Abstract
Phosphoinositides are involved in regulation of recruitment and activity of signalling proteins in cell membranes. Phosphatidylinositol (PI) 4-kinases (PI4Ks) generate PI4-phosphate the precursor of regulatory phosphoinositides. No type II PI4K research on the abiotic stress response has previously been reported in plants. A stress-inducible type II PI4K gene, named TaPI4KIIγ, was obtained by de novo transcriptome sequencing of drought-treated wheat (Triticum aestivum). TaPI4KIIγ, localized on the plasma membrane, underwent threonine autophosphorylation, but had no detectable lipid kinase activity. Interaction of TaPI4KIIγ with wheat ubiquitin fusion degradation protein (TaUDF1) indicated that it might be hydrolysed by the proteinase system. Overexpression of TaPI4KIIγ revealed that it could enhance drought and salt stress tolerance during seed germination and seedling growth. A ubdkγ7 mutant, identified as an orthologue of TaPI4KIIγ in Arabidopsis, was sensitive to salt, polyethylene glycol (PEG), and abscisic acid (ABA), and overexpression of TaPI4KIIγ in the ubdkγ7 mutant compensated stress sensitivity. TaPI4KIIγ promoted root growth in Arabidopsis, suggesting that TaPI4KIIγ might enhance stress resistance by improving root growth. Overexpression of TaPI4KIIγ led to an altered expression level of stress-related genes and changes in several physiological traits that made the plants more tolerant to stress. The results provided evidence that overexpression of TaPI4KIIγ could improve drought and salt tolerance.
Key words: Kinase activity, overexpression, PI4K, protein interaction, stress response, Triticum aestivum.
Introduction
Environmental stresses, such as drought and salt, limit crop productivity worldwide. Understanding plant responses to these stresses is essential for rational engineering of hardier crop plants. Recent studies revealed the important role of the phosphoinositide signalling pathway at multiple developmental stages in plants (Munnik and Vermeer, 2009 Ischebeck et al., 2010). Phosphatidylinositol 4-kinases (PI4Ks) at the apex of the phosphoinositide cascade are involved in many functions, including lipid- and protein-mediated signalling transductions (Carpenter and Cantley, 1996; Balla, 2001; Abraham, 2004; Wullschleger et al., 2006; Szumlanski and Nielsen, 2010). The process by which PI4Ks phosphorylate phosphatidylinositol (PI) in yeast and mammals has been thoroughly investigated: the enzymes catalyse phosphorylation of PI to PI4-phosphate, which is the most abundant of the monophosphorylated inositol phospholipids in mammalian cells (Strahl and Thorner, 2007; D’Angelo et al., 2008) and therefore represents a potentially crucial point in the regulation of phosphoinositide-dependent pathways.
Phylogenetic analysis by amino acid sequence alignment using the PI4K domain from mammals, yeast, and plant proteins revealed that PI4K-domain-containing proteins fall into major types II and III according to size and sensitivity to inhibitors. Those originally identified as type I PI4Ks were later shown to be PI3Ks. Type II PI4K is a membrane-bound, 55kDa enzyme that is readily renaturable after SDS–PAGE. Two forms of type III PI4K were detected in the membrane and soluble fractions of animal tissues. One is 110kDa in size, and the other is 200–230kDa. Type II PI4K is insensitive to wortmannin (at concentrations significantly higher than those inhibiting PI3K), whereas type III enzyme activity is inhibited (Balla, 1998).
Up to now, most of the information on PI4Ks was derived from mammals and yeast (Barylko et al., 2001; Minogue et al., 2001). The yeast genome contains three genes that encode PI4Ks (Pik1, Stt4, and Lsb6, that are functionally non-overlapping) (Han et al., 2002; Strahl and Thorner, 2007). Mammalian homologues of Pik1 and Stt4 are called PI4KIIIββ and PI4KIIIα, respectively (Wong et al., 1997; Zhao et al., 2001; Strahl and Thorner, 2007). PI4Ks are not only the enzymes catalysing PI4-phosphate, but also carry out other functions, including membrane trafficking (Strahl et al., 2005), cytoskeletal regulation (Audhya and Emr, 2002), and regulation of cell apoptosis (Chu et al., 2012).
Although PI4K activity was found in plants many years ago, the isolation and functional analysis of a full-length plant PI4K cDNA was first reported in 1999 (Xue et al., 1999). AtPI4Kβ was the first PI4K cloned from a plant, and its transcript levels were similar in all tissues. Treatment of seedlings with hormones, CaCl2, or NaCl had no effect on AtPI4Kβ mRNA levels (Xue et al., 1999). Currently, eight Arabidopsis putative type II PI4Ks, four Arabidopsis putative type III PI4Ks, and two cotton putative type II PI4Ks are reported (Mueller-Roeber and Pical, 2002; Galvão et al., 2008; Liu et al., 2012). Arabidopsis type III PI4Kβ1 and β2, upstream components of the phospholipase C pathway, were induced by cold stress. Cold-responsive genes were impaired in a PI4KIIIβ1β2 double mutant in Arabidopsis, whose seed germination was hypersensitive to low temperature (Delage et al., 2012). Cotton type II PI4K genes GbPI4K and GhPI4K were developmentally regulated in fibres and shared a similar expression pattern of ‘from high to low’ according to the fibre development stage (Liu et al., 2012). Salicylic acid (SA) addition induced a rapid decrease in the phosphoinositide pool. This decrease paralleled an increase in PI4-phosphate and PI4,5-bisphosphate. These changes were inhibited by two different inhibitors of type III PI4Ks, phenylarsine oxide and wortmannin (Krinke et al., 2007). Although there has been some progress in identification and functional analyses of plant PI4Ks, limited information is available compared with that in mammals and yeast. Except for PI4Kβ1 and β2, there is no report on the PI4K family that directly relates to abiotic stress responses.
Wheat (Triticum aestivum) is one of the most important food staples worldwide. Drought and salt affect plant growth and productivity, and reduce wheat yields worldwide (Rhoades and Loveday, 1990; Morran et al., 2011). In this study, a putative type II PI4K gene named TaPI4KIIγ was found to be up-regulated using de novo transcriptome sequencing of drought-treated wheat. TaPI4KIIγ, localized on the plasma membrane, was identified to have threonine autophosphorylation kinase activity, but not lipid kinase activity. TaPI4KIγ conferred salt and drought tolerance to Arabidopsis.
Materials and methods
Plant materials
The cultivated wheat cultivar KeNong 199 was used to amplify cDNA sequences of TaPI4KIIγ and TaUFD1. Arabidopsis Columbia-0 (WT) was used as background for overexpressing TaPI4KIIγ. By amino acid sequence alignment with Arabidopsis genes, the gene At1g13640 was found to be the closest relative to TaPI4KIIγ (identity=350/611 (57%)]. In this study, the At1G13640 (UBDKγ7) mutant (SALK_107574C) was chosen for the experiments. The ubdkγ7 mutant is a T-DNA insertion mutant (knock out), and the insertion site is in the promoter (at about –500bp). UBDKγ7 is a constitutively expressed gene (Supplementary Fig. S1 available at JXB online).
Stress treatment of wheat
Wheat seedlings were grown in Hoagland’s liquid culture at 22 °C under a 16h light/8h dark photoperiod. Ten-day-old wheat seedlings were used for dehydration, salt, and abscisic acid (ABA) treatments. For dehydration treatment, seedlings were transferred onto filter paper, and dried at 25 °C under 60% humidity conditions. For salt and ABA treatments, seedling roots were immersed in solutions containing 200mM NaCl and 1 μM ABA, respectively. The samples were harvested at 0, 0.5, 1, 3, 6, 12, 24, and 48h. RNA extraction and quantitative reverse transcription–PCR (qRT–PCR) analyses were performed. The specific primers for TaPI4KIIγ expression profiles and the primers of the wheat β-actin reference gene are listed in Supplementary Table S1 at JXB online.
Cloning and sequence analysis of the TaPI4KIIγ and TaUFD1 genes
The full-length opening reading frames of TaPI4KIIγ and TaUFD1 were obtained from wheat cDNA. The primers for cloning TaPI4KIIγ were 5′-GAGACCTCGGCGGAGAATCAAC-3′ and 5′-TCTCTGCTGCTCTATGGACCTAGTCA-3′. The primers for TaUFD1 were 5′-GTTTAACCTTCTTGTCCAT-3′ and 5′-GATAAATACATACTCTGGCAT-3′. The PCR products were cloned into pEASY-T1 vectors (TransGen) and sequenced with an ABI 3730XL 96-capillary DNA analyzer (Lifetech).
The amino acid sequences of TaPI4KIIγ homologous proteins were obtained from the NCBI using Blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequence similarity comparisons were performed using the MegAlign program in DNAStar software. The complete amino acid sequences of PI4K proteins were used to construct phylogenetic trees. Sequence alignment was performed by ClustalX using BioEdit software and adjusted manually. The Neighbor–Joining method was used to construct a phylogenetic tree by the MEGA5.1 program, and the confidence level of monophyletic groups was estimated using a bootstrap analysis of 10 000 replicates (Tamura et al., 2011).
Plasmid construction for localization analysis
TaPI4KIIγ was inserted into the subcellular localization vector p16318, which contains the 35S promoter and C-terminal green fluorescent protein (GFP) (Xu et al., 2007). The primers for TaPI4KIIγ-p16318 were 5′-TTTAAGCTTATGTCCCCCAACCTGGAG-3′ and 5′-AAACCATGGTGAAAATTTGCAGGAGGTGC-3′. For transient expression assays, ~4×104 mesophyll protoplasts were isolated from 14-day-old seedlings, and transfected with 10 μg of p16318::TaPI4KIIγ. Transfected protoplasts were incubated in darkness at 22 °C. GFP fluorescence signals were observed with a confocal laser scanning microscope (Nikon). FM4-64 dye (Molecular Probes, Carlsbad, CA, USA) was excited at 543nm, and its fluorescence was recorded using a 650nm long pass filter. All transient expression experiments were repeated three times.
Protein purification and pull-down assays
TaPI4KIIγ was inserted into the prokaryotic expression vector pCOLD (TaKaRa), and TaUFD1 was inserted into pGEX-4T-1. The primers for TaPI4KIIγ-pCOLD were 5′-TTTGGTACCATGTCCCCCAACCTGGAG-3′ and 5′-AAATC TAGAAAATTTGCAGGAGGTGCCCAG-3′. The primers for TaUFD1-pGEX were 5′-TTTGGATCCATGAATATGTATTTCG AAGGCT-3′ and 5′-TTTCTCGAGTAAATACATACTCTGGCA TTCAG-3′. TF-His-TaPI4KIIγ (TF refers to the trigger factor that improves the solubility of the protein) and GST–TaUFD1 were expressed in Escherichia coli and purified by the standard procedure using Ni and glutathione agarose beads, respectively (GE Healthcare). In brief, 100ml of BL21 cells grown overnight and expressing the desired constructs were transferred into 500ml of LB and grown at 37 °C for 3h. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1mM) was then added to the LB and left overnight at 16 °C to induce protein expression. The bacterial cells were sonicated in phosphate-buffered saline (PBS) with 1% Triton X-100 and centrifuged at 10 000 g for 10min to remove insoluble cell debris. The supernatant was incubated with PBS pre-equilibrated with Ni or glutathione agarose beads and rotated at 4 °C for 2h. After washing with PBS five times, the glutathione S-transferse (GST)-tagged and His-tagged proteins were eluted using 5mM glutathione and 125mM imidazole, respectively (Tang et al., 2008).
For GST pull-down assays, TF-His-TaPI4KIIγ was incubated with GST–TaUFD1 in Z-binding buffer [25mM HEPES (pH 7.6), 12.5mM MgCl2, 150mM KCl, 0.1% NP-40, and 20% glycerol] buffer for 2h at 4 °C. After washes in NETN buffer [100mM NaCl, 1mM EDTA, 0.5% NP-40, and 20mM TRIS (pH 8.0)], proteins were analysed on SDS–polyacrylamide gels followed by immunoblotting using anti-His antibody.
Bimolecular fluorescence complementation (BiFC) assay
TaPI4KIIγ was cloned into pSPYNE, and TaUDF1 was cloned into pSPYCE (Kerppola, 2008). The primers for TaPI4KIIγ-pSPYNE were 5′-AAATCTAGAATGTCCCCCAACCTGGAG-3′ and 5′-TT TGGTACCAAATTTGCAGGAGGTGCCCAG-3′. The primers for TaUFD1-pSPYCE were 5′-TTTTCTAGAATGAATATGTATTTCG AAGGCTAT-3′ and 5′-TTCGGTACCGCCCTTCAAAGAGTAC TTCT-3′. For the BiFC assay, the pSPYNE::TaPI4KIIγ and pSPYCE::TaUFD1 reconstruction vectors were bombarded into onion epidermal cells by a particle gun. Plasmolysis was carried out with a filter-sterilized solution prepared by dissolving the sucrose in culture medium (50% w/v). The onion epidermal cells were treated with 50% sucrose solution for 10min, and then monitored by confocal microscopy as previously described (Xu et al., 2007).
In vitro autophosphorylation assays
TF-His-TaPI4KIIγ was expressed in BL21 cells and purified using a Zi affinity gel. After elution from the beads, the protein solution was dialysed against a 1000× volume buffer containing 20mM MOPS, pH 7.5, and 1mM dithiothreitol (DTT). Autophosphorylation assays were performed by immunoblotting after transfer to polyvinylidene fluoride (PVDF) membranes using antiphosphoserine (1:500 dilution), antiphosphotyrosine (1:500), and antiphosphothreonine antibodies (1:500) (Oh et al., 2009).
Generation of transgenic Arabidopsis
The coding sequence of TaPI4KIIγ-pBI121 was amplified using primers 5′-AAATCTAGAATGTCCCCCAACCTGGAG-3′ and 5′-TCC CGGGCTCTGCTGCTCTATGGACCTAGTC-3′, and cloned into pBI121 under control of the Cauliflower mosaic virus (CaMV) 35S promoter, resulting in a 35S::TaPI4KIIγ construct. The construct was confirmed by sequencing and then transformed into wild-type (WT) plants and the ubdkγ7 mutant by the vacuum infiltration method (Bechtold and Pelletier, 1998). 35S::TaPI4KIIγ transgenic (WT background) and TaPI4KIIγ::ubdkγ7 lines (ubdkγ7 mutant background), with various expression levels of the TaPI4KIIγ gene, were obtained for further analysis.
Performance of transgenic Arabidopsis under stress treatment
Homozygous T3 seeds of the transgenic lines were used for phenotypic analysis. Seeds were surface sterilized, kept at 4 °C for 3 d, and then sown on germination medium [1/4 strength Murashige and Skoog medium with salt and vitamins (PhytoTechnology Laboratories) and further supplemented with 0.8% (w/v) agar, 30mg l–1 hygromycin B (Sigma-Aldrich), and 2% (w/v) sucrose, pH 5.7] under long-day conditions (15h light/9h dark) at 22 °C. For the germination assay, seeds were subjected to 100mM or 200mM NaCl, 5% or 10 % (w/v) polyethylene glycol (PEG) 3600 (to simulate osmotic stress), and 0.5 μM or 1 μM ABA treatments. The percentage of germinated seeds was calculated based on the number of seedlings that reached the cotyledon stage at 2 weeks (Saleki et al., 1993). For seedling phenotype analysis of the WT and the 35S::TaPI4KIIγ lines, all plants were grown in plates or soil under long-day conditions (15h light/9h dark) at 22 °C with 70% relative humidity. Seeds of WT and transgenic plants were plated on half-strength MS (Murashige and Skoog) agar, kept at 4 °C for 3 d, and then incubated in a growth chamber under continuous light at 23 °C. Six-day-old seedlings were transplanted in soil containing vermiculite. For salt treatment, 15-day-old seedlings were treated with 200mM NaCl for 20 d. For water deficit treatment, watering was withheld from 15-day-old seedlings for 20 d, by which time they were fully affected by water deficit stress.
Expression analysis of TaPI4KIIγ-regulated genes
Total RNAs from 35S::TaPI4KIIγ, ubdkγ7 mutant, and the WT were extracted using an RNAprep plant kit (TIANGEN), and first-strand cDNAs were synthesized using a PrimeScript First-Strand cDNA Synthesis kit (TaKaRa); cDNAs were combined with SYBR master mix for PCR (TIANGEN). qRT–PCRs were performed in triplicate with a Bio-Rad iCycler. The Arabidopsis β-actin gene was used as a reference. The qRT–PCRs were performed using an ABI Prism 7300 real-time PCR system (Lifetech) and repeated three times. Quantitative and data analyes were performed as previously described (Le et al., 2011). The specific primers are listed in Supplementary Table S1 at JXB online.
Measurement of malondialdehyde (MDA) and free proline contents
Seven-day-old seedlings were planted in 4cm deep rectangular plates filled with a 1:1 mixture of vermiculite and humus. After a further 7 d of growth, they were treated with 200mM NaCl and 10% (w/v) PEG, and then grown for 7 d before harvesting and measuring. All the measurements were repeated three times.
MDA contents were assayed according to Lv et al. (2007). About 0.1g of wheat leaf was used and absorbance values at 450, 532, and 600nm were determined with a spectrometer (Perkin-Elmer Lambda 25, Boston, MA, USA). The MDA content was calculated using the formula: C (1mol/l)= 6.45 (OD532–OD600)–0.56 OD450.
Wheat leaf samples (~0.1g) for the measurement of proline were treated with 3% (w/v) sulphosalicylic acid followed by boiling for 1h. The amounts of proline were measured with ninhydrin, which was detected at 520nm, and a proline standard liquid was used as a reference (Bates et al., 1973).
Results
Phylogenetic analysis and domain organization of putative wheat PI4K
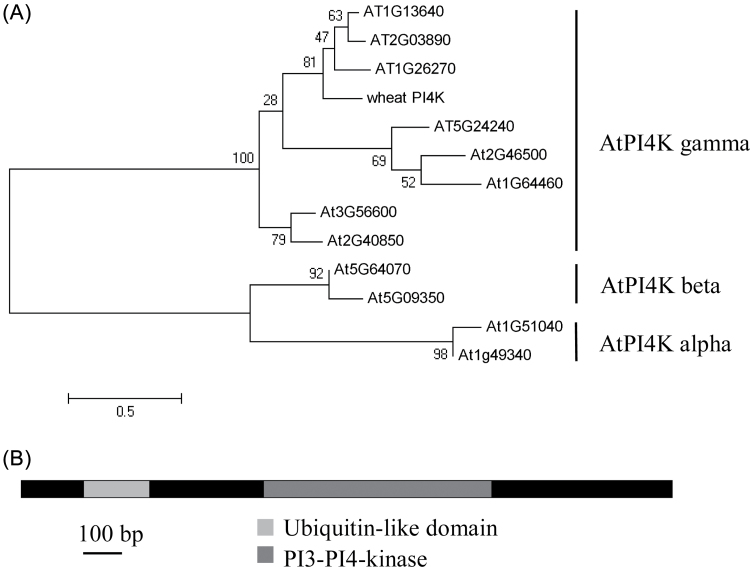
An up-regulated putative PI3/4K protein selected from the drought-treated wheat de novo transcriptome contained a predicted PI3/4K domain coupled with an N-terminal ubiquitin-like (UBL) domain (amino acids 53–120). The gene was cloned from wheat cDNA, and the full length was 1890bp encoding a 69.3kDa protein (Fig 1A). In multiple amino acid sequence alignments with 29 PI3/4K genes from human, mouse, yeast, and Arabidopsis, the wheat PI3/4K was classified as a member of the PI4K family (Supplementary Fig. S2 at JXB online). Currently, two major groups (types II and III) identified in the PI4K family occur in a wide range of tissues and cellular compartments in animals, yeast, and plants (Endemann et al., 1987; Pike, 1992; Galvão et al., 2008). Twelve Arabidopsis PI4K genes, comprising eight type II (gamma) and four type III PI4Ks (alpha and beta), together with the wheat PI4K, were aligned based on multiple sequence alignments (Fig. 1B). The wheat PI4K fell into the type II PI4K gamma cluster and was named TaPI4KIIγ.
Fig. 1.
Phylogenetic analysis and domain organization of TaPI4KIIγ. (A) Twelve Arabidopsis PI4K genes were divided into three subfamilies, type III PI4K alpha and beta and type II gamma. The wheat PI4K gene was classified as a type II PI4K gamma family member. The closest members to wheat PI4K were AT1G26270, AT2G03890, and AT1G13640, and were phosphoinositide 4-kinase gamma 7 types. (B) Domain organization of TaPI4KIIγ. Amino acid sequence alignment comparing Arabidopsis ubiquitin with UBL domains from TaPI4KIIγ.
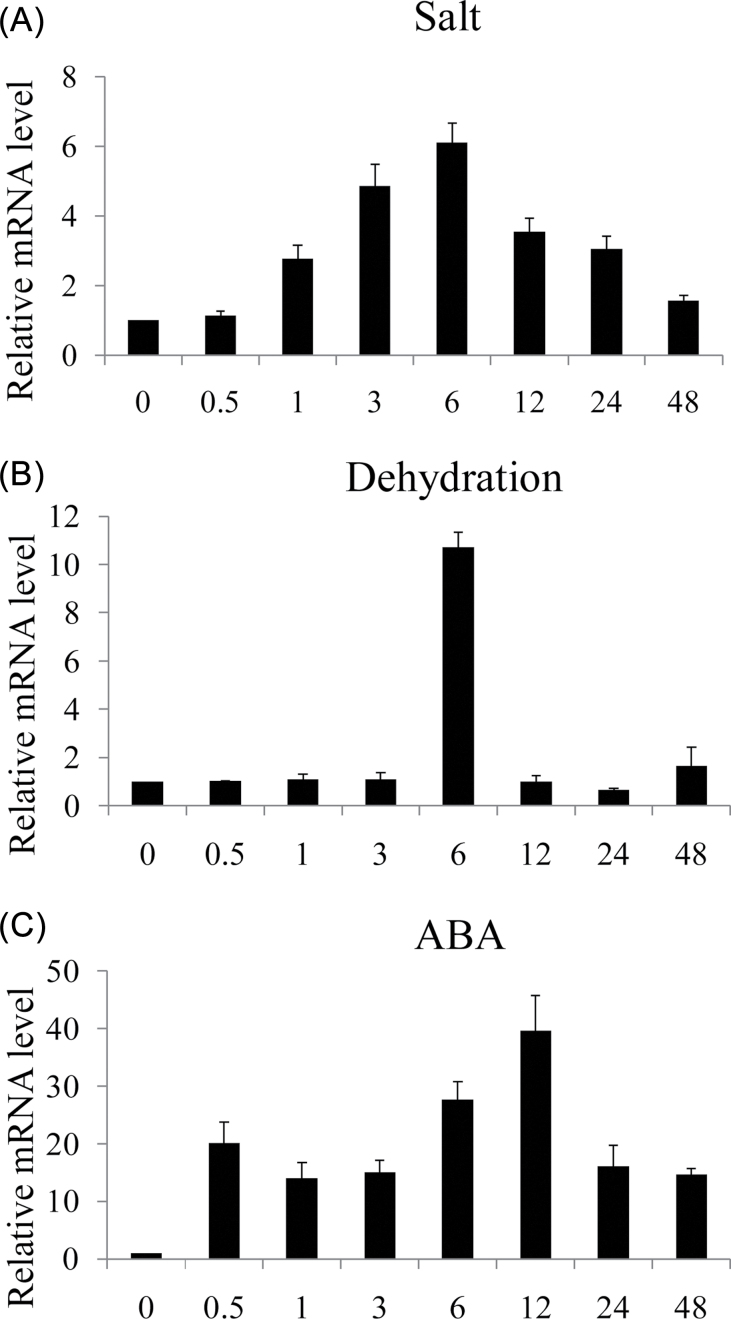
TaPI4KIIγ was induced by dehydration, salt, and ABA
The expression pattern of TaPI4KIIγ was monitored by qRT–PCR. The expression of TaPI4KIIγ increased after 1h treatment with salt and reached a peak (almost 6-fold) at 6h (Fig. 2A). TaPI4KIIγ was also induced by dehydration and ABA treatments. ABA initially induced the expression at an early phase (30min after treatment), and, after a slight decline, this increased to a peak of ~40-fold at 12h before declining again (Fig 2B). Under dehydration treatment, TaPI4KIIγ was not induced until 6h, and the peak point (~11-fold) was at 6h; followed by an abrupt decline to the normal level (Fig. 2C).
Fig. 2.
TaPI4KIIγ is a stress-responsive gene. Expression profiles of TaPI4KIIγ after salt (A), dehydration (B), and ABA (C) treatments for 0, 0.5, 1, 3, 6, 12, 24, and 48h. Ten wheat seedlings were pooled as one sample. The vertical coordinates are fold changes, and the horizontal ordinates are treatment time. The data are representative of three independent experiments.
Threonine autophosphorylation activity of TaPI4KIIγ
The coding region of TaPI4KIIγ was cloned into expression vectors, and the recombinant protein was successfully produced in E. coli as an N-terminal TF-His fusion. Autophosphorylation assays carried out using anti-phosphoamino acid antibodies revealed that TaPI4KIIγ autophosphorylated threonine residues (Fig. 3). The TF-His fusion showed no detectable cross-reaction with anti-phosphoamino acid antibodies. The failure to detect autophosphorylation of TF-His was used to eliminate non-specificity of the antibodies, and served as a crucial control to validate detection of phosphoamino acid. However, TF-His-TaPI4KIIγ showed no in vitro PIK activity in the presence of PI or Type I Folch lipid fraction and [γ-32P]ATP (results not shown).
Fig. 3.
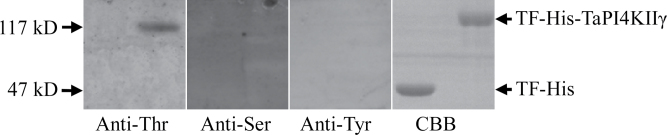
Kinase activity of TaPI4KIIγ. Autophosphorylation activity of TaPI4KIIγ detected by immunoblotting with antiphosphotyrosine (Anti-Tyr), antiphosphothreonine (Anti-Thr), and antiphosphoserine (Anti-Ser) antibodies. The molecular weights of fusion proteins TF-His-TaPI4KIIγ and TF-His are marked at the left. CBB, Coomassie brilliant blue.
Subcellular localization of TaPI4KIIγ
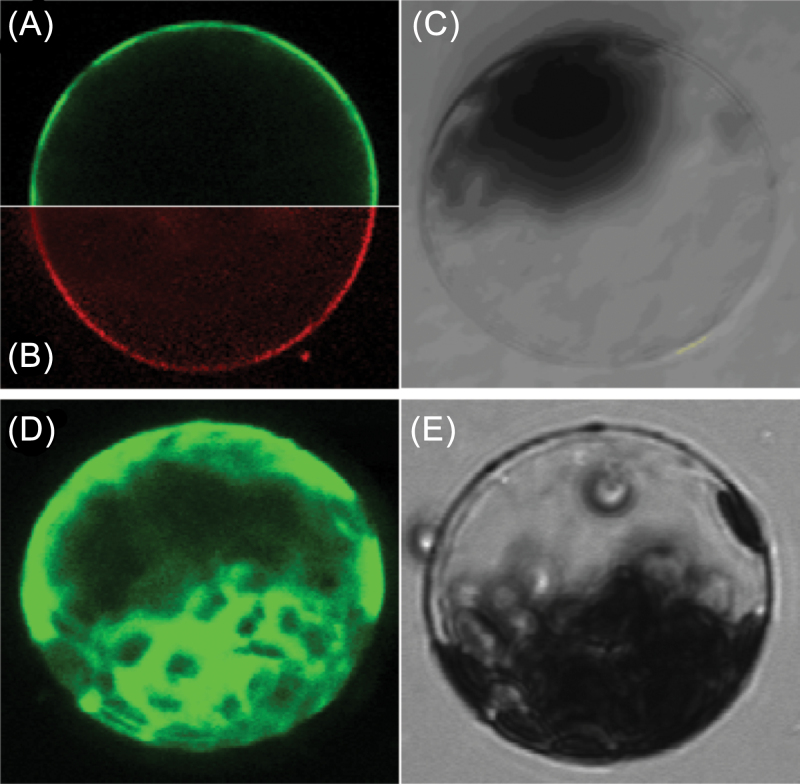
To investigate the localization of TaPI4KIIγ in cells, the gene was inserted into a subcellular localization vector. The recombinant vector p16318::TaPI4KIIγ was transformed into Arabidopsis protoplasts and, when observed by confocal microscopy, GFP fluorescence of the TaPI4KIIγ–GFP fusion protein in transformed cells occurred exclusively on the plasma membrane, whereas the GFP fluorescence of GFP-transformed cells was distributed throughout the entire cell. These data indicated that the TaPI4KIIγ protein is a plasma membrane-localized protein (Fig. 4).
Fig. 4.
Subcellular localization of TaPI4KIIγ in Arabidopsis protoplasts. The TaPI4KIIγ–GFP fusion proteins (A) or GFP alone (D) driven by the 35S promoter were transiently expressed in Arabidopsis protoplast cells and observed under a laser scanning confocal microscope. In (B), the protoplast plasma membrane was labelled with the FM4-64 steryl dye. Bright field images are shown in (C) and (E). The scale bar is 15.38 μm. (This figure is available in colour at JXB online.)
TaPI4KIIγ interacted with a component of the ubiquitin–proteasome TaUFD1 system
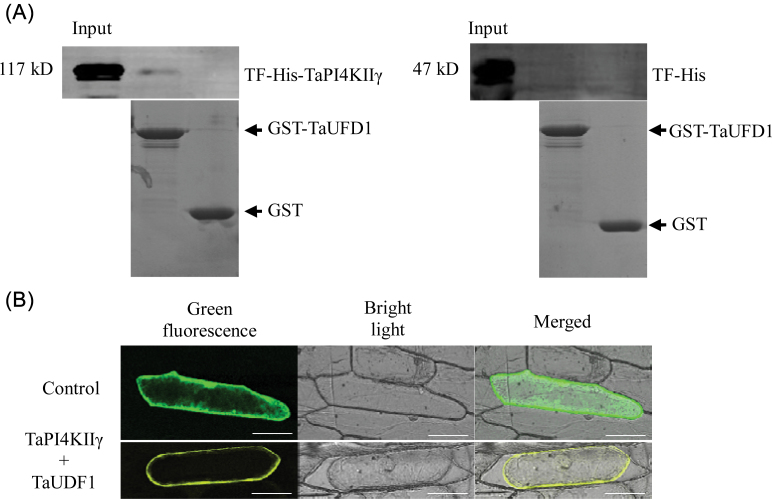
TaPI4KIIγ, as bait protein, was used to screen the wheat cDNA library by a yeast two-hybrid assay. One expressed sequence tag (EST) sequence, named TaUDF1, sharing the highest homology with Arabidoposis UDF1 was obtained. UDF1 binds to some forms of ubiquitin and is known to deliver target proteins to the 26S proteasome, which is the major degradation pathway in plants (Mayor et al., 2005). Interaction between TaPI4KIIγ and TaUDF1 was confirmed by pull-down and BiFC assays. TaPI4KIIγ and TaUDF1 were cloned into expression vectors, and the two recombinant proteins were successfully produced in E. coli as TF-His-TaPI4KIIγ and GST–TaUDF1, respectively. The in vitro pull-down assay was performed by incubating TF-His-TaPI4KIIγ and GST–TaUDF1 in combination (Fig. 5A). Interaction of the proteins was identified by BiFC. TaPI4KIIγ was cloned into pSPYNE, and TaUDF1 was cloned into pSPYCE. The TaPI4KIIγ–YFPN (N-terminal fragment of yellow fluorescent protein) and TaUDF1–YFPC (C-terminal fragment) recombination vectors were bombarded into onion epidermis, and fluorescence signals were observed. Plasmolysis was carried out to distinguish the plasma membrane from the cytoderm. Interaction of TaPI4KIIγ–YFPN and TaUDF1–YFPC occurred on the plasma membrane (Fig. 5B).
Fig. 5.
Interaction of TaPI4KIIγ and TaUDF1. (A) The in vitro pull-down assay was performed by incubating TF-His-TaPI4KIIγ with GST–TaUDF1. (B) The assay for interaction between TF-His and GST–TaUFD1 was used as the control. (C) The BiFC assay of the interaction of TaPI4KIIγ with TaUDF1. Fluorescence, bright field, and merged images of onion epidermal cells co-transfected with constructs encoding the indicated fusion proteins: TaPI4KIIγ–YFPN and TaUFD1–YFPC. Plasmolysis was carried out to distinguish the plasma membrane from the cytoderm. The scale bar is 73.61 μm. (This figure is available in colour at JXB online.)
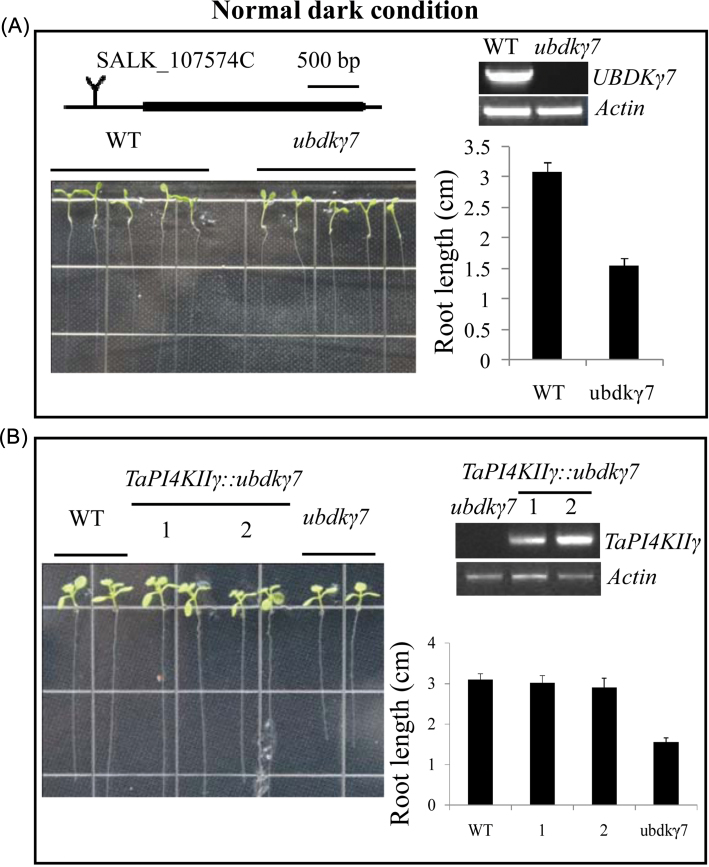
TaPI4KIIγ rescued the Arabidposis ubdkγ7 mutant and affected root growth
The phenotype of the ubdkγ7 mutant was identified as having a shorter root length (WT 3.1±0.14mm, ubdkγ7 mutant 1.56±0.1mm) (Fig. 6A). To assess whether expression of TaPI4KIIγ in Arabidopsis could rescue the ubdkγ7 mutant phenotype, TaPI4KIIγ was introduced into ubdkγ7 mutant plants under control of the CaMV35S promoter, and transgenic TaPI4KIIγ::ubdkγ7 lines were obtained. The TaPI4KIIγ::ubdkγ7 lines showed similar root lengths to the wild type (WT 3.1±0.14mm; TaPI4KIIγ::ubdkγ7 lines 3.02±0.17mm and 2.9±0.22mm) after 5 ds growth (Fig. 6B).
Fig. 6.
TaPI4KIIγ rescued the root length of the ubdkγ7 mutant. (A) The ubdkγ7 mutant displayed a shorter root length than the WT. The average root lengths of at least 30 seedlings are shown at the right. The ubdkγ7 mutant is a T-DNA insertion line, and the insertion site is in the promoter (about –500bp). The ubdkγ7 mutant is a knock out line. (B) Overexpression of TaPI4KIIγ in the ubdkγ7 mutant partially rescued the shorter root length phenotype. Gel electrophoretic map shows TaPI4KIIγ expression in the WT and the TaPI4KIIγ::UbDKγ7 lines. The histogram on the right shows average root lengths of at least 30 seedlings. The numbers 1 and 2 represent different TaPI4KIIγ::ubdkγ7 lines. (This figure is available in colour at JXB online.)
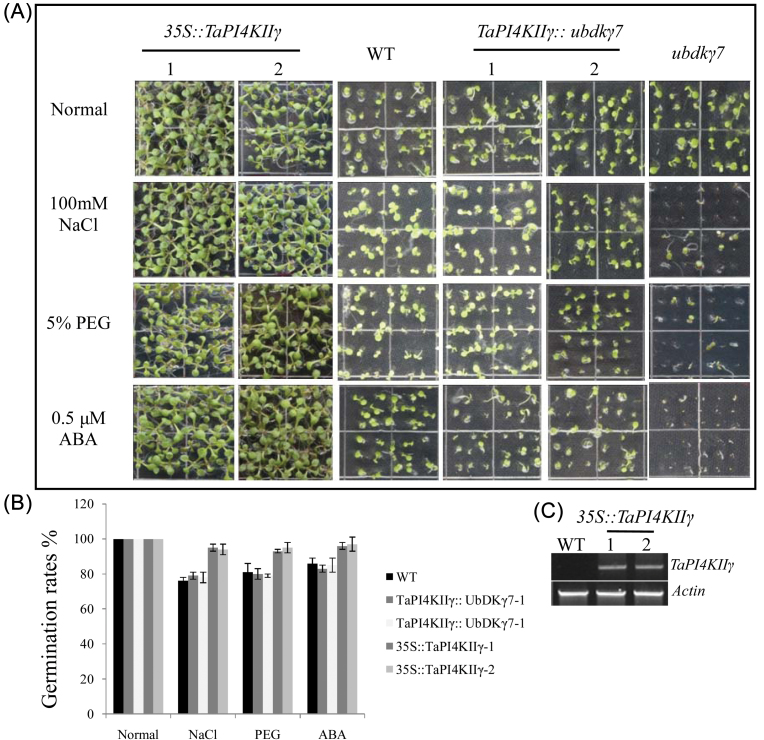
TaPI4KIIγ conferred stress tolerance to Arabidopsis
TaPI4KIIγ should not only rescue root growth, but also enhance the stress tolerances of the ubdkγ7 mutant and the WT (Fig. 7A). TaPI4KIIγ was introduced into the WT, generating several 35S::TaPI4KIIγ lines. Three lines were selected to assay for stress tolerance. Under normal conditions, nearly 100% of the seeds from the 35S::TaPI4KIIγ, WT, TaPI4KIIγ::ubdkγ7, and ubdkγ7 mutant lines germinated. However, when sown on MS medium containing PEG, NaCl, and ABA, these Arabidopsis lines displayed different germination rates (Fig. 7B). The ubdkγ7 mutant treated with PEG, NaCl, and ABA exhibited obviously lower seed germination rates than the WT (~50%). Overexpression of TaPI4KIIγ in the ubdkγ7 mutant to some extent rescued the resistance to stress treatments (the average germination rates of TaPI4KIIγ::ubdkγ7 lines were almost 80%, and similar to the WT), and overexpressing TaPI4KIIγ in the WT improved the stress tolerance (average germination rates of 35S::TaPI4KIIγ lines were ~95% compared with 80% for the WT).
Fig. 7.
Overexpression of TaPI4KIIγ improved seed germination under NaCl, PEG, and ABA. (A) The ubdkγ7 mutant was sensitive to NaCl, PEG, and ABA treatments. The TaPI4KIIγ::ubdkγ7 transformants showed a similar germination rate to the WT, and 35::TaPI4KIIγ lines displayed higher germination rates than the WT. (B) The seed germination rates of different lines. Each data point is the mean of three experiments, and each experiment comprised 30 plants. (C) Gel electrophoresis image detection of TaPI4KIIγ in the WT and 35S::TaPI4KIIγ lines. (This figure is available in colour at JXB online.)
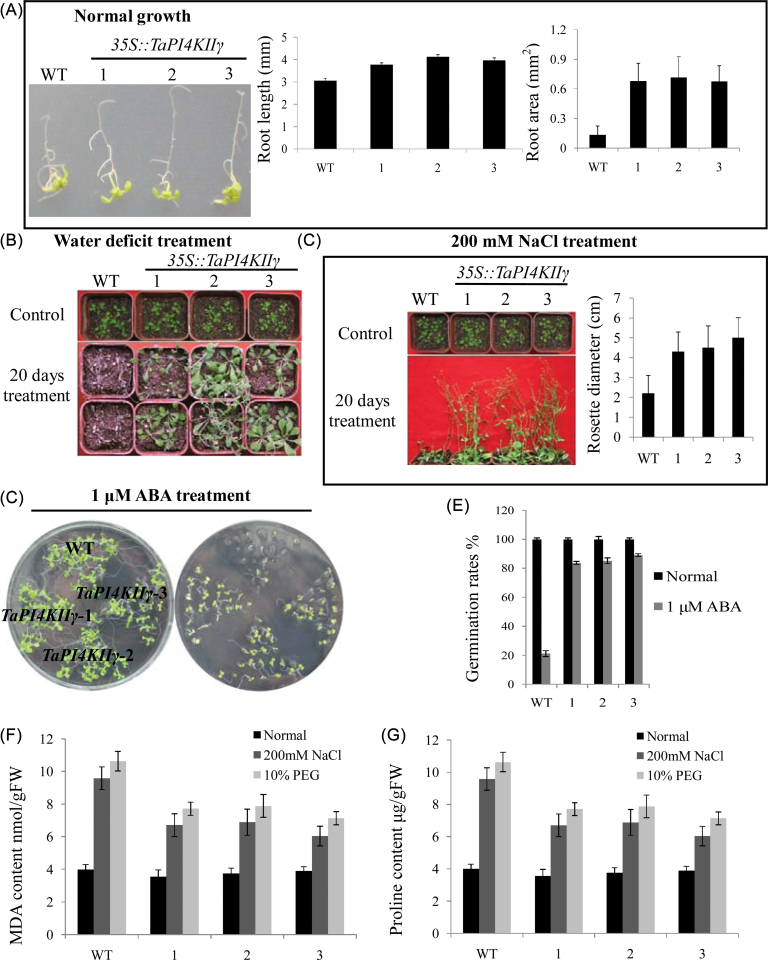
The three 35S::TaPI4KIIγ lines displayed longer root length and larger root areas than the WT under normal growth conditions, with increases of nearly 40% in root length and a 5-fold increase in root surface area (Fig. 8A). 35S::TaPI4KIIγ transgenic lines and WT seedlings were exposed to water deficit and salt treatments. After 20 d of water deficit, all WT plants exhibited severe symptoms of water loss and significant wilting. In contrast, most of the 35S::TaPI4KIIγ transgenic lines were green, and only slight wilting was evident in some of the 35S::TaPI4KIIγ transgenic leaves (Fig. 8B). After 20 d of salt treatment, the 35S::TaPI4KIIγ lines displayed a better performance than the WT based on rosette size (nearly 2-fold larger) (Fig. 8C). Improved ABA tolerance was identified by germination rates of Arabidopsis on MS medium containing ABA (Fig. 8D). The germination rates of 35S::TaPI4KIIγ lines were higher than those of the 35S::TaPI4KIIγ lines and the WT (nearly 4-fold) (Fig. 8E).
Fig. 8.
Overexpression of TaPI4KIIγ in wild-type Arabidopsis improved stress tolerance. (A) TaPI4KIIγ affected the root growth of Arabidopsis. The left image shows growth of the WT compared with the 35S::TaPI4KIIγ lines. The histograms show root length and root surface area. (B) Transgenic Arabidopsis plants overexpressing TaPI4KIIγ under water deficit treatment. The seedlings were not watered for 20 d. (C) Performance of transgenic Arabidopsis plants overexpressing TaPI4KIIγ under salt stress. The plants were treated with 200mM NaCl for 20 d. Rosette diameters of transgenic plants and the WT under salt stress are shown on the right. Each bar is the mean of 25 seedlings. (D) Arabidopsis grown on MS medium containing 1 μM ABA. Germination rates are shown in (E). (F) MDA contents in seedlings treated with 200mM NaCl and 10% PEG compared with normal growth conditions. (G) Proline contents in seedlings treated with 200mM NaCl and 10% PEG compared with seedlings grown under normal conditions. The numbers 1, 2, and 3 represent three 35S::TaPI4KIIγ lines, respectively. (This figure is available in colour at JXB online.)
The improved stress tolerance of 35S::TaPI4KIIγ transgenic plants was correlated with changes in proline and MDA contents. Under normal growing conditions, the proline and MDA contents of 35S::TaPI4KIIγ and the WT were similar. However, the MDA contents of 35S::TaPI4KIIγ lines were lower (50% and 42%, respectively) than that of the WT after NaCl and PEG treatments (Fig. 9F), and the proline contents of 35S::TaPI4KIIγ lines were much higher (70% and 38%, respectively) than those of the WT (Fig. 8G).
Fig. 9.
Expression levels of stress-responsive genes under regulation of TaPI4KIIγ. Gene-specific primers were used for detection of relative transcript levels of stress-responsive genes. The vertical coordinates are fold changes, and the horizontal ordinates are gene names. The data are means of three replicates.
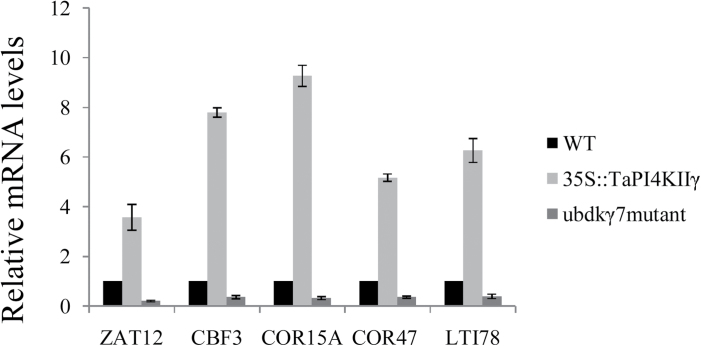
TaPI4KIIγ altered expression of stress-related genes in transgenic Arabidopsis
To elucidate the possible molecular mechanisms of TaPI4KIIγ in stress response, the expression of seven stress-response genes, namely DREB1B/CBF1, DREB1C/CBF2, DREB1A/CBF3, ZAT12, COR15A, COR47, and LTI78, was investigated in ubdkγ7 mutant, 35S::TaPI4KIIγ, and WT plants under normal growing conditions. A 2-fold change in expression was arbitrarily considered an expression induction. In three independent qRT–PCR analyses, five genes (DREB1A/CBF3, ZAT12, COR15A, COR47, and LTI78) showed altered expression in the transgenic plants and the mutant (Fig. 9). The expression of the five genes increased in the 35S::TaPI4KIIγ transformant under normal conditions, but was decreased in the ubdkγ7 mutant. The other two genes (DREB1B/CBF1 and DREB1C/CBF2) were not differentially expressed in the WT compared with the transgenic and mutant plants (data not shown).
Discussion
TaPI4KIIγ generated threonine autophosphorylation, but not lipid kinase activity
PI4K was necessary for the generation of two secondary messengers from PI4,5-bisphosphate and caused a series of cellular reactions (Jung et al., 2002). A plasma membrane-localized gene, TaPI4KIIγ, was identified to be up-regulated in drought-treated wheat de novo transcriptome sequencing. Known mammalian and yeast PI4K genes, such as Pik1, Stt4, Lsb6, PI4KIIα, PI4KIIβ, PI4KIIIα, and PI4KIIIβ, are responsible for catalysing PI to PI4-phosphate and PI4,5-bisphosphate, which possess lipid kinase activity (D’Angelo et al., 2008). However, in this study, TaPI4KIIγ generated threonine autophosphorylation, but not lipid kinase activity (Fig. 3). Several PI3/4K family members show similar kinase activities (Abraham, 2004; Galvão et al., 2008; Lovejoy and Cortez, 2009). As reported previously, phosphoinositide 3-kinase-related kinases (PIKKs) usually transduce cell signalling via phosphorylation of serine or threonine protein residues, such as ATM, ATR, mTOR, DNA-PK, and hSMG-1, and function as protein serine/threonine kinases (Lovejoy and Cortez, 2009). For instance, the protein kinase ATM (ataxia telangiectasia mutated) was homodimerized, and dissociation to monomers was stimulated by autophosphorylation, which activated ATM kinase activity (Bakkenist and Kastan, 2003). Arabidopsis type II PI4Ks UbDKγ4 and UbDKγ7 displayed the same kinase activity as TaPI4KIIγ, possessing protein kinase activity, but not lipid kinase activity (Galvão et al., 2008). It is proposed that TaPI4KIIγ is a member of a new group of protein kinases belonging to the PIKK family of atypical protein kinases (Manning et al., 2002). It is somewhat confusing in that PI4Ks are usually considered part of the phosphoinositide signalling pathway and function as key enzymes that catalyse changes of PI to PI4-phosphate. Further investigation is needed to determine how TaPI4KIIγ functions in the phosphoinositide signalling pathway.
UFD1, which was first found in a yeast mutant defective in degrading short-lived ubiquitin fusion proteins (Johnson et al., 1995), is involved in regulation of protein degradation as a ‘vehicle’, that binds some forms of ubiquitin and target proteins and delivers the complex to 26S proteasomes (Hartmann-Petersen and Gordon, 2004; Madura, 2004). In this study, TaUFD1 was identified to interact with TaPI4KIIγ (Fig. 5), suggesting a possible degradation pathway: TaPI4KIIγ interacts with TaUFD1 which is already bound to ubiquitin, and then the protein complex is transported to the 26S proteasome degradation system. The ubiquitin–26S proteasome pathway is involved in various vital processes, such as cell cycle control, programmed cell death, and signalling transduction (Vierstra, 2009). Recently, it was found that the ubiquitin–26S proteasome pathway plays significant roles in regulation of abiotic and biotic responses in plants (Dielen et al., 2010). Although there are a few reports about UFD1 regulation of stress response in plants, it was demonstrated that silencing UFD1 in Nicotiana benthamiana resulted in significantly improved biotic stress tolerance (Lai et al., 2012). These results indicated that UFD1 might act as a negative regulator in stress response by causing the degradation of interacting proteins, and in this study the function of TaPI4KIIγ in stress response might be negatively controlled by TaUFD1 via the 26S proteasome degradation system.
TaPI4KII responded to multiple stresses
Phytohormones, such as SA, jasmonic acid (JA), ethylene (ET), and ABA, primarily regulate protective responses of plants to both biotic and abiotic stresses via synergistic or antagonistic actions (Fujita et al., 2006). ABA is commonly considered an abiotic resistance-related phytohormone. Arabidopsis ABA mutants aba1, aba2, and aba3 grow relatively normally compared with the WT, but are very sensitive to drought and high salt conditions (Zhu, 2002). Many drought-inducible genes are also activated by ABA (Abe et al., 2003; Ding et al., 2009). In this study, TaPI4KIIγ was rapidly induced by ABA, and was also up-regulated by dehydration and salt (Fig. 2). However, compared with the rapid response to ABA, the TaPI4KIIγ activity induced by dehydration and salt was delayed. The different expression profiles of TaPI4KIIγ under ABA, dehydration, and salt implied that TaPI4KIIγ might function via different mechanisms under different stresses.
SA is associated with biotrophic pathogen resistance (Pieterse et al., 2009), and ABA is generally considered to be a negative regulator of SA-mediated disease resistance (Fujita et al., 2006). For example, the ABA-deficient tomato mutant sitiens had increased resistance to pathogens, and application of exogenous ABA restored susceptibility to sitiens (Audenaert et al., 2002). In the present work, TaPI4KIIγ, induced by ABA, was negatively regulated by SA, and showed decreased expression under SA treatment (Supplementary Fig. S3 at JXB online). This suggested that the ABA and SA signalling pathways affect expression of TaPI4KIIγ in antagonistic ways.
TaPI4KIIγ enhanced stress tolerance
Overexpression of stress-inducible genes in plants is an effective strategy for improving abiotic stress tolerance (Rudrabhatla and Rajasekharan, 2002; Flowers, 2004; Cattivelli et al., 2008; Xu et al., 2008, 2011). To date, there had been no report on the abiotic stress response of type II PI4Ks in plants. Overexpression of TaPI4KIIγ in the WT background led to obviously enhanced tolerance to water deficit, salt, and ABA (Figs 7, 8). One significant piece of evidence was that the 35S::TaPI4KIIγ lines exhibited high seed germination rates following salt, PEG, and ABA treatments (Fig. 7), and the seedlings exhibited higher survival rates and better growth than the WT under water deficit and salt stresses (Fig. 8B, C). In contrast, the ubdkγ7 mutant was sensitive to salt, PEG, and ABA treatments, as identified by germination rates on the culture medium (Fig. 7). These results suggested that TaPI4KIIγ may be involved in regulating plant responses to abiotic stresses. Introduction of TaPI4KIIγ into the ubdkγ7 mutant resulted in the recovery of the normal phenotype, with near-normal root growth and stress tolerance. It was reported that PI4K was required for root tip growth in Arabidopsis. Plants lacking members of the PI4K familiy displayed defective growth and morphology of root hairs (Szumlanski and Nielsen, 2010). Root length and surface area are phenotyping traits for measuring plant drought stress tolerance (Manavalan et al., 2009). In this study, the roots of 35S::TaPI4KIIγ lines were obviously longer and possessed a larger surface areas than the WT, whereas the root length of the ubdkγ7 mutant was shorter. Thus TaPI4KIIγ might enhance stress tolerance by improving root growth.
Water deficit and salt stresses induce various physiological responses in plants. Proline, which functions as an osmotic regulator, prevents water loss from cells when plants are subject to water deficit and salt stresses (Bais and Ravishankar, 2002; Urano et al., 2009). Increased proline content in 35S::TaPI4KIIγ transformants under PEG and salt stresses could be taken as evidence for stress resistance. Moreover, stresses such as water deficit and salt are accompanied by production of MDA, which damages the membrane and results in cell death (Levine et al., 1994). The lower content of MDA in 35S::TaPI4KIIγ when subjected to PEG and salt stresses could be utilized as a sign of tolerance to stress.
Possible mechanisms of TaPI4KIIγ in stress response
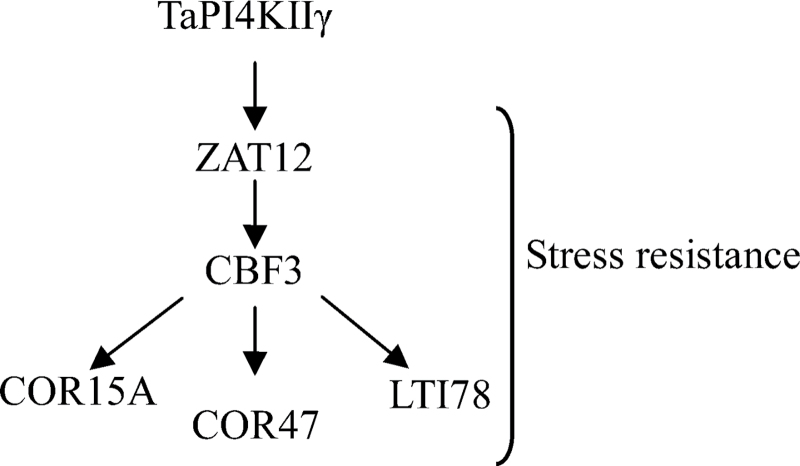
Many transcription factors and other genes responding to stress have been identified, providing the insight that plants have developed flexible molecular and cellular mechanisms to tolerate various abiotic stresses. The CBF gene family encodes ERF/AP2 transcription factors that have been described as playing important roles in drought and salt tolerance in Arabidopsis (Dubouzet et al., 2003; Shen et al., 2003; Oraby and Ahmad, 2012). In this study, it was found that the expression of CBF3 was activated in the 35S::TaPI4KIIγ plants, but decreased in the ubdkγ7 mutant. In Arabidopsis, CBF was demonstrated to control the accumulation of other stress response genes, such as COR15A, COR47, and LTI78 (Novillo et al., 2012). COR15A, COR47, and LTI78, which are CBF target genes, were reported as drought- and salt-responsive genes (Xiong et al., 2002; Cheong et al., 2011; Novillo et al., 2012; Zhang et al., 2011). The expression of COR15A, COR47, and LTI78 was significantly increased in 35S::TaPI4KIIγ plants, but was decreased in the ubdkγ7 mutant. These results indicated that TaPI4KIIγ could activate the expression of CBF3, which increased the expression of COR15A, COR47, and LTI78, and ultimately improved the salt and drought tolerances of Arabidopsis. ZAT12, a salt- and drought-responsive gene (Qin et al., 2012), controls the expression of DREB1/CBF genes (Gorsuch et al., 2010; Hirayama and Shinozaki, 2010). The present work showed that the expression of ZAT12 was changed in the 35S::TaPI4KIIγ transformant and the ubdkγ7 mutant, indicating that a ZAT12 signal pathway may exist in the DREB1/CBF gene regulatory network that was stimulated by TaPI4KIIγ (Fig 10). The present work thus suggests that TaPI4KIIγ is involved in regulating plant responses to abiotic stress, and has potential for improving stress tolerance in crop plants.
Fig. 10.
A proposed model for TaPI4KIIγ improvement of stress tolerance. TaPI4KIIγ increases the expression of ZAT12, leading to activated expression of CBF3. Stress-responsive genes, including COR15A, COR47, and LTI78, were induced to enhance plant stress tolerance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Information on the ubdkγ7 mutant.
Figure S2. Phylogeny of the PI3/4K domain.
Figure S3. Expression profile of TaPI4KIIγ under SA treatment.
Table S1. Drought- and salt-responsive genes selected from the TAIR database, and gene-specific primers used for qRT–PCR.
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (31171546) and the National 863 High-tech Project (2012AA10A309). We are grateful to Drs Rui-Lian Jing and Yong-Fu Fu (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing wheat seeds and for the BiFC system, respectively. We also thank Dr R.A. McIntosh (Plant Breeding Institute, University of Sydney) for suggestions on the manuscript.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham RT. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair 3, 883–887. [DOI] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Höfte MM. 2002. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiology 128, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Developmental Cell 2, 593–605. [DOI] [PubMed] [Google Scholar]
- Bais HP, Ravishankar GA. 2002. Role of polyamines in the ontogeny of plants and their biotechnological applications. Plant Cell, Tissue and Organ Culture 69, 1–34. [Google Scholar]
- Bakkenist CJ, Kastan MB. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506. [DOI] [PubMed] [Google Scholar]
- Balla T. 1998. Phosphatidylinositol 4-kinases. Biochimica et Biophysica Acta 1436, 69–85. [DOI] [PubMed] [Google Scholar]
- Balla T. 2001. Pharmacology of phosphoinositides, regulators of multiple cellular functions. Current Pharmaceutical Design 7, 475–507. [DOI] [PubMed] [Google Scholar]
- Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Südhof TC, Albanesi JP. 2001. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. Journal of Biological Chemistry 276, 7705–7708. [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant and Soil 39, 205–207. [Google Scholar]
- Bechtold N, Pelletier G. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods in Molecular Biology 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Carpenter CL, Cantley LC. 1996. Phosphoinositide kinases. Current Opinion in Cell Biology 8, 153–158. [DOI] [PubMed] [Google Scholar]
- Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Mare C, Tondelli A, Stanca AM. 2008. Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crops Research 105, 1–14. [Google Scholar]
- Cheong YH, Sung SJ, Kim BG, Pandey GK, Cho JS, Kim KN, Luan S. 2010. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis . Molecules and Cells 29, 159–165. [DOI] [PubMed] [Google Scholar]
- Chu KM, Minogue S, Hsuan JJ, Waugh MG. 2012. Differential effects of the phosphatidylinositol 4-kinases, PI4KIIα and PI4KIIIβ, on Akt activation and apoptosis. Cell Death and Disease 1, e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G, Vicinanza M, Di Campli A, De Matteis MA. 2008. The multiple roles of PtdIns(4)P—not just the precursor of PtdIns(4,5)P2. Journal of Cell Science 121, 1955–1963. [DOI] [PubMed] [Google Scholar]
- Delage E, Ruelland E, Guillas I, Zachowski A, Puyaubert J. 2012. Arabidopsis type-III phosphatidylinositol 4-kinases β1 and β2 are upstream of the phospholipase C pathway triggered by cold exposure. Plant and Cell Physiology 53, 565–576. [DOI] [PubMed] [Google Scholar]
- Dielen AS, Badaoui S, Candresse T, German-Retana S. 2010. The ubiquitin/26S proteasome system in plant–pathogen interactions: a never-ending hide-and-seek game. Molecular Plant Pathology 11, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Li S, An X, Liu X, Qin H, Wang D. 2009. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana . Journal of Genetics and Genomics 36, 17–29. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal 33, 751–763. [DOI] [PubMed] [Google Scholar]
- Endemann G, Dunn SN, Cantley LC. 1987. Bovine brain contains two types of phosphatidylinositol kinase. Biochemistry 26, 6845–6852. [DOI] [PubMed] [Google Scholar]
- Flowers TJ. 2004. Improving crop salt tolerance. Journal of Experimental Botany 55, 307–319. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. 2006. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology 9, 436–442. [DOI] [PubMed] [Google Scholar]
- Galvão RM, Kota U, Soderblom EJ, Goshe MB, Boss WF. 2008. Characterization of a new family of protein kinases from Arabidopsis containing phosphoinositide 3/4-kinase and ubiquitin-like domains. Biochemical Journal 409, 117–127. [DOI] [PubMed] [Google Scholar]
- Gorsuch PA, Sargeant AW, Penfield SD, Quick WP, Atkin OK. 2010. Systemic low temperature signaling in Arabidopsis . Plant and Cell Physiology 51, 1488–1498. [DOI] [PubMed] [Google Scholar]
- Han GS, Audhya A, Markley DJ, Emr SD, Carman GM. 2002. The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. Journal of Biological Chemistry 277, 47709–47718. [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Gordon C. 2004. Integral UBL domain proteins: a family of proteasome interacting proteins. Seminars in Cell and Developmental Biology 15, 247–259. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. 2010. Research on plant abiotic stress responses in the post-genome era: past, present and future. The Plant Journal 61, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Seiler S, Heilmann I. 2010. At the poles across kingdoms: phosphoinositides and polar tip growth. Protoplasma 240, 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. Journal of Biological Chemistry 270, 17442–17456. [DOI] [PubMed] [Google Scholar]
- Jung JY, Kim YW, Kwak JM, Hwang JU, Young J, Schroeder JI, Hwang I, Lee Y. 2002. Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. The Plant Cell 14, 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annual Review of Biophysics 37, 465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke O, Ruelland E, Valentová O, Vergnolle C, Renou JP, Taconnat L, Flemr M, Burketová L, Zachowski A. 2007. Phosphatidylinositol 4-kinase activation is an early response to salicylic acid in Arabidopsis suspension cells. Plant Physiology 144, 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YY, Li F, Xu YP, Cai XZ. 2012. Functional analysis of an ubiquitin fusion degradation protein gene UFD1 in regulation of plant disease and stress resistance. Journal of Zhejiang University 38, 21–27. [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. 2011. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Research 18, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. 1994. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Liu HW, Shi RF, Wang XF, Pan YX, Zang GY, Ma ZY. 2012. Cloning of a phosphatidylinositol 4-kinase gene based on fiber strength transcriptome QTL mapping in the cotton species. Gossypium barbadense. Genetics and Molecular Research 11, 3367–3378. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Cortez D. 2009. Common mechanisms of PIKK regulation. DNA Repair 8, 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Yang A, Zhang K, Wang L, Zhang J. 2007. Increase of glycine betaine synthesis improves drought tolerance in cotton. Molecular Breeding 20, 233–248. [Google Scholar]
- Madura K. 2004. Rad23 and Rpn10: perennial wallflowers join the melee. Trends in Biochemical Sciences 29, 637–640. [DOI] [PubMed] [Google Scholar]
- Manavalan LP, Guttikonda SK, Tran LS, Nguyen HT. 2009. Physiological and molecular approaches to improve drought resistance in soybean. Plant and Cell Physiology 50, 1260–1276. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. 2005. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Molecular and Cellular Proteomics 4, 741–751. [DOI] [PubMed] [Google Scholar]
- Minogue S, Anderson JS, Waugh MG, dos Santos M, Corless S, Cramer R, Hsuan JJ. 2001. Cloning of a human type II phosphatidylinositol 4-kinase reveals a novel lipid kinase family. Journal of Biological Chemistry 276, 16635–16640. [DOI] [PubMed] [Google Scholar]
- Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S. 2011. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnology Journal 9, 230–249. [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C. 2002. Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C1. Plant Physiology 130, 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Vermeer JE. 2009. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant, Cell and Environment 33, 655–669. [DOI] [PubMed] [Google Scholar]
- Novillo F, Medina J, Rodríguez-Franco M, Neuhaus G, Salinas J. 2012. Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis response to abiotic stress. Journal of Experimental Botany 63, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. 2009. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oraby H, Ahmad R. 2012. Physiological and biochemical changes of CBF3 transgenic oat in response to salinity stress. Plant Science 185–186 331–339. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Pike LJ. 1992. Phosphatidylinositol 4-kinases and the role of polyphosphoinositides in cellular regulation. Journal of Clinical Endocrinology and Metabolism 13, 692–706. [DOI] [PubMed] [Google Scholar]
- Qin Y, Wang M, Tian Y, He W, Han L, Xia G. 2012. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis . Molecular Biology Reports 39, 7183–7192. [DOI] [PubMed] [Google Scholar]
- Rhoades JD, Loveday J. 1990. Salinity in irrigated agriculture. Agronomy 30, 1089–1142. [Google Scholar]
- Rudrabhatla P, Rajasekharan R. 2002. Developmentally regulated dual-specificity kinase from peanut that is induced by abiotic stresses. Plant Physiology 130, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleki R, Young PG, Lefebvre DD. 1993. Mutants of Arabidopsis thaliana capable of germination under saline conditions. Plant Physiology 101, 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY. 2003. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theoretical and Applied Genetics 106, 923–930. [DOI] [PubMed] [Google Scholar]
- Strahl T, Hama H, DeWald DB, Thorner J. 2005. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. Journal of Biological Chemistry 171, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Thorner J. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. . Biochimica et Biophysica Acta 1771, 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlanski A, Nielsen E. 2010. Phosphatidylinositol 4-phosphate is required for tip growth in Arabidopsis thaliana. Lipid Signaling in Plants 16, 65–77. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. 2008. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, et al. 2009. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. The Plant Journal 57, 1065–1078. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2009. The ubiquitin–26S proteasome system at the nexus of plant biology. Nature Reviews Molecular Cell Biology 10, 385–397. [DOI] [PubMed] [Google Scholar]
- Wong K, Meyers DDR, Cantley LC. 1997. Subcellular locations of phosphatidylinositol 4-kinase isoforms. Journal of Biological Chemistry 272, 13236–13241. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK. 2002. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis . Journal of Biological Chemistry 277, 8588–8596. [DOI] [PubMed] [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ. 2008. Functions of the ERF transcription factor family in plants. Botany 86, 969–977. [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ. 2011. Functions and application of the AP2/ERF transcription factor family in crop improvement. Journal of Integrative Plant Biology 53, 570–585. [DOI] [PubMed] [Google Scholar]
- Xu ZS, Xia LQ, Chen M, et al. 2007. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Molecular Biology 65, 719–732. [DOI] [PubMed] [Google Scholar]
- Xue HW, Pical C, Brearley C, Elge S, Müller-Röber B. 1999. A plant 126-kDa phosphatidylinositol 4-kinase with a novel repeat structure. Cloning and functional expression in baculovirus-infected insect cells. Journal of Biological Chemistry 274, 5738–5745. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Z, Quan R, Li G, Wang R, Huang R. 2011. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis . Plant Physiology 157, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Várnai P, Tuymetova G, Balla A, Tóth ZE, Oker-Blom C, Roder J, Jeromin A, Balla T. 2001. Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. Journal of Biological Chemistry 276, 40183–40189. [DOI] [PubMed] [Google Scholar]
- Zhu JK. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.