Abstract
Despite substantial evidence on the separate roles of Arabidopsis nitric oxide-associated 1 (NOA1)-associated nitric oxide (NO) production and haem oxygenase 1 (HY1) expression in salt tolerance, their integrative signalling pathway remains largely unknown. To fill this knowledge gap, the interaction network among nitrate reductase (NIA/NR)- and NOA1-dependent NO production and HY1 expression was studied at the genetic and molecular levels. Upon salinity stress, the majority of NO production was attributed to NIA/NR/NOA1. Further evidence confirmed that HY1 mutant hy1-100, nia1/2/noa1, and nia1/2/noa1/hy1-100 mutants exhibited progressive salt hypersensitivity, all of which were significantly rescued by three NO-releasing compounds. The salinity-tolerant phenotype and the stronger NO production in gain-of-function mutant of HY1 were also blocked by the NO synthetic inhibitor and scavenger. Although NO- or HY1-deficient mutants showed a compensatory mode of upregulation of HY1 or slightly increased NO production, respectively, during 2 d of salt treatment, downregulation of ZAT10/12-mediated antioxidant gene expression (cAPX1/2 and FSD1) was observed after 7 d of treatment. The hypersensitive phenotypes and stress-related genes expression profiles were differentially rescued or blocked by the application of NO- (in particular) or carbon monoxide (CO)-releasing compounds, showing a synergistic mode. Similar reciprocal responses were observed in the nia1/2/noa1/hy1-100 quadruple mutant, with the NO-releasing compounds exhibit the maximal rescuing responses. Overall, the findings present the combination of compensatory and synergistic modes, linking NIA/NR/NOA1-dependent NO production and HY1 expression in the modulation of plant salt tolerance.
Key words: Arabidopsis, haem oxygenase-1 (HY1), NIA/NR, nitric oxide, NOA1, salt tolerance.
Introduction
Salinity stress is a major factor that significantly inhibits plant growth and decreases crop productivity (Zhu, 2000, 2001; Chinnusamy et al., 2004). Typically, it imposes ionic toxicity, osmotic stress, and even oxidative damage on plant cells. In response to salinity stress, plants that have successfully survived should reestablish, sustain, or enhance: (a) ion and osmotic homeostasis; (b) stress damage control and repair, or reactive oxygen species (ROS) detoxification; and (c) growth control. It has also been claimed that homeostasis and detoxification signalling could not only lead to salinity tolerance, but also negatively regulate the growth inhibition response (Mittler, 2002; Zhu, 2002; Krasensky and Jonak, 2012; Liu et al., 2012; Xie et al., 2012a).
Nitric oxide (NO), a redox-active gaseous molecule, plays a key signalling role in a broad spectrum of physiological and development processes throughout the whole plant life cycle (Delledonne, 2005; Gupta et al., 2011). For example, ample evidence demonstrated that NO could act as a signal molecule mediating responses to abiotic stresses, including salt stress (Uchida et al., 2002; Zhao et al., 2004, 2007), drought and dehydration (García-Mata and Lamattina, 2001; Lozano-Juste and León, 2010), and cold acclimation and freezing tolerance (Zhao et al., 2009).
In animals, NO is synthesized via the enzyme NO synthase (NOS) (Dulak and Józkowicz, 2003). Although NOS-like activity has been detected widely in plants, and inhibitors of mammalian NOS inhibit NO generation in plants, no archetypal NOS-encoding gene(s) have been isolated so far in higher plants (Crawford et al., 2006; Zemojtel et al., 2006; Van Ree et al., 2011). Arabidopsis nitric oxide associated 1 (NOA1), originally reported to encode a protein with a NOS activity, has been shown to be a GTPase, which plays a role in binding RNA/ribosomes (Sudhamsu et al., 2008). A relationship of NOA1-mediated NO production and plant responses against salinity stress (Zhao et al., 2007) and pathogens (Mandal et al., 2012) has been reported, although the reduced accumulation of NO in noa1 plants might be due to their inability to accumulate the sucrose reserve (Van Ree et al., 2011). The production of NO in plants can also be generated by nitrate reductase (NR), the critical enzyme responsible for nitrate assimilation (Rockel et al., 2002; Zhao et al., 2009; Gupta et al., 2011). In Arabidopsis, NR is encoded by two genes, NIA1 and NIA2 (Yamasaki and Sakihama, 2000). The involvement of NR-mediated NO production in stomatal closure and cold acclimation has been demonstrated genetically (Bright et al., 2006; Zhao et al., 2009). The mitochondrial electron transport chain and the non-enzymic pathway in the apoplast are also involved in NO production (Bethke et al., 2004; Gupta et al., 2005, 2011; Planchet et al., 2005).
Recently, plant haem oxygenase (HO; EC 1.14.99.3), the rate-limiting enzyme responsible for the breakdown of haem into another gaseous molecule (CO), iron, and bilirubin, has received overwhelming research attention (Shekhawat and Verma, 2010). In Arabidopsis, a small family of HOs with four members is classified into two subfamilies. The inducible HO1 subfamily includes HY1, HO3, and HO4, while HO2 is the only member of the HO2 subfamily (Gisk et al., 2010). Although HO1/CO was firstly described as a signalling system in animals (Bilban et al., 2008), new and exciting physiological roles as a bioregulator of plant developmental processes as well as environmental adaptation have been reported (Shekhawat and Verma, 2010; Bose et al., 2013). Close links among HO1/CO and auxin (Cao et al., 2011), glutathione (Han et al., 2008), NO (Noriega et al., 2007; Xuan et al., 2008), hydrogen peroxide (H2O2) and other ROS (Yannarelli et al., 2006; Chen et al., 2009; Wu et al., 2011b; Xie et al., 2011a; Xu et al., 2012; Bose et al., 2013), Ca2+ (Wu et al., 2011a), or hydrogen sulfide (Lin et al., 2012) pathways have also been suggested.
In plants, both NO and HO1/CO are integral components of osmotic signalling (Liu et al., 2010). Our data have shown that HO1/CO might confer an increased tolerance to salinity stress in wheat seedling roots, which is partially mediated by the NO signal (Xie et al., 2008), and further suggested that Arabidopsis HY1 plays an important role in salt acclimation signalling and requires the participation of RbohD-derived ROS peak II (Xie et al., 2011b; Bose et al., 2013). However, the genetic evidence for the cross talk between NO and HY1 in plant responses against salinity stress remains elusive.
Although there are some debates on the nature of AtNOA1 participating in NO biosynthesis (Crawford et al., 2006; Guo, 2006; Zemojtel et al., 2006), in most cases, Atnoa1 mutants are valuable materials for studying the physiological functions of NO (Zhao et al., 2007; Van Ree et al., 2011). Meanwhile, Lozano-Juste and Leόn (2010) have reported that the germination of nia1/2/noa mutant was hypersensitive to exogenous abscisic acid, salt, and osmotic stress (in particular). However, the corresponding detailed mechanism, such as the crosstalk with other signalling components and related transduction cascade, remains to be elucidated. The present study addressed the functions of both NO and HY1 signalling pathways upon salinity stress in Arabidopsis by generating a quadruple mutant in which both NR/NOA1 and HY1 that are severely impaired. The observed physiological responses of Arabidopsis plants were linked with complex sets of information provided by a combination of genetic and molecular analysis. Most importantly, corresponding elucidation of both NO- and HY1-deficient mutant plants led to identify a cross-talk signalling, including the compensatory and synergistic models of NIA/NR/ NOA1-dependent NO and HY1 expression, in the modulation of salt tolerance in Arabidopsis.
Materials and methods
Chemicals
All chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA) unless otherwise stated. The chemicals used for treatments were sodium nitroprusside (SNP), diethylamine NONOate (NONOate), S-nitrosoglutathione (GSNO), carbon monoxide-releasing molecule-2 (CORM-2), NG-nitro-l-arginine methyl ester hydrochloride (l-NAME), tungstate, 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide potassium salt (cPTIO) (Ederli et al., 2006; Zhao et al., 2009; Chen et al., 2010; García-Arnandis et al., 2011; Desmard et al., 2012; Xie et al., 2012b). The concentrations used in this study were determined in pilot experiments from which maximal induced responses were obtained.
Additionally, the Old SNP/NONOate/CORM-2 solutions were used as the negative controls by maintaining the separated solution of SNP/NONOate/CORM-2 (50 µM each) for at least 10 d in the light in a specific open tube to eliminate the entire NO or CO, respectively (Tossi et al., 2009).
Plant materials and growth conditions
Arabidopsis thaliana hy1-100 (CS236, Col-0), hy1 (CS67, Ler), noa1 (CS6511, Col-0), and nia1/2 (CS2356, Col-0) mutants were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc), and the homogenous HY1 overexpression line 35S:HY1-4 was previously constructed (Xie et al., 2011b). The Ler ecotype seeds were provided by Dr Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China).
Seeds were surface-sterilized for 20min and washed three times with sterile water, then cultured in Petri dishes on solid Murashige and Skoog (MS) medium (pH 5.8) with 1% sucrose. For culturing NR-related mutants, the nitrogen in the MS medium included 1mM NH4 + and 1.94mM NO3 – [Zhao et al., 2009; preliminary experiments also showed the performance of the wild-type and the nia1/2 mutant is clearly associated with the exogenous N supplement, although both NH4 + and NO3 – are essential, NR mutants could grow better with NH4 + (Supplementary Fig. S1, available at JXB online)]. Plates containing seeds were kept at 4 °C for 2 d, and then transferred into a growth chamber with a 16/8 light/dark cycle (23/18 °C) and 120 µmol m–2 s–1 irradiation.
Generation of the triple-mutant line nia1/2/noa1 and quadruple mutant line nia1/2/noa1/hy1-100
The triple-mutant line nia1/2/noa1 and the quadruple mutant line nia1/2/noa1/hy1-100 were obtained by crossing mutant lines nia1/2 with noa1, and then nia1/2/noa1 with hy1-100. F1 plants were self-pollinated and the resulting F2 individuals were genotyped using PCR primers (Supplementary Table S1) specific for the presence of the mutations (Supplementary Fig. S2). Homozygous hy1-related mutants were further identified by Western blot analysis (Supplementary Fig. S3) and the corresponding phenotypes (including yellow cotyledons, etc) were observed (Supplementary Figs. S4 and S5).
Salt-tolerance assay and phenotype analysis
For salt-tolerance analysis, 5-day-old seedlings of each genotype were cultured in MS medium with or without various chemical pretreatments for the indicated times, followed by 150mM NaCl or other treatments as indicated. Alternatively, stratified seeds of each genotype were sowed in the MS medium with or without 150mM NaCl in the presence or absence of l-NAME, tungstate, or cPTIO for the indicated times. The phenotypes, including the primary root growth, fresh weight, chlorophyll content, and germination rate, were then observed at the indicated times (Lichtenthaler, 1987; Xie et al., 2011b, 2012b). Meanwhile, representative images were photographed.
Real-time quantitative reverse-transcription PCR
Total RNA was isolated using the Trizol reagent (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer’s instructions. Real-time quantitative reverse-transcription (RT) PCR were performed using a Mastercycler ep® realplex real-time PCR system (Eppendorf, Hamburg, Germany) with SYBR® Premix Ex Taq™ (TaKaRa Bio, China) according to the manufacturer’s instructions. Using specific primers (Supplementary Table S2), relative expression levels of corresponding genes are presented as values relative to corresponding control samples at the indicated times or under the indicated conditions, after normalization to actin2/7 (At3g18780) transcript levels.
Confocal determination of NO production
Endogenous NO level was assayed by confocal microscopy using a fairly specific NO fluorescent probe 4-amino-5-methyl-amino-2′,7′-di-fluorofluorescein diacetate (DAF-FM DA). Arabidopsis seedlings were collected at the indicated times and loaded with 10 µM DAF-FM DA for 20min before washing in 20mM HEPES buffer (pH 7.8) three times for 5min each and then analysed using a TCS-SP2 laser scanning confocal microscope (LSCM; Leica, excitation at 488nm, emission at 500–530nm). All manipulations were performed at 25±1 °C. Pictures were taken based on 20 overlapping confocal planes of 2 µm each using the Leica software package. Each genotype had 6 replicates per experiment. Data are presented as relative units of pixel intensities via region of interest analysis, provided by the Leica software (Sieberer et al., 2009; Liesche and Schulz, 2012) or as the means of fluorescence intensity relative to that of the wild-type under the control condition (0h).
NO detection by electron paramagnetic resonance
Arabidopsis seedlings were homogenized and incubated in buffered solution (50mM Hepes, 1mM DTT, 1mM MgCl2, pH 7.6). After centrifugation at 12,000 g for 2min, the supernatant was added to 300 µl of freshly made Fe(II)(DETC)2 solution (2M Na2S2O4, 3.3mM DETC, 3.3mM FeSO4, 33g l–1 BSA), incubated for 2min at room temperature and frozen in liquid nitrogen until analysis (Huang et al., 2004; Sun et al., 2012). Electron paramagnetic resonance (EPR) measurements were performed on a A300 spectrometer (Bruker Instrument, Germany) under the following conditions: microwave frequency, 9.854 GHz; microwave power, 63.496 mW; modulation amplitude 4.00 G; modulation frequency, 100kHz.
Statistical analysis
Data are shown as means ± standard error (SE) of at least three independent experimental replications. The analysis of variance (ANOVA), multiple comparison, and t-test were also used for data analysis. In particular, the data for different genotype samples upon different treatments were analysed using a general linear model of a completely randomized design with the treatments as one fixed factor. Differences in the phenotypic indicators among various genotypes were analysed by a nested ANOVA, with genotypes nested within different treatments [model: phenotypic indicators = replication + treatment + sample (treatment)], followed by a multiple comparison. A threshold of P < 0.05 was taken to claim significance. All procedures were programmed to run in a SAS version 9.1 environment (SAS Institute, Cary, NC, USA).
Results
NIA1/2/NOA1-dependent NO production mediates Arabidopsis salt tolerance
To clarify how NO is synthesized in Arabidopsis, this study crossed nia1/2 double mutants with the noa1 mutant and found plants with homozygous mutations in the three genes by performing PCR-based genotyping of F2 progeny (Supplementary Fig. S2). As expected (Lozano-Juste and Leόn, 2010), stratified nia1/2, noa1, and nia1/2/noa1 mutants could fully germinate in normal growth conditions. It was further observed that the nia1/2/noa1 mutant was more sensitive to salinity stress than its parental lines, as measured by the responses of germination rate, and survival rate of germinated seeds (Supplementary Figs. S6 and S7). These results extended the observation reported by Zhao et al. (2004) and Lozano-Juste and Leόn (2010).
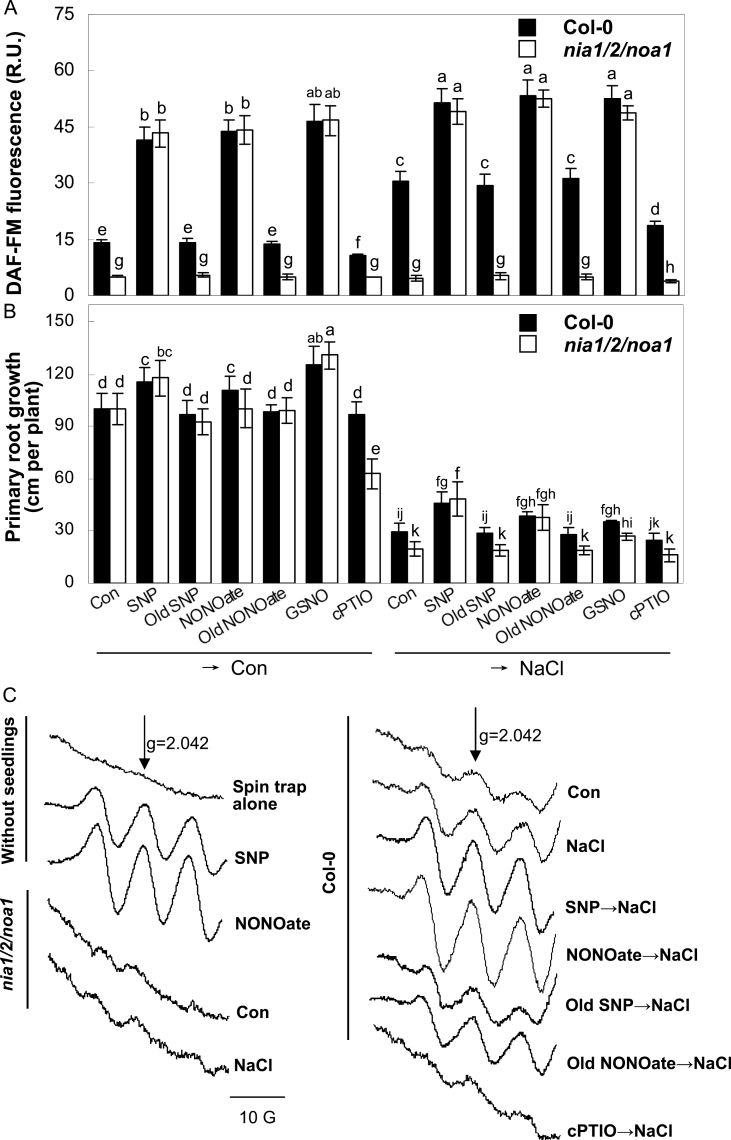
To verify the possibility that NIA1/2 and NOA1 were closely associated with NO production, the responses of endogenous NO levels in Arabidopsis roots were examined by using a fairly specific NO-fluorescent probe (DAF-FM DA). As expected, compared with the wild-type, the basal or NaCl-induced DAF-FM-associated fluorescence levels were gradually reduced in the roots of noa1, nia1/2, and even nia1/2/noa1 plants (Fig. 1A and Supplementary Fig. S8). It was further suggested that the detected fluorescence was specifically associated with the NO production because: (1) three types of NO-releasing compounds, namely SNP, NONOate, and GSNO, could obviously induce DAF-FM-associated fluorescence in both the wild-type and nia1/2/noa1; (2) Old SNP and Old NONOate, two degradation products of NO-releasing compounds, had no such inducible effects on the fluorescence intensity; and (3) the induced fluorescence detected in NaCl-treated roots was significantly reduced in the presence of (cPTIO), an NO scavenger (Fig. 1A).
Fig. 1.
NIA1/2/NOA1-dependent NO production and Arabidopsis salt tolerance. (A) DAF-FM fluorescence in wild-type and nia1/2/noa1 mutant seedling roots detected by LSCM after preincubation in the absence or presence of 1 µM SNP, Old SNP, NONOate, Old NONOate, or GSNO, or 200 µM cPTIO for 1h and then treatment with or without 150mM NaCl for another 1h. (B) Primary root growth of wild-type and nia1/2/noa1 mutant seedlings after preincubation as indicated in Fig. 1A for 5 d and then treatment with or without 150mM NaCl for 7 d. (C) NO content determined by electron paramagnetic resonance. After preincubation in the absence or presence of 10 µM SNP, Old SNP, NONOate, Old NONOate, or 400 µM cPTIO for 1h, wild-type and nia1/2/noa1 mutant seedlings were treated with or without 150mM NaCl for another 1h. A spin trap containing 10 µM SNP or 10 µM NONOate was used as positive controls. Signals were recorded at identical settings. Data are mean ± SE of three independent experiments. Means were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within different treatments [model: phenotypic indicators = replication + treatment + sample (treatment)]. Bars with different letters denote a significant difference according to multiple comparison (P < 0.05).
It has been proposed that DAF might measure other reactive species in addition to NO (Mur et al., 2011). To verify that the above-mentioned fluorescence changes were directly related to NO itself, analysis using EPR (Fig. 1C) and Griess reagent (Supplementary Fig. S9) were performed. The data presented were also in parallel with those of DAF-FM-associated fluorescence, further indicating that the DAF-FM-associated fluorescence, at least in these experimental conditions, was correlated with the NO production.
Subsequently, it was observed that the NaCl-induced growth inhibition of wild-type, noa1, nia1/2, and nia1/2/noa1 plants were significantly rescued by the pretreatment with SNP, suggesting the potential role of NO in Arabidopsis salt tolerance (Fig. 1B and Supplementary Fig. S10). Further experiments revealed that three NO-releasing compounds markedly alleviated NaCl-triggered primary root growth inhibition. In contrast, no significant rescuing responses were observed when the degradation products of SNP (Old SNP, NO2 –/NO3 –, and K3Fe(CN)6/K4Fe(CN)6) or NONOate (Old NONOate) were applied (Fig. 1B and Supplementary Fig. S11). The treatment with SNP or GSNO alone produced similar positive responses in both the wild-type and nia1/2/noa1 mutants under non-stressed conditions. A slight but no significant aggravation of NaCl-triggered primary root growth inhibition was observed by the pretreatment with cPTIO. The reversal behaviours of SNP and NONOate in the NaCl-triggered inhibition of seed germination appeared both in the wild-type and in particular the nia1/2/noa1 mutant (Supplementary Fig. S12). Together, these findings suggested that NIA1/2/NOA1-dependent NO production might mediate Arabidopsis salt tolerance. Therefore, the nia1/2/noa1 mutant line was used for further study.
The nia1/2/noa1 mutant shows the compensatory and decay modes of HY1, antioxidant gene expression after short- and long-term salt treatment
Quantitative real-time PCR was used to monitor the transcript levels of HY1, the antioxidant enzyme, and related transcription factors. As a control, expression analysis of RD29A and RAB18, two well-known stress responsive genes (Lozano-Juste and Leόn, 2010), was also performed.
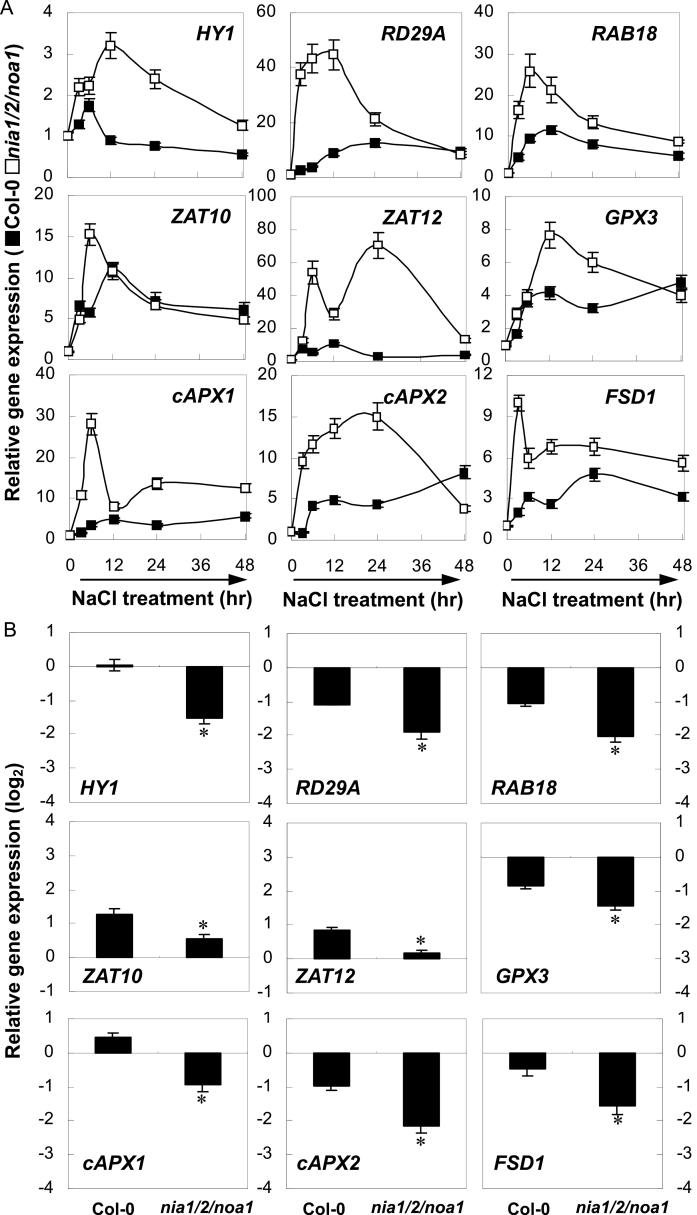
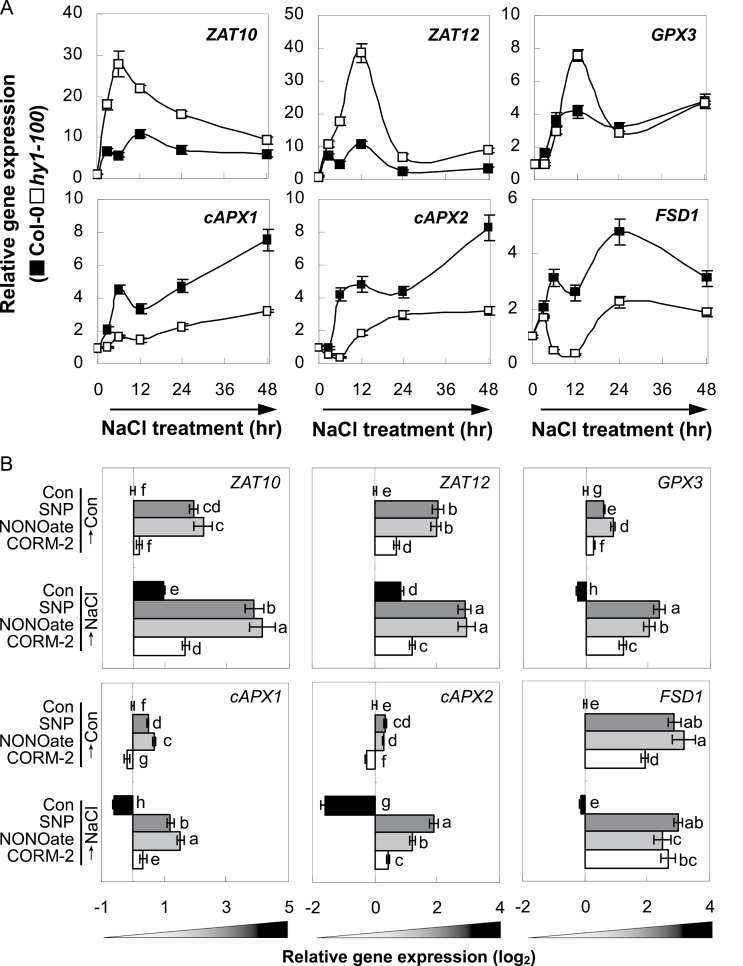
The results showed that, during a short treatment period (the first 48h of NaCl treatment; Fig. 2A), HY1 transcription in triple-mutant plants initially increased and reached a maximum level at 12h of treatment, and then decreased steadily until 48h. The upregulation of RD29A and RAB18 gene expression occurred earlier than HY1, and their induction was much higher than HY1 transcription. Further results showed that the accumulation of several antioxidant genes, including GPX3, cAPX1, cAPX2, and FSD1, were induced to different degrees, and with different kinetics, upon salt stress exposure in nia1/2/noa1 mutant plants, with strong upregulation of cAPX1 and cAPX2. In triple-mutant plants was also noticed the biphasic induction of ZAT12, which encodes a transcription factor that regulates cAPX1 expression (Miller et al., 2008), with a primary weak peak at 6h, followed by a gradual decrease and reaching another stronger peak at 24h. The transcript levels of ZAT10, which encodes a transcription factor responsible for cAPX2 and FSD1 expression (Miller et al., 2008), increased 6h after incubation in NaCl solution, followed by a decreasing tendency until 48h. However, the induction of transcription in wild-type plants was differentially delayed or blocked.
Fig. 2.
Short-term compensation (A) and long-term decay (B) of NaCl-stress-related gene expression. The expression levels of representative genes in wild-type and nia1/2/noa1 mutant seedlings over 48h (A) or at 7 d (B) of 150mM NaCl treatment were analysed by real-time RT-PCR. Expression levels were presented as values relative to corresponding untreated control samples at 0h (A), or 7 d (B) after normalization to actin2/7 levels. Data are mean ± SE of three independent experiments. Bars with asterisks are significantly different compared with the wild-type at P < 0.05 (t-test).
Contrasting results appeared after long-term salt treatment (7 d of NaCl treatment; Fig. 2B). For instance, with respect to their untreated controls, HY1 and cAPX1 expression was elevated by 2 and 39% in the the wild-type subject to salinity stress, respectively, but both correspondingly reduced by 65 and 48% in the nia1/2/noa1 mutants, indicating that NIA1/2/NOA1 might be, at least partially, required for proper expression of HY1 and cAPX1 after long-term treatment. Interestingly, the weaker induction of ZAT10 and ZAT12 was observed in nia1/2/noa1 mutants. Combined with the significant downregulation of antioxidant genes, these results supported the hypothesis that ZAT10 and ZAT12 were required for cAPX2 and FSD1, and cAPX1 expression, respectively, during salinity-induced oxidative stress.
Salinity hypersensitive responses of nia1/2/noa1 mutants are reversed by CO
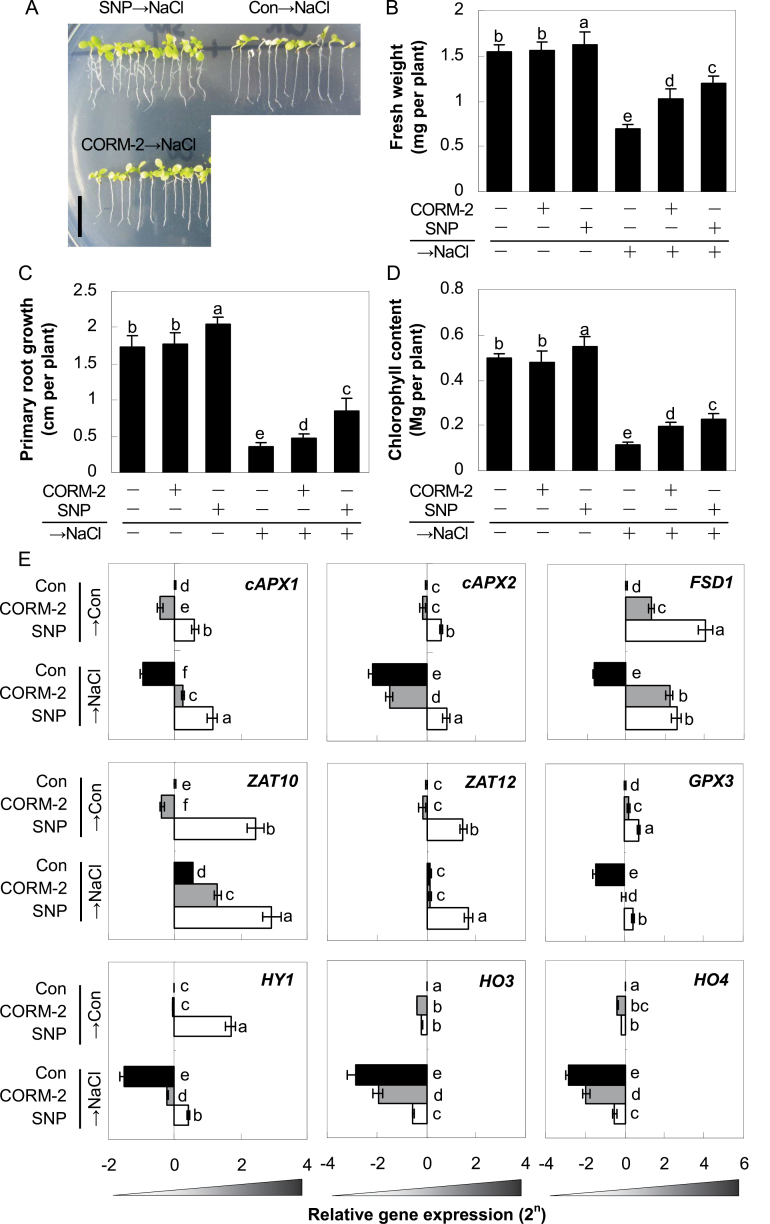
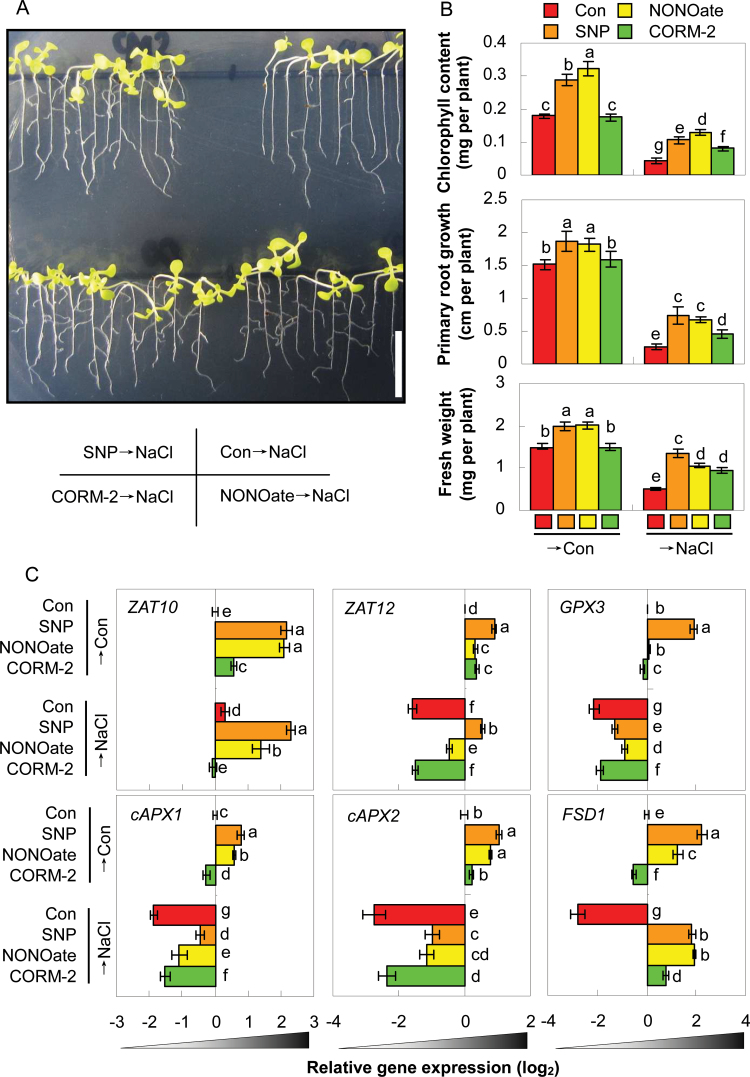
Previously, pharmacological evidence has supported interactions between NO and CO in salt tolerance (Xie et al., 2008) and the adventitious rooting process (Xuan et al., 2008). Thus, the current study characterized this possibility with respect to salinity response at the genetic and molecular levels. As expected, pretreatment with the CO-releasing compound, CORM-2, as well as SNP (in particular; as a positive control), marginally alleviated the decreases of fresh weight, primary root growth, and chlorophyll contents in nia1/2/noa1 mutants caused by the following salinity stress (Fig. 3A–D). However, there were no obvious differences in the non-stressed mutants regardless of whether CORM-2 was present or not. Interestingly, pretreatment with SNP alone brought about positive responses. It was also noticed that, with the pretreatment with Old CORM-2, the degradation product of CORM-2, which contained no CO gas, had no influence upon the growth status of nia1/2/noa1 plants in both control and the salinity stress treatment (Supplementary Fig. S13).
Fig. 3.
Effects of CO- and NO-releasing compounds on morphology (A), fresh weight (B), primary root growth (C), chlorophyll content (D), and representative stress-related genes (E) in NO-deficient mutant upon salt stress. nia1/2/noa1 mutant plants were pretreated for 5 d with or without 0.1 µM CORM-2 or 1.0 µM SNP and then grown with or without 150mM NaCl for 7 d. Bar = 1cm (A). The expression levels of representative genes analysed by real-time RT-PCR. Expression levels are presented as values relative to corresponding untreated control samples, after normalization to actin2/7 levels. Data are mean ± SE of three independent experiments. Bars with different letters denote a significant difference compared with the control according to multiple comparison (P < 0.05).
Molecular evidence showed that upon salinity stress, expression of the corresponding genes was altered compared with normal growth conditions (Fig. 3E). Moreover, the pretreatment with CORM-2 or SNP (especially) could differentially weaken or block the downregulation of cAPX1, cAPX2, FSD1, and GPX3, and strengthen the upregulation of ZAT10 gene expression in nia1/2/noa1 mutants, compared to salinity stress alone. Surprisingly, the expression of ZAT12 was unaffected by CORM-2 pretreatment, but was clearly induced by SNP. Meanwhile, NaCl treatment led to decreased HY1, HO3, and HO4 expression, all of which were weakened or blocked by the application of CO- and in particular NO-releasing compounds. Therefore, these data indicate that salinity hypersensitivity and related gene expression patterns of nia1/2/noa1 mutants were reversed or blocked differentially by CO (a synergistic mode).
Contrasting phenotypes of loss- and gain-of-function mutations of HY1 are sensitive to NO
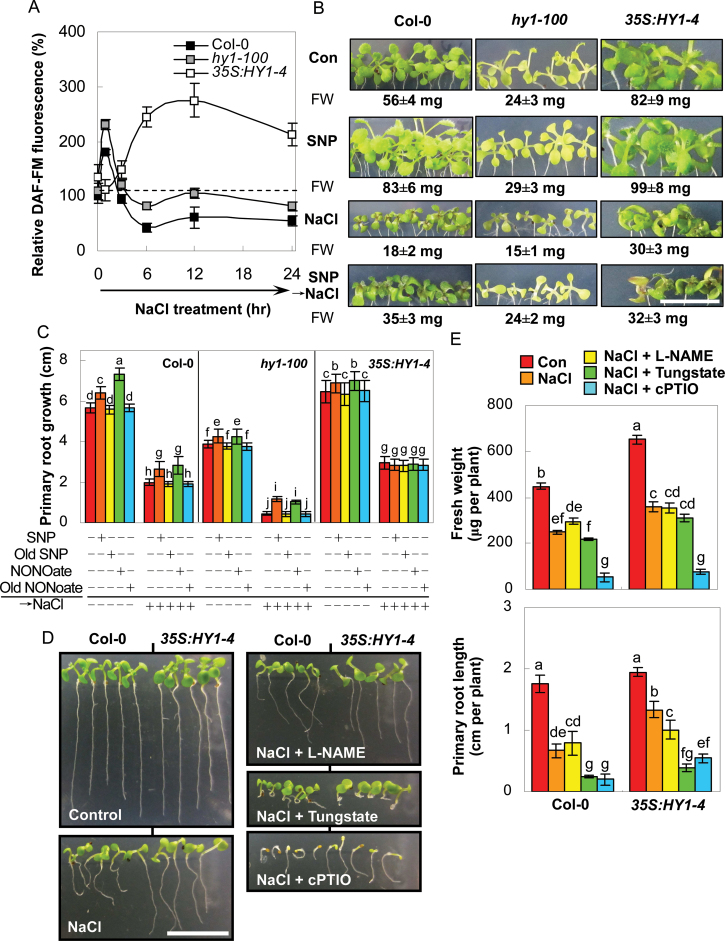
To further investigate the link between HY1 and NO during salt tolerance, HY1 loss-and-gain mutants were used. Compared with the wild-type plants, a similar but stronger induction of NO appeared after 1h of salt stress treatment in the HY1 mutant (hy1-100), as detected by the DAF-associated fluorescence and the EPR assay (Fig. 4A and Supplementary Fig. S14). Interestingly, the decreased tendency of DAF-FM level during the remainder of the monitored period was somewhat attenuated in the hy1-100 mutant, compared to the wild-type, although both were below the basal level (dashed line). Comparatively, salt stress brought about stronger but delayed fluorescence accumulation in 35S:HY1-4, a previously proven gain-of-function in a mutant of HY1 (Xie et al., 2011b).
Fig. 4.
Contrasting responses of HY1 loss-and-gain mutants were sensitive to NO. (A) Time course of 150mM NaCl-induced DAF-FM fluorescence in root tips of the wild-type, hy1-100 mutant, and HY1 overexpression line 35S:HY1-4 seedlings as detected by LSCM. (B, C) Morphology and total fresh weight (B), and primary root growth (C) of wild-type, hy1-100, and 35S:HY1-4 plants pretreated for 5 d with or without 1.0 µM SNP (B), or 1.0 µM SNP, Old SNP, NONOate, or Old NONOate (C) and then grown with or without 150mM NaCl for 7 d. Bar = 1cm (B). (D, E) Morphology (D) and fresh weight and primary root length (E) of stratified seeds of wild-type and 35S:HY1-4 plants grown in the presence or absence of 150mM NaCl with or without 200 µM l-NAME, tungstate, or cPTIO, respectively for 7 d. Bar = 1cm (D). Data are mean ± SE of three independent experiments. Means were compared by a multiple comparison on the basis of the nested ANOVA, with genotypes nested within different treatments [model: phenotypic indicators = replication + treatment + sample (treatment)]. Bars with different letters denote a significant difference according to multiple comparison (P < 0.05).
A subsequent experiment showed that compared with wild-type plants, the hy1-100 mutant exhibited salinity hypersensitivity, as validated by the observed inhibition of seedling growth and changes in primary root length. Contrasting responses were observed in the HY1 overexpression line. Pretreatment of two hy1 mutant alleles with two NO-releasing compounds, SNP or NONOate, significantly alleviated salt stress symptoms (Fig. 4B, C and Supplementary Fig. S15). However, no significant changes were observed for the pretreatment with Old SNP or Old NONOate (Fig. 4C). It should be noted that there was no obvious difference in 35S:HY1-4 plants, regardless of whether NaCl was present together with or without NO-releasing compounds.
The alleviating effects of NO-releasing compounds on salt-triggered growth inhibition of the two hy1 mutant alleles suggested that these phenotypes may result from the reductions of endogenous NO concentrations. To test this hypothesis, the effects of a NO scavenger cPTIO, a mammalian NOS inhibitor (l-NAME), and a plant NR inhibitor (tungstate) on salt tolerance of the HY1 overexpression line were evaluated. l-NAME had no obvious influence on the fresh weight of the wild-type and 35S:HY1-4 plants upon NaCl stress, whereas the application with tungstate resulted in significant reduced primary root growth (Fig. 4D, E). Meanwhile, there was a slight but non-significant decrease in fresh weight after the addition of tungstate to the stressed wild-type and overexpression line. Combined with the maximum inhibition caused by cPTIO, it was postulated that changes in endogenous NO might be involved in hy1-mediated salt hypersensitivity and that NR-mediated NO production plays a major role in this process (a synergistic mode).
hy1 mutant also shows the compensatory and decay modes of antioxidant gene expression after short- or long-term salt treatment
During the first 48h after NaCl treatment, although there was rapid induction of ZAT10, ZAT12, and GPX3 in hy1-100 mutant, the downregulation of several antioxidant genes, including cAPX1, cAPX2, and FSD1, were modulated to different degrees and with various kinetics (Fig. 5A). The data obtained indicated that ZAT10- and ZAT12-triggered transcriptional induction of corresponding antioxidant genes during a short treatment period was mostly dependent on HY1- and/or its catalytic products.
Fig. 5.
Short-term and long-term effects of NaCl on antioxidant enzymes and related transcription factors gene expression. (A) Time course analysis of gene expression in wild-type and hy1-100 mutant seedling roots upon 150mM NaCl. (B) Effect of pretreatment with SNP, NONOate, or CORM-2 on expression of antioxidant genes and related transcription factors in hy1-100 mutant seedlings upon NaCl stress. Seedling plants were pretreated for 5 d with or without 1.0 µM SNP or NONOate, or 0.1 µM CORM-2 and then grown with or without 150mM NaCl for 7 d. Gene expression was analysed by real-time RT-PCR and relative expression levels are presented as values relative to corresponding control samples at the indicated time after normalization to actin2/7 levels. Data are mean ± SE of three independent experiments. Bars with different letters denote a significant difference compared with the control according to multiple comparison (P < 0.05).
After long-term salt treatment, the upregulation of ZAT10 and ZAT12 or the downregulation of GPX3, cAPX1, cAPX2, and FSD1 were aggravated or blocked by SNP and NONOate (both in particular), and CORM-2 in hy1-100 mutants (Fig. 5B), consistent with the reversing effects of salinity hypersensitivity (Fig. 4B, C; Xie et al., 2011b). Meanwhile, under the control conditions, expression of these genes was induced differentially in mutant plants pretreated with NO- (in particular) or CO-releasing compounds, except for the downregulation of cAPX1 and cAPX2 transcripts upon CORM-2 pretreatment.
Salt hypersensitivity phenotypes of NO/HY1-deficient quadruple mutant was blocked by NO and CO
To gain insight into the compensatory and synergistic modes of NR/NOA1-dependent NO and HY1 signal transduction, a quadruple mutant nia1/2/noa1/hy1-100 was constructed and verified by PCR-based genotyping and Western blot analysis (Supplementary Figs. S2 and S3). The nia1/2/noa1/hy1-100 mutant plants displayed delayed vegetative growth but promoted reproductive growth throughout its whole life cycle (Supplementary Figs. S4 and S5), for example their true leaves developed significantly later and smaller than those in wild-type plants, but their bolting and flowering times were largely accelerated, both of which were as a result of additive effects from nia1/2/noa1 and hy1-100 mutant phenotypes. Comparatively, this mutant showed increased salinity hypersensitivity compared with its parental lines (Supplementary Table S3), which could be differentially reversed by the pretreatment with SNP or NONOate (both in particular), or CORM-2 (Fig. 6A, B). Interestingly, only NO-releasing compounds could obviously induce mutant seedling growth in the absence of NaCl. No significant changes were observed for the pretreatment with their degradation byproducts, respectively (Supplementary Figs. S13 and S16).
Fig. 6.
Rescuing effects of NO/CO-releasing compounds on quadruple mutant nia1/2/noa1/hy1-100 plants. Plants were pretreated for 5 d with or without 1.0 µM SNP or NONOate, or 0.1 µM CORM-2 and then grown with or without 150mM NaCl for 7 d. (A) Morphology; bar = 1cm. (B) Corresponding chlorophyll content, primary root growth, and fresh weight. (C) Expression levels of representative genes analysed by real-time RT-PCR, presented as values relative to corresponding control samples after normalization to actin2/7 levels. Data are mean ± SE of three independent experiments. Bars with different letters denote a significant difference compared with the control according to multiple comparison (P < 0.05).
Analysis of antioxidant gene expression in NO/HY1-deficient quadruple mutant
Further molecular evidence showed that pretreatment with SNP and NONOate could differentially weaken or block the downregulation of ZAT12, GPX3, cAPX1, cAPX2, and FSD1 or strengthen the upregulation of ZAT10 gene expression in the quadruple mutant subjected to NaCl treatment for 7 d. Comparatively, no response or only weak responses were found with CORM-2 pretreatment (except ZAT10). This finding perfectly matches the changes in corresponding phenotypes (Fig. 6A, B). Upon normal growth conditions, compared to untreated control plants, expression of the some of these genes was differentially upregulated, although GPX3, cAPX1, and FSD1 transcripts were inhibited by CORM-2.
Discussion
Arabidopsis hy1-100, nia1/2/noa1, and nia1/2/noa1/hy1-100 mutants exhibit progressive salt hypersensitivity
In this study, Arabidopsis hy1, nia1/2/noa1, and nia1/2/noa1/hy1 mutants exhibited progressive salt hypersensitivity, as evaluated by primary root growth inhibition and fresh weight loss (compared with Figs. 1, 3, 4 and, 6; also shown in Supplementary Table S3).
First, the results (Fig. 4 and Supplementary Fig. S7) confirmed the previous findings that hy1-100 and noa1 plants were more salt-sensitive than the wild-type (Zhao et al., 2007; Xie et al., 2011b). It was also observed that the nia1/2/noa1 mutant was more salinity sensitive than its parental lines, as determined by changes in seed germination and survival rates (Supplementary Figs. S6 and S7), the former of which was confirmed previously by Lozano-Juste and Leόn (2010). It was further observed that salt treatment failed to trigger NO production in nia1/2/noa1, as evaluated by the combination of DAF-FM fluorescence, EPR, and Griess reagent assays (Fig. 1A, C and Supplementary Fig. S9). Therefore, it was deduced that both NR and NOA1 were responsible for salt-induced NO production. Except for a rapid burst of stimulation, NO production in the hy1-100 mutant, however, was always below the basal level during 24h of salt stress (Fig. 4A).
Subsequent results illustrated that the salinity-induced inhibition of primary root growth was relieved by 33, 156, 141, and 185% in SNP-pretreated wild-type, hy1-100, nia1/2/noa1, and nia1/2/noa1/hy1-100 mutants, respectively (Figs. 1B, 3C, 4C, and 6B). There were similar rescuing effects triggered by the pretreatments with other NO-releasing compounds, NONOate and GSNO. Meanwhile, the degradation byproducts of SNP and NONOate produced no such effects (Figs. 1B and 4C and Supplementary Figs. S11 and S16). In contrast, the gain-of-function mutant of HY1 (35S:HY1-4) not only led to stronger but delayed NO production, but also displayed tolerance characteristics, which were reversed by the NR inhibitor tungstate and the NO scavenger cPTIO in particular, and slightly by the mammalian NOS inhibitor l-NAME (Fig. 4). These results further demonstrated that NIA1/2-dependent NO production mainly mediated Arabidopsis salt tolerance. The additive response in salt hypersensitivity of the quadruple mutant nia1/2/noa1/hy1-100 (Supplementary Table S3) also supported this proposition.
The reduced NO accumulation in the noa1 mutant has been ascribed to its inability to accumulate carbon reserve (sucrose and fumarate, etc); and application of 1% sucrose could rescue its pale green phenotypes (Van Ree et al., 2011). By contrast, a recent study showed that sucrose levels did not contribute to the noa1-mediated restoration of ssi2-triggered defence phenotypes (Mandal et al., 2012). Here, it was confirmed that, compared with wild-type seedlings, the noa1 mutant displayed severe inhibition of fresh weight under control growth conditions with 1% sucrose (Supplementary Fig. S7; 38 versus 24% compared with the results of Van Ree et al., 2011). These discrepancies may be explained by the differences in growth conditions, including temperature, light, and nitrogen supplementation. Exogenous application of nitrogen was essential for culturing NR-null mutants (Zhao et al., 2009). In this study, all ecotypes were sown in identical plates containing nitrogen. Notably, the nia1/2/noa1 mutant was severely impaired in both basal and salinity-induced NO synthesis, and additive hypersensitive phenotypes were observed (Fig. 1 and Supplementary Figs. S6 and S7). Consistently, NIA1/2- and NOA1-mediated pathways of NO biosynthesis were also additive in terms of abscisic acid-mediated inhibition of germination and seedling establishment regardless of the addition or not of sucrose (Lozano-Juste and Leόn, 2010). Strikingly, cold acclimation induced greater freezing tolerance of the noa1 mutant than wild-type plants (Zhao et al., 2009). Thus, the detailed mechanism of NOA1 in the regulation of NO synthesis and its physiological roles are yet to be fully investigated.
Compensatory and synergistic modes between NIA/NR/NOA1-dependent NO production and HY1 expression
The relationship between NO and HO1/CO, in a number of animal responses, was usually synergistic, complementary, or independent (Otterbein et al., 2003; Bauer et al., 2008; Pae et al., 2010). Since NO and HO1/CO in plants usually have similar physiological roles (Noriega et al., 2007; Xie et al., 2008; Xuan et al., 2008; Santa-Cruz et al., 2010), it is most likely that decreased NO production in plants subjected to environmental stimuli may be complemented by HO1/CO, and vice versa.
In this study’s subsequent experiment, although the nia1/2/noa1 mutant exhibited a more severe hypersensitive response than its parental lines (Supplementary Fig. S3, S4), this triple mutant displayed significantly increased ability to maintain the redox homeostasis – indicated by the upregulation of HY1, GPX3, cAPX1/2, FSD1, and ZAT10/12 transcription after short-term salt treatment (Fig. 2A). Therefore, it was deduced that although the systemic signalling role of NO is associated with plant salt tolerance, the NO-deficient mutants could increase expression of HY1 and other specific defence genes, at least temporarily, to compensate for the lack of NO. This conclusion was confirmed by the fact that NaCl failed to trigger NO accumulation in nia1/2/noa1 roots (Fig. 1A, C). A similar compensatory mode was observed for ZAT10, ZAT12, and GPX3 expression in the hy1-100 mutant (Fig. 5A). Bonifacio et al. (2011) also reported a compensatory response, showing that rice mutants double-silenced for cytosolic ascorbate peroxidase (APx1/2s) upregulated other peroxidases, which enabled the rice plants to cope with a number of stresses. Similarly, our previous reports demonstrated that cadmium-induced upregulation of HO1 gene expression was associated with glutathione depletion, leading to transiently enhanced antioxidant capability (Cui et al., 2011).
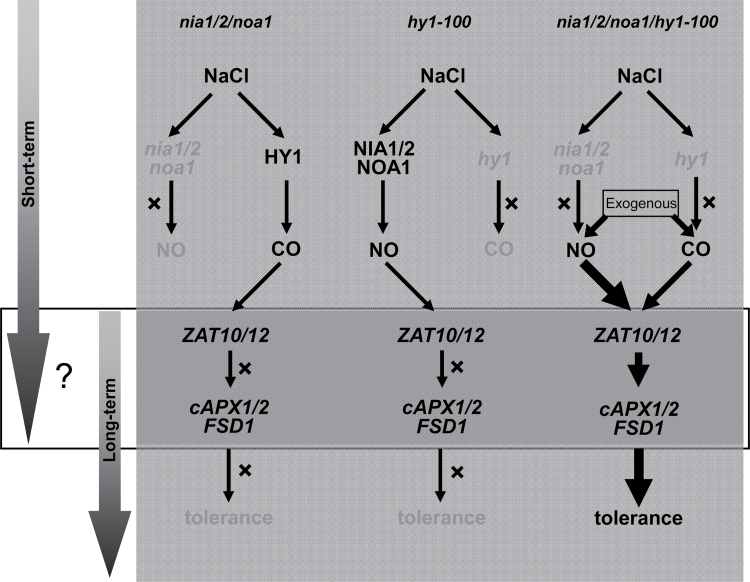
Normally, plants contain a highly efficient antioxidant defence system that can scavenge/detoxify ROS, thereby increase the salt tolerance (Ashraf, 2009). After long-term salt treatment, the majority of antioxidant gene transcripts were diminished significantly (Figs. 2B and 5B), consistent with corresponding salt hypersensitivity phenotypes (Figs. 1B and 4B, C). These results further illustrated that the temporary compensatory mode of antioxidant defence induction could not make up for the deficiency of NO or HY1 to withstand long-term saline conditions (Fig. 7). Thus, the results confirmed that NO and HY1 were beneficial and essential for plant adaptation to salt stress and that disturbed NO production and HY1 expression reduced plant’s long-term adaptation to salt stress. Similarly, previous studies showed that most antioxidants transcripts/activities were increased or decreased upon short- or long-term zinc stress, separately (Xu et al., 2010); the content of a series of amino acids, also increased during short-term NaCl stress, but decreased with long-term treatment periods (Kim et al., 2007).
Fig. 7.
Schematic representation of the proposed compensatory and synergistic modes of NIA/NR/NOA1-dependent NO production and HY1 expression in salt-tolerance signalling pathway. The relay of short-term signalling and long-term salt adaptation was mediated by unknown mechanisms/factors (question mark). The ZAT10/12-mediated antioxidant defence might be involved in the overlapping. The mutation of NIA/NR/NOA1 or HY1 was accompanied with the compensated upregulation of HY1 expression or NIA/NR/NOA1-associated NO production. However, the short-term compensatory mode followed by the downregulation of antioxidant defence system (cAPX1/2 and FSD1, etc.) was not sufficient for long-term salt tolerance. Hypersensitivity and corresponding parameters could be partially reversed by NO- (in particular) or CO-releasing compounds, a synergistic mode. In the NO/CO-deficient quadruple mutant, the application of NO- (in particular) or CO-releasing compounds could differentially trigger ZAT10/12-mediated long-term antioxidant defence, thus leading to the salt-tolerance phenotype.
There was also a synergistic mode, for example pretreatment of nia1/2/noa1 mutants with the CORM-2 significantly alleviated salt hypersensitivity phenotypes (Fig. 3A–D). Molecular evidence further showed that CORM-2 could differentially rescue the downregulation the expression of HO1 subfamily, antioxidant defence genes, and related transcription factors (Fig. 3E). There were similar rescuing responses in hy1-100 mutant plants with the addition of NO-releasing compounds (Figs. 4B, C, and 5B). Meanwhile, the salt-tolerant phenotype of 35S:HY1-4 was significantly arrested when NO production was scavenged or inhibited, whereas no additive effects were found when SNP was added (Fig. 4B–E). Finally, the aggravated hypersensitivity symptom and changes in corresponding genes of quadruple mutant nia1/2/noa1/hy1-100 could be differentially rescued by NO- and CO-releasing compounds, with SNP displaying the maximum responses (Fig. 6). Meanwhile, the degradation byproducts of NO- and CO-releasing compounds produced no such effects (Supplementary Figs. S13 and S16). These results, together with the observations that SNP pretreatment exhibiting the maximal rescuing responses in all mutant ecotypes, suggests that NO might occupy the dominant position in the interaction between NIA/NR/NOA1-dependent NO production and HY1 expression in mediating salt-tolerance signalling (Fig. 7).
How do NIA/NR/NOA1-dependent NO and HY1 work in mediating salt-tolerance signalling?
Transcriptional factor ZAT10/12 was shown to play a key role in modulating plant defence responses (Mittler, 2002; Davletova et al., 2005b; Mittler et al., 2006). The ZAT10/12-dependent increase of Arabidopsis tolerance to abiotic stresses could be attributed to the specific activation of ROS-related defence genes, including cAPX1, cAPX2, and FSD1 (Miller et al., 2008). Additionally, glutathione peroxidase3 (GPX3) was found to act as a ROS scavenger and specifically relay the H2O2 signal as an oxidative signal transducer (Miao et al., 2006). Molecular evidence illustrated that expression levels of the majority of ZAT10/12 and related ROS-scavenging enzymes in nia1/2/noa1, hy1-100, and nia1/2/noa1/hy1-100 mutants were positively regulated by pretreatment with NO/CO-releasing compounds – consistent with the alleviation symptoms (Figs. 1B and 3–6). These findings supported that ZAT10/12-associated ROS metabolism, together with the GPX3, were correlated with the NO/HY1-mediated long-term salt tolerance (Fig. 7).
The opposite induction mode of ZAT10/12 and cAPX1/2 in the hy1-100 mutant during short-term salt stress is noteworthy (Fig. 5A). These results were consistent with this study group’s previous reports showing that HY1 is involved in the induction of cAPX1 expression during salt acclimation (Xie et al., 2011b). In fact, ZAT12 was shown to be required for cAPX1 expression in response to oxidative stress (Rizhsky et al., 2004; Davletova et al., 2005a). Overexpression of ZAT12 did not induce cAPX1 transcript level, further indicating a complex relationship between ZAT12 and cAPX1, and an additional factor(s) may be required for the cAPX1 expression (Rizhsky et al., 2004). Interestingly, the expression of ZAT12, controlling a regulon of 42 different genes involved in abiotic stress, was elevated in a cAPX1-knockout mutant during several abiotic stresses (Davletova et al., 2005a). This study further hypothesizes that ZAT12 might be crucial in relaying short-term compensation to long-term tolerance through a complex signalling network, and HY1 as well as other unknown factor(s) might be involved in the ZAT12-regulated cAPX1 expression (Fig. 7). Further genetic evidence is required to confirm this hypothesis.
In summary, this report provides, as far as is known, the first mechanistic description of the link between Arabidopsis HY1 and NIA/NR/NOA1-dependent NO and their functions in salt tolerance and antioxidant defence gene expression. Thus, the ongoing analysis of the role of other HY1/NO target genes should open new window in the understanding of salt signalling.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Morphology, chlorophyll content, and survival rate of stratified seeds of wild-type and nia1/2 mutant plants grown with or without nitrate and ammonium for 20 d.
Supplementary Fig. S2. Genotyping of the nia1/2/noa1 and nia1/2/noa1/hy1-100 mutants.
Supplementary Fig. S3. Detection of HY1 protein level in 5-d-old seedlings of wild-type, hy1-100, Ler, hy1, noa1, nia1/2, nia1/2/noa1, and nia1/2/noa1/hy1-100 by Western blot analysis.
Supplementary Fig. S4. Developmental phenotypes of wild-type, hy1-100, nia1/2/noa1, and nia1/2/noa1/hy1-100 plants at the early seedling stage.
Supplementary Fig. S5. Developmental phenotypes of wild-type, nia1/2/noa1, hy1-100, and nia1/2/noa1/hy1-100 plants at the vegetative and reproductive growth stages.
Supplementary Fig. S6. Dose-dependent inhibition of seed germination in wild-type, noa1, nia1/2, and nia1/2/noa1 induced by increasing NaCl concentrations for 8 d.
Supplementary Fig. S7. Fresh weight and survival rate of stratified seeds of wild-type, noa1, nia1/2, and nia1/2/noa1 with or without 150mM NaCl treatment for 20 d.
Supplementary Fig. S8. DAF-FM-associated fluorescence in seedling roots of wild-type, noa1, nia1/2, and nia1/2/noa1.
Supplementary Fig. S9. Determination of NO content in wild-type and nia1/2/noa1 seedlings by Griess reagent assay.
Supplementary Fig. S10. Effect of SNP on growth of wild-type, noa1, and nia1/2 plants.
Supplementary Fig. S11. Effect of NO2 –/NO3 –, K3Fe(CN)6/K4Fe(CN)6 and NaCl on growth of wild-type seedlings.
Supplementary Fig. S12. Germination rate of wild-type and nia1/2/noa1 mutant seeds with or without 150mM NaCl and with or without 1 µM SNP or 1 µM NONOate for 3 d.
Supplementary Fig. S13. Effects of CORM-2 and Old CORM-2 on salt hypersensitivity phenotypes of nia1/2/noa1 and nia1/2/noa1/hy1-100 mutant plants.
Supplementary Fig. S14. Determination of NO content in wild-type, hy1-100, and 35S:HY1-4 seedlings by electron paramagnetic resonance.
Supplementary Fig. S15. Effects of NONOate and SNP on growth and chlorophyll contents of hy1 mutant plants.
Supplementary Fig. S16. Effects of fresh and old NO-releasing compounds on salt hypersensitivity phenotypes of nia1/2/noa1/hy1-100 mutant plants.
Supplementary Table S1. Primers for genotyping.
Supplementary Table S2. Primers for real-time RT-PCR.
Supplementary Table S3. Comparative analysis of seedling growth inhibition of wild-type, hy1-100, nia1/2/noa1, and nia1/2/noa1/hy1-100 mutants grown with 150mM NaCl stress for 7 d.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant no. 31170241) and the key International S&T Cooperation Programme between Italy and China (2010). The authors sincerely thank Prof Da Xing and Dr Aizhen Sun from South China Normal University, China, for their kind suggestions in EPR assay. They also thank Dr Evan Evans from the University of Tasmania, Australia for his kind help in editing the manuscript.
References
- Ashraf M. 2009. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnology Advances 27, 84–93. [DOI] [PubMed] [Google Scholar]
- Bauer M, Huse K, Settmacher U, Claus RA. 2008. The heme oxygenase–carbon monoxide system: regulation and role in stress response and organ failure. Intensive Care Medicine 34, 640–648. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Badger MR, Jones RL. 2004. Apoplastic synthesis of nitric oxide by plant tissues. The Plant Cell 16, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilban M, Haschemi A, Wegiel B, Chin BY, Wagner O, Otterbein LE. 2008. Heme oxygenase and carbon monoxide initiate homeostatic signaling. Journal of Molecular Medicine 86, 267–279. [DOI] [PubMed] [Google Scholar]
- Bonifacio A, Martins MO, Ribeiro CW, Fontenele AV, Carvalho FE, Margis-Pinheiro M, Silveira JA. 2011. Role of peroxidases in the compensation of cytosolic ascorbate peroxidase knockdown in rice plants under abiotic stress. Plant, Cell and Environment 34, 1705–1722. [DOI] [PubMed] [Google Scholar]
- Bose J, Xie Y, Shen W, Shabala S. 2013. Haem oxygenase modifies salinity tolerance in Arabidopsis by controlling K+ retention via regulation of the plasma membrane H+-ATPase and by altering SOS1 transcript levels in roots. Journal of Experimental Botany 64, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. 2006. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. The Plant Journal 45, 113–122. [DOI] [PubMed] [Google Scholar]
- Cao ZY, Geng BB, Xu S, Xuan W, Nie L, Shen WB, Liang YC, Guan RZ. 2011. BnHO1, a haem oxygenase-1 gene from Brassica napus, is required for salinity and osmotic stress-induced lateral root formation. Journal of Experimental Botany 62, 4675–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ. 2010. Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis . Plant Physiology 154, 810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Ding X, Xu S, Wang R, Xuan W, Cao ZY, Chen J, Wu HH, Ye MB, Shen WB. 2009. Endogenous hydrogen peroxide plays a positive role in the upregulation of heme oxygenase and acclimation to oxidative stress in wheat seedling leaves. Journal of Integrative Plant Biology 51, 951–960. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK. 2004. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. Journal of Experimental Botany 55, 225–236. [DOI] [PubMed] [Google Scholar]
- Crawford NM, Galli M, Tischner R, Heimer YM, Okamoto M, Mack A. 2006. Response to Zemojtel et al: plant nitric oxide synthase: back to square one. Trends in Plant Science 11, 526–527. [Google Scholar]
- Cui W, Fu G, Wu H, Shen W. 2011. Cadmium-induced heme oxygenase-1 gene expression is associated with the depletion of glutathione in the roots of Medicago sativa . Biometals 24, 93–103. [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. 2005a. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis . The Plant Cell 17, 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. 2005b. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis . Plant Physiology 139, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M. 2005. NO news is good news for plants. Current Opinion in Plant Biology 8, 390–396. [DOI] [PubMed] [Google Scholar]
- Desmard M, Foresti R, Daqoussat M, et al. 2012. Differential antibacterial activity against pseudomonas aeruginosa by carbon monoxide-releasing molecules. Antioxidants and Redox Signaling 16, 153–163. [DOI] [PubMed] [Google Scholar]
- Dulak J, Józkowicz A. 2003. Carbon monoxide – a ‘new’ gaseous modulator of gene expression. Acta Biochimica Polonica 50, 31–47. [PubMed] [Google Scholar]
- Ederli L, Morettini R, Borgogni A, Wasternack C, Miersch O, Reale L, Ferranti F, Tosti N, Pasqualini S. 2006. Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiology 142, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arnandis I, Guillén MI, Gomar F, Casteión MA, Alcaraz MJ. 2011. Control of cell migration and inflammatory mediators production by CORM-2 in osteoarthritic synoviocytes. PLoS One 6, e24591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L. 2001. Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiology 126, 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinke N. 2010. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. The Biochemical Journal 425, 425–434. [DOI] [PubMed] [Google Scholar]
- Guo FQ. 2006. Response to Zemojtel et al: plant nitric oxide synthase: AtNOS1 is just the beginning. Trends in Plant Science 11, 527–528. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. 2011. On the origins of nitric oxide. Trends in Plant Science 16, 160–168. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Stoimenovem M, Kaiser WM. 2005. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ . Journal of Experimental Botany 420, 2601–2609. [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang J, Chen XY, Gao ZZ, Xuan W, Xu S, Ding X, Shen WB. 2008. Carbon monoxide alleviates cadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa . New Phytologist 177, 155–166. [DOI] [PubMed] [Google Scholar]
- Huang X, Stettmaier K, Michel C, Hutzler P, Mueller MJ, Durner J. 2004. Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana . Planta 218, 938–946. [DOI] [PubMed] [Google Scholar]
- Kim JK, Bamba T, Harada K, Fukusaki E, Kobayashi A. 2007. Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. Journal of Experimental Botany 58, 415–424. [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. 2012. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Journal of Experimental Botany 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382. [Google Scholar]
- Liesche J, Schulz A. 2012. In vivo quantification of cell coupling in plants with different phloem-loading strategies. Plant Physiology 159, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Li MY, Cui WT, Lu W, Shen WB. 2012. Haem oxygenase-1 is involved in hydrogen sulfide-induced cucumber adventitious root formation. Journal of Plant Growth Regulation 31, 519–528. [Google Scholar]
- Liu C, Li S, Wang M, Xia G. 2012. A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line. Plant Molecular Biology 78, 159–169. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xu S, Ling T, Xu L, Shen W. 2010. Heme oxygenase/carbon monoxide system participates in regulating wheat seed germination under osmotic stress involving the nitric oxide pathway. Journal of Plant Physiology 167, 1371–1379. [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J, León J. 2010. Enhanced abscisic acid-mediated responses in nia1nia2nia1–2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis . Plant Physiology 152, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal MK, Chandra-Shekara AC, Jeong RD, Yu K, Zhu S, Chanda B, Navarre D, Kachroo A, Kachroo P. 2012. Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis . The Plant Cell 24, 1654–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. 2006. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. The Plant Cell 18, 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. 2008. Reactive oxygen signaling and abiotic stress. Physiologia Plantarum 133, 481–489. [DOI] [PubMed] [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler R, Kim YS, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. 2006. Gain- and loss-of-function mutations of Zat10 enhance the tolerance of plants to abiotic stress. FEBS Letters 580, 6537–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LAJ, Mandon J, Cristescu SM, Harren FJM, Prats E. 2011. Methods of nitric oxide detection in plants: a commentary. Plant Science 181, 509–519. [DOI] [PubMed] [Google Scholar]
- Noriega GO, Yannarelli GG, Balestrasse KB, Batlle A, Tomaro ML. 2007. The effect of nitric oxide on heme oxygenase gene expression in soybean leaves. Planta 226, 1155–1163. [DOI] [PubMed] [Google Scholar]
- Otterbein LE, Soares MP, Yamashita K, Bach FH. 2003. Heme oxygenase-1: unleashing the protective properties of heme. Trends in Immunology 24, 449–455. [DOI] [PubMed] [Google Scholar]
- Pae H, Son Y, Kim NH, Jeong HJ, Chang KC, Chung HT. 2010. Role of heme oxygenase in preserving vascular bioactive NO. Nitric oxide 23, 251–257. [DOI] [PubMed] [Google Scholar]
- Planchet E, Gupta KJ, Sonoda M, Kaiser WM. 2005. Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. The Plant Journal 41, 732–743. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. 2004. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis . Journal of Biological Chemistry 279, 11736–11743. [DOI] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. 2002. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro . Journal of Experimental Botany 53, 103–110. [PubMed] [Google Scholar]
- Santa-Cruz DM, Pacienza NA, Polizio AH, Balestrasse KB, Tomaro ML, Yannarelli GG. 2010. Nitric oxide synthase-like dependent NO production enhances heme oxygenase up-regulation in ultraviolet-B-irradiated soybean plants. Phytochemistry 71, 1700–1707. [DOI] [PubMed] [Google Scholar]
- Shekhawat GS, Verma K. 2010. Haem oxygenase (HO): an overlooked enzyme of plant metabolism and defence. Journal of Experimental Botany 61, 2255–2270. [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. 2009. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiology 151, 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhamsu J, Lee GI, Klessig DF, Crane BR. 2008. The structure of YqeH. An AtNOS1/AtNOA1 ortholog that couples GTP hydrolysis to molecular recognition. Journal of Biological Chemistry 283, 32968–32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Nie S, Xing D. 2012. Nitric oxide-mediated maintenance of redox homeostasis contributes to NPR1-dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiology 160, 1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi V, Lamattina L, Cassia R. 2009. An increase in the concentration of abscisic acid is critical for nitric oxide-mediated plant adaptive responses to UV-B irradiation. New Phytologist 181, 871–879. [DOI] [PubMed] [Google Scholar]
- Uchida A, Jagendorf AT, Hibino T, Takabe T, Takabe T. 2002. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Science 163, 515–523. [Google Scholar]
- Van Ree K, Gehl B, Chehab EW, Tsai YC, Braam J. 2011. Nitric oxide accumulation in Arabidopsis is independent of NOA1 in the presence of sucrose. The Plant Journal 68, 225–233. [DOI] [PubMed] [Google Scholar]
- Wu JY, Qu HY, Shang ZL, Tao ST, Xu GH, Wu J, Wu HQ, Zhang SL. 2011a. Reciprocal regulation of Ca2+-activated outward K+ channels of Pyrus pyrifolia pollen by heme and carbon monoxide. New Phytologist 189, 1060–1068. [DOI] [PubMed] [Google Scholar]
- Wu MZ, Huang JJ, Xu S, Ling TF, Xie YJ, Shen WB. 2011b. Haem oxygenase delays programmed cell death in wheat aleurone layers by modulation of hydrogen peroxide metabolism. Journal of Experimental Botany 62, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Cui WT, Yuan XX, Shen WB, Yang Q. 2011a. Heme oxygenase-1 is associated with wheat salinity acclimation by modulating reactive oxygen species homeostasis. Journal of Integrative Plant Biology 53, 653–670. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Ling TF, Han Y, et al. 2008. Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defence in wheat seedling roots. Plant, Cell and Environment 31, 1864–1881. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Mao Y, Lai Diwen, Zhang Wei, Shen WB. 2012a. H2 enhances Arabidopsis salt tolerance by manipulating ZAT10/12-mediated antioxidant defence and controlling sodium exclusion. PLoS One 7, e49800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Xu DK, Cui WT, Shen WB. 2012b. Mutation of Arabidopsis HY1 causes UV-C hypersensitivity by impairing carotenoid and flavonoid biosynthesis and the down-regulation of antioxidant defence. Journal of Experimental Botany 63, 3869–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DK, Yang Q, Shen WB. 2011b. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. The Plant Journal 66, 280–292. [DOI] [PubMed] [Google Scholar]
- Xu J, Yin H, Li Y, Liu X. 2010. Nitric oxide is associated with long-term zinc tolerance in Solanum nigrum . Plant Physiology 154, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wang L, Zhang B, et al. 2012. RNAi knockdown of rice SE5 gene is sensitive to the herbicide methyl viologen by the down-regulation of antioxidant defense. Plant Molecular Biology 80, 219–235. [DOI] [PubMed] [Google Scholar]
- Xuan W, Zhu FY, Xu S, Huang BK, Ling TF, Qi JY, Ye MB, Shen WB. 2008. The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiology 148, 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y. 2000. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Letters 468, 89–92. [DOI] [PubMed] [Google Scholar]
- Yannarelli GG, Noriega GO, Batlle A, Tomaro ML. 2006. Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta 224, 1154–1162. [DOI] [PubMed] [Google Scholar]
- Zemojtel T, Fröhlich A, Palmieri MC, et al. 2006. Plant nitric oxide synthase: a never-ending story? Trends in Plant Science 11, 524–525. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang F, Guo J, Yang Y, Li B, Zhang L. 2004. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiology 134, 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Chen L, Zhang LL, Zhang WH. 2009. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis . Plant Physiology 151, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Tian QY, Zhang WH. 2007. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis . Plant Physiology 144, 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2000. Genetic analysis of plant salt tolerance using Arabidopsis . Plant Physiology 124, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2001. Plant salt tolerance. Trends in Plant Science 6, 66–71. [DOI] [PubMed] [Google Scholar]
- Zhu JK. 2002. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.