Helix 69 (H69) of 23S ribosomal RNA serves as a unique model system for studying the impact of modified bases on RNA structure and function, and for screening potential antibiotics. H69 is located at the functionally important core domain of the bacterial ribosome, participates in key intersubunit bridge B2a interactions, and plays important roles in translation.[1] This helix exists in multiple conformational states,[1] and interacts with a number of translation factors[2] at different stages of protein synthesis. Chemical probing analyses revealed that H69 undergoes structural rearrangements upon ribosome association, particularly at positions A1913 and A1918, with the various conformational states being influenced by solution conditions (e.g., [Mg2+], pH, and temperature) as well as pseudouridine (Ψ) modifications.[3] Residue A1913 is proposed to be important for high-fidelity translation[2c,4] and efficient termination.[2b] Moreover, flexibility of the H69 stem region may help accommodate the twisting energy from subunit rotation.[5] Since H69 is a highly dynamic RNA domain, altering or regulating these important conformational states with small molecules could be a promising way to disrupt bacterial ribosome translation. Development of a method for easily monitoring these changes is therefore important for understanding H69 dynamics in solution, as well as discovering H69-targeting ligands.
To establish a method that enables H69 conformational states to be monitored in solution and to detect preferential binding of small molecules to these states, a 19-nucleotide RNA containing an adenine analogue, 2-aminopurine (2AP), at position 1913 was designed (Figure 1a). The 2AP modification has been widely used to monitor ligand-induced RNA structural changes.[6] Model RNAs containing either Ψ or U at the naturally modified positions 1911, 1915, and 1917 (2AP-ΨΨΨ or 2AP-UUU, respectively) were generated. The 2AP analogue is sensitive to changes in environment and undergoes significant decreases in fluorescence intensity when base stacked.[7] Previous studies suggested that A1913 is stacked with a neighboring base in 50S subunits, but exposed and extended from the H69 loop in 70S ribosomes.[3b,8] Solution studies on related small RNAs representing H69 are consistent with this model.[9] Therefore, residue 1913 was an ideal candidate for substitution by 2AP and monitoring local conformational changes in the H69 loop. Fluorescence of 2AP was expected to diminish due to increased base stacking if nucleotide 1913 forms a local conformation similar to that observed in 50S subunits, but increase if 2AP1913 becomes more solvent exposed as seen in 70S ribosomes.
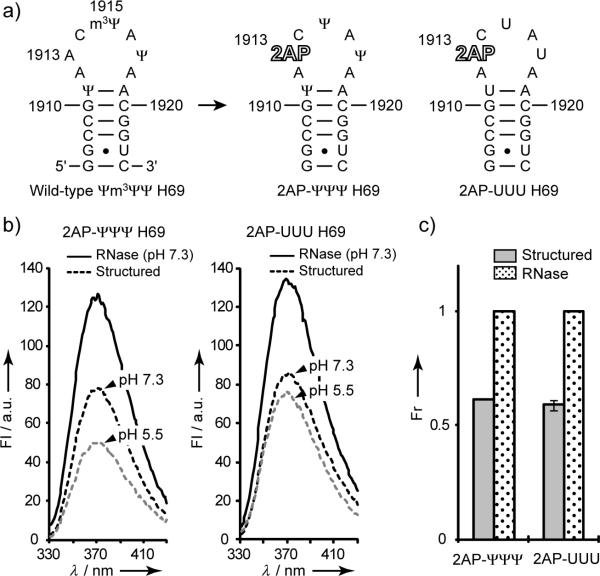
Figure 1.
(a) Secondary structure of E. coli H69 and 2AP-H69 RNAs. (b) Fluorescence data for 2AP-ΨΨΨ and 2AP-UUU before (dashed) (pH 5.5 and 7.3) and after (solid) (pH 7.3) RNase A treatment. (c) Relative fluorescence intensities (normalized to RNase A data) at the emission maximum (370 nm) (1.5 μm RNA in 20 mm K+-cacodylate, 70 mm NH4Cl, and 30 mm KCl; FI is fluorescence intensity (arbitrary units), and Fr is relative fluorescence intensity).
Comparison of fluorescence intensities between RNase-digested and renatured 2AP-H69 reveals a 40% decrease for both modified (2AP-ΨΨΨ) and unmodified (2AP-UUU) RNAs, indicating disruption of 2AP1913 base stacking upon RNA degradation (Figure 1b and c). Similar fluorescence intensities of the digested RNAs at pH 7.3 indicate that 2AP-ΨΨΨ and 2AP-UUU concentrations are similar (the extinction coefficient of unmodified H69 was used due to unavailability of nearest-neighbor extinction coefficients for 2AP and Ψ-modified RNAs[10]). Similar fluorescence intensities of the folded RNAs suggest that they exist in similar average conformational states at pH 7.3. In contrast, spectra at pH 5.5 reveal differences in the conformational states between modified and unmodified RNAs, which is consistent with thermodynamic differences of H69 at pH 5.5 and 7.0.[9a]
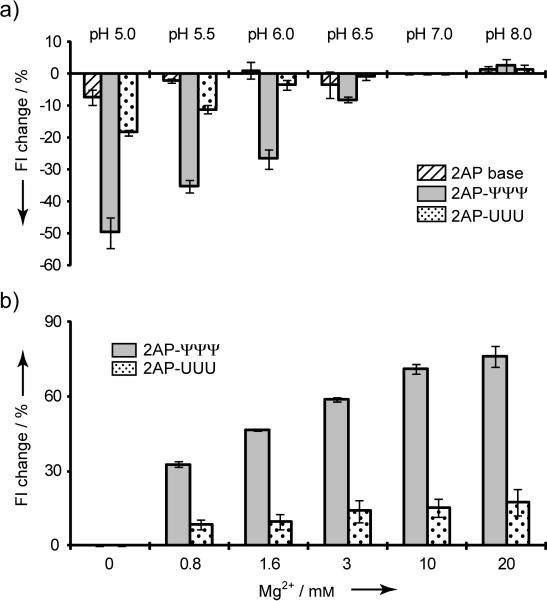
Previous model studies indicated that conformational transitions of the modified H69 loop can be induced by changes in solution pH, but the effect is dependent on the presence of Ψs.[9a] Fluorescence intensities of 2AP-ΨΨΨ and 2AP-UUU were determined from pH 5.0 to 8.0. As shown in Figure 2a, the fluorescence intensity of 2AP itself is not influenced by changes in pH.[7b] The 2AP-UUU RNA shows an 18% reduction in fluorescence intensity between pH 7.0 and 5.0, whereas, the 2AP-ΨΨΨ RNA exhibits a 50% reduced intensity (Figure 2a). This change is attributed to enhanced base-stacking interactions of 2AP1913 in the modified RNA construct (2AP-ΨΨΨ) as the pH is decreased. These results are consistent with conformational exchange of H69 between an open and closed loop conformation with respect to position 1913, as shown in previous model studies, and the equilibrium between the two conformational states can be shifted by changes in pH.[9a]
Figure 2.
(a) Percent change in fluorescence intensity (Fl) upon change in solution pH. Fluorescence intensities at 370 nm were normalized relative to the value at pH 7.0 (20 mm K+-cacodylate, 70 mm NH4Cl, 30 mm KCl, pH 5–8; measurements were done in triplicate). (b) Percent change in fluorescence intensity (Fl) upon addition of Mg2+. Fluorescence intensities at 370 nm were normalized relative to no Mg2+ (1.5 μm RNA, 20 mm HEPES, 70 mm NH4Cl, 30 mm KCl, pH 7.3, at 22 °C).
To test whether 2AP-H69 RNA can be used to monitor ligand-induced conformational changes, Mg2+ titrations were performed. Magnesium was shown previously to have different effects on H69 conformational states within the context of 70S ribosomes, depending on the presence of Ψ. Specifically, high [Mg2+] facilitated increased exposure of A1913 in modified RNA.[3b] For 2AP-ΨΨΨ, the fluorescence intensity increases 60% from 0 to 3 mm Mg2+ (pH 7.0). In contrast, the fluorescence intensity of 2AP-UUU increases by less than 20% upon addition of 3 mm Mg2+ (Figure 2b). These results are consistent with chemical probing studies on 70S ribosomes.[3b] Further increases in fluorescence are observed up to 20 mm Mg2+ for 2AP-ΨΨΨ, whereas, no additional changes are observed for 2AP-UUU (Figure 2b). Notably, the overall increase in 2AP-ΨΨΨ fluorescence at saturating amounts of Mg2+ is 75%, similar to the extent of increase observed with RNase digestion, suggesting high exposure of 2AP1913 to solvent or possibly direct interactions with Mg2+.
Our results reveal a different impact of Mg2+ on the loop conformation of unmodified RNA. Loop modifications (Ψ) in combination with metal ions appear to play a role in organizing H69 structure and/or regulating its flexibility. These results along with previous structure and stability studies suggest that at least three different structural ensembles exist for modified H69, which are influenced by both pH and Mg2+. The 2AP results show that the loop region is more sensitive to Mg2+ than pH. Specifically, addition of Mg2+ stabilizes the H69 loop structure with A1913 being more exposed, while also stabilizing the stem region.[11] A decrease in pH induces opposing changes in the stem (decreased order) and loop (increased stacking) of H69, as observed by NMR spectroscopy; whereas, a less compact loop and more ordered stem are observed at higher pH.[9a] The next goal was to determine whether other ligands could influence the equilibrium between H69 conformational states.
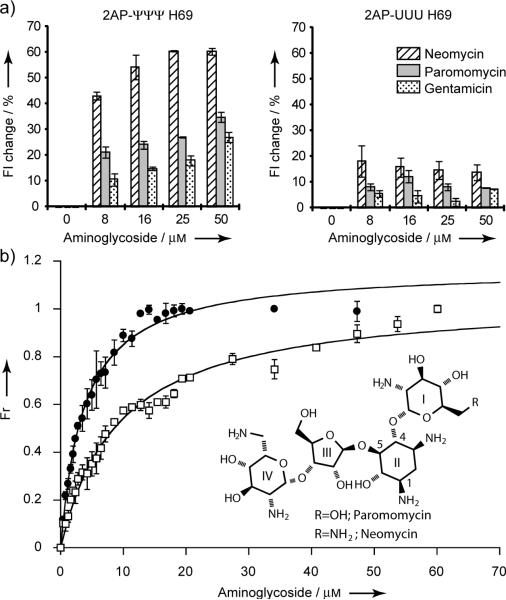
Previous crystallography and model studies showed that aminoglycoside antibiotics interact with the H69 stem region with moderate (μM) binding affinity.[12] It was suggested that H69-aminoglycoside interactions stabilize bridge B2a and promote H69 conformational arrangements that inhibit ribosome recycling factor (RRF)-mediated subunit dissociation.[12a] RRF has been proposed to induce conformational changes in H69, which lead to its movement away from the subunit surface.[12a] Thus, aminoglycosides likely influence the dynamic behavior of H69. To reveal the influence of aminoglycoside binding on H69 conformational states, neomycin, paromomycin, and gentamicin were titrated with 2AP-ΨΨΨ and 2AP-UUU in the absence of Mg2+, and 2AP fluorescence was monitored. The 2AP-ΨΨΨ fluorescence increases by 45% upon addition of neomycin (8 μm); whereas, the same concentration of paromomycin and gentamicin causes only 20 and 10% increases, respectively (Figure 3a). The 2AP-UUU RNA exhibits only a 10–20% increase of fluorescence upon addition of aminoglycosides (8 μm). A further increase in 2AP-ΨΨΨ fluorescence is observed up to 25 μm neomycin; however, no further changes are observed in the case of 2AP-UUU. For paromomycin and gentamicin, 2AP-ΨΨΨ fluorescence increases up to 50-60 μm drug and then reaches saturation (Figure 3a). These data reveal the ability of neomycin to induce or stabilize a conformational state of H69 in which residue 1913 is more exposed, similar to the effects of Mg2+, a behavior that is unique to the modified RNA.
Figure 3.
(a) Percent changes in fluorescence intensities of 2AP-H69 in the presence of various drug concentrations (20 mm HEPES, 70 mm NH4Cl, 30 mm KCl, pH 7.3 at 22 °C). Fluorescence intensities at 370 nm were normalized to spectra in the absence of drug. (b) Binding curves of neomycin and paromomycin to 2AP-ΨΨΨ were fit using non-linear curve fitting to a quadratic equation.[18] The apparent Kd was measured from three independent experiments.
Apparent dissociation constants (Kds) of neomycin and paromomycin were determined from the 2AP fluorescence data. Figure 3b and Table 1 show Kd values of 2.5 and 2.0 μm for neomycin binding to 2AP-ΨΨΨ and 2AP-UUU, respectively. For paromomycin, the apparent Kd is 7.1 μm with 2AP-ΨΨΨ; however, a value with 2AP-UUU could not be obtained because of the small fluorescence changes (<10%). These Kd values are consistent with those obtained with ΨΨΨ, Ψm3ΨΨ (the wild-type E. coli H69, with Ψ at positions 1911 and 1917, and 3-methylpseudouridine, m3Ψ, at 1915), and UUU constructs without 2AP using electrospray ionization mass spectrometry (ESI-MS) (Table 1), indicating the usefulness of the 2AP-H69 system for binding studies. More importantly, however, is the ability to monitor conformational states of H69 RNA by using 2AP1913 fluorescence, which is not revealed by ESI-MS. The RNA structural changes induced by aminoglycosides are most apparent with the modified RNA.
Table 1.
Comparison of dissociation constants (Kds) for H69
| Ligand | Method[a] | RNA type | Kd [μM] |
|---|---|---|---|
| neomycin | Fluorescence | 2AP-ψψψ | 2.5 ± 0.2 |
| ESI-MS | 2AP-ψψψ | 1.6 ± 0.7 | |
| ESI-MS | ψ ψ ψ | 1.2 ± 0.4 | |
| ESI-MS | ψm3ψψ | 3.0 ± 0.6 | |
| Fluorescence | 2AP-UUU | 2.0 ± 0.4 | |
| ESI-MS | 2AP-UUU | 1.8 ± 0.3 | |
| ESI-MS | UUU | 1.4 ± 0.6 | |
| paromomycin | Fluorescence | 2AP-ψψψ | 7 ± 1 |
| ESI-MS | 2AP-ψψψ | 11 ± 3 | |
| ESI-MS | ψ ψ ψ | 10 ± 3 | |
| ESI-MS | ψm3ψψ | 11 ± 3 | |
| Fluorescence | 2AP-UUU | ND[b] | |
| ESI-MS | 2AP-UUU | 9 ± 3 | |
| ESI-MS | UUU | 8 ± 2 | |
Experimental conditions: Fluorescence; 20 mm HEPES, 70 mm NH4CI, 30 mm KCl, pH 7.3, ESI-MS; 150 mm NH4OAc, pH 7.0.
ND; not determined due to too small FI changes.
Binding studies of aminoglycosides with 2AP-H69 suggest that structurally similar aminoglycosides, particularly neomycin and paromomycin, have different effects on the H69 loop conformation, even though their binding modes are similar in crystal structures.[12a] ESI-MS binding analysis of the neomycin and paromomycin complexes with H69 reveals 1:1 binding stoichiometries (Supporting Information), although higher stoichiometries are observed at higher concentrations due to non-specific binding. Previous studies showed binding of aminoglycosides in the H69 stem region (G1906/G1921-U1923).[12] If the same binding mode occurs in the 2AP model RNAs, then the fluorescence results are consistent with neomycin binding to the H69 stem region and stabilizing a conformational state of the H69 loop with residue 1913 exposed to solvent. In order for this to occur, a conformational change in the stem must be propagated to the H69 loop. This effect is also influenced by Ψ modifications, as revealed by differences in 2AP1913 fluorescence enhancement. Crosstalk between the H69 stem and loop was observed in previous circular dichroism studies, in which H69 RNAs with different stem sequences had varying loop conformations.[13] Thus, an altered H69 stem conformation induced or stabilized by neomycin could in turn alter the H69 loop state in a similar manner. Considering the biological roles of H69, an altered loop structure could promote changes in the ribosome functional state. This idea is consistent with proposed roles of another ribosomal motif, helix 44 in 16S rRNA, based on crystal structures and fluorescence studies with 2AP, in which conformational changes that are stabilized by aminoglycoside binding cause defects in translation.[2c,6c] Indeed, a recent study indicates binding of neomycin to H69 in 70S ribosomes and subsequent allosteric perturbations of global ribosome dynamics.[14]
Feldman and coworkers observed that under low neomycin concentrations (<100 nm) the ribosome has a classical configuration, whereas, higher concentrations induce a hybrid configuration.[15] Paromomycin and gentamicin do not show such effects. Toe-printing analysis also revealed that neomycin induces a stronger back-translocation of ribosomes than paromomycin.[16] Moreover, neomycin was found to have a stronger inhibition effect of EF-G dissociation from ribosomes compared to paromomycin.[17] These differences were suggested to be due to a secondary binding site, and H69 is a likely candidate. The different binding activities and varying effects on conformational states of H69 could be the reason for different influences of aminoglycosides on ribosome function.
In conclusion, structural information from X-ray crystallography, chemical probing, and synthetic RNA model studies was used to design a small fluorescent H69 that allows monitoring of loop conformational states in solution. The fluorescence method is convenient and rapid, and only requires pmol amounts of RNA. The 2AP fluorescence method allows binding information to be obtained for a variety of ligands, such as metal ions and small molecules. In this study, H69 loop conformational states were shown to vary with factors such as [Mg2+], pH, and Ψ modifications, consistent with previous observations on 70S ribosomes.[3,9a] Structurally similar aminoglycoside antibiotics have different impacts on the conformational states of the H69 loop, which is also influenced by modified nucleotides. The 2AP-H69 results also suggest communication between the H69 stem and loop. These observations demonstrate the utility of the 2AP system, which will allow specific types of H69-binding ligands to be identified, namely, those that alter the H69 loop conformation and possibly its function, as observed with neomycin. Combined with ESI-MS assays to obtain binding affinities and stoichiometries, the 2AP method provides an opportunity to screen large libraries of small molecules with the potential for binding to H69 with high affinity as well as the ability to stabilize certain H69 conformational states. This information will be helpful for identification of novel H69-targeting ligands that have a higher potential to inhibit different phases of ribosome translation.
Supplementary Material
Acknowledgments
We thank D. Rueda for providing access to the fluorometer. This project is supported by the NIH (GM087596).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 2.a Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]; c Selmer M, Dunham CM, Murphy F. t., Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]; d Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 3.a Sakakibara Y, Chow CS. J. Am. Chem. Soc. 2011;133:8396–8399. doi: 10.1021/ja2005658. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Sakakibara Y, Chow CS. ACS Chem. Biol. 2012;7:871–878. doi: 10.1021/cb200497q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Jenner L, Demeshkina N, Yusupova G, Yusupov M. Nat. Struct. Mol. Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]; b Ortiz-Meoz RF, Green R. J. Biol. Chem. 2011;286:25604–25610. doi: 10.1074/jbc.M111.256255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Souliere MF, Haller A, Rieder R, Micura R. J. Am. Chem. Soc. 2011;133:16161–16167. doi: 10.1021/ja2063583. [DOI] [PubMed] [Google Scholar]; b Tam VK, Kwong D, Tor Y. J. Am. Chem. Soc. 2007;129:3257–3266. doi: 10.1021/ja0675797. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kaul M, Barbieri CM, Pilch DS. J. Am. Chem. Soc. 2006;128:1261–1271. doi: 10.1021/ja056159z. [DOI] [PubMed] [Google Scholar]; d Bradrick TD, Marino JP. RNA. 2004;10:1459–1468. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Lacourciere KA, Stivers JT, Marino JP. Biochemistry. 2000;39:5630–5641. doi: 10.1021/bi992932p. [DOI] [PubMed] [Google Scholar]
- 7.a Guest CR, Hochstrasser RA, Sowers LC, Millar DP. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]; b Ward DC, Reich E, Stryer L. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 8.a Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]; b Zhang W, Dunkle JA, Cate JH. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Abeysirigunawardena SC, Chow CS. RNA. 2008;14:782–792. doi: 10.1261/rna.779908. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Desaulniers JP, Chang YC, Aduri R, Abeysirigunawardena SC, SantaLucia J, Jr., Chow CS. Org. Biomol. Chem. 2008;6:3892–3895. doi: 10.1039/b812731j. [DOI] [PubMed] [Google Scholar]
- 10.Richards EG. Handbook of Biochemistry and Molecular biology. CRC Press; Cleveland, OH: 1975. pp. 596–599. [Google Scholar]
- 11.Sumita M, Desaulniers JP, Chang YC, Chui HM, Clos L, 2nd, Chow CS. RNA. 2005;11:1420–1429. doi: 10.1261/rna.2320605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]; b Scheunemann AE, Graham WD, Vendeix FA, Agris PF. Nucleic Acids Res. 2010;38:3094–3105. doi: 10.1093/nar/gkp1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumita M, Jiang J, SantaLucia J, Jr., Chow CS. Biopolymers. 2012;97:94–106. doi: 10.1002/bip.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Doudna Cate JH, Blanchard SC. Nat. Struct. Mol. Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman MB, Terry DS, Altman RB, Blanchard SC. Nat. Chem. Biol. 2010;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoji S, Walker SE, Fredrick K. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campuzano S, Vazquez D, Modolell J. Biochemistry. 1979;18:1570–1574. doi: 10.1021/bi00575a029. [DOI] [PubMed] [Google Scholar]
- 18.Llano-Sotelo B, Azucena EF, Jr., Kotra LP, Mobashery S, Chow CS. Chem. Biol. 2002;9:455–463. doi: 10.1016/s1074-5521(02)00125-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.