1. Introduction
Lipids spontaneously form bilayered structures when brought into an aqueous environment. This is the foundation in the architecture of biological cell membranes. However, lipid bilayers do not lend themselves easily to common biophysical studies; be it of the bilayer itself or of embedded membrane proteins. Detergents, on the other hand, form small aggregates known as micelles that readily solubilize membrane proteins and are well-suited for numerous biophysical methods. However, they are not excellent models of biological membranes as they may denature the structure of a protein and the curvature of the micelle may impose a non-native protein folding. When lipid and detergent meet in an aqueous environment, entities with wholly different properties are formed: lipid bicelles. Bicelles are made of patches of lipid bilayers that are either encircled or perforated by detergent ‘rims’. They combine the advantages of both components alone (micelle and lipid bilayer), namely being good models for a biological membrane and having advantageous properties for biophysical experiments. An additional advantage of certain bicelle preparations is their tendency to macroscopically align when brought into a magnetic field. This fact has been exploited not only in the highresolution structural and dynamics studies of membrane proteins, but also for globular proteins using nuclear magnetic resonance (NMR) experiments.
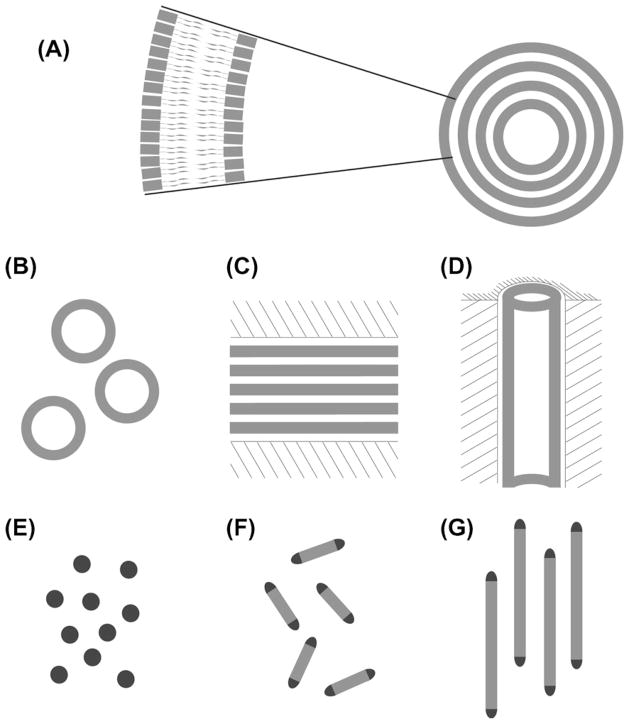
Fig. 1 gives a graphical introduction to the two types of bicellar phases most commonly employed. At a high detergent concentration and low temperatures, isotropically tumbling disk-like aggregates are formed, the so-called isotropic bicelles (Fig. 1B). At a high lipid concentration and in certain temperature ranges, extended bilayered lamellae are formed that are perforated or delimited by detergent, and have the potential for magnetic alignment (Fig. 1D). Cryo-transmission electron microscopy (TEM) micrographs (A, C) of bicelles taken from the literature [1] are also included in Fig. 1.
Fig. 1.
Lipid bicelles are supramolecular aggregates that are formed when appropriate amounts of lipids and detergents are mixed in an aqueous environment. The size and phase of bicellar aggregates depend on the [lipid]:[detergent] ratio as well as on the temperature. Two fundamentally different phases of bicellar preparations have proven highly useful in the study of protein structure using NMR spectroscopy: isotropic bicelles rapidly tumble freely and are formed at a high detergent concentration (A and B). At low detergent concentrations extended bilayered lamellae are formed (C and D), that spontaneously align macroscopically in a magnetic field. Cryo-TEM micrographs (A and C) are reproduced from the literature [1]. Micrograph (A) contains arrows marked A and B that point to disk-like bicelles viewed from the side and the top, respectively.
Since their first description in 1988, the great potential of bicelles in the study of membrane proteins and proteins in general has been realized. A steady stream of remarkable insights and applications has emerged that is still growing in size. In the present contribution, we will give an introduction to the properties of lipid bicelle phases with an emphasis on NMR experimental measurements. In addition, we will discuss some of the most exciting recent applications of bicelles in the structural and dynamic studies of membrane proteins.
2. Different types of model membranes used in NMR studies
2.1. Vesicles
Lipid membranes and membrane proteins have been investigated by NMR spectroscopy for more than 40 years. Numerous types of membrane samples and preparation protocols have been developed. An overview of the most popular ones is depicted in Fig. 2. The choice of a certain type of sample depends on the task in hand. The simplest type of lipid bilayer sample is formed spontaneously when pure lipids are mixed with a buffer. In this case, multilamellar vesicles (MLVs) are formed, which are approximately spherical aggregates up to tens or thousands of μm in diameter where large numbers of lipid bilayers are stacked in the fashion of an onion. Fig. 2A gives a simple schematic idea. By means of sonication, or by extrusion through the pores of suitable membrane filters, MLV samples can be converted into small unilamellar vesicles (SUVs, Fig. 2B) made up of small spheres consisting of only a single lipid bilayer. The size or size distribution of SUVs is governed by the preparation method employed and is usually much more homogeneous when filter extrusion is employed [2–4].
Fig. 2.
Different types of model membrane samples suitable for studying membrane proteins by NMR spectroscopy: (A) lipid bilayers (inset) are present in multilamellar vesicles; (B) small unilamellar vesicles and macroscopically-aligned bilayer samples using either (C) glass plates or (D) cylindrical nanopores in anodic aluminum oxide. (E) Micelles are formed from pure detergents. When lipid and detergent mix, isotropic bicelles (F) and magnetically-aligned bicelles (G) are formed. Light gray color indicates a lipid bilayer, dark gray is detergent, and hatching represents either a glass plate (C) or aluminum oxide (D) as solid supporting material.
For use in conventional NMR spectroscopy, vesicle samples have a drawback: they do not reorient fast on the NMR time scale, hence the anisotropic NMR interactions (chemical shift anisotropy, dipolar coupling, quadrupolar coupling) dominate the spectra. This is in stark contrast to systems usually investigated in solution-state NMR spectroscopy, where fast molecular reorientation makes anisotropic interactions collapse to an average isotropic value. A situation of fast isotropic tumbling can be recreated in detergent micelles (Fig. 2E) which do not form bilayers and, hence, give unreliable environments for mimicking membrane conditions and may not always preserve the membrane protein structure and function. Alternatively, anisotropic NMR interactions can be suppressed by rapid spinning of a vesicle sample around a certain angle with respect to the external magnetic field. This angle (54°74′) is dubbed as the ‘magic angle’ in solid-state NMR, and magic-angle spinning (MAS) is one of the most commonly used approaches in solid-state NMR today. MAS techniques are also used in the specialized area of membrane proteins in vesicle samples, where MAS has been successfully applied for a long time. MAS at suitable intermediate speeds can also be used to determine the tensorial quantities of anisotropic NMR interactions by analysis of spinning sidebands [5]. Since the tensors of anisotropic NMR interactions observed in peptides or proteins embedded in lipid bilayers are dependent on molecular alignment, information on the global orientation of such molecules can be extracted from these tensors [6,7].
2.2. Mechanically-aligned lipid bilayers
In an alternative experimental approach, anisotropic interactions are not suppressed but are put to good use. By a number of preparation protocols it is possible to generate macroscopically oriented samples of lipid bilayers. In a well-oriented sample, all lipid bilayers are parallel to each other and enclose a single, well-defined angle between their bilayer normal and the external magnetic field of an NMR spectrometer. As a result, proteins or peptides embedded in the lipid bilayers are also partially oriented with respect to the external magnetic field. The result is a dramatically improved spectral resolution, and the possibility to infer geometrical information from the observed values of anisotropic spin interactions [8]. Most commonly, anisotropic interactions are thus used for angle measurements and therefore allow imaging molecules at atomic-level resolution.
The conceptually most straightforward preparation method for macroscopically oriented lipid bilayer samples involves stacks of glass plates. The space between two adjacent glass plates in the stack is filled with the lipid preparation of interest [9]. The lipid preparation is applied to the glass plate in dehydrated form and is then hydrated in an environment of suitable temperature and humidity. Relative humidity is controlled over saturated solutions of a salt [10]. During the hydration process, the lipids form well-hydrated bilayers that orient parallel to the glass plates of the stack. Since thousands of lipid bilayers are typically enclosed between each pair of glass plates, the data measured in NMR spectroscopic experiments are not influenced by the glass plates that provide the mechanical support. A (smaller) number of lipid bilayers oriented between two glass plates is shown schematically in Fig. 2C. Initial studies on a membrane-embedded fd coat protein [11], gramicidin A [12,13], and the retinal of bacteriorhodopsin [14] have established the usefulness of the technique. The Ramamoorthy laboratory has made numerous contributions to the development and application of macroscopically oriented glass plate samples. It was shown that the preparation is feasible over a wide range of temperatures [15] and that the quality of orientation can be decisively improved by including sublimable solids, such as naphthalene or para-dichlorobenzene, in the preparation process [16]. The mechanism of membrane disruption by antimicrobial peptides has been investigated in stacked glass plate samples [17–20], as well as the action of cell signaling peptides [21] and the membrane interaction of myelin basic protein [22].
2.3. Anodic aluminum oxide nanodisks
Anodic aluminum oxide (AAO) is another viable support material for macroscopic alignment. AAO is a porous material that is perforated by highly parallel hollow cylinders ranging in diameter between several nm and several hundred nm [23]. It is commercially available in flat disks of approximately 60 μm height commonly used as a filter material [24], but is equally suitable as an orienting medium for bilayer samples. A fully hydrated liposome preparation can be applied to an AAO disk using an ordinary pipette. Upon contact with the disk material, the lipid immediately covers the surface of the pores, giving macroscopically oriented cylinders or ‘nanotubes’ of lipid bilayers which may contain embedded or attached protein or peptide (Fig. 2D) [25]. The geometrical and dynamic properties of lipid bilayers in AAO have been carefully characterized [26,27]; the latter contribution includes a detailed description of the preparation protocol.
The incorporation of the transmembrane (TM) domain TM-A of the membrane protein acetylenase CREP-1 was a first proof that integral membrane proteins can be studied in AAO-immobilized lipid bilayers [28]. Similarly, a transmembrane domain of the M2 protein from influenza virus was investigated in lipid nanotubes, including the use of two-dimensional 1H-15N-PISEMA spectroscopy [24]. This contribution also discusses the high conductivity of AAO for heat, which may potentially be advantageous in dissipating the heating effects of sophisticated NMR pulse sequences. Rhodopsin, a full-length integral membrane protein with seven transmembrane α-helices, has been successfully aligned in nanotubes [29]. Most remarkably, it was found that rhodopsin retains its binding affinity for G protein in the nanotube environment. In the Ramamoorthy laboratory, immediate alignment in AAO was employed to study membrane damage caused by amyloid-forming peptides [30]. Such a study is not possible using the glass plate samples where the peptide aggregation is much faster than the hydration step necessary in preparing stacked glass plate samples.
Other innovative media examined for the incorporation of membrane proteins include lipoprotein particles [31–34] and amphipols [35].
2.4. Bicelles
Bicelles, the topic of this review article, are a type of lipid sample that combines the advantages of most of the sample preparations introduced so far. Bicelles are formed when bilayer-forming long-chain lipids are mixed with detergent molecules. Under certain conditions, the two components mix to form aggregates, but are spatially separated into a central portion forming an actual lipid bilayer, surrounded or interspersed with ‘rims’ of detergent molecules. Fig. 2F shows the bilayer patch in light gray and the detergent rims in dark gray. At low concentration of a long-chain lipid, the aggregates tumble at a rate that is fast on the NMR time scale, almost comparable to the tumbling rate of detergent micelles (Fig. 2E). In addition, they offer a bilayer environment for embedded membrane proteins, making them a far more realistic membrane mimetic than detergent micelles. The term ‘bicelles’ was coined to denote such bilayer-containing mixed micelle-like aggregates, and the fact that diacylglycerol kinase retains enzymatic activity in bicelles but not in micelles was a proof of their advantageous properties [36].
When the concentration of a long-chain lipid is increased beyond a certain threshold, another advantageous property of bicelles is observed: they assume a well-defined orientation with respect to the magnetic field, see Fig. 2G for a graphic representation. This behavior is rooted in the geometric shape and the anisotropic magnetic susceptibility of bicelles. Hence, macroscopically ordered bicelle samples can be prepared with a degree of alignment that is typically much higher than what is possible in glass plate samples (Fig. 2C) and anodic aluminum oxide material (Fig. 2D). Magnetically-aligned bicelles are often used in solid-state NMR experiments, but can easily be converted into the isotropic bicelles introduced above, which are more amenable for solution-state NMR experiments. Hence, lipid bicelles form an important bridge linking solid- and solution-state NMR approaches. Today, lipid bicelles are a frequently utilized tool in NMR studies of protein structure in both soluble and membrane-bound proteins [8,150,172,236].
It needs to be mentioned that a suitable choice of lipid environment is equally important in crystallization assays of membrane proteins [37,38], and that bicellar lipid samples have also found application in this field. In particular, crystallization of bacteriorhodopsin (bR) from bicelle preparations at 37 °C yielded a novel monomeric structure for bR [39]. A similar result was later obtained at room temperature using different bicelle formulations [40]. For bR, crystallized from bicelle formulations, characteristic properties in optical spectroscopy were reported [41]. A mutant of bR was crystallized from bicelles to investigate structural differences of the proton pump bR and the homologous phototaxis receptor sensory rhodopsin II [42]. LeuT, a neurotransmitter sodium symporter, was crystallized in different crystal forms from bicelles in the presence of a substrate molecule [43]. Membrane protein crystallization using bicelles has recently received an exclusive review that reports a list of protein structures solved by bicelle crystallization [44]. A step-by-step protocol for using bicelles in high-throughput crystallization assays is available in video form [45].
Bicelle formation and morphology can be rationalized by the overall geometry of the involved molecules, depicted in Fig. 3. All involved molecules have hydrophilic head groups and hydrophobic acyl chains. In detergent molecules, on the one hand, the hydrophobic portion tends to be small when compared to the headgroup, giving the detergent molecule a conical overall shape (Fig. 3A). This geometry explains the tendency for detergents to form micelles in an aqueous environment (Fig. 2E). In lipid molecules, on the other hand, the number and size of the acyl chains tend to be larger giving the molecule a cylindrical overall shape (Fig. 3B), and resulting in the formation of bilayered structures in aqueous environments (Fig. 2A). The difference in overall shape results in a low miscibility of lipid and detergent. Hence, lipid and detergent phase-separate into bilayer and rim portions, which causes the formation of flat bilayer patches, i.e. bicelles.
Fig. 3.
The overall geometrical shape of detergents tends to be conical (A), while phospholipid molecules have mostly cylindrical overall geometry (B). The geometry of pegylated phospholipids again tends to be conical (C).
A similar conical molecular geometry results when a polyethyleneglycol (PEG) strand is attached to the head groups of a lipid (Fig. 3C). PEGylated lipids insert readily into lipid bilayers. Mixed micelles of lipid and PEGylated lipid are formed at very high concentration of PEGylated lipids. In between both regimes, the formation of flat bilayer disks similar to isotropic bicelles was observed in mixtures of cholesterol, lipids and PEGylated lipids [46]. These discoidal aggregates do not show a propensity for magnetic- alignment and are not referred to as bicelles; no specific name has been introduced for them. Discoidal aggregates can be prepared without cholesterol [47,48]. Their phase behavior has been characterized [49,50]. The influence of the preparation path [51] and the effect of phospholipid hydrolysis [52] on aggregate structure have been studied. Special fluorophores were developed to study the highly curved regions of disks formed by dipalmitoylphosphatidylcholine (DPPC) and PEGylated distearoylposphatidylethanolamine (DSPE-PEG2000) disks by fluorescence resonance energy transfer [53]. Discoidal assemblies of PEGylated lipids have been investigated in a coarse-grained molecular dynamics simulation [54]. Biotin-derivatized discoidal aggregates can be used as model membrane biosensors in surface plasmon resonance [55]. A potential pharmaceutical application has been demonstrated for PEGylated discoidal aggregates, since tight binding of comparably large quantities of melittin to the rim of the stable and well-defined PEG-stabilized disks may be exploited for drug delivery purposes [56]. Similar assemblies were observed when PEGylated lipids were replaced by the more common detergents octaethylene glycol monododecyl ether (C12E8), hexadecyltrimethylammonium bromide (CTAB), and sodium dodecyl sulfate (SDS) [57]. In a similar approach, the lipid dimyristoylphosphatidylethanolamine (DMPE) extended by a diethylenetriaminepentaacetate (DTPA) group chelating a paramagnetic Tm3+ or La3+ lanthanide ion was used instead of PEGylated lipids. Formation of flat discoidal aggregates was observed, which in the case of Tm3+-doping were slightly orientable in a field of 8 T [58]. The presence of 16 mol% cholesterol in discoidal aggregates of DMPC, DMPE-DTPA, and Tm3+ increases the disk size and the propensity for magnetic-alignment [318]. Linear peptide copolymers with hydrophilic and hydrophobic portions of different length have been designed that assemble into vesicle-like structures-dubbed as ‘polymersomes’-and bicelle-like flat discoidal structures [59].
3. What are bicelles?
3.1. General description
Bicelles are formed when long-chain lipids are brought in contact with detergent molecules. Long-chain lipids alone form lipid bilayers, while detergent molecules on their own form detergent micelles. When they are mixed, lyotropic mesophases are observed that combine the properties of both bilayers and micelles. In the simplest case, such a bicellar phase is made up of disk-like aggregates where a central bilayer patch is enclosed by a ‘rim’ of detergent molecules. However, this simple picture does not apply to all bicellar phases, see Section 5 on bicelle morphology. We want to stress three general traits of the bicelle preparations that are in general use today. First, they contain lipid bilayers with very similar properties as found in biological membranes. Second, these bilayers form flat patches rather than having a more or less pronounced curvature found in vesicles. Third, they can potentially be macroscopically aligned by an external magnetic field. This last trait is especially relevant for NMR studies.
3.2. Some landmarks in the development of bicelles
There are numerous systems that can be seen as bicelle precursors. For example, mixtures of sodium decyl sulfate, decanol, sodium sulfate, and water, form disk-like aggregates that orient in an external magnetic field [60]. Numerous other mixtures of hydrophobic, hydrophilic, and amphipathic molecules show similar behavior that has been extensively investigated in the context of lyotropic liquid crystals [61,62]. However, these systems do not contain the lipids that are the constituents of biological membranes. At high water content of at least 80 wt.%, lipid vesicles can be macroscopically ordered by a strong magnetic field [63]. However, being vesicles, they do not contain flat bilayers. The same objection is true for small unilamellar vesicles that form spontaneously when long- and short-chain phospholipids are mixed in certain conditions. (Obviously, these conditions are different from the conditions favoring the formation of bicelles.) Spontaneously formed short-chain long-chain unilamellar vesicles (SLUVs) have been introduced by Gabriel and Roberts [64] and have non-spherical shapes [65]. SLUVs are still of considerable interest today, and recent contributions have investigated the relationship between SLUVs and bicelles [67,68]. SLUV and bicellar phases can be inter-converted, and the interconversion can be used to generate small unilamellar vesicles of path-dependent size distribution and characteristic shape [68]. Kinetically trapped unilamellar vesicles of uniform size were prepared by passing through a metastable bicellar phase at 10 °C [69].
Coexistence of spherical and disk-like aggregates has been found in mixtures of bile salt and phospholipids [70], and in 1988 magnetic field induced order was reported for such mixtures [71]. This result by the Prestegard laboratory was the first to meet all three criteria for bicelles that were postulated in the preceding section. A bile salt analog, 3-(cholamidopropyl)dimethylammonio-2-hydroxy-1-propanesulfonate (CHAPSO), forms similar bicellar phases with lipids and gives advantageous experimental properties [72]. Lipid/CHAPSO complexes were structurally characterized [73] and used to study the conformation of glyco- and sulfo- lipids and their interaction with membrane binding proteins [74–76]. In 1992, it was demonstrated by the Sanders laboratory that the short-chain phospholipid dihexanoylphosphatidylcholine (DHPC) can be used to replace CHAPSO, eliminating the need for a potentially disadvantageous non-lipid detergent [77]. In the same contribution it was shown that long-chain phospholipids and DHPC in bicelles are spatially separated, presumably into bilayered patches and detergent rims. A first comprehensive review on bicelles, especially with regard to their potential in the study of membrane proteins, was published in 1994 [78] and has since been cited by most studies employing bicelles. Remarkably, the term “bicelles” was not introduced until a year later [36] to denote “binary, bilayered, mixed micelles bearing a resemblance to the classical model for bile-salt phosphatidylcholine aggregates (see Müller 1981)”, citing the article that is reference [70] in the current contribution.
3.3. Bicelle preparation
Most preparation protocols for bicelle samples are straightforward, highly reproducible, and do not require much time or effort. Protocols usually start with mixing of long-chain and short-chain lipids. The most popular choice is dimyristoyl-phosphatidylcholine (DMPC) as a long-chain lipid and dihexanoyl-phosphatidylcholine (DHPC) as a short-chain lipid (or detergent) component. Section 3.4 below gives an overview of other choices. Care must be taken to completely remove any residual organic solvent from the ingredients. The dry lipid-detergent is hydrated by adding a suitable amount of buffer, and after several cycles of cooling/heating (or freeze/thawing, if possible) the bicelle sample should be ready. It is also possible, but less convenient and common, to add detergent to pre-formed vesicular samples, giving the option for a titration with a short-chain lipid component. Preparation protocols have been reviewed [79,4], and Mäler and Gräslund [4] include a comparison to protocols to prepare MLVs and SUVs.
At least two parameters are necessary to describe the composition of bicelles and are needed to establish phase diagrams. The first one is the molar ratio of lipid molecules over detergent molecules. This ratio is usually denoted with the letter q. In the most common case of DMPC as a lipid component and DHPC as a detergent, it is
It has to be noted that q does not reflect the microscopic ratio in bicellar aggregates with full accuracy. That is because detergents typically have a noticeable solubility in buffer, better known as the critical micelle concentration (CMC). The CMC of DHPC is around 7 mM. Since the presence of ‘free’ detergent in micelles may reduce the amount of detergent available for bicelle formation, an ‘effective-q′-ratio was suggested to take this effect into account [80]. Similarly, a corrected ratio q* was used to describe the effect of non-negligible amounts of short-chain phospholipid invading the bilayer fraction [81]. The second important parameter is the level of hydration. It is usually given as the ratio between the added lipid and detergent weight with respect to the total weight (or volume) of the sample.
If the ratio of lipid over detergent is raised above a certain threshold, macroscopic alignment of the bilayer patches in a strong external magnetic field can be observed (Fig. 4). Most commonly, the normal of the bilayer patches aligns perpendicularly with respect to the external magnetic field. See Section 3.4 for modifications of this behavior of bicelles. Magnetic field-induced macroscopic alignment is observed for a magnetic field strength above about 1 T [82], which is well below the field strength in modern NMR spectrometers. It has been demonstrated that alignment can be achieved in a weaker field of 0.63 T as used in X-band EPR spectroscopy [83]. Alignment is possible even without an external magnetic field, as demonstrated by shear forces in a Couette flow cell, enabling linear dichroism experiments on embedded molecules [84]. It was shown that the presence of protein, namely the antimicrobial peptide gramicidin A, changes the alignment as a function of q, probably because the embedded protein increases the area of the bilayer patch [85]. Magnetic-alignment can remain present for days after the removal of the external magnetic field [86]. Bicelles with a high concentration of a long-chain lipid or a low hydration level tend to be very viscous. However, they become much more fluid at lower temperatures, which can be used for easy handling using common pipettes.
Fig. 4.
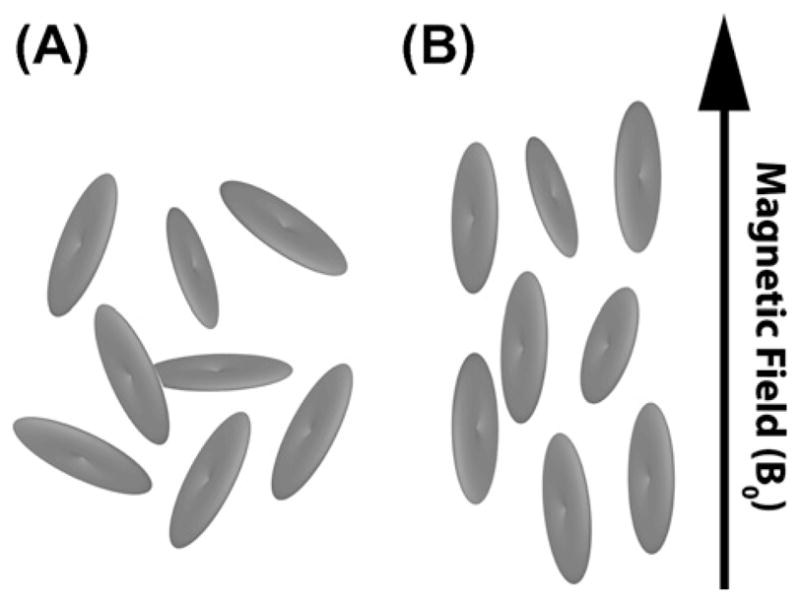
(A) In the absence of a magnetic field, bicelles assume random orientations. (B) Anisotropy in magnetic susceptibility causes macroscopic alignment of bicelles when an external magnetic field is applied.
3.4. Popular bicelle modifications
Numerous other constituents have been used to make bicelles different from the most common choice, DMPC and DHPC. Most bicelle preparations that show magnetic field-induced alignment are oriented with the bilayer normal perpendicular to the applied magnetic field (Figs. 4B and 5B). In a number of cases, a parallel alignment would be better since it can give increased spectral resolution in NMR spectra. Paramagnetic lanthanide ions, especially ytterbium ions, Yb3+, were found to bind to lipid bilayers and reverse the sign of the anisotropy in their magnetic susceptibility. This results in bicelles with parallel magnetic-alignment, often called “flipped” bicelles (Fig. 5A) [87,88]. Lipid-bound chelating agents can sequester the lanthanide ions and protect embedded proteins from possible disadvantageous effects [89]. By means of the chelating agent DTPA attached to DMPE lipids, it was possible to dope bicelles with Cu2+ ions [90]. The paramagnetic ions greatly increase spin–lattice relaxation, thus reducing T1 by a factor of 10 and speeding up NMR experiments accordingly. On the other hand, no significant change in the line width was observed suggesting that the change in the spin–spin relaxation is negligible. Using relaxation enhancement by Cu2+ ions, it was possible to record SO-FAST- HMQC spectra within 1 h from a bicelle-embedded antimicrobial peptide that had no isotopic labeling [91]. A special lipid was synthesised that carries a biphenyl group in one of its acyl chains. It forms bicellar phases over a wide range of temperatures, but only for a fairly limited range of q-values. Because the biphenyl group causes the anisotropy of magnetic susceptibility to change its sign, these bicelles have their bilayer normals oriented in parallel to the magnetic field without the need for added lanthanide ions [92,93].
Fig. 5.
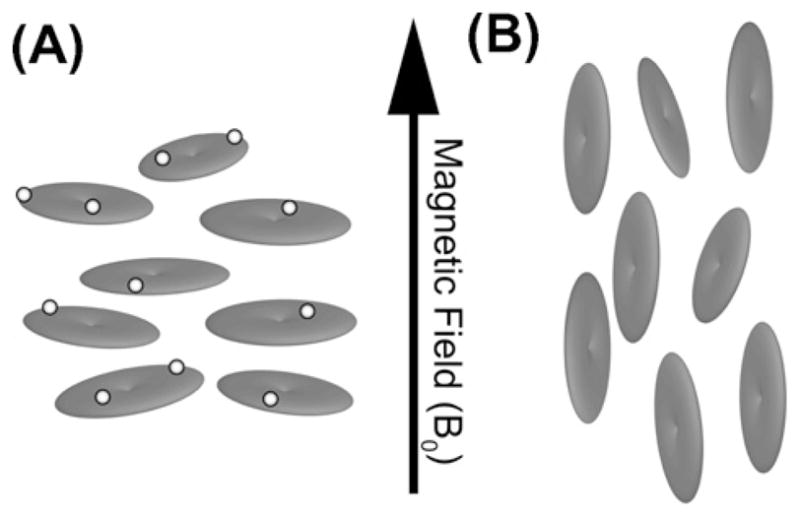
In the presence of lanthanide ions (light spheres), lipid bicelles adopt an orientation where the bilayer normal is parallel to the applied magnetic field (A). This is termed as “flipped” bicelle with respect to the perpendicular orientation adopted by undoped bicelles (B).
Bicellar phases change with the presence of ions and their concentration, with cations having the stronger effect [94]. Lipids with more stable ether-linkages between their acyl chains and the glycerol backbone have been used to increase the long-term stability of bicelles [95]. Phase diagrams for such ether-linked lipid bicelles have been established with respect to q-ratio, hydration level, and temperature [96]. Ether-lipid bicelles have also been applied to residual dipolar coupling (RDC) studies of globular proteins [97]. However, the structure of the antimicrobial peptide novicidin was found to be altered in ether-lipid bicelles when compared to DMPC/DHPC bicelles [98]. Small amounts of added PEGylated lipids are another choice to make bicelle samples more stable [99], but their use is not as wide-spread as that of ether-lipids. Rather, PEGylated lipids have been extensively used in the study of translational lipid diffusion (see Section 6). Doping of dilute oriented bicelle preparations with charged amphiphiles improves the stability and the degree of alignment [100]. The influences of lipid unsaturation and chain length on bicelle stability have been studied [101]. The addition of cholesterol and especially cholesterol sulfate was reported to stabilize bicelles thermally, with magnetic-alignment possible in an extended temperature range [102]. The influence of divalent cations, which are required by many classes of biomolecules for optimal activity, on the formation and alignment of DMPC/DHPC bicelles has been studied [319]. It was found that higher concentrations of Zn2+ and Cd2+ disrupt the magneticallyaligned phase, while Ca2+ and Mg2+ result in more strongly oriented phases.
A number of bicelle modifications have been designed to make bicelles resemble biological membranes more closely. One important characteristic of biological membranes is their bilayer thickness, which affects embedded proteins by means of hydrophobic mismatch [103,21]. Bicelles with different bilayer thickness have successfully been prepared [103]. The effect of varying chain-length in both lipid and detergent components has been investigated systematically [101]. Bicelles formed by DHPC and the phospholipid 1-palmitoyl-2-stearoyl-phosphatidylcholine (PSPC), that is 16:0–18:0-PC, have been investigated [104]. Another important characteristic of biological membranes is their composition with respect to head group charge and cholesterol content. Acidic bicelles with the addition of the charged phospholipid dimyristoylphosphatidylglycerol (DMPG) have been prepared and their stability has been investigated [105,106]. Other researchers have investigated the effect of cholesterol [107–110], of the unsaturated phospholipid 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) [100], and of polyunsaturated phosphatidylcholine [110] added to bicelles. It was shown that sphingomyelin, a common sphingo-lipid in mammalian membranes, and DHPC can form isotropic as well as aligned bicelles [111]. Morphological effects caused by the addition of ceramide to DMPC/DHPC bicelles were studied [112]. Cyclofos-6 as detergent formed bicelles with DMPC; since the critical micelle concentration of Cyclofos-6 is very low (and lower than that of DHPC), bicelle formation is observed down to and below 0.5% total lipid weight per volume [113]. The T-shaped molecule A6/6, which forms closed vesicles in water, was shown to mix with DPPC to form bicelles [114]. In studies of membrane proteins, it is practical that the detergent which is present in the end products of expression, purification, and refolding protocols may in some cases also be used as the short-chain or detergent component in aligned bicelles, as demonstrated for the detergents Triton X-100 [115] and dodecylphosphocholine (DPC) [116] in combination with DMPC.
4. Bicelles in electron paramagnetic resonance (EPR) spectroscopy
A large body of work, mainly by the Lorigan laboratory, has been dedicated to establish aligned bicelles as a membrane mimetic for electron paramagnetic resonance (EPR) spectroscopy studies. The first successful preparations of aligned bicelles were reported on bicellar samples doped with paramagnetic lanthanide ions [117,118]. As described above and shown in Fig. 5A, bicelles doped in this way orient with their normal parallel to the external magnetic field. This effect is caused by the large positive anisotropies in the magnetic susceptibilities caused by suitable paramagnetic lanthanide ions (Eu3+, Er3+, Tm3+, Yb3+) in the lipid bilayer, which itself has a negative anisotropy in magnetic susceptibility. In the comparably weak magnetic field of 0.63 T used in X-band EPR, no spontaneous alignment of undoped bicelles was observed. However, using Dy3+ lanthanide ions which enhance the negative anisotropic magnetic susceptibility of the bilayer, it was possible to achieve alignment of bicelles with their normals perpendicular to the magnetic field (Fig. 5B) [119,83]. EPR studies require the addition of a spin label to the investigated bicelles to detect their orientation; doxyl-labeled cholestane and doxyl-labeled stearic acid were used for this purpose in the studies mentioned so far. Doxyl spin labels attached to different positions along the acyl chain of stearic acid have been used as a probe to study the dynamic properties of the bilayer portions of bicelles and have shown that they agree well with other biological and model membrane systems [120]. Bilayer dynamics and the effect of temperature were studied in more detail using spin-labeled phosphocholines [121]. In addition to the mentioned X-band studies at 9 GHz EPR frequency, bicelles were also introduced for the higher field strength used in Q-band [122] and W-band [123] spectroscopy at 35 and 94 GHz EPR frequency, respectively. At these higher magnetic field strengths of 1.25 T and 3.4 T, respectively, reduced concentrations of lanthanide ions were needed to achieve magneticalignment of bicelles. The magnetic-alignment of bicelles at high q was investigated by EPR and NMR spectroscopy [124].
EPR spectroscopy was applied to study the effect of cholesterol on bicelle model membranes using phosphocholine [125] and cholestane [126] with doxyl spin labels. At Q-band, higher order was observed in cholesterol bicelles than at X-band [126]. Another study on cholesterol in bicelles compared results from EPR and NMR spectroscopy [108]. Nitroxide spin labels can be introduced into proteins to study their properties by EPR spectroscopy, but labeling can potentially perturb protein properties. The sidechain conformation of a nitroxide spin label has been studied in the homodimeric protein CylR2 by comparing results from X-ray crystallography, EPR and NMR spectroscopy [127]. Structural and dynamic properties of the transmembrane protein phospholamban were determined by EPR of aligned bicelles [128]. The helical tilt of the M2δ transmembrane peptide of the nicotinic acetylcholine receptor in aligned bicelles was determined [129]. The analysis method, which is similar to the dipolar waves [130] used in solid-state NMR, is described in detail elsewhere [131]. Similar results could be obtained in unoriented samples [132]. The quenching of EPR spin labels by water-soluble reducing agents can be monitored in real-time to determine details of membrane immersion, as demonstrated for the M2δ peptide [133]. It was shown by EPR that α-synuclein in bicelles forms an extended α-helix rather than a helix-turn-helix structure [134].
5. Phase diagrams and morphology of bicelles
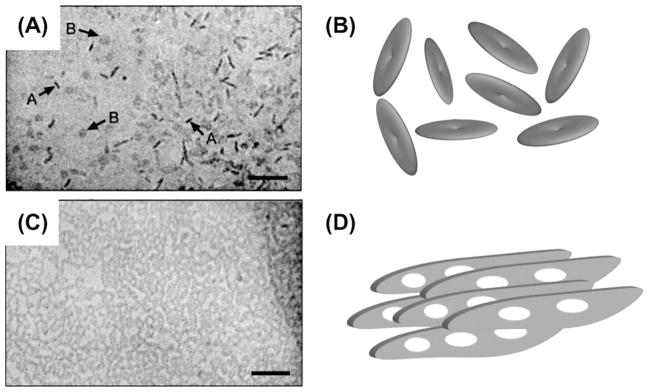
The morphology of bicelles is most often described as microscopic disks of lipid bilayers where the detergent covers the ‘rims’ (see Fig. 2F,G for schematic representations). This picture holds true only in a limited range of conditions, especially for low q-ratios and fast-tumbling bicelles, as seen in small-angle neutron scattering (SANS) [135] and electron microscopy (EM) [94]. In other conditions, more complex models are necessary. To derive unambiguous conclusions and models of complex morphologies, usually complementary techniques such as SANS or EM are needed in combination with NMR spectroscopy.
The influence of the ratio q between DMPC and DHPC on the formation of bicellar phases is shown in Fig. 6. At low q, small isotropically tumbling aggregates are observed, and no magnetically orientable phase is formed. Only in a limited range of q values between ~2.5 and 7.5 is it possible to obtain magnetic alignment. Above a q of 7.5, morphology similar to multilamellar vesicles is present, where the detergent gathers in nanopore defects [136]. At higher temperatures, annealing of the nanopore defects is observed [137]. 31P- and 2H NMR are the methods of choice to study the structure and dynamics of lipid phases and to quickly map out phase diagrams [138–140]. Partial magnetic-orientation can be determined by the first spectral moments of 31P NMR spectra [141]. The deconvolution of global orientational distributions of lipid bilayers and local order parameters of embedded molecular reporters has been demonstrated [142].
Fig. 6.
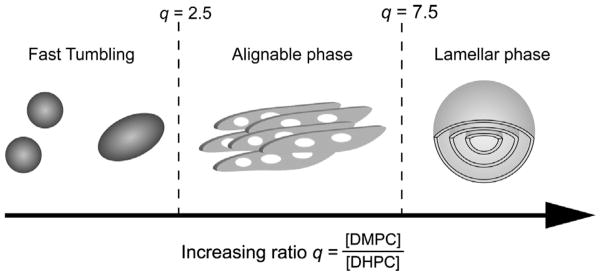
The morphology of bicellar preparations is dependent on the ratio q between a long-chain lipid and a detergent. Fast tumbling bicelles exist when q < 2.5 (left panel), macroscopically aligned bicelles exist between q ratios of 2.5 and 7.5 (middle), and at high q > 7.5, multilamellar vesicles are formed (right).
The phase behavior of bicelles, including phase diagrams, has been the objective of numerous studies. The limited miscibility of long- and short-chain lipid components dictates the separation into bilayer and ‘rim’ proportions and is thus the key to the phase behavior of bicelles [143,144]. It was possible to predict phase diagrams from miscibility properties [144]. Based on this insight, lipid mixtures forming bicellar phases in specified concentration and temperature ranges can be rationally designed [101]. As a general rule, it is observed that oriented bicellar phases are formed at temperatures above the main lipid phase transition between the gel state and the liquid–crystalline phase of the long-chain lipid component [145].
Phase diagrams have been established for pure phospholipids [146] as well as for lanthanide-doped bicelle mixtures [147]. Phase diagrams for DMPC/DHPC bicelles at various q-values have been reported [148]. Phase diagrams for ether-lipid bicelles have been established [96]. A review article dedicated to morphology and phase diagrams of bicellar phases is available [149]. A very broad general review on bicelles [150] offers a good focus on morphology. The feasibility of magnetic-alignment of dilute bicellar solutions has been investigated [151].
The model of flat, microscopic disks for bicelle morphology (Fig. 2F and G) is based on the fact that DHPC and DMPC are phase separated into bilayer and ‘rim’ regions in oriented bicelles [77]. A first geometrical model was developed for ideal disk-shaped bicelles [152]. This study also found a new proof in addition to the ones reported [77] that DMPC and DHPC are indeed spatially separated in oriented bicelles, and mentions the need for an experimental proof in the case of isotropic bicelles. This need was again stressed in a work on Mastoparan X in isotropic bicelles [153]. The flat disk model was proven to apply to isotropic bicelles by 31P NMR, dynamic light scattering, and electron microscopy [154]. An independent proof was given by small angle neutron scattering and Eu3+-doping [135]. Further support comes from a recent study of a protein embedded in isotropic lipid bicelles [155].
It has to be noted that from the beginning, the flat disk model has been seen as only tentative [77]. The existence of DHPC pores in DMPC lamellae (Fig. 7C) was clearly seen as an alternative structural model for aligned bicelles, and was termed the “Swiss cheese model” [88]. The high diffusion rate measured for tetramethylsilane (TMS) in oriented bicelles would require extensive transient edge-to-edge contacts in the disk model [156]. Measurements of high viscosity in orientable bicelles [157,158] contradict the theory of individual disks and suggest some entangling. The use of optical microscopy and SANS found yet another bicelle morphology, described as “wormlike micelles” and depicted in Fig. 7B [159]. The use of SANS experiments has in all cases been able to identify the morphology that is present in the bicellar phase [149]. It was possible to obtain very clear cryo-TEM micrographs for isotropic as well as oriented bicellar phases [1,160,161]. The kinetic pathway of the phase transition from bilayered micelles to perforated lamellae has been characterized [162]. Stimulated echo-pulsed field gradient (STE-PFG) experiments were used to measure water diffusion in bicelles [81]. In this study, the fraction of rim-located and bilayer-embedded DHPC was determined as a function of q and T. As an additional parameter, q*, the fraction between edge and planar phospholipid, was introduced.
Fig. 7.
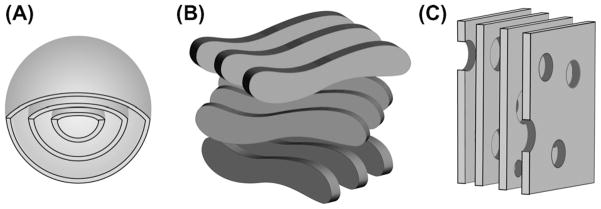
Bicelles exhibit a wide range of morphologies. Beside the simple nanodisk morphology, multilamellar vesicles (A), chiral nematic ribbons (B), and perforated lamellae (C) are found under different sample conditions.
A single contribution has claimed the observation of tight stacks of disk-like bicelles at very high hydration levels [163], similar to the elongated stacks reported for disk-like high-density lipoprotein (HDL) particles [31]. However, the stacking effect claimed for dilute bicelles has not been reported for other systems.
Molecular dynamics (MD) simulations have in a few instances been carried out on bicelle systems. Most studies to date have used coarse-grained models. Structures resembling bicelles were found in the spontaneous aggregation of DPPC into small unilamellar vesicles [164]. A special coarse-grained force field was developed to model zwitterionic lipid assemblies; in coarse-grained MD runs, bicelle-like invaginations referred to as ‘buds’ were observed to form from DPPC monolayers [165]. A coarse-grained force field called MARTINI was used to simulate discoidal aggregates of PEGylated lipids [54]. A coarse-grained simulation showed that functionalized carbon nanotubes and lipids form bicellar assemblies [166]. The influence of line tension on length and shape of bilayer edges was investigated by course-grained MD of lipid bilayer ribbons of different tail lengths [167]. Only recently, two atomistic simulations on bilayer ribbons were reported which provide insight into lipid behavior in bicelles: the effect of bilayer edge and curvature on the partitioning of lipids by tail lengths was investigated [168]. A two-step semi-grand-canonical mixed Monte Carlo/molecular dynamics approach found a possible mechanism for attraction and merging of DHPC pores [169]. The three-dimensional structure of glycolipids embedded in bilayers starts to be investigated by combined use of isotropic bicelle NMR experiments and molecular dynamics simulations [320].
6. Diffusion studies on bicelles
Molecular diffusion, particularly translational diffusion, is the most fundamental transport process in nature. Importantly, Brownian motion in lipid bilayers governs a variety of important biological processes that ranges from signal transduction to the transport of nutrients across cell membranes such that a significant body of literature is devoted to this subject matter. However, Brownian motion in lipid membranes can be extremely complex due to the heterogeneity of most biological systems; only a single elegant coefficient, namely the lateral diffusion coefficient, i.e. the component of the diffusion tensor that is perpendicular to the bilayer normal, is required to describe this complicated process. The elegance of this coefficient lies in the depth of information it holds, particularly the relationship between the diffusant and its environment. Lateral diffusion coefficients in a cell membrane vary by orders of magnitude: from the rapidly diffusing phospholipids to the slowly moving multi-helix membrane proteins. For species whose sizes are comparable to that of a lipid, their diffusion coefficients seem to follow the free volume model whereas larger species diffuse according to the hydrodynamic model. Currently, most diffusion coefficients are obtained via fluorescence recovery after photobleaching (FRAP) while single molecule tracking is increasingly used to study diffusion of molecules in situ. These optical techniques are valuable in providing detailed information on molecular diffusion; yet, these techniques only work if the molecules are inherently fluorescent. Therefore, an alternative approach is required for molecules that lack such an optical property and where the introduction of a fluorescent tag is not an option.
NMR spectroscopy provides an alternative means of measuring diffusion coefficients in a model membrane system. In particular, the pulsed field gradient (PFG) NMR technique introduced by Stejskal and Tanner [66] has evolved into a powerful tool capable of determining the diffusion coefficients for a wide range of macro-molecular systems. PFG NMR allows for the rapid and simultaneous determination of multiple diffusion coefficients from different species, provided that their resonances are resolvable. The use of the stimulated echo (STE) was subsequently found to be advantageous for the measurement of diffusion coefficients [170]. The seminal publications [66,170] provide the basic theoretical framework for PFG NMR diffusion measurements. Therefore, only a brief description will be given here. The diffusion coefficient of a species, D, under isotropic condition, can be extracted from a STE-PFG NMR experiment by measuring the signal attenuation as a function of gradient duration (δ), gradient amplitude (g) and diffusion time (Δ) as indicated in the pulse sequence of Fig. 9. The observed NMR intensity is attenuated according to
Fig. 9.
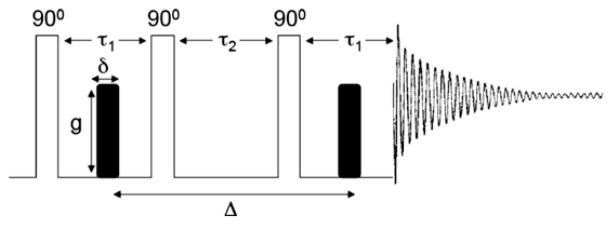
STE-PFG NMR pulse sequence composed of three 90° radio frequency pulses and two gradient pulses of identical amplitude and duration. The STE-PFG NMR experiment is arranged such that the decay of the echo intensity, as a function of δ, g or Δ, is proportional to the diffusion coefficient of the species of interest.
| (1) |
where γ is the gyromagnetic ratio of the observed nucleus and I and I0 are the observed and the initial signal intensity, respectively [66,170].
Lateral diffusion in a lipid bilayer environment is anisotropic such that the diffusion coefficient is represented as a diffusion tensor according to
| (2) |
There is uniaxial symmetry about the bilayer normal and so only two principal components are required to represent the system. These two components correspond to molecular diffusion along perpendicular (D⊥) and parallel (D||) directions to the bilayer normal as illustrated in Fig. 8.
Fig. 8.
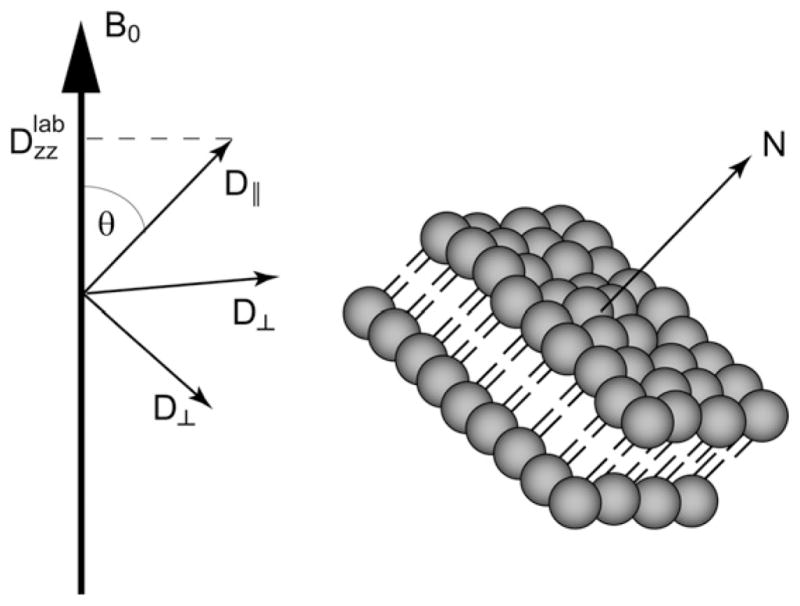
In a lipid bilayer system, only two components are needed to describe the diffusion tensor. The relative orientation of the bilayer normal and hence the principal components of the diffusion with respect to the external magnetic field is described by the angle θ. It determines the component of the diffusion tensor that lines up with the magnetic field and can be measured.
If the gradient is applied along the laboratory z-axis, chosen along the direction of the external magnetic field B0 (see Fig. 8), then only the Dzz component of the diffusion tensor in the laboratory frame is measured. The relationship between Dzz and the principal components in the molecular frame, which align parallel and perpendicular with respect to the bilayer normal as described above, is given by
| (3) |
If the molecules forming the phase have a low critical micelle concentration (CMC), as is typical for lipids in general, then there is no detectable diffusion parallel to the normal of the bilayer, D|| = 0. Thus, only the term Dzz = D⊥sin2 θ remains in Eq. (3). Interestingly, for the case where the bilayer normal is perpendicular to the applied gradient, the observed diffusion coefficient in the laboratory frame (Dzz) is equal to that of the lateral diffusion (D⊥) in the lipid bilayer. Therefore, in order to measure the diffusion coefficient of a molecule in a lipid bilayer system, an aligned sample is required and bicelles provide a suitable medium for the purpose. Recently, Soong and Macdonald [171] demonstrated the feasibility of measuring diffusion of a polymer-grafted lipid, namely DMPEPEG2000, in magnetically-aligned bicelles using the STE-PFG NMR technique; the pulse sequence is shown in Fig. 9.
The diffusion coefficient is measured by monitoring the decay of the 1H signal intensity of polymer-grafted lipids as a function of the gradient duration. The rapid internal motion of PEG yields a narrow 1H resonance, which made the measurement feasible. The measured diffusion coefficient was found to be comparable in magnitude to the FRAP measurements of DMPC in the liquid crystalline phase [171]. Therefore, this illustrates the viability of bicelles as a medium for the lateral diffusion studies of membrane-associated amphiphiles in a bilayer environment. The results also demonstrate the possibility of extracting the diffusion coefficient of a membrane protein via STE-PFG NMR, which is important to our understanding of protein trafficking in lipid bilayers. Therefore, this illustrates that bicelles are more than just a mere reconstitution medium for membrane proteins; in fact, they can be used as a platform for lateral diffusion studies via NMR. An interesting result regarding bicelle morphology was also obtained in these diffusion measurements: the molecular constituents exhibit free diffusion over micron distances. This observation is consistent with the perforated lamellae morphology of bicelles [172] and also corroborates with SANS data [159] and tetramethylsilane diffusion studies in dilute bicelle solutions [156].
Diffusion studies can also be done on bicelles with low q ratios (0.5 ≤ q ≤ 1). Due to their small size, these bicelles tumble isotropically, hence are suitable for high-resolution studies of membrane proteins. Interestingly, bicelles with a low q ratio exist as disk-like aggregates and are relatively monodisperse in their diameter and thickness. At q = 0.5, their estimated diameter is about 8 nm, double the assumed bilayer thickness [152]. The diameter of these fast-tumbling bicelles changes in the presence of membrane associated peptides. Recently, these bicelles have been used for the measurement of lateral diffusion coefficients of membrane binding peptides and the results are comparable to the literature values. While these experiments demonstrate the feasibility of using bicelles for diffusion studies, caution needs to be taken since the lateral diffusion coefficient is measured in a relative sense as all the components in the sample diffuse at different rates. In particular, the lipid bicelle as a whole does not provide a stationary reference frame, since it itself undergoes substantial lateral diffusion. Nevertheless, these bicelles are excellent model membrane systems for the investigation of binding kinetics of membrane-associated peptides and amphiphiles.
Diffusion measurements by PFG NMR were used to investigate the morphology of three media that are commonly used in the study of residual dipolar couplings, namely oriented lipid bicelles, cetylpyridinium bromide, and a mixture of PEG and n-hexanol [156]. The hydrodynamic radius of micelles and isotropic bicelles has been measured by bipolar pulsed gradients [173]. In these measurements, lysozyme was used as a reference compound to correct for the differences in hydrodynamic volume between dry and hydrated protein, bicelle, or micelle. The motion of constituent lipids in isotropic bicelles was studied by 13C relaxation, PFG NMR, and EPR [174]. Local mobility was found to depend much more strongly on the detergent used than on bicelle size. Different peptides were found to have an impact on apparent dynamics. In a later study, diffusion measurements were used to study the size and shape of bicelles of low q ratio which tumble isotropically [175]. Three different long-chain lipid components, namely dilauroylphosphatidylcholine (DLPC), DMPC, and DPPC, were used to study lipid dynamics as a function of bilayer thickness [176].
Rotational diffusion concerns a molecule’s rotational degrees of freedom in contrast to translational motion discussed so far. Lipids as well as proteins in a bilayer sample experience rotational diffusion, which occurs most freely around the bilayer normal. It is usually fast on the NMR time scale, as long as proteins and peptides of moderate molecular size are studied. In NMR spectra, rotational diffusion becomes evident from the observed NMR lineshape as the averaging of anisotropic spin interactions along the axis of rotation alters the spectral lineshape and the magnitude of the interaction. As a consequence, multilamellar vesicles (MLVs) can potentially give the same information as macroscopically oriented samples [6,7,177,178], and the choice is mostly determined by ease of preparation and sensitivity issues. However, it was reported that structural measurements for the antimicrobial peptide PGLa may be influenced by different hydration levels present in MLVs compared with data obtained from macroscopically oriented samples made of stacks of glass plates [7]. For disk-like bicelles, rotational diffusion about the bilayer normal was found to be fast enough to average the cylindrical distribution about that axis, even in the rare cases where the embedded protein itself does not undergo rotational diffusion fast enough for the required averaging [179]. As a result, even very large proteins and protein complexes are amenable to studies in unflipped bicelles.
7. Separated local field (SLF) NMR studies on bicelles
7.1. Separated local field spectroscopy
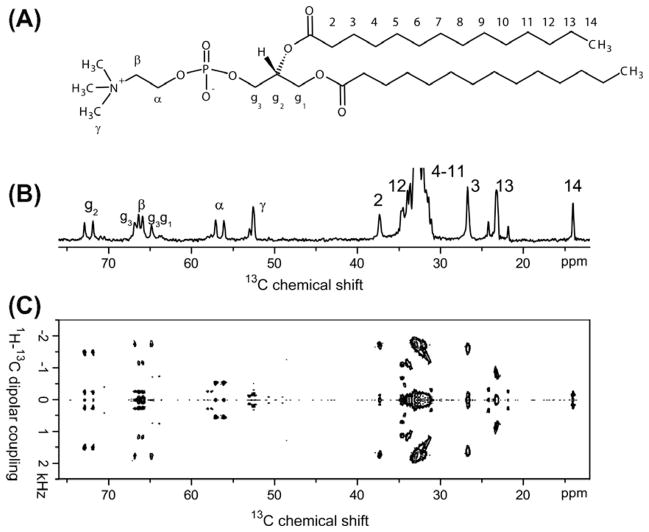
Though one-dimensional 31P, 1H and 13C NMR experiments are commonly used to characterize magnetically-aligned bicelles [180], sophisticated two-dimensional (2D) experiments are essential to probe the order/disorder of lipid and detergent molecules in bicelles as well as to measure the interaction of ligand or peptide with hydrophilic and hydrophobic domains of bicelles. Experiments correlating short-range heteronuclear dipolar couplings with the chemical shift of a given nucleus, referred to as separated local field (SLF) experiments, are powerful in providing insights into the atomic-level structure of bicelles. Magnetically-aligned bicelles were found to be readily accessible objects in 2D SLF studies [181–183]. Fig. 10 demonstrates how 2D SLF gives piercing insights into bicelle properties. The chemical structure of a DMPC lipid molecule is shown in Fig. 10A. The commonly used labeling scheme is indicated: Cα, Cβ, Cγ denote the carbon positions in the choline headgroup, Cg1, Cg2, Cg3 make up the glycerol backbone, and C1 to C14 the myristoyl chains. The very high concentration of DMPC molecules in typical bicelle preparations means that natural abundance 13C NMR of DMPC carbons is sufficient for one- and two-dimensional NMR experiments.
Fig. 10.
13C NMR spectroscopy of magnetically-aligned DMPC/DHPC bicelles, q = 3.5. At this ratio of long- to short-chain component, resonances from the long-chain component dominate the spectra. (A) Molecular structure of DMPC, including the commonly employed nomenclature. (B) The natural-abundance 13C NMR chemical shift spectrum on a 400 MHz NMR spectrometer shows clearly resolved resonances for most carbon sites in DMPC. (C) A 400 MHz 2D SLF-spectrum correlates 13C chemical shift to 1H–13C dipolar coupling of carbon nuclei to adjacent hydrogen nuclei and yields information on the local structure and mobility of lipid molecules in bicelles.
The one-dimensional 13C chemical shift spectrum in Fig. 10B shows well-resolved resonances for almost all positions; assignments are available for all resolved resonances and are given in Fig. 10B. In an SLF experiment, an incrementable time delay is added to evolve the transverse magnetization under heteronuclear dipolar couplings. The heteronuclear dipolar couplings (also known as the local field) associated with each chemically distinct carbon nucleus in the indirect dimension of the 2D spectrum are separated by the 13C-chemical shift frequency; hence the name ‘separated local field’. The SLF spectrum in Fig. 10C shows well-resolved dipolar multiplets for each carbon group. A large splitting corresponds to a large molecular order parameter of the respective 1H–13C bond, whereas a small splitting indicates a small order parameter. The magnitude of the dipolar splitting can be plotted versus the carbon position in the lipid molecule, yielding an order parameter profile over the whole lipid bilayer. In this fashion the dynamics of the whole lipid molecule, from the headgoup to the acyl chains, can be thoroughly mapped out via one single 2D experiment. This is similar to order parameter profiles acquired by 2H NMR spectroscopy [138,139], but does not need deuterated lipids and gives unambiguous site-specific information. Static and/or macroscopically oriented samples are not a necessary prerogative for SLF experiments; SLF is equally possible under MAS conditions by re-introducing dipolar interactions [184,185].
Spectroscopically, SLF experiments can be divided into two categories: laboratory-frame and rotating-frame experiments. In a rotating-frame SLF, the heteronuclear dipolar couplings are evolved under the influence of a spin-lock matched at the appropriate Hartmann–Hahn condition. Prominent examples include polarization inversion spin exchange at the magic angle (PISEMA) [186], heteronuclear isotropic mixing spin exchange via local field (HIMSELF) or heteronuclear rotating-frame spin exchange via local field (HERSELF) [181], and magic-sandwich PISEMA (SAMMY) [187]. The PISEMA experiment and its improved derivatives are commonly used to investigate the structure and dynamics of membrane proteins in aligned lipid bilayers as they provide ultra-high spectral resolution; since those are not the main focus of this article, they will not be discussed further. Readers can consult a review article on PISEMA spectroscopy [188].
In the second type of SLF experiment, laboratory-frame SLF, the heteronuclear dipolar couplings are evolved through the protons and are subsequently transferred, by cross polarization, to the dilute spin such as 13C or 15N for detection. An example is proton evolved local field spectroscopy (PELF) [321]. Rotating-frame SLF is preferable particularly when large dipolar couplings are measured by efficiently suppressing large homonuclear proton–proton dipolar couplings, as in a single crystal or in transmembrane proteins. Laboratory-frame SLF, on the other hand, is suitable when motionally averaged dipolar couplings are present. The latter case is found in magnetically-aligned bicelles. Consequently, laboratory- frame SLF experiments have recently been successfully applied to a number of biological problems.
7.2. Application of SLF to study bicelle properties
As a proof of feasibility, our laboratory has recorded 2D 1H-13C-PELF spectra on oriented q = 3.5 DMPC/DHPC-bicelles [189]. In addition, a 2D correlation spectrum of 13C chemical shift and 13C–31P dipolar couplings was acquired, which reports on the conformation of the phosphocholine headgroup region of DMPC. Similar results were obtained with a solution-state NMR spectrometer using a SAMMY experiment [182]. In a subsequent study, experimental parameters were optimized to maximize sensitivity and resolution [180]. Ramped cross-polarization [190] and the FLOP-SY- 8 [191] scheme were found to be the best choices for polarization transfer and heteronuclear decoupling, respectively. By carefully calibrating the sample temperature and minimizing sample heating, spectral quality could be further improved. A direct experimental comparison of 2D SLF, PELF, and rotating-frame SLF schemes proved the superior quality of the PELF approach [183]. The method was further developed by reintroducing proton-spin diffusion to aid in resonance assignment of embedded molecules [192].
1H–13C-PELF spectra were used to characterize bilayer properties in bicelles for a wide range of conditions [193]. For different q-values, temperatures, and hydration levels, 1H-13C-PELF spectra were recorded. The extracted order parameter profiles are reproduced in Fig. 11. The order parameter profiles show that at higher q values (higher DMPC content, Fig. 11B), the bilayered regions become more rigid. The same is true for lower temperatures (Fig. 11C). More surprising is the fact that mobility seems to be fairly constant over a large range of hydration levels (Fig. 11D).
Fig. 11.
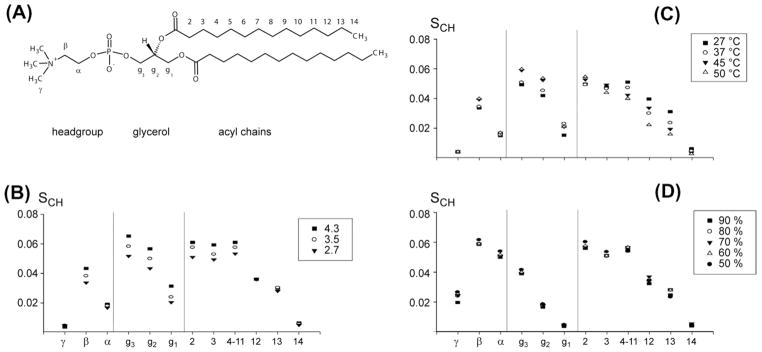
Order parameter profiles determined from 2D 1H–13C-PELF NMR spectra of magnetically-aligned DMPC/DHPC bicelles [193]. (A) DMPC structure with nomenclature and indication of distinct molecular regions. Order parameter profiles were determined for different values of (B) composition q, (C) temperature and (D) hydration level. These results demonstrate that experimentally measured 13C–1H dipolar couplings can be utilized in measuring the changes in the order/disorder associated with various regions of the lipid and detergent in bicelles without the need for isotopic enrichment. Such measurements have been shown to provide insights into the mechanism of membrane disruption by antimicrobial peptides and amyloid peptides/proteins.
It was mentioned above that the order parameter profiles of lipid bilayers can be—and traditionally have been—recorded using 2H NMR. A tremendous number of results has been achieved from 2H NMR studies of lipid vesicles [138,139]. The determination of order parameters of lipid C–C-bonds from 2H quadrupolar splittings has been thoroughly investigated, and a number of conformational details could be determined [194]. It has been shown that structural and dynamical details of DMPC in bicelles can be studied by deuteration [195]. However, there are serious drawbacks to this technique: a study [195] found that progressive deuteration has an effect on thermotropic behavior. The fact that partially deuterated molecules were employed, rather than single-site deuteration, makes their assignments ambiguous. Earlier results on DMPC conformation in crystals [196] or MLVs [177] were confirmed and refined for the bicelle environment. As described, the 1H–13C-PELF method gives equivalent results and does not require isotope labeling and does not suffer from the sensitivity and assignment problems imposed by 2H-labeled phospholipids. A promising extension would be the use of order parameters as orientational angular constraints in molecular dynamics simulations [197].
The DMPC choline head group in DMPC/DHPC bicelles has found particular interest. It was found that a deuterated head group can be employed as molecular voltmeter, since its conformation reacts almost linearly to the presence of negatively charged DMPG and positively charged 1,2-dimyristoyl-3-trimethylammoniumpropane (DMTAP) [198,199]. POPC with site-specific 2H- and 13C-labels was used to record 2H quadrupolar as well as 13C–31P dipolar splittings, which can be used to provide torsional constraints on conformation [200]. Again, carbon dipolar couplings with 1H and 31P nuclei can be detected in 2D HIMSELF or HERSELF experiments without the need for isotopic labeling. In addition, quadrupolar splittings of the naturally most abundant nitrogen isotope, 14N, can be employed to report on head group conformation [180,201]. The immersion depth and orientation of the headgroup of ganglioside GM1, a glycosphingolipid, was investigated in isotropic DMPC/CHAPSO bicelles by paramagnetic relaxation enhancement experiments [202].
The application of 1H–13C-PELF to membrane-associated peptides and small molecules has already yielded interesting results. The antimicrobial peptide MSI-78 (also known as pexiganan) was found to reduce the order parameter profile smoothly in each position, indicating that MSI-78 fragments the bicelles into smaller, more dynamic segments [189]. The antidepressant desipramine was found to be localized in the glycerol backbone and head group regions in DMPC/DHPC bicelles [183]. A 1H–13C-PELF study on the interaction between curcumin and membranes shows an ordering effect of the lipid acyl chain at low curcumin concentrations but at a high concentration of curcumin, a disordering of the acyl chains was observed [203]. The latter two studies dealt with the interaction of small molecules with lipid bicelles. This topic has also found interest in other types of studies, and will be further discussed in Section 9.
8. Bicelles under magic-angle spinning (MAS)
Magic-angle spinning (MAS) experiments on macroscopically aligned samples have been established which used solid supports, such as stacks of round glass plates [204], or a polymer film wrapped into a cylinder [205]. Similarly, lipid bicelles have been investigated under magic-angle spinning. It was found by variable- angle sample spinning (VASS) that at spinning angles smaller than the magic-angle the bilayer normal will align perpendicular to the rotation axis [206]. It was demonstrated that this fact can be used to determine the relative signs of dipolar and scalar (or J-) couplings. Perpendicular alignment at spinning angles smaller than the magic-angle was confirmed by 2H NMR [207]. For spinning angles larger than 54.7°, the same study found parallel alignment with respect to the spinning axis. At the magic-angle itself, no preferential orientation is present, and the bilayer normals are distributed isotropically. In addition, the study described metastable phases with different alignment behavior whose presence depends on spinning speed and can be manipulated by switched-angle spinning to yield arbitrary angles between bilayer normal and applied magnetic field. The width of the distribution of the bilayer normal around its average value, known as mosaic spread, was the subject of another study [208]. In this study, mosaic spread in spinning bicelles was investigated by 31P NMR and found to be small at spinning angles far away from the magic-angle. As the spinning axis approaches the magic-angle, mosaic spread becomes larger until an isotropic distribution is reached at the magic-angle.
It was shown that studies of peptides and proteins under MAS can benefit from using bicelles rather than MLVs [209]. Both isotropic and aligned spectra for the study of RDCs in soluble proteins can be collected from the same sample using bicelles and MAS [210]. Precise values of chemical shift anisotropy (CSA) were determined for a transmembrane segment of an acetylcholine receptor in bicelles under MAS [211]. Two-dimensional MAS experiments on bicelles containing a membrane-associated cytochrome b5 have been reported and the results suggest that bicelles form better model membranes than MLVs or SUVs [212].
9. Interaction of small molecules with bicelles
Bicelles have in a few cases been used to study the interaction of small organic molecules with lipid bilayers. Both isotropically tumbling and magnetically-aligned bicelles were used. Tea catechins interact with isotropic bicelles according to the partition coefficient and their amphiphilic properties, with attachment mostly to the lipid headgroup region [213]. Erythromycin A, a macrolide antibiotic, showed shallow insertion into isotropic bicelles in a paramagnetic relaxation enhancement (PRE) study [214]. Salinomycin, an ionophore antibiotic, showed different conformation in presence and absence of sodium ions in isotropic bicelles [215]. The conformation found for the salinomycin-sodium complex is significantly different from the structure found in solution or in a crystal. The conformation and a model of insertion were established for amphidinol 3, a potent antifungal agent, in isotropic bicelles [216]. The study of small molecules in isotropic bicelles has been reviewed [217]. The polyene antibiotic amphotericin B was investigated in aggregates formed with dioctadecyl dimethylammonium bromide that are presumed to have discoidal geometry similar to bicelles [218].
Magnetically-aligned bicelles have also been used to study small molecules. RDCs were measured for ethanol [219]. The orientation and motion of ethanol in its membrane bound state could be inferred from these measurements. About 4% of ethanol is bound to the membrane; the lifetime of ethanol association is on the order of a few nanoseconds. Structural and dynamic properties of stearic acid-d35 in bicelles containing cholesterol have been studied by 2H NMR [220] and later compared to EPR results [221]. Two cannabinoids were investigated in the presence of oriented bicelles [222]. In this study, two deuterium labeled sites yielded orientational constraints from spectra obtained on a solution-state NMR spectrometer, and were analyzed in terms of structure and dynamics. In an earlier parallel study, the same two cannabinoids were investigated in isotropic bicelles of high DMPC content, q = 2.0, which was necessary in order to observe a bilayer-bound conformation [223]. Another cannabinoid was investigated by 2H NMR in bicelles of different lipid composition [224]. Glycosidic torsional motions were determined in a bicelle-associated disaccharide using RDCs [225]. Single-site deuterium labeled epigallocatechin gallate was found to interact with aligned bicelles [226]. Three fullerene derivatives in magnetically-alignable bicelles were investigated by EPR spectroscopy and were found to reside as single molecules just below the hydrophilic/hydrophobic interface with a preferential orientation [227]. Our own studies on membrane distortion by the antidepressant desipramine [183] are presented in Section 7.
In biopartitioning chromatography (BPC), a biomembrane-mimetic (e.g. liposomes, phospholipid monolayers, micelles, or bicelles) is introduced into a chromatographic system to study drug-membrane interactions [228]. Pure and highly stable phases of PEG-stabilized bilayer disks (described in Section 2.4) were developed as model membranes for drug partition studies, and have the potential to give more accurate results than liposomes [229]. Formulations designed to model porcine brush border membrane [230] and plant tissue [231] in drug partition studies were described. Discoidal aggregates of PEGylated lipids and lipids were used as pseudostationary phases in capillary electrophoresis to study drug partitioning [232,233] and the results verified by using a quartz crystal microbalance [233].
Numerous studies on peptides in bicelles have been conducted, for example on antimicrobial peptides [234] and neuropeptides [235]. A recent review [236] gives a comprehensive overview of proteins and peptides studied in bicelles and therefore we do not report on peptide studies here.
10. Magic touch added to studies of protein structure
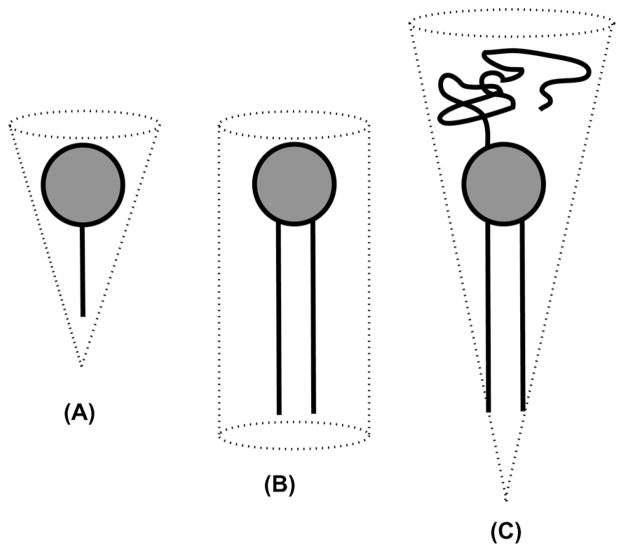
The most exciting and fruitful area of bicelle application is without doubt found in structural biology of membrane proteins. Bicelles can be used in four fundamentally different ways to study membrane protein structure, as illustrated in Fig. 12. The difference is found in the proteins studied and in the type of bicelle chosen for their study. Integral membrane proteins (Fig. 12A and B), soluble proteins (Fig. 12C), and membrane-interacting proteins (Fig. 12D) have all been studied in bicelles. Both aligned (Fig. 12A and C) and isotropically tumbling (Fig. 12B and D) bicelles are in common use. In addition, a variation in the lipid:detergent ratio (the so called q titration) converting one type into the other can be performed and may be useful for differentiating between structured and mobile residues of membrane proteins [237]. In this section, a very short and highly subjective overview of the most exciting results on proteins is presented. For a comprehensive presentation, the reader is referred to our recent review article [236]. Several other, excellent review contributions on proteins studied in bicelles are available [8,150,172].
Fig. 12.
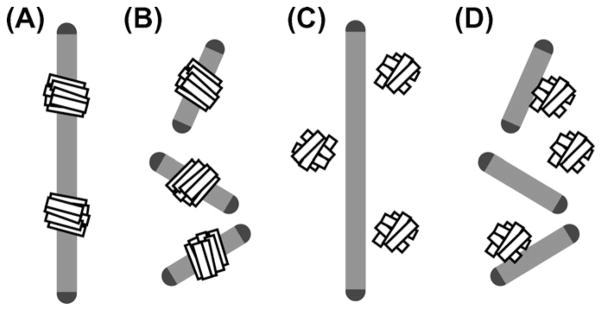
Graphic representation of the four fundamentally different ways in which bicelles are used in protein structure and protein–membrane interaction studies. Integral membrane proteins can be studied in aligned (A) as well as isotropically tumbling bicelles (B). Aligned bicelles can be used to impose a residual orientation on soluble proteins for RDC measurements (C). Isotropic as well as aligned bicelles can be used to study membrane interaction of soluble proteins (D).
The propensity of bicelles for magnetic-alignment opened a completely new route for the preparation of macroscopically oriented membrane samples. Typically, proteins embedded in magnetically- aligned bicelles (Figs. 2G and 12A) reach a much higher quality of alignment than is possible for example in stacks of glass plates [15,16] or anodic aluminum disks (see Section 2 and Fig. 2C and D). Solid-state NMR of macroscopically aligned, static samples is a well-established and very productive branch of solid-state NMR spectroscopy [238]. It offers a wide array of well-tested custom-tailored pulse sequences, such as the separated local field experiments that were described in Section 7.1. Magneticallyaligned bicelles have given fresh impetus to this field, especially in the study of incorporated membrane proteins.
The largest integral membrane protein studied in this fashion to date is CXCR1, a human chemokine receptor that consists of seven transmembrane α-helices. CXCR1 was successfully incorporated in aligned bicelles [239]. Local and global dynamics of CXCR1 were studied by a combination of solid- and solution-state NMR experiments [240]. The interaction of CXCR1 with its ligand interleukin- 8 was characterized in aligned as well as isotropic bicelles [241]. For OmpX, outer membrane porin X which forms an 8-stranded β-barrel, the absolute orientation with respect to the surrounding bicelle bilayer was determined [242]. Functional (α4)2(β2)3 pentamers of transmembrane α-helices from the α4β2 neuronal nicotinic acetylcholine receptor were prepared in aligned bicelles and the effect of anesthetics was investigated by observing a selectively 15N-Leu labeled α4 peptide [243]. Results on the viral proteins p7 and Vpu in aligned bicelles have recently been reviewed [244]. Although both proteins belong to the same family of viroporins, p7 of HCV passes the membrane in two α-helical stretches, while Vpu of HIV-1 consists of only one transmembrane α-helix plus two α-helices that are attached peripherally to the membrane. The structure of MerFt was determined, a truncated version of a bacterial mercury transporter which has two transmembrane α-helices [245]. Several fragments of TatA, twin-arginine translocase consisting of a transmembrane α-helix and two membrane-associated α-helices, were investigated in aligned bicelles [246,247]. Bacteriophage Pf1 coat protein has a single transmembrane α-helix and was studied in bicelles with biphenyl lipid side-chains that align with their bilayer normal parallel to the external magnetic field [248] (Section 3.4). The feasibility of de novo sequential assignments in oriented bicelles has been demonstrated on sarcolipin, where 26 out of 31 amide 15N-resonances could be unambiguously assigned [249].
The Ramamoorthy laboratory has studied the integral membrane protein cytochrome b5 in aligned bicelles [250]. Cytochrome b5 consists of a transmembrane α-helix that serves as a membrane anchor, and a water soluble, globular domain. It was found to give high quality of orientation in aligned bicelles, which resulted in solid-state NMR spectra of very high resolution [251]. This result suggests that bicelles with their high degree of hydration are especially useful in the common case where membrane proteins have large globular domains in addition to their transmembrane portions. It was possible to reconstitute a complex of cytochrome b5 with its binding partner, the 56 kDa enzyme cytochrome P450 [250]. Innovative pulse sequences were developed on cytochrome b5 in bicelles that used proton-evolved local-field experiments to distinguish transmembrane and soluble domains [252], and used insensitive nuclei enhancement by polarization transfer (INEPT)-type magnetization transfer to study side-chain dynamics [253]. When cytochrome b5 in aligned bicelles was studied under magic-angle spinning conditions, where macroscopic orientation vanishes, spectra of very high quality could be observed, confirming the favorable conditions that bicelles offer for membrane proteins [212].
The more traditional field of solution-state NMR has also made rapid progress in the study of integral membrane proteins, and bicelles have made a significant contribution. Solution-state NMR offers a wide range of sophisticated tools to study proteins that tumble quickly on the NMR time scale [254–258]. Integral membrane proteins that are natively embedded in membranes typically do not meet the criterion of fast reorientation. For this reason, they are regularly solubilized in detergent micelles that do show fast tumbling. The detergent used for solubilization has been humorously described as “French swimwear” for the protein [259]. Naturally, smaller protein–detergent aggregates will result in faster reorientation and better solution-state NMR spectra. However, it has been found that not all proteins retain their functional form in protein–detergent micelles. Isotropic bicelles are especially attractive as an alternative in this context, as they possess a bilayered portion that offers a native-like environment for the embedded protein (Fig. 12B). For example, the functional form of Smr, staphylococcal small multidrug-resistance pump, is stabilized in bicelles [260]. A review on solubilizing agents presents Smr as prototypical case and stresses the importance of an assay to verify that a protein is present in its active form [261]. This publication states with respect to protein–detergent aggregates that “small is beautiful, but sometimes bigger is better”. A complete backbone assignment for the functional form of Smr in isotropic bicelles has been reported [262]. Several other contributions have reviewed the importance of detergent systems in the study of membrane proteins [263–266]. Two review contributions present comprehensive galleries of integral membrane proteins solved by solution-state NMR spectroscopy [267,268].
The structure determination of sensory rhodopsin II, a seven transmembrane helix receptor, in detergent micelles (pdb-id 2KSY) [269] has shown that these proteins are now within reach of solution-state NMR. The same study used spectra recorded in isotropic bicelles to show that environment-specific effects are minimal when using mild detergents. A complex of rhodopsin and transducin was prepared in isotropic bicelles and displayed dramatically increased stability [270], and it was possible to generate and purify a rhodopsin/transducin complex with constitutively active, recombinant rhodopsin [271]. A bicelle-associated structure was determined for full-length myristoylated ARF1, ADP ribosylation factor 1, which consists of an N-terminal bicelle-associated and a C-terminal catalytic domain connected by a flexible linker (pdb-id 2KSQ) [272]. EmrE, a small multidrug-resistance transporter, forms homodimers where each monomeric unit consists of four transmembrane α-helices. EmrE is highly sensitive to its environment but could successfully be incorporated into isotropic bicelles [273]. It was then shown that asymmetric antiparallel EmrE exchanges between inward- and outward-facing states that are identical except that they have opposite orientation in the membrane [274]. Homo- and hetero-dimers of single-pass transmembrane α-helices are of special interest since they play central roles in a large class of transmembrane signaling receptors [275]. A structure determination has been reported for the heterodimer of the αIIb and β3 transmembrane helices of integrin (pdb-id 2K9 J) [276]. Similarly, the spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases was reported (pdb-id 2KS1) [277]. Phospholamban, a single-pass α-helical transmembrane protein, has found special interest because of its role in regulating SERCA, sarcoplasmic reticulum Ca2+-ATPase [278]. Magic angle spinning of phospholamban in isotropic bicelles was used to probe equilibria between ground and excited states [279]. For OmpX, a bacterial outer membrane porin that forms an 8-stranded β-barrel, it was possible to observe intermolecular NOE contacts to DMPC lipids in the surrounding isotropic bicelle, but not to DHPC detergent, proving that the protein is embedded solely in the bilayered portion of the bicelle [155].
When soluble, globular proteins are introduced into dilute samples of strongly aligned bicelles (Fig. 12C), they experience a weak macroscopic alignment that gives rise to small anisotropic NMR interactions, so-called residual dipolar couplings or RDCs, on the order of several Hz. The transferred partial orientation of the soluble protein is caused by interaction with the aligned medium which forms an obstacle to the free re-orientation that would be observed in solution. Numerous alignment media are used besides bicelles in the study of RDCs; tabulated overviews of alignment media are available [280,281]. The alignment that a molecule will experience in a certain environment can be predicted on the basis of steric interaction alone [282], or additionally taking into account electrostatic interaction [283]; the software PALES is available for this task [284].
RDC measurements were first reported on ubiquitin partially ordered in q = 2.9 DMPC/DHPC bicelles [285]. Their role in structure determination of biomolecules has been reviewed repeatedly; we mention a contribution that includes an overview of pulse sequences for recording RDCs [280]. Other review contributions have focused on RDCs as probes of biomolecular dynamics [281], and on RDC studies on proteins [286] and RNA [287]. It has to be noted that the interaction with the alignment medium may change the properties of the studied protein. For example, the putatively inactive dimeric state of the chemokine SDF-1/CXCL12 is favored when bicelles are used as alignment medium [288]. Since RDCs report on global orientation, they are especially useful for determining the overall orientation of protein domains [289]. Three independent sets of RDCs in aligned bicelles of three different conditions were determined for the three N-terminal domains of human factor H and analyzed to yield their relative orientation [290]. For ubiquitin, 18 independent ordering media including bicelles gave sufficient RDC data to observe protein recognition dynamics on a microsecond time scale [291].
An especially intriguing situation occurs when bicelles in RDC studies are not used as alignment medium but are themselves subjected to partial alignment. Isotropic bicelles containing the HIV-1 Env peptide were introduced into a stretched hydrogel and RDCs could be recorded that showed considerable differences when compared to RDCs recorded on the same peptide in detergent micelles [292]. Similarly, DNA tetrads can be used as alignment medium to measure RDCs on molecules embedded in isotropic bicelles [293].
The interaction of soluble, globular proteins with lipid bilayers can be studied in the presence of isotropic bicelles (Fig. 12D). This is different from the separated local field (SLF) studies described in Section 7, where strong binding to aligned bicelles is a prerequisite. In the case of globular protein and isotropic bicelle, neither strong nor partial alignment of the protein is observed and protein-lipid interactions can be investigated. For example, details of membrane binding of myristoylated and non-myristoylated ARF1, ADP-ribosylation factor 1 (pdb-id 2K5U), was compared in isotropic bicelles [294]. Spontaneous insertion of the apoptotic repressor BclXL into q = 0.5 DMPC/DHPC bicelles was found to cause a dramatic conformational change that inhibits binding with its BH3 ligand [295]. Isotropic bicelles were used to supply lipid substrate to cobra venom phospholipase A2 for kinetic assays [296]. It has also been observed that amyloid formation is modulated in bicellar solutions [297].
11. New and notable
A number of recent publications have presented new bicelle designs and novel bicelle applications that do not fit easily into the categories of this review article, but that feel particularly noteworthy. A new bicelle was designed that contains both cholesterol and unsaturated lipid sidechains and forms separated domains that can potentially be used as models for physiological lipid rafts [298]. Bicelles were stabilized by encompassing them into a layer of siloxane ceramics [299]. Newly designed peptide copolymers form flat discoidal structures similar to bicelles [59]. The use of special detergents extends the possible range of bicelle concentrations to below 0.5% lipid weight per volume [113]. Even lower concentrations can be reached by encapsulating bicelles in vesicles; such formulations have been introduced under the name ‘bicosomes’ [300].
More traditional bicelle formulations have been applied in completely novel ways. The use of bicelles in the crystallization of membrane proteins [38,44] has been discussed in detail in Section 2.4. Bicelles as detergents in the cell-free expression of membrane proteins offer distinct advantages [34,301,302]. The effects of applying bicellar preparations to the skin have received broad attention [303–308] and were recently reviewed [309]. Of special interest seems to be the prospect of delivering drug substances to the skin using bicelles, as demonstrated for diclofenac [310]. The voltage gated potassium channel modulatory membrane protein KCNE3 was delivered into oocytes in functional form using bicelles [311].
In the field of nanotechnology, bicelles were used as a temporary scaffold to synthesize rigid organic nanodisks by polymerization [312] and to generate platinum nanowheels [313,314]. Bicelles were used to build assemblies of multiple highly fluid free lipid bilayers that are tethered together by cholesterol-anchored double-stranded DNA [315]. Supported and suspended lipid bilayers have been generated on silicon chips using bicelles [316].
12. Summary and conclusion
Bicelles have quickly emerged as another amazing possibility in the host of lipid morphologies [317]. The present contribution has focused on the studies that generated today’s comprehensive understanding of bicelle properties. NMR experiments along with scattering and diffusion measurements can give quick and unambiguous characterizations of bicelle morphology and phase diagrams. Bicelles quickly took on an important role in the context of lipid bilayer samples for NMR and EPR spectroscopy. SLF and MAS experiments on bicelles are regularly used to study bilayer properties. The interaction of small molecules with bilayers can also be studied in bicelle samples. The study of membrane protein structure forms the most prominent practical application of bicelles.
The application of bicelles in the structural studies of membrane proteins has a number of unique advantages. Bicelles can embed membrane proteins into truly flat, bilayered lipid patches with lipid compositions that can be chosen from a wide array of options. Bicelles form only in limited ranges within the phase diagram, but these ranges are continuously expanded and typically coincide with ranges of conditions that are desirable for the study of membrane proteins. By the ratio q of lipid over detergent, the size of bicelles can be freely chosen. Bicelles with small q values can be used for high-throughput solution-state NMR studies while those with large q values are ideal for solid-state NMR studies. Even q-variations ranging from solid-state into solution-state conditions are possible. Since bicelles contain bulk water, they enable natural folding of even those membrane-associated proteins that contain large soluble domains and therefore make physiologically relevant structural studies possible. In summary, bicelles are the most versatile model membrane and as a consequence they are increasingly and routinely used in studies ranging from crystallography to NMR spectroscopy.
Today, the field extends so widely that it is virtually impossible to cover the phenomenon “bicelle” in a single review article. Insights on protein structure gained with the help of bicellar formulations are emerging at an amazing pace. At the same time, research that is focused on bicelles in their own right has elucidated most of their basic structural and dynamic properties. The most exciting activity is found on the fringes of the field, where new imaginative variations and applications keep emerging at a similar pace.
Acknowledgments
This study was supported by funds from NIH (GM084018 and GM095640 to A.R.).
Glossary of abbreviations
- AAO
anodic aluminum oxide
- BPC
biopartitioning chromatography
- bR
bacteriorhodopsin
- C12E18
octaethylene glycol monododecyl ether
- CHAPSO
3-(cholamidopropyl)dimethylammonio-2-hydroxy-1-propanesulfonate
- CMC
critical micelle concentration
- CSA
chemical shift anisotropy
- CTAB
hexadecyltrimethylammonium bromide
- DHPC
dihexanoylphosphatidylcholine
- DLPC
dilauroylphosphatidylcholine
- DMPC
dimyristoylphosphatidylcholine
- DMPE
dimyristoylphosphatidylethanolamine
- DMPG
dimyristoylphosphatidylglycerol
- DMTAP
1,2-dimyristoyl-3-trimethylammonium-propane
- DPC
dodecylphosphocholine
- DPPC
dipalmitoylphosphatidylcholine
- DSPE
distearoylphosphatidylethanolamine
- DTPA
diethylenetriaminepentaacetate
- EM
electron microscopy
- EPR
electron paramagnetic resonance
- FRAP
fluorescence recovery after photobleaching
- HDL
high-density lipoprotein
- HERSELF
heteronuclear rotating-frame spin exchange via local field
- HIMSELF
heteronuclear isotropic mixing spin exchange via local field
- INEPT
insensitive nuclei enhancement by polarization transfer
- MAS
magic-angle spinning
- MD
molecular dynamics
- MLV
multilamellar vesicle
- NMR
nuclear magnetic resonance
- PEG
polyethyleneglycol
- PELF
proton evolved local field spectroscopy
- PFG
pulsed field gradient
- PISEMA
polarization inversion spin exchange at the magic angle
- POPC
1-palmitoyl-2-oleoyl-phosphatidylcholine
- PRE
paramagnetic relaxation enhancement
- PSPC
1-palmitoyl-2-stearoyl-phosphatidylcholine
- RDC
residual dipolar coupling
- SAMMY
magic-sandwich PISEMA
- SANS
small-angle neutron scattering
- SDS
sodium dodecyl sulfate
- SLF
separated local field
- SLUV
short-chain long-chain unilamellar vesicle
- STE
stimulated echo
- SUV
small unilamellar vesicle
- TEM
transmission electron microscopy
- TM
transmembrane
- TMS
tetramethylsilane
- VASS
variable-angle sample spinning
References
- 1.van Dam L, Karlsson G, Edwards K. Morphology of magnetically aligning DMPC/DHPC aggregates-perforated sheets, not disks. Langmuir. 2006;22:3280–3285. doi: 10.1021/la052988m. [DOI] [PubMed] [Google Scholar]
- 2.Winterhalter M, Lasic DD. Liposome stability and formation: experimental parameters and theories on the size distribution. Chem Phys Lipids. 1993;64:35–43. doi: 10.1016/0009-3084(93)90056-9. [DOI] [PubMed] [Google Scholar]
- 3.Da Costa G, Mouret L, Chevance S, Le Rumeur E, Bondon A. NMR of molecules interacting with lipids in small unilamellar vesicles. Eur Biophys J. 2007;36:933–942. doi: 10.1007/s00249-007-0186-7. [DOI] [PubMed] [Google Scholar]
- 4.Mäler L, Gräslund A. Artificial membrane models for the study of macromolecular delivery. Meth Mol Biol. 2009;480:129–139. doi: 10.1007/978-1-59745-429-2_9. [DOI] [PubMed] [Google Scholar]
- 5.Herzfeld J, Berger AE. Sideband intensities in NMR spectra of samples spinning at the magic angle. J Chem Phys. 1980;73:6021–6030. [Google Scholar]
- 6.Prongidi-Fix L, Bertani P, Bechinger B. The membrane alignment of helical peptides from non-oriented 15N chemical shift solid-state NMR spectroscopy. J Am Chem Soc. 2007;129:8430–8431. doi: 10.1021/ja072668k. [DOI] [PubMed] [Google Scholar]
- 7.Tremouilhac P, Strandberg E, Wadhwani P, Ulrich AS. Conditions affecting the re-alignment of the antimicrobial peptide PGLa in membranes as monitored by solid-state 2H-NMR. Biochim Biophys Acta. 2006;1758:1330–1342. doi: 10.1016/j.bbamem.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 8.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moll F, III, Cross TA. Optimizing and characterizing alignment of oriented lipid bilayers containing gramicidin D. Biophys J. 1990;57:351–362. doi: 10.1016/S0006-3495(90)82536-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winston PW, Bates DH. Saturated solutions for the control of humidity in biological research. Ecology. 1960;41:232–237. [Google Scholar]
- 11.McDonnel PA, Shon K, Kim Y, Opella SJ. Fd coat protein structure in membrane environments. J Mol Biol. 1993;233:447–463. doi: 10.1006/jmbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- 12.Ketchem RR, Hu W, Cross TA. High-resolution conformation of gramicidin A in lipid bilayer by solid-state NMR. Science. 1993;261:1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- 13.Separovic F, Killian JA, Cotten M, Busath DD, Cross TA. Modeling the membrane environment for membrane proteins. Biophys J. 2011;100:2073–2074. doi: 10.1016/j.bpj.2011.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulrich AS, Heyn MP, Watts A. Structure determination of the cyclohexene ring of retinal in bacteriorhodopsin by solid-state deuterium NMR. Biochemistry. 1992;31:10390–10399. doi: 10.1021/bi00157a029. [DOI] [PubMed] [Google Scholar]
- 15.Lee D-K, Henzler Wildman KA, Ramamoorthy A. Solid state NMR spectroscopy of aligned lipid bilayers at low temperatures. J Am Chem Soc. 2004;126:2318–2319. doi: 10.1021/ja039077o. [DOI] [PubMed] [Google Scholar]
- 16.Hallock KJ, Henzler Wildman KA, Lee D-K, Ramamoorthy A. An innovative procedure using a sublimable solid to align lipid bilayers for solid-state NMR studies. Biophys J. 2002;82:2499–2503. doi: 10.1016/S0006-3495(02)75592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallock KJ, Lee DK, Omnaas J, Mosberg HI, Ramamoorthy A. Membrane composition determines pardaxin’s mechanism of lipid bilayer disruption. Biophys J. 2002;83:1004–1013. doi: 10.1016/S0006-3495(02)75226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallock KJ, Lee DK, Ramamoorthy A. MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys J. 2003;84:3052–3060. doi: 10.1016/S0006-3495(03)70031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henzler Wildman KA, Lee D-K, Ramamoorthy A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 2003;42:6545–6558. doi: 10.1021/bi0273563. [DOI] [PubMed] [Google Scholar]
- 20.Henzler Wildman KA, Martinez GV, Brown MF, Ramamoorthy A. Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry. 2004;43:8459–8469. doi: 10.1021/bi036284s. [DOI] [PubMed] [Google Scholar]
- 21.Ramamoorthy A, Kandasamy SK, Lee DK, Kidambi S, Larson RG. Topology and tilt of cell-signaling peptides containing nuclear localization sequences in membrane bilayers determined by solid-state NMR and molecular dynamics simulation studies. Biochemistry. 2007;46:965–975. doi: 10.1021/bi061895g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pointer-Keenan CD, Lee DK, Hallock KJ, Tan A, Zand R, Ramamoorthy A. Investigation of the interaction of myelin basic protein with phospholipid bilayers using solid-state NMR spectroscopy. Chem Phys Lipids. 2004;132:47–54. doi: 10.1016/j.chemphyslip.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Furneaux RC, Rigby WR, Davidson AP. The formation of controlled-porosity membranes from anodically oxidized aluminum. Nature. 1989;337:147–149. [Google Scholar]
- 24.Chekmenev EY, Hu J, Gorkov PL, Brey WW, Cross TA, Ruuge A, Smirnov AI. 15N and 31P solid-state NMR study of transmembrane domain alignment of M2 protein of influenza A virus in hydrated cylindrical lipid bilayers confined to anodic aluminum oxide nanopores. J Magn Reson. 2005;173:322–327. doi: 10.1016/j.jmr.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Smirnov AI, Poluektov OG. Substrate-supported lipid nanotube arrays. J Am Chem Soc. 2003;125:8434–8435. doi: 10.1021/ja0349406. [DOI] [PubMed] [Google Scholar]
- 26.Gaede HC, Luckett KM, Polozov IV, Gawrisch K. Multinuclear NMR studies of single lipid bilayers supported in cylindrical aluminum oxide nanopores. Langmuir. 2004;20:7711–7719. doi: 10.1021/la0493114. [DOI] [PubMed] [Google Scholar]
- 27.Karp ES, Newstadt JP, Chu S, Lorigan GA. Characterization of lipid bilayer formation in aligned nanoporous aluminium oxide nanotube arrays. J Magn Reson. 2007;187:112–119. doi: 10.1016/j.jmr.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorigan GA, Dave PC, Tiburu EK, Damodaran K, Abu-Baker S, Karp ES, Gibbons WJ, Minto RE. Solid-state NMR spectroscopic studies of an integral membrane protein inserted into aligned phosopholipid bilayer nanotube arrays. J Am Chem Soc. 2004;126:9504–9505. doi: 10.1021/ja047317+. [DOI] [PubMed] [Google Scholar]
- 29.Soubias O, Polozov IV, Teague WE, Yeliseev AA, Gawrisch K. Functional reconstitution of rhodopsin into tubular lipid bilayers supported by nanoporous material. Biochemistry. 2006;45:15583–15590. doi: 10.1021/bi061416d. [DOI] [PubMed] [Google Scholar]
- 30.Brender JR, Dürr UHN, Heyl D, Budarapu MB, Ramamoorthy A. Membrane fragmentation by an amyloidogenic fragment of human islet amyloid polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes. Biochim Biophys Acta. 2007;1768:2026–2029. doi: 10.1016/j.bbamem.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brouillette CG, Jones JL, Ng TC, Kercret H, Chung BH, Segrest JP. Structural studies of apolipoprotein A-I/phosphatidylcholine recombinants by high-field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry. 1984;23:359–367. doi: 10.1021/bi00297a027. [DOI] [PubMed] [Google Scholar]
- 32.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shenkarev ZO, Lyukmanova EN, Solozhekin OI, Gagnidze IE, Nekrasova OV, Chupin VV, Tagaev AA, Yakimenko ZA, Ovchinnikova TV, Kirpichnikov MP, Arseniev AS. Lipid–protein nanodiscs: possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry (Moscow) 2009;74:756–765. doi: 10.1134/s0006297909070086. [DOI] [PubMed] [Google Scholar]
- 34.Lyukmanova EN, Shenkarev ZO, Khabibullina NF, Kopeina GS, Shulepko MA, Paramonov AS, Mineev KS, Tikhonov RV, Shingarova LN, Petrovskaya LE, Dolgikh DA, Arseniev AS, Kirpichnikov MP. Lipid–protein nanodisks for cell-free production of integral membrane proteins in a soluble and folded state: Comparison with detergent micelles, bicelles and liposomes. Biochim Biophys Acta. 1818;2011:349–358. doi: 10.1016/j.bbamem.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 35.Popot JL, Berry EA, Charvolin D, Creuzenet C, Ebel C, Engelman DM, Flötenmeyer M, Giusti F, Gohon Y, Hervé P, Hong Q, Lakey JH, Leonard K, Shuman HA, Timmins P, Warschawski DE, Zito F, Zoonens M, Pucci B, Tribet C. Amphipols: polymeric surfactants for membrane biology research. Cell Mol Life Sci. 2003;60:1559–1574. doi: 10.1007/s00018-003-3169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders CR, Landis GC. Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry. 1995;34:4030– 4040. doi: 10.1021/bi00012a022. [DOI] [PubMed] [Google Scholar]
- 37.Caffrey M. Membrane protein crystallization. J Struct Biol. 2003;142:108– 132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 38.Johansson LC, Wöhri AB, Katona G, Engström S, Neutze R. Membrane protein crystallization from lipidic phases. Curr Opin Struct Biol. 2009;19:372–378. doi: 10.1016/j.sbi.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 40.Faham S, Boulting GL, Massey EA, Yohannan S, Yang D, Bowie JU. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Prot Sci. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanii LS, El-Sayed MA. Partial dehydration of the retinal binding pocket and proof for photochemical deprotonation of the retinal Schiff base in bicelle bacteriorhodopsin. Photochem Photobiol. 2005;81:1356–1360. doi: 10.1562/2005-03-09-RA-458. [DOI] [PubMed] [Google Scholar]
- 42.Spudich EN, Ozorowski G, Schow EV, Tobias DJ, Spudich JL, Luecke H. A transporter converted into a sensor, a phototaxis signaling mutant of bacteriorhodopsin at 3.0 Å. J Mol Biol. 2011;415:455–463. doi: 10.1016/j.jmb.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Elferich J, Gouaux E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol. 2012;19:212–219. doi: 10.1038/nsmb.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ujwal R, Abramson J. High-throughput crystallization of membrane proteins using the lipidic bicelle method. J Vis Exp. 2012;59:e3383. doi: 10.3791/3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards K, Johnsson M, Karlsson G, Silvander M. Effect of polyethyleneglycol-phospholipids on aggregate structure in preparations of small unilamellar liposomes. Biophys J. 1997;73:258–266. doi: 10.1016/S0006-3495(97)78066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnsson M, Edwards K. Phase behavior and aggregate structure in mixtures of dioleoylphosphatidylethanolamine and poly(ethylene glycol)-lipids. Biophys J. 2001;80:313–323. doi: 10.1016/S0006-3495(01)76016-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ickenstein LM, Arfvidsson MC, Needham D, Mayer LD, Edwards K. Disc formation in cholesterol-free liposomes during phase transition. Biochim Biophys Acta. 2003;1614:135–138. doi: 10.1016/s0005-2736(03)00196-2. [DOI] [PubMed] [Google Scholar]
- 49.Johnsson M, Edwards K. Liposomes, disks, and spherical micelles: aggregate structure in mixtures of gel phase phosphatidylcholines and poly(ethylene glycol)-phospholipids. Biophys J. 2003;85:3839–3847. doi: 10.1016/S0006-3495(03)74798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandström MC, Johansson E, Edwards K. Structure of mixed micelles formed in PEG-lipid/lipid dispersions. Langmuir. 2007;23:4192–4198. doi: 10.1021/la063501s. [DOI] [PubMed] [Google Scholar]
- 51.Sandström MC, Johansson E, Edwards K. Influence of preparation path on the formation of discs and threadlike micelles in DSPE-PEG2000/lipid systems. Biophys Chem. 2008;132:97–103. doi: 10.1016/j.bpc.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 52.Ickenstein LM, Sandström MC, Mayer LD, Edwards K. Effects of phospholipid hydrolysis on the aggregate structure in DPPC/DSPE-PEG2000 liposome preparations after gel to liquid crystalline phase transition. Biochim Biophys Acta. 2006;1758:171–180. doi: 10.1016/j.bbamem.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 53.Šachl R, Mikhalyov I, Gretskaya N, Olzynska A, Hof M, Johansson LB-Å. Distribution of BODIPY-labelled phosphatidylethanolamines in lipid bilayers exhibiting different curvatures. Phys Chem Chem Phys. 2011;13:11694– 11701. doi: 10.1039/c1cp20608g. [DOI] [PubMed] [Google Scholar]
- 54.Lee H, Pastor RW. Coarse-grained model for PEGylated lipids: effect of PEGylation on the size and shape of self-assembled structures. J Phys Chem B. 2011;115:7830–7837. doi: 10.1021/jp2020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundquist A, Hansen SB, Nordström H, Danielson UH, Edwards K. Biotinylated lipid bilayer disks as model membranes for biosensor analyses. Anal Biochem. 2010;405:153–159. doi: 10.1016/j.ab.2010.06.030. [DOI] [PubMed] [Google Scholar]
- 56.Lundquist A, Wessman P, Rennie AR, Edwards K. Melittin–lipid interaction: a comparative study using liposomes, micelles and bilayer disks. Biochim Biopyhs Acta. 2008;1778:2210–2216. doi: 10.1016/j.bbamem.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Johansson E, Sandström MC, Bergström M, Edwards K. On the formation of discoidal versus threadlike micelles in dilute aqueous surfactant/lipid systems. Langmuir. 2008;24:1731–1739. doi: 10.1021/la702637h. [DOI] [PubMed] [Google Scholar]
- 58.Beck P, Liebi M, Kohlbrecher J, Ishikawa T, Rüegger H, Fischer P, Walde P, Windhab E. Novel type of bicellar disks from a mixture of DMPC and DMPE-DTPA with complexed lanthanides. Langmuir. 2010;26:5382–5387. doi: 10.1021/la903806a. [DOI] [PubMed] [Google Scholar]
- 59.Marsden HR, Handgraaf JW, Nudelman F, Sommerdijk NAJM, Kros A. Uniting polypeptides with sequence-designed peptides: synthesis and assembly of poly(γ-benzyl L-glutamate)-β-coiled-coil peptide copolymers. J Am Chem Soc. 2010;132:2370–2377. doi: 10.1021/ja909540a. [DOI] [PubMed] [Google Scholar]
- 60.Amaral LQ, Pimentel CA, Tavares MR, Vanin JA. Study of magnetically oriented lyotropic mesophase. J Chem Phys. 1979;71:2940–2945. [Google Scholar]
- 61.Forrest BJ, Reeves LW. New lyotropic liquid crystals composed of finite nonspherical micelles. Chem Rev. 1981;81:1–14. [Google Scholar]
- 62.Ramamoorthy A. Thermotropic Liquid Crystals: Recent Advances. Springer; Dordrecht, The Netherlands: 2007. [Google Scholar]
- 63.Seelig J, Borle F, Cross TA. Magnetic ordering of phospholipid membranes. Biochim Biophys Acta. 1985;814:195–198. [Google Scholar]
- 64.Gabriel NE, Roberts MF. Spontaneous formation of stable unilamellar vesicles. Biochemistry. 1984;23:4011–4015. doi: 10.1021/bi00313a001. [DOI] [PubMed] [Google Scholar]
- 65.Gabriel NE, Roberts MF. Interaction of short-chain lecithin with long-chain phospholipids: characterization of vesicles that form spontaneously. Biochemistry. 1986;25:2812–2821. doi: 10.1021/bi00358a012. [DOI] [PubMed] [Google Scholar]
- 66.Stejskal EO, Tanner J. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42:288– 292. [Google Scholar]
- 67.Nieh MP, Harroun TA, Raghunathan VA, Glinka CJ, Katsaras J. Spontaneously formed monodisperse biomimetic unilamellar vesicles: the effect of charge, dilution, and time. Biophys J. 2004;86:2615–2629. doi: 10.1016/S0006-3495(04)74316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nieh MP, Raghunathan VA, Kline SR, Harroun TA, Huang CY, Pencer J, Katsaras J. Spontaneously formed unilamellar vesicles with path-dependent size distribution. Langmuir. 2005;21:6656–6661. doi: 10.1021/la0508994. [DOI] [PubMed] [Google Scholar]
- 69.Nieh MP, Dolinar P, Kucerka N, Kline SR, Debeer-Schmitt LM, Littrell KC, Katsaras J. The formation of kinetically-trapped nanoscopic unilamellar vesicles from metastable nanodiscs. Langmuir. 2011;27:14308–14316. doi: 10.1021/la2023314. [DOI] [PubMed] [Google Scholar]
- 70.Müller K. Structural dimorphism of bile salt/lecithin mixed micelles. A possible regulatory mechanism for cholesterol solubility in bile? X-ray structure analysis. Biochemistry. 1981;20:404–414. doi: 10.1021/bi00505a028. [DOI] [PubMed] [Google Scholar]
- 71.Ram P, Prestegard JH. Magnetic field induced ordering of bile salt/phospholipid micelles: new media for NMR structural investigations. Biochim Biophys Acta. 1988;940:289–294. doi: 10.1016/0005-2736(88)90203-9. [DOI] [PubMed] [Google Scholar]
- 72.Sanders CR, Prestegard JH. Magnetically orientable phospholipid bilayers containing small amounts of a bile salt analogue, CHAPSO. Biophys J. 1990;58:447–460. doi: 10.1016/S0006-3495(90)82390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hare BJ, Prestegard JH, Engelman DM. Small angle X-ray scattering studies of magnetically oriented lipid bilayers. Biophys J. 1995;69:1891–1896. doi: 10.1016/S0006-3495(95)80059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aubin Y, Ito Y, Paulson JC, Prestegard JH. Structure and dynamics of the sialic acid moiety of GM3-ganglioside at the surface of a magnetically oriented membrane. Biochemistry. 1993;32:13405–13413. doi: 10.1021/bi00212a005. [DOI] [PubMed] [Google Scholar]
- 75.Hare BJ, Rise F, Aubin Y, Prestegard JH. 13C NMR studies of wheat germ agglutinin interactions with N-acetylglucosamine at a magnetically oriented bilayer surface. Biochemistry. 1994;33:10137–10148. doi: 10.1021/bi00199a042. [DOI] [PubMed] [Google Scholar]
- 76.Howard KP, Prestegard JH. Conformation of sulfoquinovosyldiacylglycerol bound to a magnetically oriented membrane system. Biophys J. 1996;71:2573–2582. doi: 10.1016/S0006-3495(96)79449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanders CR, Schwonek JP. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry. 1992;31:8898–8905. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- 78.Sanders CR, Hare BJ, Howard KP, Prestegard JH. Magnetically oriented phospholipid micelles as a tool for the study of membrane associated molecules. Prog Nucl Magn Reson Spectrosc. 1994;26:421–444. [Google Scholar]
- 79.de Angelis AA, Opella SJ. Bicelle samples for solid-state NMR of membrane proteins. Nat Prot. 2007;2:2332–2338. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- 80.McKibbin C, Farmer NA, Jeans C, Reeves PJ, Khorana HG, Wallace BA, Edwards PC, Villa C, Booth PJ. Opsin stability and folding: modulation by phospholipid bicelles. J Mol Biol. 2007;374:1319–1332. doi: 10.1016/j.jmb.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 81.Soong R, Macdonald PM. Water diffusion in bicelles and the mixed bicelle model. Langmuir. 2009;25:380–390. doi: 10.1021/la801739a. [DOI] [PubMed] [Google Scholar]
- 82.Katsaras J, Donaberger RL, Swainson IP, Tennant DC, Tun Z, Vold RR, Prosser RS. Rarely observed phase transitions in a novel lyotropic liquid crystal system. Phys Rev Lett. 1997;78:899–902. [Google Scholar]
- 83.Cardon TB, Tiburu EK, Lorigan GA. Magnetically aligned phospholipid bilayers in weak magnetic fields: optimization, mechanism, and advantages for X-band EPR studies. J Magn Reson. 2003;161:77–90. doi: 10.1016/s1090-7807(02)00109-x. [DOI] [PubMed] [Google Scholar]
- 84.Kogan M, Beke-Somfai T, Nordén B. Flow-alignment of bicellar lipid mixtures: orientations of probe molecules and membrane-associated biomacromolecules in lipid membranes studied with polarized light. Chem Commun. 2011;47:7356–7358. doi: 10.1039/c1cc12313k. [DOI] [PubMed] [Google Scholar]
- 85.Marcotte I, Bélanger A, Auger M. The orientation effect of gramicidin A on bicelles and Eu3+-doped bicelles as studied by solid-state NMR and FT-IR spectroscopy. Chem Phys Lip. 2006;139:137–149. doi: 10.1016/j.chemphyslip.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 86.Loudet-Courreges C, Nallet F, Dufourc EJ, Oda R. Unprecedented observation of days-long remnant orientation of phospholipid bicelles: a small-angle X-ray scattering and theoretical study. Langmuir. 2011;27:9122–9130. doi: 10.1021/la1050817. [DOI] [PubMed] [Google Scholar]
- 87.Prosser RS, Hunt SA, DiNatale JA, Vold RR. Magnetically aligned membrane model systems with positive order parameter: switching the sign of Szz with paramagnetic ions. J Am Chem Soc. 1996;118:269–270. [Google Scholar]
- 88.Prosser RS, Hwang JS, Vold RR. Magnetically aligned phospholipids with positive ordering: a new model membrane system. Biophys J. 1998;74:2405–2418. doi: 10.1016/S0006-3495(98)77949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prosser RS, Volkov VB, Shiyanovskaya IV. Novel chelate-induced magnetic alignment of biological membranes. Biophys J. 1998;75:2163–2189. doi: 10.1016/S0006-3495(98)77659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamamoto K, Xu J, Kawulka KE, Vederas JC, Ramamoorthy A. Use of copper-chelated lipid speeds up NMR measurements from membrane proteins. J Am Chem Soc. 2010;132:6929–6931. doi: 10.1021/ja102103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamamoto K, Vivekanandan S, Ramamoorthy A. Fast NMR data acquisition from bicelles containing a membrane-associated peptide at natural-abundance. J Phys Chem B. 2011;115:12448–12455. doi: 10.1021/jp2076098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loudet C, Manet S, Gineste S, Oda R, Achard MF, Dufourc EJ. Biphenyl bicelle disks align perpendicular to magnetic fields on large temperature scales. A study combining synthesis, solid state NMR, TEM and SAXS. Biophys J. 2007;92:3949–3959. doi: 10.1529/biophysj.106.097758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park SH, Loudet C, Marassi FM, Dufourc EJ, Opella SJ. Solid-state NMR spectroscopy of a membrane protein in biphenyl phospholipid bicelles with the bilayer normal parallel to the magnetic field. J Magn Reson. 2008;193:133–138. doi: 10.1016/j.jmr.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arnold A, Labrot T, Oda R, Dufourc EJ. Cation modulation of bicelle size and magnetic alignment as revealed by solid-state NMR and electron microscopy. Biophys J. 2002;83:2667–2680. doi: 10.1016/S0006-3495(02)75276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cavagnero S, Dyson HJ, Wright PE. Improved low pH bicelle system for orienting macromolecules over a wide temperature range. J Biomol NMR. 1999;13:387–391. doi: 10.1023/a:1008360022444. [DOI] [PubMed] [Google Scholar]
- 96.Aussenac F, Lavigne B, Dufourc EJ. Toward bicelle stability with ether-linked phospholipids: temperature, composition, and hydration diagrams by 2H and 31P solid-state NMR. Langmuir. 2005;21:7129–7135. doi: 10.1021/la050243a. [DOI] [PubMed] [Google Scholar]
- 97.Ottiger M, Bax A. Bicelle-based liquid crystals for NMR-measurement of dipolar couplings at acidic and basic pH values. J Biomol NMR. 1999;13:187–191. doi: 10.1023/a:1008395916985. [DOI] [PubMed] [Google Scholar]
- 98.Bertelsen K, Vad B, Nielsen EH, Hansen SK, Skrydstrup T, Otzen DE, Vosegaard T, Nielsen NC. Long-term-stable ether-lipid vs conventional ester-lipid bicelles in oriented solid-state NMR: altered structural information in studies of antimicrobial peptides. J Phys Chem B. 2011;115:1767–1774. doi: 10.1021/jp110866g. [DOI] [PubMed] [Google Scholar]
- 99.King V, Parker M, Howard KP. Pegylation of magnetically oriented lipid bilayers. J Magn Reson. 2000;142:177–182. doi: 10.1006/jmre.1999.1928. [DOI] [PubMed] [Google Scholar]
- 100.Losonczi JA, Prestegard JH. Improved dilute bicelle solutions for high-resolution NMR of biological macromolecules. J Biomol NMR. 1998;12:447. doi: 10.1023/a:1008302110884. [DOI] [PubMed] [Google Scholar]
- 101.Triba MN, Devaux PF, Warschawski DE. Effects of lipid chain length and unsaturation on bicelles stability. A phosphorus NMR study. Biophys J. 2006;91:1357–1367. doi: 10.1529/biophysj.106.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shapiro RA, Brindley AJ, Martin RW. Thermal stabilization of DMPC/DHPC bicelles by addition of cholesterol sulfate. J Am Chem Soc. 2010;132:11406–11407. doi: 10.1021/ja1041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Whiles JA, Glover KJ, Vold RR, Komives EA. Methods for studying transmembrane peptides in bicelles: consequences of hydrophobic mismatch and peptide sequence. J Magn Reson. 2002;158:149–156. doi: 10.1016/s1090-7807(02)00068-x. [DOI] [PubMed] [Google Scholar]
- 104.Tiburu EK, Moton DM, Lorigan GA. Development of magnetically aligned phospholipid bilayers in mixtures of palmitoylstearoylphosphatidylcholine and dihexanoylphosphatidylcholine by solid-state NMR spectroscopy. Biochim Biophys Acta. 2001;1512:206–214. doi: 10.1016/s0005-2736(01)00320-0. [DOI] [PubMed] [Google Scholar]
- 105.Struppe J, Komives EA, Taylor SS, Vold RR. 2H NMR studies of a myristoylated peptide in neutral and acidic phospholipid bicelles. Biochemistry. 1998;37:15523–15527. doi: 10.1021/bi981326b. [DOI] [PubMed] [Google Scholar]
- 106.Struppe J, Whiles JA, Vold RR. Acidic phospholipid bicelles: a versatile model membrane system. Biophys J. 2000;78:281–289. doi: 10.1016/S0006-3495(00)76591-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sasaki H, Fukuzawa S, Kikuchi J, Yokoyama S, Hirota H, Tachibana K. Cholesterol doping induced enhanced stability of bicelles. Langmuir. 2003;19:9841–9844. [Google Scholar]
- 108.Lu JX, Caporini MA, Lorigan GA. The effects of cholesterol on magnetically aligned phospholipid bilayers: a solid-state NMR and EPR spectroscopy study. J Magn Reson. 2004;168:18–30. doi: 10.1016/j.jmr.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 109.Tiburu EK, Dave PC, Lorigan GA. Solid-state 2H NMR studies of the effects of cholesterol on the acyl chain dynamics of magnetically aligned phospholipid bilayers. Magn Reson Chem. 2004;42:132–138. doi: 10.1002/mrc.1324. [DOI] [PubMed] [Google Scholar]
- 110.Minto RE, Adhikari PR, Lorigan GA. A 2H solid-state NMR spectroscopic investigation of biomimetic bicelles containing cholesterol and polyunsaturated phosphatidylcholine. Chem Phys Lip. 2004;132:55–64. doi: 10.1016/j.chemphyslip.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 111.Yamaguchi T, Suzuki T, Yasuda T, Oishi T, Matsumori N, Murata M. NMR-based conformational analysis of sphingomyelin in bicelles. Bioorg Med Chem. 2012;20:270–278. doi: 10.1016/j.bmc.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 112.Barbosa-Barros L, de la Maza A, Walther P, Estelrich J, López O. Morphological effects of ceramide on DMPC/DHPC bicelles. J Microsc. 2008;230:16–26. doi: 10.1111/j.1365-2818.2008.01950.x. [DOI] [PubMed] [Google Scholar]
- 113.Lu Z, Van Horn WD, Chen J, Mathew S, Zent R, Sanders CR. Bicelles at low concentrations. Mol Pharm. 2012;9:752–761. doi: 10.1021/mp2004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scholtysek P, Achilles A, Hoffmann CV, Lechner BD, Meister A, Tschierske C, Saalwächter K, Edwards K, Blume A. A T-shaped amphiphilic molecule forms closed vesicles in water and bicelles in mixtures with a membrane lipid. J Phys Chem B. 2012;116:4871–4878. doi: 10.1021/jp207996r. [DOI] [PubMed] [Google Scholar]
- 115.Park SH, Opella SJ. Triton X-100 as the ‘short chain lipid’ improves the magnetic alignment and stability of membrane proteins in phosphatidylcholine bilayers for oriented sample (OS) solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:12552–12553. doi: 10.1021/ja1055565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nolandt OV, Walther TH, Grage SL, Ulrich AS. Magnetically oriented dodecylphosphocholine bicelles for solid-state NMR structure analysis. Biochim Biophys Acta. 1818;2012:1142–1147. doi: 10.1016/j.bbamem.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 117.Garber SM, Lorigan GA, Howard KP. Magnetically oriented phospholipid bilayers for spin label EPR studies. J Am Chem Soc. 1999;121:3240–3241. doi: 10.1021/ja005574i. [DOI] [PubMed] [Google Scholar]
- 118.Mangels ML, Cardon TB, Harper AC, Howard KP, Lorigan GA. Spectroscopic characterization of spin-labeled magnetically oriented phospholipid bilayers by EPR spectroscopy. J Am Chem Soc. 2000;122:7052–7058. [Google Scholar]
- 119.Cardon TB, Tiburu EK, Padmanabhan A, Howard KP, Lorigan GA. Magnetically aligned phospholipid bilayers at the parallel and perpendicular orientations for X-band spin-label EPR studies. J Am Chem Soc. 2001;123:2913–2914. doi: 10.1021/ja005574i. [DOI] [PubMed] [Google Scholar]
- 120.Nusair NA, Lorigan GA. Investigating the structural and dynamic properties of n-doxylstearic acid in magnetically-aligned phospholipid bilayers by X-band EPR spectroscopy. Chem Phys Lip. 2005;133:151–164. doi: 10.1016/j.chemphyslip.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 121.Dave PC, Inbaraj JJ, Lorigan GA. Electron paramagnetic resonance studies of magnetically aligned phospholipid bilayers utilizing a phospholipid spin label. Langmuir. 2004;20:5801–5808. doi: 10.1021/la036377a. [DOI] [PubMed] [Google Scholar]
- 122.Inbaraj JJ, Nusair NA, Lorigan GA. Investigating magnetically aligned phospholipid bilayers with EPR spectroscopy at Q-band (35 GHz): Optimization and comparison with X-band (9 GHz) J Magn Reson. 2004;171:71–79. doi: 10.1016/j.jmr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 123.Mangels ML, Harper AC, Smirnov AI, Howard KP, Lorigan GA. Investigating magnetically aligned phospholipid bilayers with EPR spectroscopy at 94 GHz. J Magn Reson. 2001;151:253–259. doi: 10.1006/jmre.2001.2368. [DOI] [PubMed] [Google Scholar]
- 124.Cardon TB, Dave PC, Lorigan GA. Magnetically aligned phospholipid bilayers with large q ratios stabilize magnetic alignment with high order in the gel and Lα phases. Langmuir. 2005;21:4291–4298. doi: 10.1021/la0473005. [DOI] [PubMed] [Google Scholar]
- 125.Dave PC, Nusair NA, Inbaraj JJ, Lorigan GA. Electron paramagnetic resonance studies of magnetically aligned phospholipid bilayers utilizing a phospholipid spin label: the effect of cholesterol. Biochim Biophys Acta. 2005;1714:141–151. doi: 10.1016/j.bbamem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 126.Ghimire H, Inbaraj JJ, Lorigan GA. A comparative study of cholesterol on bicelle model membranes using X-band and Q-band EPR spectroscopy. Chem Phys Lip. 2009;160:98–104. doi: 10.1016/j.chemphyslip.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gruene T, Cho M-K, Karyagina I, Kim H-Y, Grosse C, Giller K, Zweckstetter M, Becker S. Integrated analysis of the conformation of a proteinlinked spin label by crystallography, EPR and NMR spectroscopy. J Biomol NMR. 2011;49:111–119. doi: 10.1007/s10858-011-9471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ghimire H, Abu-Baker S, Sahu ID, Zhou A, Mayo DJ, Lee RT, Lorigan GA. Probing the helical tilt and dynamic properties of membrane-bound phospholamban in magnetically aligned bicelles using electron paramagnetic resonance spectroscopy. Biochim Biophys Acta. 1818;2011:645–650. doi: 10.1016/j.bbamem.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Inbaraj JJ, Cardon TB, Laryukhin M, Grosser SM, Lorigan GA. Determining the topology of integral membrane peptides using EPR spectroscopy. J Am Chem Soc. 2006;128:9549–9554. doi: 10.1021/ja0622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mesleh MF, Lee S, Veglia G, Thiriot DS, Marassi FM, Opella SJ. Dipolar waves map the structure and topology of helices in membrane proteins. J Am Chem Soc. 2003;125:8928–8935. doi: 10.1021/ja034211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Newstadt JP, Mayo DJ, Inbaraj JJ, Subbaraman N, Lorigan GA. Determining the helical tilt of membrane peptides using electron paramagnetic resonance spectroscopy. J Magn Reson. 2009;198:1–7. doi: 10.1016/j.jmr.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mayo D, Zhou A, Sahu I, McCarrick R, Walton P, Ring A, Troxel K, Coey A, Hawn J, Emwas AH, Lorigan GA. Probing the structure of membrane proteins with electron spin echo envelope modulation spectroscopy. Prot Sci. 2011;20:1100–1104. doi: 10.1002/pro.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nusair NA, Mayo DJ, Dorozenski TD, Cardon TB, Inbaraj JJ, Karp ES, Newstadt JP, Grosser SM, Lorigan GA. Time-resolved EPR immersion depth studies of a transmembrane peptide incorporated into bicelles. Biochim Biophys Acta. 1818;2011:821–828. doi: 10.1016/j.bbamem.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Luchette PA, Vetman TN, Prosser RS, Hancock REW, Nieh MP, Glinka CJ, Krueger S, Katsaras J. Morphology of fast-tumbling bicelles: a small angle neutron scattering and NMR study. Biochim Biophys Acta. 2001;1513:83–94. doi: 10.1016/s0005-2736(01)00358-3. [DOI] [PubMed] [Google Scholar]
- 136.Yamada NL, Hishida M, Torikai N. Nanopore formation on unilamellar vesicles of long- and short-chain lipids. Phys Rev E. 2009;79:032902. doi: 10.1103/PhysRevE.79.032902. [DOI] [PubMed] [Google Scholar]
- 137.Nieh MP, Raghunathan VA, Pabst G, Harroun T, Nagashima K, Morales H, Katsaras J, Macdonald P. Temperature driven annealing of perforations in bicellar model membranes. Langmuir. 2011;27:4838–4847. doi: 10.1021/la104750x. [DOI] [PubMed] [Google Scholar]
- 138.Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q Rev Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 139.Davis JH. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim Biophys Acta. 1983;737:117–171. doi: 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- 140.Watts A, Spooner PJR. Phospholipid phase transitions as revealed by NMR. Chem Phys Lip. 1991;57:195–211. doi: 10.1016/0009-3084(91)90076-n. [DOI] [PubMed] [Google Scholar]
- 141.Picard F, Paquet MJ, Levesque J, Bélanger A, Auger M. 31P NMR first sprectral moment study of the partial magnetic orientation of phospholipid membranes. Biophys J. 1999;77:888–902. doi: 10.1016/S0006-3495(99)76940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sternin E, Schäfer H, Polozov IV, Gawrisch K. Simultaneous determination of orientational and order parameter distributions from NMR spectra of partially oriented model membranes. J Magn Reson. 2001;149:110–113. doi: 10.1006/jmre.2000.2278. [DOI] [PubMed] [Google Scholar]
- 143.Sternin E, Nizza D, Gawrisch K. Temperature dependence of DMPC/DHPC mixing in bicellar solution and its structural implications. Langmuir. 2001;17:2610–2616. [Google Scholar]
- 144.Triba MN, Warschawski DE, Devaux PF. Reinvestigation by phosphorus NMR of lipid distribution in bicelles. Biophys J. 2005;88:1887–1901. doi: 10.1529/biophysj.104.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ottiger M, Bax A. Characterization of magnetically oriented phospholipid micelles for measurement of dipolar couplings in macromolecules. J Biomol NMR. 1998;12:361–372. doi: 10.1023/a:1008366116644. [DOI] [PubMed] [Google Scholar]
- 146.Raffard G, Steinbruckner S, Arnold A, Davis JH, Dufourc EJ. Temperature-composition diagram of dimyristoylphosphatidylcholine-dicaproylphosphatidylcholine “bicelles” self-orienting in the magnetic field. A solid state 2H and 31P NMR study. Langmuir. 2000;16:7655–7662. [Google Scholar]
- 147.Nieh MP, Glinka CJ, Krueger S, Prosser RS, Katsaras J. SANS study of the structural phases of magnetically alignable lanthanide-doped phospholipid mixtures. Langmuir. 2001;17:2629–2638. [Google Scholar]
- 148.Harroun TA, Koslowsky M, Nieh MP, de Lannoy CF, Raghunathan VA, Katsaras J. Comprehensive examination of mesophases formed by DMPC and DHPC mixtures. Langmuir. 2005;21:5356–5361. doi: 10.1021/la050018t. [DOI] [PubMed] [Google Scholar]
- 149.Katsaras J, Harroun TA, Pencer J, Nieh MP. “Bicellar” lipid mixtures as used in biochemical and biophysical studies. Naturwissenschaften. 2005;92:355–366. doi: 10.1007/s00114-005-0641-1. [DOI] [PubMed] [Google Scholar]
- 150.Marcotte I, Auger M. Bicelles as model membranes for solid- and solution-state NMR studies of membrane peptides and proteins. Conc Magn Reson. 2005;24A:17–37. [Google Scholar]
- 151.Struppe J, Vold RR. Dilute bicellar solutions for structural NMR work. J Magn Reson. 1998;135:541–546. doi: 10.1006/jmre.1998.1605. [DOI] [PubMed] [Google Scholar]
- 152.Vold RR, Prosser RS. Magnetically oriented phospholipid bilayered micelles for structural studies of polypeptides. Does the ideal bicelle exist? J Magn Reson B. 1996;113:267–271. [Google Scholar]
- 153.Vold RR, Prosser RS, Deese AJ. Isotropic solutions of phospholipid bicelles: a new membrane mimetic for high-resolution NMR studies of polypeptides. J Biomol NMR. 1997;9:329–335. doi: 10.1023/a:1018643312309. [DOI] [PubMed] [Google Scholar]
- 154.Glover KJ, Whiles JA, Wu G, Yu NJ, Deems R, Struppe JO, Stark RE, Komives EA, Vold RR. Structural evaluation of phospholipid bicelles for solution-state studies of membrane-associated biomolecules. Biophys J. 2001;81:2163–2171. doi: 10.1016/s0006-3495(01)75864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee D, Walter KFA, Brückner AK, Hilty C, Becker S, Griesinger C. Bilayer in small bicelles revealed by lipid–protein interactions using NMR spectroscopy. J Am Chem Soc. 2008;130:13822–13823. doi: 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- 156.Gaemers S, Bax A. Morphology of three lyotropic liquid crystalline biological NMR media studied by translational diffusion anisotropy. J Am Chem Soc. 2001;123:12343–12352. doi: 10.1021/ja011967l. [DOI] [PubMed] [Google Scholar]
- 157.Rowe BA, Neal SL. Fluorescence probe study of bicelle structure as a function of temperature: developing a practical bicelle structure model. Langmuir. 2003;19:2039–2048. [Google Scholar]
- 158.Hwang JS, Oweimreen GA. Anomalous viscosity behavior of a bicelle system with various molar ratios of short- and long-chain phospholipids. Arab J Sci Eng. 2003;28:43–49. [Google Scholar]
- 159.Nieh MP, Raghunathan VA, Glinka CJ, Harroun TA, Pabst G, Katsaras J. Magnetically alignable phase of phospholipid “bicelle” mixtures is a chiral nematic made up of wormlike micelles. Langmuir. 2004;20:7893–7897. doi: 10.1021/la048641l. [DOI] [PubMed] [Google Scholar]
- 160.van Dam L, Karlsson G, Edwards K. Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochim Biophys Acta. 2004;1664:241–256. doi: 10.1016/j.bbamem.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 161.Barbosa-Barros L, de la Maza A, Walther P, Linares AM, Feliz M, Estelrich J, López O. Use of high-pressure freeze fixation and freeze fracture electron microscopy to study the influence of the phospholipid molar ratio in the morphology and alignment of bicelles. J Microsc. 2009;233:35–41. doi: 10.1111/j.1365-2818.2008.03093.x. [DOI] [PubMed] [Google Scholar]
- 162.Wang H, Nieh MP, Hobbie EK, Glinka CJ, Katsaras J. Kinetic pathway of the bilayered-micelle to perforated-lamellae transition. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67:060902. doi: 10.1103/PhysRevE.67.060902. [DOI] [PubMed] [Google Scholar]
- 163.Bolze J, Fujisawa T, Nagao T, Norisada K, Saito H, Naito A. Small-angle X-ray scattering and 31P-NMR studies on the phase behaviour of phospholipid bilayered mixed micelles. Chem Phys Lett. 2000;329:215–220. [Google Scholar]
- 164.Marrink SJ, Mark AE. Molecular dynamics simulation of the formation, structure, and dynamics of small phospholipid vesicles. J Am Chem Soc. 2003;125:15233–15242. doi: 10.1021/ja0352092. [DOI] [PubMed] [Google Scholar]
- 165.Shinoda W, DeVane R, Klein ML. Zwitterionic lipid assemblies: molecular dynamics studies of monolayers, bilayers, and vesicles using a new coarse grain force field. J Phys Chem B. 2010;114:6836–6849. doi: 10.1021/jp9107206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dutt M, Kuksenok O, Nayhouse MJ, Little SR, Balazs AC. Modeling the self-assembly of lipids and nanotubes in solution: forming vesicles and bicelles with transmembrane nanotube channels. ACS Nano. 2011;5:4769–4782. doi: 10.1021/nn201260r. [DOI] [PubMed] [Google Scholar]
- 167.Jiang Y, Kindt JT. Simulations of edge behavior in a mixed-lipid bilayer: fluctuation analysis. J Chem Phys. 2007;126:045105. doi: 10.1063/1.2430714. [DOI] [PubMed] [Google Scholar]
- 168.Wang H, de Joannis J, Jiang Y, Gaulding JC, Albrecht B, Yin F, Khanna K, Kindt JT. Bilayer edge and curvature effects on partitioning of lipids by tail lengths: atomistic simulations. Biophys J. 2008;95:2647–2657. doi: 10.1529/biophysj.108.131409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jiang Y, Wang H, Kindt JT. Atomistic simulations of bicelle mixtures. Biophys J. 2010;98:2895–2903. doi: 10.1016/j.bpj.2010.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tanner JE. Use of the stimulated echo in NMR diffusion studies. J Chem Phys. 1970;52:2523–2526. [Google Scholar]
- 171.Soong R, Macdonald PM. Lateral diffusion of PEG-lipid in magnetically aligned bicelles measured using stimulated echo pulsed field gradient 1H NMR. Biophys J. 2005;88:255–268. doi: 10.1529/biophysj.104.043620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 173.Chou JJ, Baber JL, Bax A. Characterization of phospholipid mixed micelles by translational diffusion. J Biomol NMR. 2004;29:299–308. doi: 10.1023/B:JNMR.0000032560.43738.6a. [DOI] [PubMed] [Google Scholar]
- 174.Andersson A, Mäler L. Magnetic resonance investigations of lipid motion in isotropic bicelles. Langmuir. 2005;21:7702–7709. doi: 10.1021/la0513003. [DOI] [PubMed] [Google Scholar]
- 175.Andersson A, Mäler L. Size and shape of fast-tumbling bicelles as determined by translational diffusion. Langmuir. 2006;22:2447–2449. doi: 10.1021/la053177l. [DOI] [PubMed] [Google Scholar]
- 176.Lind J, Nordin J, Mäler L. Lipid dynamics in fast-tumbling bicelles with varying bilayer thickness: effect of model transmembrane peptides. Biochim Biophys Acta. 2008;1778:2526–2534. doi: 10.1016/j.bbamem.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 177.Hong M, Schmidt-Rohr K, Zimmermann H. Conformational constraints on the headgroup and sn-2 chain of bilayer DMPC from NMR dipolar couplings. Biochemistry. 1996;35:8335–8341. doi: 10.1021/bi953083i. [DOI] [PubMed] [Google Scholar]
- 178.Cady SD, Goodman C, Tatko CD, DeGrado WF, Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: a 2H, 13C, and 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 179.Nevzorov AA, De Angelis AA, Park SH, Opella SJ. Uniaxial motional averaging of the chemical shift anisotropy of membrane proteins in bilayer environments. In: Ramamoorthy A, editor. NMR Spectroscopy of Biological Solids. CRC Press; New York: 2006. pp. 177–190. [Google Scholar]
- 180.Dvinskikh SV, Yamamoto K, Dürr UHN, Ramamoorthy A. Sensitivity and resolution enhancement in solid-state NMR spectroscopy of bicelles. J Magn Reson. 2007;184:228–235. doi: 10.1016/j.jmr.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dvinskikh SV, Yamamoto K, Ramamoorthy A. Heteronuclear isotropic mixing separated local field NMR spectroscopy. J Chem Phys. 2006;125:034507. doi: 10.1063/1.2212939. [DOI] [PubMed] [Google Scholar]
- 182.Lu JX, Damodaran K, Lorigan GA. Probing membrane topology by high-resolution 1H–13C heteronuclear dipolar solid-state NMR spectroscopy. J Magn Reson. 2006;178:283–287. doi: 10.1016/j.jmr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 183.Dvinskikh SV, Dürr UHN, Yamamoto K, Ramamoorthy A. High-resolution 2D NMR spectroscopy of bicelles to measure the membrane interaction of ligands. J Am Chem Soc. 2007;129:794–802. doi: 10.1021/ja065536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schmidt-Rohr K, Spiess HW. Multidimensional NMR and Polymers. Academic Press; San Diego, CA: 1994. [Google Scholar]
- 185.Yamamoto K, Ermakov VL, Lee DK, Ramamoorthy A. PITANSEMA-MAS, a solid-state NMR method to measure heteronuclear dipolar couplings under MAS. Chem Phys Lett. 2005;408:118–122. doi: 10.1016/j.cplett.2005.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu CH, Ramamoorthy A, Opella SJ. High-resolution heteronuclear dipolar solid-state NMR spectroscopy. J Magn Reson A. 1994;109:270–272. [Google Scholar]
- 187.Nevzorov AA, Opella SJ. A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J Magn Reson. 2003;164:182–186. doi: 10.1016/s1090-7807(03)00240-4. [DOI] [PubMed] [Google Scholar]
- 188.Ramamoorthy A, Wei Y, Lee DK. PISEMA solid-state NMR spectroscopy. Annu Rep NMR Spectrosc. 2004;52:1–52. [Google Scholar]
- 189.Dvinskikh SV, Dürr UHN, Yamamoto K, Ramamoorthy A. A high-resolution solid-state NMR approach for the structural studies of bicelles. J Am Chem Soc. 2006;128:6326–6327. doi: 10.1021/ja061153a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Metz G, Wu X, Smith SO. Ramped-amplitude cross polarization in magic-angle- spinning NMR. J Magn Reson Ser A. 1994;110:219–227. [Google Scholar]
- 191.Mohebbi A, Shaka AJ. Improvements in carbon-13 broadband homonuclear cross-polarization for 2D and 3D NMR. Chem Phys Lett. 1991;178:374– 378. [Google Scholar]
- 192.Xu J, Smith PES, Soong R, Ramamoorthy A. A proton spin diffusion based solid-state NMR approach for structural studies in aligned samples. J Phys Chem B. 2011;115:4863–4871. doi: 10.1021/jp201501q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yamamoto K, Soong R, Ramamoorthy A. Comprehensive analysis of lipid dynamics variation with lipid composition and hydration of bicelles using nuclear magnetic resonance (NMR) spectroscopy. Langmuir. 2009;25:7010– 7018. doi: 10.1021/la900200s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Douliez JP, Léonard A, Dufourc EJ. Restatement of order parameters in biomembranes: calculation of C–C bond order parameters from C–D quadrupolar splittings. Biophys J. 1995;68:1727–1739. doi: 10.1016/S0006-3495(95)80350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Aussenac F, Laguerre M, Schmitter JM, Dufourc EJ. Detailed structure and dynamics of bicelle phospholipids using selectively deuterated and perdeuterated labels. 2H NMR and molecular mechanics study. Langmuir. 2003;19:10468–10479. [Google Scholar]
- 196.Pearson RH, Pascher I. The molecular structure of lecithin dihydrate. Nature. 1979;281:499–501. doi: 10.1038/281499a0. [DOI] [PubMed] [Google Scholar]
- 197.Sternberg U, Witter R, Ulrich AS. All-atom molecular dynamics simulations using orientational constraints from anisotropic NMR samples. J Biomol NMR. 2007;38:23–39. doi: 10.1007/s10858-007-9142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Crowell KJ, Macdonald PM. Surface charge response of the phosphatidylcholine head group in bilayered micelles from phosphorus and deuterium nuclear magnetic resonance. Biochim Biophys Acta. 1999;1416:21–30. doi: 10.1016/s0005-2736(98)00206-5. [DOI] [PubMed] [Google Scholar]
- 199.Porcelli F, Buck B, Lee DK, Hallock KJ, Ramamoorthy A, Veglia G. Structure and orientation of pardaxin determined by NMR experiments in model membranes. J Biol Chem. 2004;279:45815–45823. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Semchyschyn DJ, Macdonald PM. Conformational response of the phosphatidylcholine headgroup to bilayer surface charge: torsion angle constraints from dipolar and quadrupolar couplings in bicelles. Magn Reson Chem. 2004;42:89–104. doi: 10.1002/mrc.1333. [DOI] [PubMed] [Google Scholar]
- 201.Santos JS, Lee DK, Ramamoorthy A. Effects of antidepressants on the conformation of phospholipid headgroups studied by solid-state NMR. Magn Reson Chem. 2004;42:105–114. doi: 10.1002/mrc.1327. [DOI] [PubMed] [Google Scholar]
- 202.DeMarco ML, Woods RJ, Prestegard JH, Tian F. Presentation of membrane-anchored glycosphingolipids determined from molecular dynamics simulations and NMR paramagnetic relaxation rate enhancement. J Am Chem Soc. 2010;132:1334–1338. doi: 10.1021/ja907518x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Barry J, Fritz M, Brender JR, Smith PES, Lee DK, Ramamoorthy A. Determining the effects of lipophilic drugs on membrane structure by solid-state NMR spectroscopy: the case of the antioxidant curcumin. J Am Chem Soc. 2009;131:4490–4498. doi: 10.1021/ja809217u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Glaubitz C, Watts A. Magic angle-oriented sample spinning (MAOSS): a new approach toward biomembrane studies. J Magn Reson. 1998;130:305–316. doi: 10.1006/jmre.1997.1344. [DOI] [PubMed] [Google Scholar]
- 205.Sizun C, Bechinger B. Bilayer sample for fast or slow magic angle oriented sample spinning solid-state NMR spectroscopy. J Am Chem Soc. 2002;124:1146–1147. doi: 10.1021/ja016571o. [DOI] [PubMed] [Google Scholar]
- 206.Tian F, Losonczi JA, Fischer MWF, Prestegard JH. Sign determination of dipolar couplings in field-oriented bicelles by variable angle sample spinning (VASS) J Biomol NMR. 1999;15:145–150. doi: 10.1023/a:1008356210291. [DOI] [PubMed] [Google Scholar]
- 207.Zandomeneghi G, Tomaselli M, van Beek JD, Meier BH. Manipulation of the director in bicellar mesophases by sample spinning: a new tool for NMR spectroscopy. J Am Chem Soc. 2001;123:910–913. doi: 10.1021/ja0019326. [DOI] [PubMed] [Google Scholar]
- 208.Zandomeneghi G, Tomaselli M, Williamson PTF, Meier BH. NMR of bicelles: orientation and mosaic spread of the liquid-crystal director under sample rotation. J Biomol NMR. 2003;25:113–123. doi: 10.1023/a:1022236217018. [DOI] [PubMed] [Google Scholar]
- 209.Carlotti C, Aussenac F, Dufourc EJ. Towards high-resolution H-1-NMR in biological membranes: magic angle spinning of bicelles. Biochim Biophys Acta. 2002;1564:156–164. doi: 10.1016/s0005-2736(02)00446-7. [DOI] [PubMed] [Google Scholar]
- 210.Kurita J, Shimahara H, Utsunomiya-Tate N, Tate S. Measurement of 15N chemical shift anisotropy in a protein dissolved in a dilute liquid crystalline medium with the application of magic angle sample spinning. J Magn Reson. 2003;163:163–173. doi: 10.1016/s1090-7807(03)00080-6. [DOI] [PubMed] [Google Scholar]
- 211.Williamson PTF, Zandomeneghi G, Barrantes FJ, Watts A, Meier BH. Structural and dynamic studies of the γ-M4 trans-membrane domain of the nicotinic acetylcholine receptor. Mol Membr Biol. 2005;22:485–496. doi: 10.1080/09687860500370653. [DOI] [PubMed] [Google Scholar]
- 212.Xu J, Dürr UHN, Im SC, Gan Z, Waskell L, Ramamoorthy A. Bicelle-enabled structural studies on a membrane-associated cytochrome b5 by solid-state MAS NMR spectroscopy. Angew Chem Int Ed. 2008;47:7864– 7867. doi: 10.1002/anie.200801338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Uekusa Y, Kamihira M, Nakayama T. Dynamic behavior of tea catechins interacting with lipid membranes as determined by NMR spectroscopy. J Agric Food Chem. 2007;55:9986–9992. doi: 10.1021/jf0712402. [DOI] [PubMed] [Google Scholar]
- 214.Matsumori N, Morooka A, Murata M. Detailed description of the conformation and location of membrane-bound erythromycin A using isotropic bicelles. J Med Chem. 2006;49:3501–3508. doi: 10.1021/jm051210v. [DOI] [PubMed] [Google Scholar]
- 215.Matsumori N, Morooka A, Murata M. Conformation and location of membrane-bound salinomycin–sodium complex deduced from NMR in isotropic bicelles. J Am Chem Soc. 2007;129:14989–14995. doi: 10.1021/ja075024l. [DOI] [PubMed] [Google Scholar]
- 216.Houdai T, Matsumori N, Murata M. Structure of membrane-bound amphidinol 3 in isotropic small bicelles. Org Lett. 2008;10:4191–4194. doi: 10.1021/ol8016337. [DOI] [PubMed] [Google Scholar]
- 217.Matsumori N, Murata M. 3D structures of membrane-associated small molecules as determined in isotropic bicelles. Nat Prod Rep. 2010;27:1480–1492. doi: 10.1039/c0np00002g. [DOI] [PubMed] [Google Scholar]
- 218.Oliveira TR, Benatti CR, Lamy MT. Structural characterization of the interaction of the polyene antibiotic amphotericin B with DODAB bicelles and vesicles. Biochim Biophys Acta. 1808;2011:2629–2637. doi: 10.1016/j.bbamem.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 219.Koenig BW, Gawrisch K. Lipid–ethanol interaction studied by NMR on bicelles. J Phys Chem B. 2005;109:7540–7547. doi: 10.1021/jp044980b. [DOI] [PubMed] [Google Scholar]
- 220.Dave PC, Tiburu EK, Nusair NA, Lorigan GA. Calculating order parameter profiles utilizing magnetically aligned phospholipid bilayers for 2H solid-state NMR studies. Solid State Nucl Magn Reson. 2003;24:137–149. doi: 10.1016/S0926-2040(03)00052-3. [DOI] [PubMed] [Google Scholar]
- 221.Nusair NA, Tiburu EK, Dave PC, Lorigan GA. Investigating fatty acids inserted into magnetically aligned phospholipid bilayers using EPR and solid-state NMR spectroscopy. J Magn Reson. 2004;168:228–237. doi: 10.1016/j.jmr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 222.Guo J, Yang DP, Chari R, Tian X, Pavlopoulos S, Lu D, Makriyannis A. Magnetically aligned bicelles to study the orientation of lipophilic ligands in membrane bilayers. J Med Chem. 2008;51:6793–6799. doi: 10.1021/jm800766x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Guo J, Pavlopoulos S, Tian X, Lu D, Nikas SP, Yang DP, Makriyannis A. Conformational study of lipophilic ligands in phospholipid model membrane systems by solution NMR. J Med Chem. 2003;46:4838–4846. doi: 10.1021/jm020385r. [DOI] [PubMed] [Google Scholar]
- 224.Tiburu EK, Bass CE, Struppe JO, Lorigan GA, Avraham S, Avraham HK. Structural divergence among cannabinoids influences membrane dynamics: a 2H solid-state NMR analysis. Biochim Biophys Acta. 2007;1768:2049– 2059. doi: 10.1016/j.bbamem.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 225.Yi X, Venot A, Glushka J, Prestegard JH. Glycosidic torsional motions in a bicelle-associated disaccharide from residual dipolar couplings. J Am Chem Soc. 2004;126:13636–13638. doi: 10.1021/ja045697t. [DOI] [PubMed] [Google Scholar]
- 226.Kajiya K, Kumazawa S, Naito A, Nakayama T. Solid-state NMR analysis of the orientation and dynamics of epigallocatechin gallate, a green tea polyphenol, incorporated into lipid bilayers. Magn Reson Chem. 2008;46:174–177. doi: 10.1002/mrc.2157. [DOI] [PubMed] [Google Scholar]
- 227.Bortolus M, Parisio G, Maniero AL, Ferrarini A. Monomeric fullerenes in lipid membranes: effect of molecular shape and polarity. Langmuir. 2011;27:12560–12568. doi: 10.1021/la202524r. [DOI] [PubMed] [Google Scholar]
- 228.Sun J, Wu X, Liu J, Wang Y, He Z. Profiling drug membrane permeability and activity via biopartitioning chromatography. Curr Drug Metab. 2008;9:152–166. doi: 10.2174/138920008783571800. [DOI] [PubMed] [Google Scholar]
- 229.Johannson E, Engvall C, Arfvidsson M, Lundahl P, Edwards K. Development and initial evaluation of PEG-stabilized bilayer disks as novel model membranes. Biophys Chem. 2005;113:183–192. doi: 10.1016/j.bpc.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 230.Johansson E, Lundquist A, Zuo S, Edwards K. Nanosized bilayer disks: attractive model membranes for drug partition studies. Biochim Biophys Acta. 2007;1768:1518–1525. doi: 10.1016/j.bbamem.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 231.Boija E, Lundquist A, Edwards K, Johansson G. Evaluation of bilayer disks as plant cell membrane models in partition studies. Anal Biochem. 2007;364:145–152. doi: 10.1016/j.ab.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 232.Boija E, Lundquist A, Nilsson M, Edwards K, Isaksson R, Johansson G. Bilayer disk capillary electrophoresis: a novel method to study drug partitioning into membranes. Electrophoresis. 2008;29:3377–3383. doi: 10.1002/elps.200700682. [DOI] [PubMed] [Google Scholar]
- 233.Vainikka K, Reijmar K, Yohannes G, Samuelsson J, Edwards K, Jussila M, Riekkola ML. Polyethylene glycol-stabilized lipid disks as model membranes in interaction studies based on electrokinetic capillary chromatography and quartz crystal microbalance. Anal Biochem. 2011;414:117–124. doi: 10.1016/j.ab.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 234.Marcotte I, Wegener KL, Lam YH, Chia BCS, de Planque MRR, Bowie JH, Auger M, Separovic F. Interaction of antimicrobial peptides from Australian amphibians with lipid membranes. Chem Phys Lip. 2003;122:107–120. doi: 10.1016/s0009-3084(02)00182-2. [DOI] [PubMed] [Google Scholar]
- 235.Marcotte I, Separovic F, Auger M, Gagné SM. A multidimensional 1H NMR investigation of methionine–enkephalin in fast-tumbling bicelles. Biophys J. 2004;86:1587–1600. doi: 10.1016/S0006-3495(04)74226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dürr UHN, Gildenberg M, Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chem Rev. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Son WS, Park SH, Nothnagel HJ, Lu GJ, Wang Y, Zhang H, Cook GA, Howell SC, Opella SJ. ‘q-Titration’ of long-chain and short-chain lipids differentiates between structured and mobile residues of membrane proteins studied in bicelles by solution NMR spectroscopy. J Magn Reson. 2012;214:111–118. doi: 10.1016/j.jmr.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ramamoorthy A. NMR Spectroscopy of Biological Solids. CRC Press, Taylor & Francis Group; New York: 2006. [Google Scholar]
- 239.Park SH, Prytulla S, de Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. J Am Chem Soc. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Park SH, Casagrande F, Das BB, Albrecht L, Chu M, Opella SJ. Local and global dynamics of the G-protein-coupled receptor CXCR1. Biochemistry. 2011;50:2371–2380. doi: 10.1021/bi101568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Park SH, Casagrande F, Cho L, Albrecht L, Opella SJ. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. J Mol Biol. 2011;414:194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mahalakshmi R, Marassi FM. Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-state NMR. Biochemistry. 2008;47:6531–6538. doi: 10.1021/bi800362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Cui TX, Canlas CG, Xu Y, Tang P. Anesthetic effects on the structure and dynamics of the second transmembrane domains of nAChR α4β2. Biochim Biophys Acta. 2010;1798:161–166. doi: 10.1016/j.bbamem.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cook GA, Zhang H, Park SH, Wang Y, Opella SJ. Comparative NMR studies demonstrate profound differences between two viroporins: p7 of HCV and Vpu of HIV-1. Biochim Biophys Acta. 1808;2011:554–560. doi: 10.1016/j.bbamem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.de Angelis AA, Howell SC, Nevzorov AA, Opella SJ. Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128:12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Müller SD, De Angelis AA, Walther TH, Grage SL, Lange C, Opella SJ, Ulrich AS. Structural characterization of the pore forming protein TatAd of the twin-arginine translocase in membranes by solid-state 15N-NMR. Biochem Biophys Acta. 2007;1768:3071–3079. doi: 10.1016/j.bbamem.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 247.Walther TH, Grage SL, Roth N, Ulrich AS. Membrane alignment of the pore-forming component TatAd of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:15945–15956. doi: 10.1021/ja106963s. [DOI] [PubMed] [Google Scholar]
- 248.Park SH, Marassi FM, Black D, Opella SJ. Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly. Biophys J. 2010;99:1465–1474. doi: 10.1016/j.bpj.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mote KR, Gopinath T, Traaseth NJ, Kitchen J, Gor’kov PL, Brey WW, Veglia G. Multidimensional oriented solid-state NMR experiments enable the sequential assignment of uniformly 15N-labeled integral membrane proteins in magnetically aligned lipid bilayers. J Biomol NMR. 2011;51:339–346. doi: 10.1007/s10858-011-9571-8. [DOI] [PubMed] [Google Scholar]
- 250.Dürr UHN, Waskell L, Ramamoorthy A. The cytochromes P450 and b5 and their reductases—Promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim Biophys Acta. 2007;1768:3235–3259. doi: 10.1016/j.bbamem.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 251.Dürr UHN, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J Am Chem Soc. 2007;129:6670– 6671. doi: 10.1021/ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Soong R, Smith PES, Xu JD, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Proton-evolved local-field solid-state NMR studies of cytochrome b5 embedded in bicelles, revealing both structural and dynamical information. J Am Chem Soc. 2010;132:5779–5788. doi: 10.1021/ja910807e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Xu JD, Soong R, Im SC, Waskell L, Ramamoorthy A. INEPT-based separated-local-field NMR spectroscopy: a unique approach to elucidate side-chain dynamics of membrane-associated proteins. J Am Chem Soc. 2010;132:9944–9947. doi: 10.1021/ja103983f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 255.Wider G. Technical aspects of NMR spectroscopy with biological macromolecules and studies of hydration in solution. Prog Nucl Magn Reson Spectrosc. 1998;32:193–275. [Google Scholar]
- 256.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- 257.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles & Practice. Academic Press Inc; San Diego: 2006. [Google Scholar]
- 258.Keeler J. Understanding NMR Spectroscopy. Wiley; Chichester: 2010. [Google Scholar]
- 259.Sanders CR, Hoffmann AK, Grayn DN, Keyes MH, Ellis CD. French swimwear for membrane proteins. Chembiochem. 2004;5:423–426. doi: 10.1002/cbic.200300830. [DOI] [PubMed] [Google Scholar]
- 260.Poget SF, Cahill SM, Girvin ME. Isotropic bicelles stabilize the functional form of a small multidrug-resistance pump for NMR structural studies. J Am Chem Soc. 2007;129:2432–2433. doi: 10.1021/ja0679836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Poget SF, Girvin ME. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim Biophys Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Poget SF, Harris R, Cahill SM, Girvin ME. 1H, 13C, 15N backbone NMR assignments of the Staphylococcus aureus small multidrug-resistance pump (Smr) in a functionally active conformation. Biomol NMR Assign. 2010;4:139–142. doi: 10.1007/s12104-010-9228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 2004;1666:105– 117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 264.Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 265.Wang G. NMR of membrane-associated peptides and proteins. Curr Prot Pept Sci. 2008;9:50–69. doi: 10.2174/138920308783565714. [DOI] [PubMed] [Google Scholar]
- 266.Raschle T, Hiller S, Etzkorn M, Wagner G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr Opin Struct Biol. 2010;20:471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Prog Nucl Magn Reson Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Qureshi T, Goto NK. Contemporary methods in structure determination of membrane proteins by solution NMR. Top Curr Chem. 2012;326:123–185. doi: 10.1007/128_2011_306. [DOI] [PubMed] [Google Scholar]
- 269.Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat Struct Biol. 2010;17:768–774. doi: 10.1038/nsmb.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kaya AI, Thaker TM, Preininger AM, Iverson TM, Hamm HE. Coupling efficiency of rhodopsin and transducin in bicelles. Biochemistry. 2011;50:3193–3203. doi: 10.1021/bi200037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Xie G, D’Antona AM, Edwards PC, Fransen M, Standfuss J, Schertler GFX, Oprian DD. Preparation of an activated rhodopsin/transducin complex using a constitutively active mutant of rhodopsin. Biochemistry. 2011;50:10399– 10407. doi: 10.1021/bi201126r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Liu YZ, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf.GTP. Nat Struct Biol. 2010;17:876–882. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Morrison EA, Henzler-Wildman KA. Reconstitution of integral membrane proteins into isotropic bicelles with improved sample stability and expanded lipid composition profile. Biochim Biophys Acta. 1818;2012:814–820. doi: 10.1016/j.bbamem.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T, Henzler-Wildman KA. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2011;481:45–50. doi: 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bocharov EV, Volynsky PE, Pavlov KV, Efremov RG, Arseniev AS. Structure elucidation of dimeric transmembrane domains of bitopic proteins. Cell Adh Migr. 2010;4:284–298. doi: 10.4161/cam.4.2.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Mineev KS, Bocharov EV, Pustovalova YE, Bocharova OV, Chupin VV, Arseniev AS. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J Mol Biol. 2010;400:231–243. doi: 10.1016/j.jmb.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 278.Traaseth NJ, Ha KN, Verardi R, Shi L, Buffy JJ, Masterson LR, Veglia G. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca2+-ATPase. Biochemistry. 2008;47:3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Gustavsson M, Traaseth NJ, Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochim Biophys Acta. 1818;2012:146–153. doi: 10.1016/j.bbamem.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chem Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 281.Tolman JR, Ruan K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem Rev. 2006;106:1720–1736. doi: 10.1021/cr040429z. [DOI] [PubMed] [Google Scholar]
- 282.Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J Am Chem Soc. 2000;122:3791–3792. [Google Scholar]
- 283.Zweckstetter M, Hummer G, Bax A. Prediction of charge-induced molecular alignment of biomolecules dissolved in liquid-crystalline phases. Biophys J. 2004;86:3444–3460. doi: 10.1529/biophysj.103.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat Prot. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 285.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 286.Bouvignies G, Markwick PRL, Blackledge M. Simultaneous definition of high resolution protein structure and backbone conformational dynamics using NMR residual dipolar couplings. ChemPhysChem. 2007;8:1901–1909. doi: 10.1002/cphc.200700353. [DOI] [PubMed] [Google Scholar]
- 287.Getz M, Sun X, Casiano-Negroni A, Zhang Q, Al-Hashimi HM. NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 288.Veldkamp CT, Ziarek JJ, Su JD, Basnet H, Lennertz R, Weiner JJ, Peterson FC, Baker JE, Volkman BF. Monomeric structure of the cardioprotective chemokine SDF-1/CXCL12. Prot Sci. 2009;18:1359–1369. doi: 10.1002/pro.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Fischer MWF, Losonczi JA, Weaver JL, Prestegard JH. Domain orientation and dynamics in multidomain proteins from residual dipolar couplings. Biochemistry. 1999;38:9013–9022. doi: 10.1021/bi9905213. [DOI] [PubMed] [Google Scholar]
- 290.Maciejewski M, Tjandra N, Barlow PN. Estimation of interdomain flexibility of N-terminus of factor H using residual dipolar couplings. Biochemistry. 2011;50:8138–8149. doi: 10.1021/bi200575b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lange OF, Lakomek NA, Farès C, Schröder GF, Walter KFA, Becker S, Meiler J, Grubmüller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 292.Chou JJ, Kaufmann JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement on stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 293.Lorieau J, Yao L, Bax A. Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins. J Am Chem Soc. 2008;130:7536– 7537. doi: 10.1021/ja801729f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Liu Y, Kahn RA, Prestegard JH. Structure and membrane interaction of myristoylated ARF1. Structure. 2009;17:79–87. doi: 10.1016/j.str.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bhat V, McDonald CB, Mikles DC, Deegan BJ, Seldeen KL, Bates ML, Farooq A. Ligand binding and membrane insertion compete with oligomerization of the BclXL apoptotic repressor. J Mol Biol. 2012;416:57–77. doi: 10.1016/j.jmb.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Whiles JA, Deems R, Vold RR, Dennis EA. Bicelles in structure-function studies of membrane-associated proteins. Bioorg Chem. 2002;30:431–442. doi: 10.1016/s0045-2068(02)00527-8. [DOI] [PubMed] [Google Scholar]
- 297.Lührs T, Zahn R, Wüthrich K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J Mol Biol. 2006;357:833–841. doi: 10.1016/j.jmb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 298.Cho HS, Dominick JL, Spence MM. Lipid domains in bicelles containing unsaturated lipids and cholesterol. J Phys Chem B. 2010;114:9238–9245. doi: 10.1021/jp100276u. [DOI] [PubMed] [Google Scholar]
- 299.Yasuhara K, Miki S, Nakazono H, Ohta A, Kikuchi JI. Synthesis of organic– inorganic hybrid bicelles—lipid bilayer nanodiscs encompassed by siloxane surfaces. Chem Commun. 2011;47:4691–4693. doi: 10.1039/c1cc10254k. [DOI] [PubMed] [Google Scholar]
- 300.Rodríguez G, Soria G, Coll E, Rubio L, Barbosa-Barros L, López-Iglesias C, Planas AM, Estelrich J, de la Maza A, López O. Bicosomes: bicelles in dilute systems. Biophys J. 2010;99:480–488. doi: 10.1016/j.bpj.2010.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Park KH, Billon-Denis E, Dahmane T, Lebaupain F, Pucci B, Breyton C, Zito F. In the cauldron of cell-free synthesis of membrane proteins: playing with new surfactants. New Biotechnol. 2011;28:255–261. doi: 10.1016/j.nbt.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 302.Uhlemann EM, Pierson HE, Fillingame RH, Dmitriev OY. Cell-free synthesis of membrane subunits of ATP synthase in phospholipid bicelles: NMR shows subunit fold similar to the protein in the cell membrane. Prot Sci. 2012;21:279–288. doi: 10.1002/pro.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Barbosa-Barros L, Barba C, Cócera M, Coderch L, López-Iglesias C, de la Maza A, López O. Effect of bicellar systems on skin properties. Int J Pharm. 2008;352:263–272. doi: 10.1016/j.ijpharm.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 304.Barbosa-Barros L, de la Maza A, Estelrich J, Linares AM, Feliz M, Walther P, Pons R, López O. Penetration and growth of DPPC/DHPC bicelles inside the stratum corneum of the skin. Langmuir. 2008;24:5700–5706. doi: 10.1021/la703732h. [DOI] [PubMed] [Google Scholar]
- 305.Barbosa-Barros L, Barba C, Rodríguez G, Cócera M, Coderch L, López-Iglesias C, de la Maza A, López O. Lipid nanostructures: self-assembly and effect on skin properties. Mol Pharm. 2009;6:1237–1245. doi: 10.1021/mp9000734. [DOI] [PubMed] [Google Scholar]
- 306.Rodríguez G, Barbosa-Barros L, Rubio L, Cócera M, Díez A, Estelrich J, Pons R, Caelles J, de la Maza A, López O. Conformational changes in stratum corneum lipids by effect of bicellar systems. Langmuir. 2009;25:10595– 10603. doi: 10.1021/la901410h. [DOI] [PubMed] [Google Scholar]
- 307.Rodríguez G, Rubio L, Cócera M, Estelrich J, Pons R, de la Maza A, López O. Application of bicellar systems on skin: diffusion and molecular organization effects. Langmuir. 2010;26:10578–10584. doi: 10.1021/la100691m. [DOI] [PubMed] [Google Scholar]
- 308.Rodríguez G, Barbosa-Barros L, Rubio L, Cócera M, López-Iglesias C, de la Maza A, López O. Bicellar systems as modifiers of skin lipid structure. Coll Surf B: Biointerf. 2011;84:390–394. doi: 10.1016/j.colsurfb.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 309.Barbosa-Barros L, Rodríguez G, Barba C, Cócera M, Rubio L, Estelrich J, López-Iglesias C, de la Maza A, López O. Bicelles: lipid nanostructured platforms with potential dermal applications. Small. 2012;8:807–818. doi: 10.1002/smll.201101545. [DOI] [PubMed] [Google Scholar]
- 310.Rubio L, Rodríguez G, Barbosa-Barros L, Coderch L, de la Maza A, Parra JL, López O. Bicellar systems for in vitro percutaneous absorption of diclofenac. Int J Pharm. 2010;386:108–113. doi: 10.1016/j.ijpharm.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 311.Kang C, Vanoye CG, Welch RC, Van Horn WD, Sanders CR. Functional delivery of a membrane protein into oocyte membranes using bicelles. Biochemistry. 2010;49:653–655. doi: 10.1021/bi902155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Tekobo S, Pinkhassik E. Directed covalent asssembly of rigid organic nanodisks using self-assembled temporary scaffolds. Chem Commun. 2009:1112–1114. doi: 10.1039/b817598e. [DOI] [PubMed] [Google Scholar]
- 313.Song Y, Dorin RM, Garcia RM, Jiang YB, Wang H, Li P, Qiu Y, van Swol F, Miller JE, Shelnutt JA. Synthesis of platinum nanowheels using a bicellar template. J Am Chem Soc. 2008;130:12602–12603. doi: 10.1021/ja8047464. [DOI] [PubMed] [Google Scholar]
- 314.Garcia RM, Song Y, Dorin RM, Wang H, Moreno AM, Jiang YB, Tian Y, Qiu Y, Medforth CJ, Coker EN, van Swol F, Miller JE, Shelnutt JA. Templated growth of platinum nanowheels using the inhomogeneous reaction environment of bicelles. Phys Chem Chem Phys. 2011;13:4846–4852. doi: 10.1039/c0cp01930e. [DOI] [PubMed] [Google Scholar]
- 315.Tabaei SR, Jönsson P, Brändén M, Höök F. Self-assembly formation of multiple DNA-tethered lipid bilayers. J Struct Biol. 2009;168:200–206. doi: 10.1016/j.jsb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 316.Zeineldin R, Last JA, Slade AL, Ista LK, Bisong P, O’Brien MJ, Brueck SRJ, Sasaki DY, Lopez GP. Using bicellar mixtures to form supported and suspended lipid bilayers on silicon chips. Langmuir. 2006;22:8163–8168. doi: 10.1021/la060817r. [DOI] [PubMed] [Google Scholar]
- 317.Epand RM. Membrane lipid polymorphism: relationship to bilayer properties and protein function. Meth Mol Biol. 2007;400:15–26. doi: 10.1007/978-1-59745-519-0_2. [DOI] [PubMed] [Google Scholar]
- 318.Liebi M, Kohlbrecher J, Ishikawa T, Fischer P, Walde P, Windhab EJ. Cholesterol increases the magnetic aligning of bicellar disks from an aqueous mixture of DMPC and DMPE-DTPA with complexed thulium ions. Langmuir. 2012;28:10905–10915. doi: 10.1021/la3019327. [DOI] [PubMed] [Google Scholar]
- 319.Brindley AJ, Martin RW. Effect of divalent cations on DMPC/DHPC bicelle formation and alignment. Langmuir. 2012;28:7788–7796. doi: 10.1021/la300885u. [DOI] [PubMed] [Google Scholar]
- 320.DeMarco ML. Three-dimensional structure of glycolipids in biological membranes. Biochemistry. 2012;51:5725–5732. doi: 10.1021/bi3003633. [DOI] [PubMed] [Google Scholar]
- 321.Schmidt-Rohr K, Nanz D, Emsley L, Pines A. NMR measurement of resolved heteronuclear dipole couplings in liquid crystals and lipids. J Phys Chem. 1994;98:6668–6670. [Google Scholar]