Summary
CD4+ T follicular helper (Tfh) cells provide the required signals to B cells for germinal center reactions that are necessary for long-lived antibody responses. However, it remains unclear whether there are CD4+ memory T cells committed to the Tfh cell lineage after antigen clearance. Using adoptive transfer of antigen-specific memory CD4+ T cell subpopulations in the LCMV infection model, we found that there are distinct memory CD4+ T cell populations with commitment to either Tfh- or Th1-cell lineages. Our conclusions are based on gene expression profiles, epigenetic studies, and phenotypic and functional analyses. Our findings indicate that CD4+ memory T cells “remember” their previous effector lineage after antigen clearance, being poised to reacquire their lineage-specific effector functions upon antigen reencounter. These findings have important implications for rational vaccine design, where improving the generation and engagement of memory Tfh cells could be used to enhance vaccine-induced protective immunity.
Introduction
Naïve pathogen-specific CD4+ T cells respond to acute infections through robust proliferation and differentiation to generate effector cells with the capacity to provide help to the many and diverse branches of the immune system. Following antigen clearance, the majority of antigen-specific effector cells undergo apoptosis, leaving behind a population of memory CD4+ T cells. In addition to their ability to survive and undergo homeostatic proliferation in the absence of antigen, memory T cells retain the capacity to rapidly recall effector function, traffic to a wide range of tissues, and exist at much higher frequencies than naïve cells specific for the same antigen. These features provide the host with a protective network of pathogen-specific memory T helper cells that are poised to swiftly respond upon a secondary challenge (Sallusto et al., 2010).
Naive CD4+ T cells have multiple fates and upon activation can develop into a variety of specialized subsets, such as T helper 1 (Th1), Th2, Th17, and Treg cells. Each of these lineages has distinct gene expression programs that are regulated by specific STATS, transcription factors, and epigenetic mechanisms (O’Shea and Paul, 2010). More recently, an additional subset known as T follicular helper (Tfh) cells have been identified as the CD4+ T cell subset that provides help for antibody responses. Tfh cells provide the necessary signals to antigen-specific B cells to generate and maintain the germinal center reaction, thus facilitating efficient class switching and affinity maturation of antibodies, and the generation of long-lived antibody-secreting plasma cells (Crotty, 2011). Tfh cells were first characterized in humans by their expression of the B cell follicle homing receptor CXCR5 (Breitfeld et al., 2000; Kim et al., 2001; Schaerli et al., 2000), high ICOS and PD-1 expression, and the transcription factor Bcl6 (Crotty et al., 2010). Tfh cells can localize to the B cell follicle by sensing CXCL13 through CXCR5 (Ansel et al., 1999; Kim et al., 2001). Bcl6 has recently been identified as a Tfh lineage regulator (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009), and shares a reciprocal relationship with the transcriptional repressor Blimp-1, which suppresses Tfh differentiation (Crotty et al., 2010; Johnston et al., 2009). However, it remains unclear whether Tfh cells possess the capacity to further differentiate into the resting memory CD4+ T cell pool and retain their Tfh lineage commitment after antigen clearance (Crotty, 2011; Fazilleau et al., 2007; Liu et al., 2012; Luthje et al., 2012; Marshall et al., 2011; Pepper et al., 2011; Weber et al., 2012).
To address whether Tfh memory cells exist within the pool of memory CD4+ T cells, we studied virus-specific CD4+ T cells throughout the primary, memory, and secondary effector phases of the immune response following acute LCMV infection. We report here that a distinct CXCR5+ subset of antigen-specific CD4+ T cells preferentially recalled a Tfh cell secondary response following transfer and challenge with virus, while CXCR5− memory cells generated a Th1 cell secondary response. Based on these findings, we propose a model in which Th1 and Tfh cells differentiate to become respectively Th1 and Tfh memory cells, poised to preferentially recall their previously programmed lineage-associated gene expression patterns and effector functions upon antigen rechallenge. These findings have important implications for vaccine design, where adjuvants and strategies that promote a higher quantity and quality of memory Tfh cells may enable enhanced humoral immunity following prime and boost vaccination.
Results
Phenotypic heterogeneity of virus-specific CD4+ T cells is maintained during effector and memory differentiation
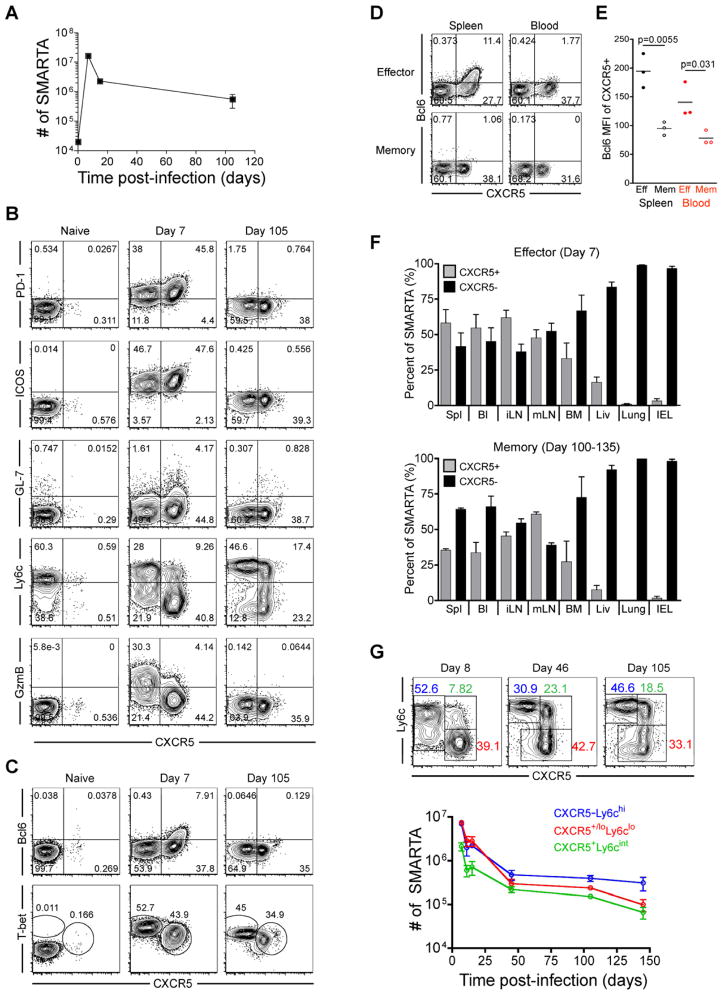
To determine whether heterogeneity in the effector CD4+ T cell population persists during memory development, we performed a longitudinal analysis of Th1- and Tfh-cell phenotypic marker expression on LCMV-specific CD4+ T cells following acute LCMV infection. Congenically-marked (CD45.1) naïve SMARTA transgenic (Tg) CD4+ T cells specific for the LCMV GP66-77 epitope were adoptively-transferred into recipient mice, and donor SMARTA cells (CD4+CD45.1+ gated) were analyzed at effector and memory timepoints following infection (Figure 1a). We observed that approximately 45% of virus-specific effector (day 7 post-infection) SMARTA cells expressed CXCR5, high amounts of PD-1 and ICOS, and contained a subpopulation of GL-7hi germinal center Tfh cells (Yusuf et al., 2010), consistent with a Tfh phenotype (Figure 1b). Further, the majority of CXCR5+ effector cells downregulated Ly6c expression and expressed low levels of granzyme B, while the CXCR5− effector cells displayed higher Ly6c and granzyme B expression (Figure 1b). In addition, CXCR5+ effector SMARTA cells expressed Bcl6 and low amounts of T-bet, while CXCR5− effector cells were Bcl6 negative and expressed high amounts of T-bet (Figure 1c). Similar to LCMV-specific SMARTA Tg cells, endogenous LCMV GP66-77 specific (tetramer+) effector CD4+ T cells in LCMV-infected B6 mice exhibited the same dichotomy of CXCR5+ and CXCR5− cells with similar expression patterns of PD-1, Ly6c, Bcl6, and T-bet (Figure S1a and S1b). These data demonstrate the generation of both Tfh and Th1 virus-specific effector cells during LCMV infection, which could be generally distinguished by Ly6c and CXCR5 expression. In agreement with this, Ly6clo SMARTA effector cells localized predominantly within the B cell follicle and germinal centers, while Ly6chi effector cells were generally outside of B cell follicles and germinal centers (Figure S1f).
Figure 1. Phenotypic heterogeneity of virus-specific CD4+ T cells is maintained during effector and memory differentiation.
2×105 CD45.1+ LCMV-specific naive SMARTA transgenic CD4+ T cells were adoptively transferred into CD45.2+ naïve recipients that were then infected with 2×105 PFU of LCMV Armstrong. FACS plots are gated on CD4+CD45.1+ SMARTA cells at the indicated timepoints relative to infection. A) Kinetics of splenic SMARTA CD4+ T cells. B) CXCR5, PD-1, ICOS, GL-7, Ly6c, and granzyme B analysis of naïve, effector, and memory SMARTA CD4+ T cells. C) Analysis of T-bet and Bcl6 expression. D) Analysis of Bcl6 and CXCR5 on effector (Day 8) and memory (Day 100–133) SMARTA cells in blood and spleen. E) Bcl6 MFI of CXCR5+ gated effector and memory SMARTA cells. Statistically significant p values are shown, and were determined using a two-tailed unpaired Students t test. F) The frequency of effector and memory SMARTA cells that are CXCR5+ and CXCR5− in the following tissues: Spl=spleen; Bl=blood; iLN=inguinal lymph node; mLN=mesenteric lymph node; BM=bone marrow; Liv=liver; Lung, and IEL=intestinal intraepithelial lymphocytes. Graphs show the mean and s.e.m. N=3 mice at each timepoint. G) Plots of CD4+CD45.1+ gated SMARTA cells with gates indicating the Ly6c+CXCR5− (blue), CXCR5+Ly6clo (red), and CXCR5+Ly6cint (green) subsets. Chart indicates the number of each subset in spleen following infection (N≥3 at each timepoint). Error bars represent the SEM. See also Figure S1 and Table S1.
Consistent with a previous report (Poholek et al.), we observed that CXCR5+ Tfh effector cells were lower for Psgl1 expression than CXCR5− Th1 effector cells (Figure S1c). A recent study also using the LCMV infection model used the markers Psgl1 and Ly6c to identify effector and memory subsets of LCMV-specific CD4 T cells. Their study described the Psgl1loLy6clo effector subset as Tfh cells, the Psgl1hiLy6chi (Tbethi) effector subset as Th1 cells, and the Psgl1hiLy6clo effector cells as less terminally-differentiated Th1 cells. However, we observed that the Psgl1loLy6clo population did not account for all CXCR5+ Tfh effector cells, and the Psgl1hi-intLy6clo effector population was comprised of a similar proportion of both CXCR5+ Tfh and CXCR5− Th1 effector cells (Figure S1d and S1e). Thus, Psgl1 used in combination with Ly6c (Marshall et al., 2011) does not clearly distinguish between Tfh and Th1 lineage effector cells (Figure S1d and S1e). Furthermore, unlike at effector timepoints, the Psgl1hi-intLy6clo memory subset had higher CXCR5 expression than the Psgl1loLy6clo memory subset (Figure S1d and S1e), indicating that the combination of Psgl1 and Ly6c markers is not useful for subsetting cells with the most Tfh-like qualities at both effector and memory timepoints.
Interestingly, while PD-1, ICOS, GL-7, and Bcl6 expression were absent on virus-specific memory cells (Day 105), approximately 40% of SMARTA cells maintained CXCR5 expression, albeit at decreased surface expression relative to effector Tfh cells (Figure 1b and 1c), a pattern which was also observed in endogenous GP66-77 tetramer+ memory cells (Figure S1b). Antigen-specific CXCR5+ memory cells were also observed in the blood, but had reduced Bcl6 expression relative to their effector counterparts (Figure 1d and 1e). CXCR5+ effector cells were abundant in blood and secondary lymphoid organs; however, unlike CXCR5− Th1 effector cells, CXCR5+ cells were almost entirely excluded from non-lymphoid tissues such as lung, liver, and IEL (Figure 1f). As expected, the total number of memory SMARTA CD4+ T cells in each tissue was reduced compared to the effector stage (Table S1), and coincided with a similar pattern in the tissue distribution of CXCR5+ and CXCR5− cells at the memory timepoint (Figure 1f). Thus, antigen-specific CXCR5+ cells are mostly found in lymphoid organs and excluded from non-lymphoid tissues at both effector and memory timepoints.
At memory timepoints, CXCR5+ memory cells fall into two subsets: Ly6clo and Ly6cint. Following the contraction phase of antigen-specific Th1 and Tfh effector cells, the CXCR5−Ly6chi, CXCR5+Ly6clo, and CXCR5+Ly6cint CD4+ SMARTA subsets are maintained into the memory phase in stable numbers (Figure 1g). Together, these data show that distinct effector T helper subsets (Th1 and Tfh) develop from a clonal population of virus-specific cells and suggest that these populations are maintained as distinct lineages within the heterogeneous memory pool.
Transcriptional profiling suggests lineage relationships between Tfh effector and memory cells and between Th1 effector and memory cells
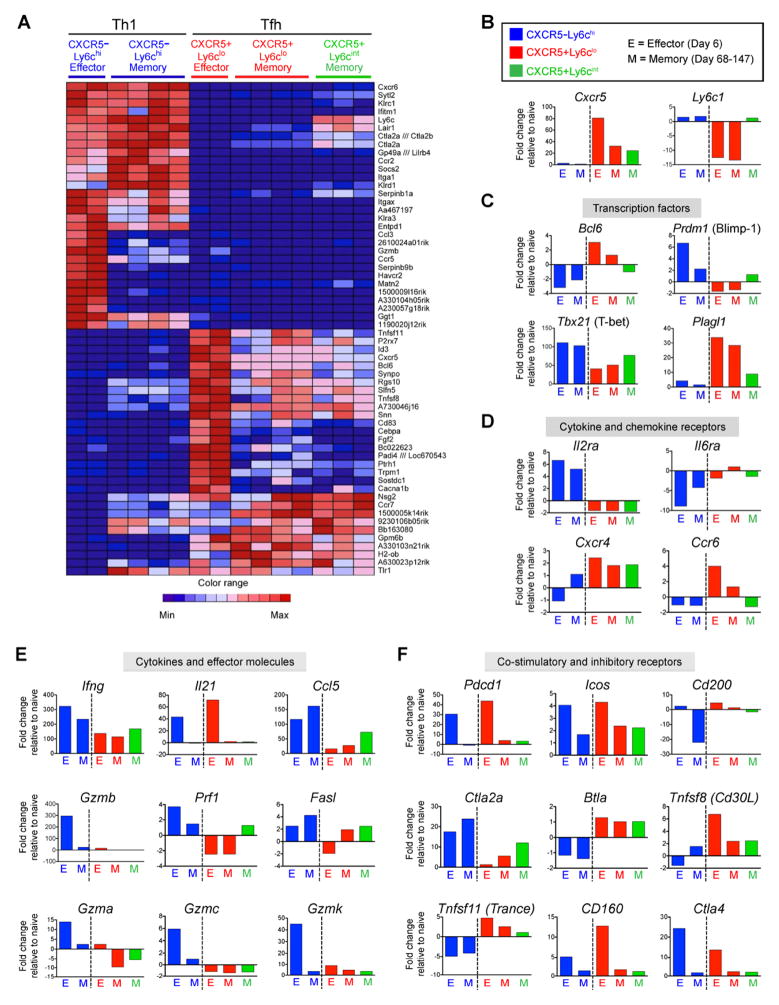
To examine potential relationships between effector and memory cells of the Tfh and Th1 cell lineages, we performed gene expression profiling of sorted CXCR5−Ly6chi, CXCR5+Ly6clo, and CXCR5+Ly6cint SMARTA subsets at effector and memory time points. These analyses revealed many genes that were commonly regulated between the Th1 effector and CXCR5−Ly6chi Th1-like memory cells, and that distinguished them from Tfh effector and CXCR5+ Tfh-like memory cells (Figure 2a). Similarly, many genes that were expressed in Tfh effector cells were also expressed in the CXCR5+Ly6clo and CXCR5+Ly6cint memory subsets while expression of these same genes was downregulated in Th1 effector and memory cells (Figure 2a). We performed gene set enrichment analysis (GSEA) of the microarray datasets to further evaluate the degree that gene expression patterns were shared between these various subsets. These analyses revealed that gene expression patterns by CXCR5−Ly6chi Th1 memory cells were enriched in the effector Th1 cell expression profile (Enrichment Score 0.78) (Figure S2a), while the upregulated gene set in CXCR5+Ly6clo memory cells was enriched in the Tfh effector profile (Enrichment score:0.70) (Figure S2b).
Figure 2. Transcriptional profiling suggests a lineage relationship between Tfh effector and memory cells and between Th1 effector and memory cells.
Microarray analysis of CD4+ SMARTA CXCR5−Ly6chi (blue), CXCR5+Ly6clo (red), and CXCR5+Ly6cint (green) subsets sorted from chimeric mice at effector (day 6 post-infection) and memory time points (day 68–147 p.i.). A) The 30 most up-regulated and 30 most down-regulated genes in the Day 6 Ly6hiCXCR5− (Th1) relative to the day 6 CXCR5+Ly6clo (Tfh) SMARTA CD4+ T cells were identified and their expression in all subsets at effector and memory stages is shown as a heatmap. B–F) Expression of select genes as determined by microarray analysis, including: B) Cxcr5 and Ly6c; C) transcription factors; D) cytokine and chemokine receptors; E) cytokines and cytotoxic effector molecules; and F) costimulatory and inhibitory receptors. Data are shown as fold change relative to naïve SMARTA CD4+ T cells. Bars show the average value for each indicated population; effector CXCR5−Ly6chi (n=2), effector CXCR5+Ly6clo (n=2), memory CXCR5−Ly6chi (n=4), memory CXCR5+Ly6clo (n=4), and memory CXCR5+Ly6cint (n=3). See also Figure S2.
Expression analysis of specific genes by CXCR5−Ly6chi memory cells revealed a similar pattern as the CXCR5−Ly6chi Th1 effector subset, showing higher levels of Tbx21 (T-bet), Prdm1 (Blimp-1), and Ly6c and no Cxcr5 expression compared to the CXCR5+Ly6clo memory subset (Figure 2b and 2c). In contrast, CXCR5+Ly6clo memory cells maintained Cxcr5 expression, intermediate levels of T-bet gene expression, and did not express Prdm1 Blimp-1, a pattern similar to CXCR5+Ly6clo Tfh effector cells (Figure 2b and 2c). Furthermore, Bcl6 transcript levels remained slighltly upregulated in the CXCR5+Ly6clo memory cells (relative to naïve cells). Similar patterns of Bcl6 and Prdm1 (Blimp-1) gene expression were confirmed by real-time PCR (Figure S2c). Together, these data indicate that the gene expression patterns of Cxcr5 and these transcription factors that define the Th1 and Tfh lineages in CD4+ T cells are maintained (although at lower expression levels compared to effector cells) in subsets of resting memory cells. In addition, we found that gene expression of the transcription factor Plagl1 (Abdollahi, 2007), a gene with no previously reported role in CD4+ T cell differentiation, distinguished the Th1 and Tfh cell populations at both the effector and memory phases of the immune response (Figure 2c).
We next examined the expression of genes that encode cytokine and chemokine receptors, costimulatory and inhibitory receptors, cytokines, and cytotoxic molecules. Expression of Th1 associated genes including Il-2ra, Ifnγ, Ccl5, and Gzmb were more highly expressed in the CXCR5−Ly6chi compared to the CXCR5+Ly6clo and CXCR5+Ly6cint memory populations (Figure 2d–e). In contrast, genes involved in Tfh function and differentiation (Crotty, 2011), including Cxcr5, Cxcr4, Il6ra, Pdcd1 (PD-1), Cd200, and Sh2d1a (SAP) were more highly expressed in memory CXCR5+Ly6clo compared to memory CXCR5−Ly6chi cells (Figure 2b and 2d–2f and Figure S2d). Of note, several genes related to the cytotoxic potential of effector cells, including those that code for Granzyme B and other granzymes, perforin, and Fas ligand were preferentially increased in the CXCR5−Ly6chi SMARTA effector cells (Figure 2e). Interestingly, the CXCR5+Ly6cint memory population in many cases displayed a gene expression pattern that was intermediate between that of the CXCR5−Ly6chi and CXCR5+Ly6clo memory populations (Cxcr5, Bcl6, Prdm1, Tbx21, Plagl1, Ccr6, Ccl5, Prf1, and Ctla2a), suggesting more heterogeneity and/or lineage pluripotency within this population (Figure 2b–f). Collectively, these data show that memory CXCR5−Ly6chi and CXCR5+Ly6clo CD4+ T cell populations are enriched for Th1 and Tfh associated gene expression programs, respectively, and may thus be poised to preferentially recall distinct T helper effector responses upon antigen re-exposure.
Th1 and Tfh memory CD4+ T cells are committed for recall of lineage-specific functions
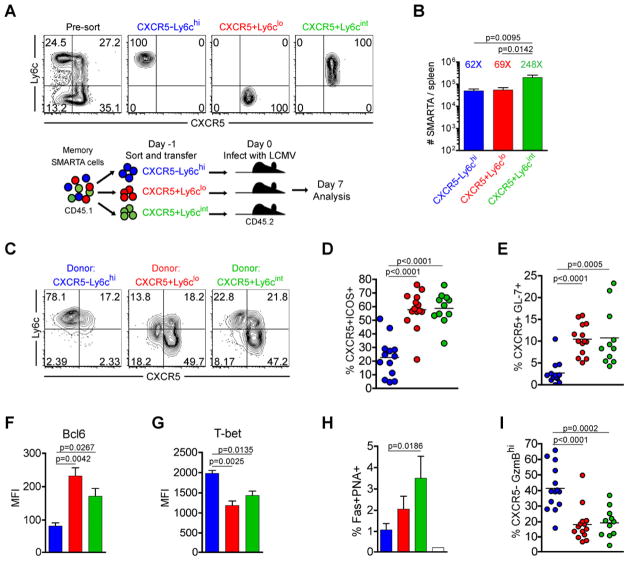
To determine whether the generation of secondary Th1 versus Tfh effector CD4+ T cells arise from these distinct memory populations, we sorted memory SMARTA cells into CXCR5−Ly6chi, CXCR5+Ly6clo, and CXCR5+Ly6cint subsets and transferred them into naïve recipient mice, then challenged the chimeric mouse with an acute LCMV infection (Figure 3a). The expansion of the CXCR5−Ly6chi and CXCR5+Ly6clo responding memory populations was relatively similar in the spleen 7 days post-infection (between 62–69 fold assuming a 10% take in the spleen following adoptive transfer), while the CXCR5+Ly6cint subset expanded more extensively (approximately 248 fold) (Figure 3b). Interestingly, CXCR5+Ly6clo and CXCR5+Ly6cint memory SMARTA cells preferentially recalled a Tfh phenotype following rechallenge, becoming mostly CXCR5+ effector cells, while the majority of secondary effectors derived from the transferred CXCR5−Ly6chi memory cells remained Ly6chi and CXCR5− (Figure 3c). Furthermore, both the CXCR5+Ly6clo and CXCR5+Ly6cint memory cells gave rise to significantly increased frequencies of CXCR5+ ICOShi Tfh effector cells and contained significantly more GL-7+ germinal center Tfh cells relative to those derived from CXCR5−Ly6chi memory cells (Figure 3d and 3e and Figure S3a). Secondary effector cells derived from the CXCR5+Ly6clo and CXCR5+Ly6cint memory populations displayed significantly higher Bcl6+ and lower T-bet expression relative to those derived from CXCR5−Ly6chi memory CD4+ T cells (Figure 3f and 3g and Figure S3d). Importantly, the transfer of CXCR5+ memory SMARTA cells promoted the rapid appearance (within seven days post-infection) of Fas+PNA+ germinal center B cells compared to the transfer of CXCR5−Ly6chi memory cells (Figure 3h). Together, these data indicate that CXCR5+ memory cells are biased toward a Tfh cell recall response.
Figure 3. Th1 and Tfh memory CD4+ T cells are committed for recall of lineage-specific functions.
CD45.1 congenically marked CXCR5−Ly6chi (blue), CXCR5+Ly6clo (red), and CXCR5+Ly6cint (green) memory CD4+ subsets (between days 56 and 101 post-infection) were FACS purified to >97%. 8×103 sorted cells were adoptively transferred into naïve CD45.2 recipients, and recipient mice were then infected with LCMV Armstrong 16–20 hours later. In (C–I), the phenotype and function of transferred SMARTA cells was analyzed 7 days post-infection. A) Representative post-sort analysis of memory SMARTA subsets and cartoon of experimental setup. B) Absolute number of transferred CD4+ CD45.1+ SMARTA splenocytes 7 days following rechallenge with LCMV. The relative fold increase of each population, assuming a 10% take of transferred SMARTA cells, is shown above each bar. C) Representative CXCR5 and Ly6c analysis. D) Chart shows the percent of transferred SMARTA cells that are CXCR5+ICOS+ Tfh cells. E) The percent of SMARTA cells that are CXCR5+GL-7+ germinal center Tfh cells. F) Bcl6 MFI of SMARTA cells. G) T-bet MFI of SMARTA cells. H) Chart shows the percent of B220+ gated splenic B cells that are Fas+PNA+ germinal center B cells. Data were combined from 2 experiments. I) Chart shows the percent of transferred SMARTA cells that are CXCR5−GranzymeBhi cells. Data in B, D, E and I are combined from 4 independent experiments for a total of N=11–14 mice per group. Data in F and G were from a single experiment (N=3 per group) and were representative of 2–4 independent experiments. Statistically significant p values are shown, and were determined using a two-tailed unpaired Students t test. Error bars represent the SEM. See also Figure S3.
To further define the characteristics of CXCR5−Ly6chi, CXCR5+Ly6clo, CXCR5+Ly6cint memory SMARTA cells, we evaluated their capacity to generate a Th1 cell secondary response. Effector cells derived from CXCR5−Ly6chi memory cells had significantly increased levels of T-bet expression (Figure 3g and Figure S3d). In addition, the majority of these cells were CXCR5−, and had an enhanced capacity for granzyme B expression in both the spleen (Figure 3i and Figure S3a) and in lung (Figure S3g). In contrast, while the majority of effector cells generated from CXCR5+Ly6clo and CXCR5+Ly6cint subsets were CXCR5+, those that became CXCR5− cells had a diminished capacity to express granzyme B (Figure 3i and Figure S3a), even in the lung, a non-lymphoid tissue that enriches for Th1 effector cells (Figure S3g). Thus, CXCR5−Ly6chi Th1 memory cells efficiently produce Th1 effector cells following antigen challenge, while CXCR5+Ly6clo and CXCR5+Ly6cint Tfh memory cells have a cell-intrinsic restriction that limits their capacity to express granzyme B. In addition, significantly fewer effector cells derived from CXCR5+Ly6clo and CXCR5+Ly6cint Tfh subsets (compared to those from CXCR5− Ly6chi memory cells) produced IFNγ and had lower expression of this cytokine, being more consistent with the IFNγ expression levels of Tfh cells (Figure S3h).
To determine whether the CXCR5−Ly6chi, CXCR5+Ly6clo, and CXCR5+Ly6cint memory subsets are committed to their respective Th1 and Tfh cell lineages, we compared their responses to the primary effector response generated from uncommitted (naïve) SMARTA CD4+ T cells. CXCR5−Ly6chi memory cells exhibited Th1 lineage commitment, producing a significantly higher frequency of Granzyme B+ and IFNγ+ effector cells, while CXCR5+Ly6clo and CXCR5+Ly6cint memory cells generated a significantly higher frequency of Tfh cells and fewer Th1-like cells compared to the primary effector response (Figure S3b, S3f, and S3h). We thus conclude that the CXCR5−Ly6chi subset are Th1 memory cells, and that both CXCR5+Ly6clo and CXCR5+Ly6cint subsets are Tfh memory cells.
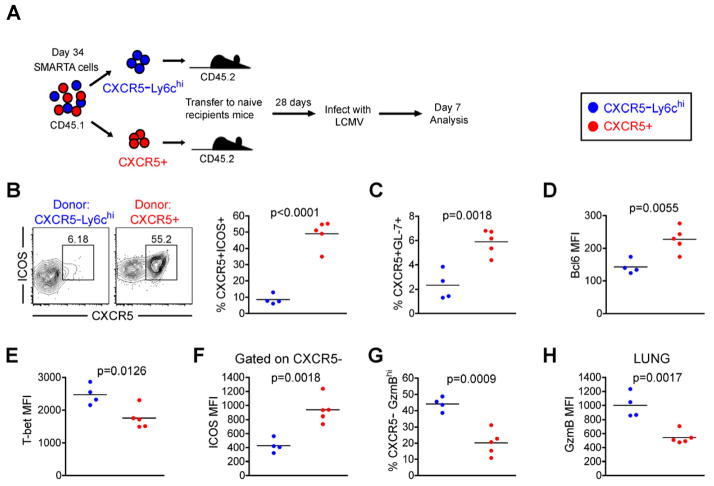
To determine whether Th1 and Tfh memory cells maintain their respective lineage biases in the absence of antigen, sorted Th1 memory and Tfh memory SMARTA cells were adoptively-transferred into naive recipient mice and rested for 28 days before rechallenging these recipients with acute LCMV infection (Figure 4a). Consistent with our previous results (Figure 3), CXCR5+ Tfh memory cells preferentially generated Tfh effector cells (Figure 4b) and GL7+ GC Tfh cells (Figure 4c). Furthermore, these effector cells expressed significantly more Bcl6 and less T-bet (Figure 4d and 4e). The CXCR5− effector cells that were generated from the CXCR5+ Tfh memory population also had significantly higher ICOS expression (Figure 4f) and exhibited an impairment for Granzyme B expression compared to those generated from CXCR5− Th1 memory cells (Figure 4g and 4h). After challenge, memory Th1 cells generated Th1 effector cells with high T-bet and Granzyme B expression (Figure 4). Thus, the capacity of CXCR5− Th1 and CXCR5+ Tfh memory cells to recall their lineage-specific responses is not dependent on the continued presence of antigen during the maintenance phase of memory differentiation.
Figure 4. Lineage commitment of Th1 and Tfh memory subsets is maintained in the absence of antigen.
CD45.1 congenically marked CXCR5−Ly6chi (blue) and CXCR5+ (red) memory SMARTA CD4+ T cell subsets 34 days post-infection were FACS purified to >97%. 8×104 purified cells were adoptively transferred into naïve CD45.2 recipients. 28 days following cell transfer, naïve recipients were infected with 2×105 p.f.u. LCMV Armstrong 16–20 hours later. The phenotype (B–H) of transferred CD4+CD45.1+ gated SMARTA cells were analyzed 7 days post-infection. A) Cartoon of experimental setup. B) CXCR5 and ICOS analysis. Chart shows the percent of SMARTA cells that are CXCR5+ICOS+ Tfh cells. C) The percent of transferred SMARTA cells that are CXCR5+GL-7+ germinal center Tfh cells. D) Bcl6 MFI of SMARTA cells. E) T-bet MFI of SMARTA cells. F) ICOS MFI on CXCR5− gated effector SMARTA cells 7 days post rechallenge. G) Chart shows the percent of transferred SMARTA cells that are CXCR5−Granzyme Bhi cells 7 days post rechallenge. H) Chart shows the Granzyme B MFI of SMARTA cells in lung 7 days post rechallenge. For A–H: CXCR5−Ly6chi (N=4); CXCR5+ (N=5). Statistically significant p values of <0.05 are indicated, and were determined using a two-tailed unpaired Students t test. See also Figure S4.
Cells with a central memory phenotype exist within the Tfh and Th1 memory populations
We next evaluated whether a central memory phenotype (by CD62L and CCR7 expression) was associated with CXCR5 expression by memory cells. As expected, within 5 days post-LCMV infection, SMARTA cells had downregulated CD62L, followed by the gradual re-expression by memory cells (Figure S4a). Compared to Day 8 effector cells that were almost entirely CD62L− and had downregulated CCR7, all memory SMARTA cells expressed some surface CD62L and CCR7, being either low-intermediate or intermediate-high for each of these surface markers (Figure S4b). Further analysis showed that cells expressing high levels of surface CD62L and CCR7 existed within both CXCR5+ and CXCR5− memory populations (Figure S4c). Thus, cells of a central memory phenotype exist within both Th1 and Tfh memory populations (Figure S4c). To determine whether CD62L+ compared to CD62L− Th1 and Tfh memory cells exhibited different T helper lineage recall potential following rechallenge, we sorted CD62L− and CD62L+ subsets of CXCR5+ and CXCR5− memory cells, transferred into congenic recipient mice, and challenged these mice with LCMV (Figure S4d). Both CD62L+ and CD62L− Th1 memory cells preferentially generated CXCR5− Th1 effector cells to similar degrees, while CXCR5+ CD62L− and CD62L+ memory cells generated substantial Tfh effector cells (Figure S4e). These data suggest that while CD62L+ central memory phenotype cells exist within both Th1 and Tfh memory populations, the CXCR5− Th1 memory and CXCR5+ Tfh memory cells maintain and recall their relative lineage-associated phenotypes regardless of CD62L expression.
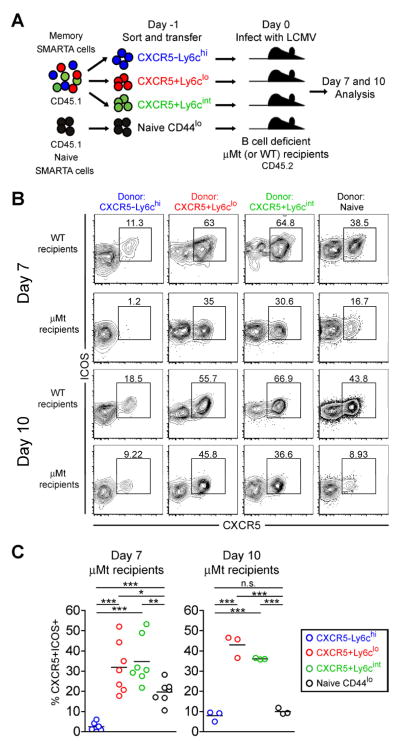
Tfh memory CD4+ T cells recall a Tfh-like response even in B cell deficient recipient mice
Interaction with cognate B cells and continued ICOS signaling are required for stabilizing the Tfh phenotype and the maintenance of primary effector Tfh cells (Crotty, 2011). We next evaluated whether memory Tfh cells could recall aspects of the Tfh effector program when reactivated in the absence of B cells by transferring them into B cell deficient recipients prior to LCMV infection (Figure 5a). As expected, GL-7+ (germinal center phenotype) Tfh-like effector cells were nearly undetectable in B cell deficient recipient mice following infection (Figure S5). Similar to what has been previously reported (Johnston et al., 2009), naïve SMARTA cells transferred into B cell deficient mice generated a significantly reduced frequency of CXCR5+ Tfh-like effector cells at day 7 post-infection, which continued to decrease to less than 10 percent by day 10 (Figure 5b and 5c). In contrast, approximately 30 percent of effectors derived from CXCR5+Ly6clo and CXCR5+Ly6cint memory cells were CXCR5+ Tfh-like cells at day 7, and surprisingly this frequency was maintained at approximately 35–45% at day 10 in the absence of B cells (Figure 5b and 5c). Thus, compared to naïve CD4+ T cells, CXCR5+Ly6clo and CXCR5+Ly6cint memory cells have a cell-intrinsic capacity to recall and maintain aspects of the Tfh phenotype such as CXCR5 expression, even in the absence of B cells. These results clearly demonstrate the persistence of memory CD4+ T cells with commitment to the Tfh lineage.
Figure 5. Tfh memory CD4+ T cells recall a Tfh-like response even in B cell deficient recipient mice.
CD45.1 congenically marked CXCR5−Ly6chi (blue), CXCR5+Ly6clo (red), and CXCR5+Ly6cint (green) SMARTA memory CD4+ T cell subsets (days 68 and 88 post-infection), and naïve SMARTA cells sorted as CD44lo (black) were FACS purified to >97%. 8×103 purified cells were adoptively transferred into naïve CD45.2+ WT and μMT (B cell deficient) B6 recipients, and recipient mice were then infected with LCMV Armstrong. The phenotype and function of SMARTA cells were analyzed 7 and 10 days post-infection. A) Cartoon of experimental setup. B) Representative CXCR5 and ICOS analyses of CD4+ CD45.1+ gated SMARTA cells 7 and 10 days post-infection. C) Charts show the frequency of SMARTA cells that are CXCR5+ICOS+ Tfh-like cells in B cell deficient mice at each time point following infection with LCMV. Data for day 7 were combined from 2 independent experiments (N=7 per experimental group) and day 10 were from one experiment (N=3 per group). Statistically significant p values were determined using a two-tailed unpaired Students t test (*** p<0.001; ** p<0.01; * p<0.05). See also Figure S5.
Epigenetic modifications of the granzyme B locus distinguish Tfh memory from Th1 memory CD4+ T cells
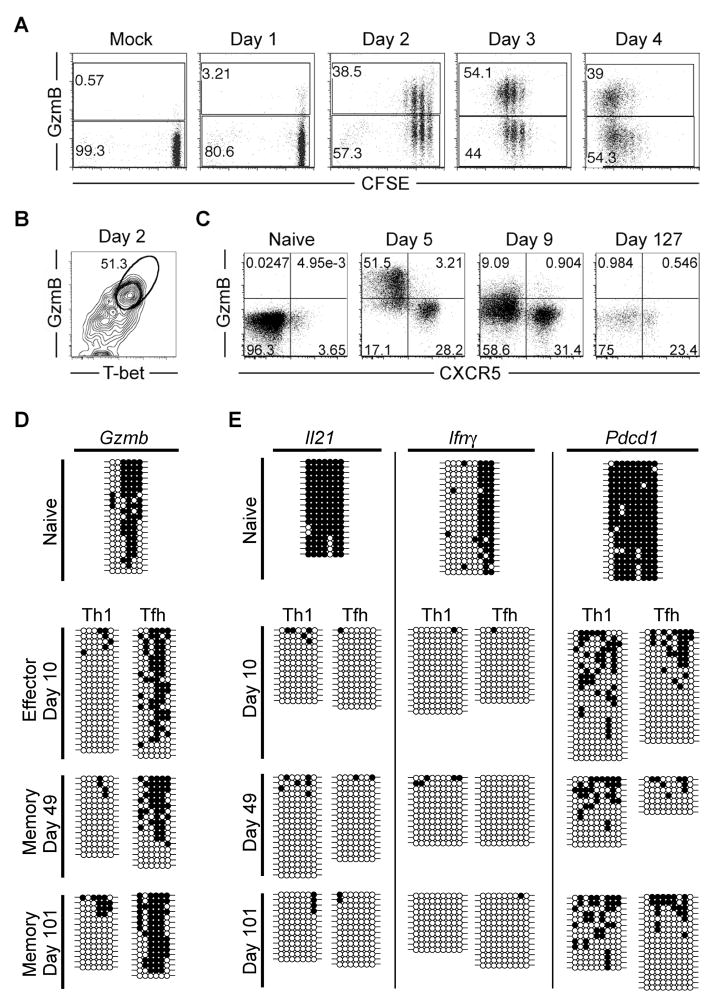
We have shown that granzyme B expression is strikingly different between Th1 and Tfh cells at an effector timepoint (Figures 1, 2, and S3), and is most efficiently re-expressed by CXCR5−Ly6chi Th1 memory cells following viral rechallenge (Figure 3 and S3). We next tested whether the difference in granzyme B expression occurs very early in CD4+ lineage differentiation following activation of naïve CD4+ T cells. Rapidly following activation, naive SMARTA cells had diverged into distinct granzyme B+ and granzyme B− populations, even in undivided cells, that persisted as separate populations while undergoing proliferation during the first 4 days of LCMV infection (Figure 6a). Granzyme B expression correlated with high T-bet expression as early as day 2 post-infection (Figure 6b), demonstrating that very early following activation (within only a few rounds of cell division), high granzyme B expression is restricted to Th1 cells. Day 5 CXCR5− Th1 cells expressed high levels of granzyme B, while expression was mostly diminished by day 9, and absent in CXCR5− SMARTA memory cells (Figure 6c and Figure 1b).
Figure 6. Epigenetic modifications of the granzyme B locus distinguish Tfh memory from Th1 memory CD4+ T cells.
A–B) 1-2×106 CFSE-labeled SMARTA cells were transferred into host mice and infected one day later. A) Plots show granzyme B expression and CFSE dilution analysis of SMARTA cells at the indicated time points post-infection. B) Plot shows Granzyme B and T-bet expression analysis of SMARTA cells 2 days post-infection. C–E) 2×105 SMARTA cells were transferred into host mice that were infected 1 day later. C) Flow cytometry plots show granzyme B and CXCR5 expression of SMARTA cells at the indicated time points post-infection. D and E) Sorted antigen-specific SMARTA CD4+ Tfh and Th1 cells were isolated from the spleen and purified by FACS at naïve, effector, and memory stages of differentiation. Tfh subsets were defined as CXCR5+ Ly6clo, while Th1 subsets were defined as CXCR5−Ly6chi. DNA methylation status of D) Gzmb and E) Il21, Ifnγ, and Pdcd1 loci were determined by bisulfite sequencing the genomic DNA from the purified cells. Each horizontal line corresponds to the sequence of an individual clone. Filled circles = methylated cytosine. Open circles = non-methylated cytosine. Representative dot plots from one complete data set are shown. Data are representative of at least 3 indepentently isolated populations of naïve (uninfected CD44lo), effector (days 5–10), and memory (days 49–101) SMARTA cells. See also Figure S6.
The bimodal distribution of granzyme B expression during effector CD4+ T cell differentiation, in conjunction with our observation that granzyme B transcript is significantly upregulated only in CD4+ Th1 effector cells, suggests that CD4+ Tfh cells retain a transcriptional regulatory mechanism for repressing granzyme B expression. Epigenetic modifications serve as a mechanism for a dividing cell population to “remember” the transcriptional status of the parental cell population (Reik, 2007). To determine whether the restriction of granzyme B expression is coupled to repressive epigenetic modifications in effector and memory Tfh cells, we analyzed the DNA methylation status in the Gzmb transcriptional regulatory region in naïve, Th1 and Tfh effector, and Th1 and Tfh memory cells following LCMV infection. We found that the Gzmb locus becomes unmethylated exclusively in Th1 effector cells and remains unmethylated in Th1 memory cells, while CD4+ Tfh cells retain the naïve DNA methylation program at the Gzmb locus throughout effector and memory differentiation (Figure 6d). Thus, our data show that methylation of the Gzmb locus can be used to distinguish CD4+ Th1 cells from Tfh cells, and further confirm the lineage relationship between effector and memory Tfh cells. Furthermore, the repression of cytotoxic molecule expression such as granzyme B and perforin (Figure 2e) in Tfh cells may be essential for preventing the unwanted destruction of antigen-presenting B cells.
We also examined the methylation status of several other loci in Th1 and Tfh lineage cells. Surprisingly, the Il21 locus was demethylated in both Tfh and Th1 cells, and remained unmethylated in both memory populations (Figure 6e). Although IL-21 is characterized as a cytokine primarily relevant to Tfh function, Th1 cells can express it, albeit with reduced transcript levels (Figure 2) (Fahey et al., 2011; Spolski and Leonard, 2010). Similarly, the Ifnγ locus became demethylated in both Tfh and Th1 cells and remained unmethylated in memory cells (Figure 6e). These data suggest that both Th1 and Tfh memory CD4+ T cells transitioned through an effector stage of differentiation, and that the Ifnγ and Il21 loci may be similarly poised for potential rapid reexpression in both subsets of memory cells following antigen reencounter. The Pdcd1 (PD-1) locus was demethylated following infection in both Tfh and Th1 cells, although there may be slightly more methylation in Th1 compared to Tfh memory cells (Figure 6e).
Discussion
There is considerable interest in understanding the generation of CD4+ Tfh memory cells. However, this topic remains controversial since several recent studies have reported different conclusions regarding the existence of a committed population of memory Tfh cells (Liu et al., 2012; Luthje et al., 2012; MacLeod et al., 2011; Marshall et al., 2011; Pepper et al., 2011; Weber et al., 2012). In this study using the mouse model of acute LCMV infection, we show that there are distinct populations of virus-specific memory CD4+ T cells with commitment to either the Tfh or Th1 cell lineages. Our findings indicate that antigen-specific memory CD4+ T cells that maintain CXCR5 expression are biased towards the recall of a Tfh effector response, while CXCR5−Ly6chi memory cells efficiently recall Th1 effector cells. These findings are consistent with a model in which the concomitant generation of a pool of resting memory Th1 or Tfh cells arise from the corresponding effector Th1 and Tfh cells, respectively. Commitment of memory CD4+ T cells to Th1 or Tfh lineages provides cells that are poised for the lineage-specific reexpression of effector molecules upon reexposure to antigen (see model in Figure S6).
In contrast to our findings, using a Listeria monocytogenes infection model, Pepper et al. suggested that memory CD4+ T cells that express CXCR5 and CCR7 are a central memory population with a capacity to fully reconstitute Th1 and Tfh effector populations (Pepper et al., 2011). Wheras our study also shows that there is more pluripotency among the Tfh compared to Th1 memory population, there is still clear evidence of Tfh lineage commitment in CXCR5+ memory CD4+ T cells. The differing results seen in recall responses from CXCR5+ memory cells between our study and the report by Pepper et al. may result from the different infection systems used (LCMV versus Listeria monocytogenes). The report by Pepper et al. suggests that Tfh cells do not enter the memory pool based on the absence of CXCR5hiPD-1hi cells at memory timepoints (Pepper et al., 2011). In our study, LCMV-specific cells CD4+ T cells (both SMARTA and endogenous GP66-77 tetramer+) at memory timepoints show decreased CXCR5 and relatively absent PD-1, ICOS, and Bcl6 expression levels by FACS staining. We interpret this observation (the lack of CXCR5hiPD-1hi memory cells) to signify that activation molecules such as PD-1 (and to some extent CXCR5) are downregulated on memory Tfh cells following antigen clearance, rather than indicate the disappearance of Tfh lineage cells altogether from cells entering the memory pool.
A recent study by Luthje et al. using an Il21 GFP reporter system showed that both GFP+ and GFP− subsets of CXCR5+ Tfh cells efficiently enter the memory pool of CD4+ T cells following adoptive transfer. In contrast with our observations, they observed a higher frequency of CXCR5− effectors generated from Tfh-derived memory cells following influenza rechallenge (Luthje et al., 2012). Clear interpretation of their study is limited by the lack of analysis of antigen-specific cells at effector versus memory and recall timepoints (either by MHC tetramer staining or using TCR transgenic cells). Their study also showed that the majority of GFP+ (Il21+) effector cells were CXCR5+ Tfh effector cells (Luthje et al., 2012). Our data show that both Tfh and Th1 effector cells express Il21 following LCMV infection, and that the Il21 locus becomes demethylated in both Tfh and Th1 effector subsets. Therefore, IL-21 expression in itself is not a clear universal indicator of T helper lineage. While our data indicate that some CXCR5+Ly6clo and CXCR5+Ly6cint memory cells become CXCR5 negative following reactivation, we observed that such CXCR5− (Th1-like) effector cells do not express granzyme B to the same extent as secondary effector cells derived from Th1 memory cells. Our findings suggest that while there is some degree of heterogeneity and/or plasticity within the Tfh (CXCR5+Ly6clo and CXCR5+Ly6cint) memory population, these cells do not yield secondary effector cells with the same capacity for Th1 cell function as memory Th1 (CXCR5−Ly6chi) cells.
Several observations within our study clearly show that memory CD4+ T cells with commitment to the Tfh lineage exist within the memory pool following acute LCMV infection. First, the Tfh and Th1 cell lineage dichotomy that is established very early following LCMV infection based on both CXCR5 surface expression and the restriction of the capacity for Granzyme B expression can be tracked into the memory pool. Indeed, similar to their effector precursors that do not express Granzyme B, CXCR5+ memory cells carry repressive DNA methylation marks at the Gzmb locus, and the CXCR5− minority cell population that emerges from the CXCR5+ memory cells following reexposure to antigen shows a delayed or impaired ability to express Granzyme B. It has been shown that maintenance of repressive DNA methylation programs at the Il4 and Foxp3 loci are essential for preventing aberrant expression of these molecules in non-Th2 and non-Treg CD4+ T cell lineages, respectively (Josefowicz et al., 2009; Makar et al., 2003). Thus, the maintenance of DNA methylation at the Gzmb locus (and other Th1-specific loci) in Tfh memory cells may reinforce their Tfh lineage commitment by repressing genes used by Th1 lineage cells (Wilson et al., 2009). Therefore, differential DNA methylation at the Gzmb locus can be used as an additional criterion for distinguishing CD4+ Tfh memory from Th1 memory cells. Second, compared to the primary response generated from uncommitted naïve CD4+ T cells of the identical TCR specificity (SMARTA transgenic), CXCR5+ Tfh memory cells are more biased toward generating Tfh cells. Third, transfer experiments to B cell deficient mice demonstrate that a large fraction of memory CXCR5+ Tfh cells have acquired cell-intrinsic programs that allow them to recapitulate portions of the Tfh phenotype (including CXCR5 expression) following reactivation in the absence of bidirectional signals from B cells that are required for the generation and maintenance of primary effector Tfh cells.
In agreement with our findings, several recent reports provide evidence supporting the existence of memory Tfh cells. Marshall et al observed that a subset of memory cells with similar phenotypic features as Tfh cells existed within the memory cell pool (Marshall et al., 2011). MacLeod et al. demonstrated that CXCR5+ memory cells enhance the kinetics of B cell expansion and class switching, and suggested that such CXCR5 expression on memory cells promotes migration to areas where encounter with congnate B cells can occur following reintroduction of antigen (MacLeod et al., 2011). Similarly, findings by Liu et al. are suggestive that effector Tfh cells enter the memory T cell pool and retain their preference for recall of Tfh cells, although whether the cells are truly memory cells is unclear based on the early timepoints of cells used for transfer (Liu et al., 2012). Another study suggested that rechallenge of memory cells derived from adoptively-transferred Tfh effector cells exhibited a Tfh phenotype 2.5 days after reimmunization, however, this phenotype was diminished 6 days post reactivation (Weber et al., 2012). Morita et al. reported that CXCR5+CD4+ T cells found in human blood are functional counterparts to Tfh cells found in lymphoid organs, and suggest that these may be memory Tfh cells (Morita et al.).
The repression of cytotoxic potential appears to be a hallmark of Tfh cells, and expression of key cytolytic molecules such as Granzyme B and perforin was inhibited in Tfh but not Th1 effector cells. Together, these data suggest that Tfh differentiation is coupled to the restriction in cytotoxic potential by preventing expression of Th1 cell-related killing apparatus components. The epigenetic repression of cytotoxic molecules by Tfh cells may be essential to prevent the unwanted destruction of antigen-presenting germinal center B cells which rely on cell contact dependent interactions (including CD40L and SAP) with Tfh cells for essential signals (Cannons et al., 2010; Crotty, 2011; Crotty et al., 2003; Qi et al., 2008).
Our findings demonstrate that acute LCMV infection induces a strikingly balanced response of both Tfh and Th1 effector CD4+ T cells and robust long-lived memory populations for both of these lineages. This raises the question as to whether such Tfh and Th1 balance and memory generation is achieved by other viral infections, vaccine vectors, and protein immunization strategies. In addition, balancing the generation of Tfh memory and other lineages of CD4+ memory T cells may be essential for providing secondary responses that provide both optimal CD4+ T cell help for antibody responses, as well as CD4+ T cells with effector function. Thus, future studies are needed to determine how different adjuvants, inflammatory cytokines, costimulatory molecules and interactions with different types of antigen presenting cells can be used to specifically induce either Tfh and Th1 memory cells.
The majority of our successful vaccines rely on neutralizing antibody and long-lived humoral responses for protective immunity (Plotkin et al., 2013). Gaining a better understanding of the development, function, and contribution of Tfh memory cells within the context of prime/boost vaccination and pathogen challenge will provide avenues for rational vaccine design (D’Argenio and Wilson). Developing effective vaccines for challenging pathogens such as HIV and malaria will likely require the generation of high titer broadly neutralizing antibody responses (Burton et al.). Similarly, efforts to develop universal influenza vaccines by targeting conserved epitopes (Sette and Rappuoli) may possibly be improved by optimally inducing memory Tfh cells. It is likely that vaccine strategies that fail to induce the generation of primary Tfh cells will subsequently leave a gap in the pool of antigen-specific memory CD4+ T cells, resulting in suboptimal boosting of antibody responses. Conversely, immunogens, adjuvants, or strategies that promote robust Tfh responses and drive commitment and maintenance of high numbers of antigen-specific memory Tfh cells may enhance the quality and/or quantity of antibody production following antigen boost or pathogen encounter. Thus, it is critical to design rational prime and boost strategies for optimal generation of Tfh memory cells.
Experimental Procedures
Mice and Adoptive Transfers
Congenically marked (CD45.1) CD4+ T cell splenocytes specific to the GP66-77 epitope of LCMV obtained from naïve SMARTA TCR transgenic mice (Oxenius et al., 1998) were intravenously transferred into naïve C57BL/6 (CD45.2) mice (Jackson Laboratory, Bell Harbor, ME). In order to generate SMARTA chimeric mice with a high number of memory SMARTA cells, thus facilitating memory cell isolation for adoptive transfer studies, we transfered 2×105 naïve SMARTA CD4+ T cells. ~24 hours post-transfer, chimeric mice were infected i.p. with 2×105 PFU of LCMV Armstrong. For adoptive transfer of memory SMARTA cells, CD4+ splenocytes from chimeric mice (day 56–101 post-infection) were enriched using a MACS CD4+ T cell Isolation Kit II (Miltenyi). Enriched CD4+ T cells were stained and sorted to isolate CD45.2− (CD45.1 SMARTA) memory subsets based on CXCR5, Ly6c, or CD62L to greater than 98% purity. For rechallenge experiments, 8×103 sorted memory (or Naïve) SMARTA cells were adoptively transferred into naïve C57BL/6 mice (CD45.2) or B cell deficient μMT mice that were subsequently infected 24 hours later with 2×105 PFU of LCMV Armstrong. Animal experiments were conducted in accordance with Emory University IACUC protocols.
FACS Analysis and sorting
Cell were stained as described previously (Youngblood et al., 2011) with fluorochrome-conjugated antibodies (purchased from BD, eBiosciences, BioLegend, Vector Laboratories Inc., and Invitrogen). A three step CXCR5 staining was performed as described by (Johnston et al., 2009) using purified rat anti-mouse CXCR5 (BD), a secondary Biotin-SP-conjugated Affinipure F(Ab′)2 Goat anti-rat IgG (Jackson Immunoresearch) and finally with streptavidin-APC or streptavidin-PeCy7 (Invitrogen). For Bcl6 and T-bet staining, cells were first stained for surface antigens, followed by permeabilization, fixation and staining using the Foxp3 Permeabilization/Fixation kit and protocol (eBioscience). Intracellular cytokine staining was done by standard techniques following 5 hour stimulation with GP61-80 peptide (Murali-Krishna et al., 1998). Cell sorting was performed using a FACS Aria II (BD) and flow cytometry data were collected on a FACS Canto II (BD). FACS data were analyzed using FlowJo software (TreeStar Inc.)
Microscopy
Spleens were frozen in OCT compound (Fisher Scientific, Waltham, MA) and sectioned. Sections were fixed in cold acetone for 10 min, dried, and stained with the indicated antibodies. Images were acquired using a Zeiss Axioskop 2 Plus microscope with 10×/0.25 and 20×/0.50 (magnification/aperature) objectives and a Zeiss Axiocam MRc5 camera. Image overlays were performed with ImageJ software (NIH, Bethesda, MD).
RNA isolation and Microarray analysis
RNA from sorted cells was purified (Qiagen), linearly amplified (NuGEN), and hybridized to Affymetrix mouse 430 2.0 arrays (Memorial Sloan-Kettering Cancer Center, Genomics Core Facility). Gene pattern 3.4 and the associated modules were used to analyze the microarray data. Gene set enrichment analysis (GSEA) was performed as described previously (Subramanian et al., 2005).
Genomic DNA Methylation Analysis
Bisulfite modification of genomic DNA from FACS purified cells was performed using the Zymo Research EZ DNA methylation kit. Bisulfite modified DNA was PCR amplified with locus-specific primers (Table S2) as previously described (Youngblood et al., 2010).
Statistical Analysis
All experiments were analyzed using Prism 4. Statistically significant p values of <0.05 are indicated, and were determined using a two-tailed unpaired Students t test.
Accession numbers
The microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE43863.
Supplementary Material
Highlights.
Tfh effector and Tfh memory cells share phenotypic and transcriptional similarities.
CXCR5+ Tfh memory cells show commitment to the Tfh-cell lineage during recall.
Granzyme B locus methylation distinguishes Tfh memory from Th1 memory cells.
Acknowledgments
We thank Dr. Marion Pepper (University of Washington) and Dr. Marc Jenkins (University of Minnesota) for the generous gift of I:Ab Gp66-77 tetramer. We thank R. Karaffa and S. Durham for FACS sorting at the Emory University School of Medicine Flow Cytometry Core Facility. This work was supported by the National Institutes of Health (NIH) grant P01 A1080192 (to R.A.), grant RO1 AI030048 (to R.A.), Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery UM1AI100663 (to R.A), the American Cancer Society (ACS) postdoctoral fellowship PF-09-134-01-MPC (to B.A.Y.), and the National Institute of Allergy and Infectious Diseases Training Grant T32AI074492 and F32 A1096709-01A1 (to J.S.H.).
Footnotes
There are no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdollahi A. LOT1 (ZAC/PLAGL1) and its family members: mechanisms and functions. Journal of Cellular Physiology. 2007;210:16–25. doi: 10.1002/jcp.20835. [DOI] [PubMed] [Google Scholar]
- Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. Journal of Experimental Medicine. 1999;190:1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. Journal of Experimental Medicine. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, et al. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodrigquez J, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 2010;32:253–265. doi: 10.1016/j.immuni.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular Helper CD4 T Cells (Tfh) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentation. Nature Immunology. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. SAP is required for generating long-term humoral immunity. Nature. 2003;421:282–287. doi: 10.1038/nature01318. [DOI] [PubMed] [Google Scholar]
- D’Argenio DA, Wilson CB. A decade of vaccines: Integrating immunology and vaccinology for rational vaccine design. Immunity. 33:437–440. doi: 10.1016/j.immuni.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. Journal of Experimental Medicine. 2011;208:987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N, Eisenbraun MD, Malerbe L, Ebright JN, Pogue-Caley RR, McHeyzer-Williams LJ, McHeyzer-Williams MG. Lymphoid reservoirs of antigen-specific memory T helper cells. Nature Immunology. 2007;8:753–761. doi: 10.1038/ni1472. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, Ditoro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J Immunol. 2009;182:6648–6652. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. Journal of Experimental Medicine. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. Journal of Experimental Medicine. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthje K, Kallies A, Shimohakamada Y, Tbelz GT, Light A, Tarlington DM, Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nature Immunology. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- MacLeod MK, David A, McKee AS, Crawford F, Kappler JW, Marrack P. Memory CD4 T cells that express CXCR5 provide accelerated help T B cells. Journal of Immunology. 2011;186:2889–2896. doi: 10.4049/jimmunol.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nature Immunology. 2003;4:1183–1190. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, Kaech SM. Differential expression of Ly6c and T-bet distinguish effector and memory Th1 CD4+ cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-speciifc MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. European Journal of Immunology. 1998;28:390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA, Orenstein WA, Offit PA. Vaccines. 6. Elsevier Inc; 2013. [Google Scholar]
- Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. Journal of Experimental Medicine. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, Leonard WJ. IL-21 and T follicular helper cells. International Immunology. 2010;22:7–12. doi: 10.1093/intimm/dxp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JP, Fuhrmann F, Hutloff A. T-follicular helper cells survive as long-term memory cells. European Journal of Immunology. 2012;42:1981–1988. doi: 10.1002/eji.201242540. [DOI] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T helper cell differentiation. Nature Reviews Immunology. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- Youngblood B, Davis CW, Ahmed R. Making memories that last a lifetime: heritable functions of self-renewing memory CD8 T cells. Internation Immunology. 2010;22:797–803. doi: 10.1093/intimm/dxq437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8+ T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) Journal of Immunology. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.