions between heparan sulfate and proteins: The concept of specificity. J. Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KA, et al. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- Liotta LA, et al. New method for preparing large surfaces of intact human basement membrane for tumor invasion studies. Cancer Lett. 1980;11:141–152. doi: 10.1016/0304-3835(80)90105-6. [DOI] [PubMed] [Google Scholar]
- Longley RL, et al. Control of morphology, cytoskeleton and migration by syndecan-4. J. Cell Sci. 1999;112(Pt 20):3421–3431. doi: 10.1242/jcs.112.20.3421. [DOI] [PubMed] [Google Scholar]
- Malinda KM. In vivo matrigel migration and angiogenesis assay. Methods Mol. Biol. 2009;467:287–294. doi: 10.1007/978-1-59745-241-0_17. [DOI] [PubMed] [Google Scholar]
- Matthews H, et al. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development. 2008;135:1771–1780. doi: 10.1242/dev.017350. [DOI] [PubMed] [Google Scholar]
- Mochizuki S, et al. Role of hyaluronic acid glycosaminoglycans in shear-induced endothelium-derived nitric oxide release. Am. J. Physiol. Heart Circ. Physioi. 2003;285:H722–H726. doi: 10.1152/ajpheart.00691.2002. [DOI] [PubMed] [Google Scholar]
- Moon JJ, et al. Role of cell surface heparan sulfate proteoglycans in endothelial cell migration and mechanotransduction. J. Cell. Physiol. 2005;203:166–176. doi: 10.1002/jcp.20220. [DOI] [PubMed] [Google Scholar]
- Morgan MR, et al. Synergistic control of cell adhesion by integrins and syndecans. Nat. Rev. Mol. Cell Biol. 2007;8:957–969. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, et al. Derailed endocytosis: An emerging feature of cancer. Nat. Rev. Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- Munevar S, et al. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys. J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, et al. Non-canonical fibroblast growth factor signaling in angiogenesis. Cardiovasc. Res. 2008;78:223–231. doi: 10.1093/cvr/cvm086. [DOI] [PubMed] [Google Scholar]
- Mythreye K, Blobe GC. Proteoglycan signaling co-receptors: Roles in cell adhesion, migration and invasion. Cell. Signal. 2009;21:1548–1558. doi: 10.1016/j.cellsig.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache SN, et al. Binding of internalized receptors to the PDZ domain of GIPC/synectin recruits myosin VI to endocytic vesicles. Proc. Natl. Acad. Sci. USA. 2006;103:12735–12740. doi: 10.1073/pnas.0605317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, et al. Analysis of the spatiotemporal activation of rho GTPases using Raichu probes. Methods Enzymol. 2006;406:315–332. doi: 10.1016/S0076-6879(06)06023-X. [DOI] [PubMed] [Google Scholar]
- Nugent MA, Iozzo RV. Fibroblast growth factor-2. Int. J. Biochem. Cell Biol. 2000;32:115–120. doi: 10.1016/s1357-2725(99)00123-5. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, et al. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell. Biol. 1992;12:240–247. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R, et al. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovian C, et al. Syndecan-4 regulates subcellular localization of mTOR Complex2 and Akt activation in a PKCalpha-dependent manner in endothelial cells. Mol. Cell. 2008;32:140–149. doi: 10.1016/j.molcel.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge MA, Marcantonio EE. Initiation of attachment and generation of mature focal adhesions by integrin-containing filopodia in cell spreading. Mol. Biol. Cell. 2006;17:4237–4248. doi: 10.1091/mbc.E06-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce ML. Tube formation: An in vitro matrigel angiogenesis assay. Methods Mol. Biol. 2009;467:183–188. doi: 10.1007/978-1-59745-241-0_10. [DOI] [PubMed] [Google Scholar]
- Rauch U, et al. Neurocan: A brain chondroitin sulfate proteoglycan. Cell. Mol. Life Sci. 2001;58:1842–1856. doi: 10.1007/PL00000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksela O, et al. Endothelial cell-derived heparan sulfate binds basic fibroblast growth factor and protects it from proteolytic degradation. J. Cell Biol. 1988;107:743–751. doi: 10.1083/jcb.107.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Schaefer RM. Proteoglycans: From structural compounds to signaling molecules. Cell Tissue Res. 2010;339:237–246. doi: 10.1007/s00441-009-0821-y. [DOI] [PubMed] [Google Scholar]
- Tkachenko E, Simons M. Clustering induces redistribution of syndecan-4 core protein into raft membrane domains. J. Biol. Chem. 2002;277:19946–19951. doi: 10.1074/jbc.M200841200. [DOI] [PubMed] [Google Scholar]
- Tkachenko E, et al. Fibroblast growth factor 2 endocytosis in endothelial cells proceed via syndecan-4-dependent activation of Rac1 and a Cdc42-dependent macro-pinocytic pathway. J. Cell Sci. 2004;117:3189–3199. doi: 10.1242/jcs.01190. [DOI] [PubMed] [Google Scholar]
- Tkachenko E, et al. Syndecans: New kids on the signaling block. Circ. Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- Tkachenko E, et al. Syndecan-4 clustering induces cell migration in a PDZ-dependent manner. Circ. Res. 2006;98:1398–1404. doi: 10.1161/01.RES.0000225283.71490.5a. [DOI] [PubMed] [Google Scholar]
- Tolosa E, Shaw S. A fluorogenic assay of endogenous phosphatase for assessment of cell adhesion. J. Immunol. Methods. 1996;192:165–172. doi: 10.1016/0022-1759(96)00042-7. [DOI] [PubMed] [Google Scholar]
- Turley EA. Proteoglycans and cell adhesion. Their putative role during tumor-igenesis. Cancer Metastasis Rev. 1984;3:325–339. doi: 10.1007/BF00051458. [DOI] [PubMed] [Google Scholar]
- Valdembri D, et al. Neuropilin-1/GIPC1 signaling regulates alpha5beta1 integrin traffic and function in endothelial cells. PLoS Biol. 2009;7:e25. doi: 10.1371/journal.pbio.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A. Function and interactions of integrins. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, et al. Cell migration at a glance. J. Cell Sci. 2005;118:4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- Yayon A, et al. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc. Natl. Acad. Sci. USA. 2007;104:3919–3924. doi: 10.1073/pnas.0608360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicha D, et al. A new direct-viewing chemotaxis chamber. J. Cell Sci. 1991;99(Pt 4):769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Orientation chamber in chemotaxis. Methods Enzymol. 1988;162:65–72. doi: 10.1016/0076-6879(88)62064-7. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, et al. Syndecan recycling [corrected] is controlled by syntenin-PIP2 interaction and Arf6. Dev. Cell. 2005;9:377–388. doi: 10.1016/j.devcel.2005.07.011. [DOI] [PubMed] [Google Scholar]