Abstract
Grafting is particularly important to the cultivation of perennial crops such as grapevine (Vitis vinifera) because rootstocks can provide resistance to soil-borne pests and diseases as well as improve tolerance to some abiotic stresses. Successful grafting is a complex biochemical and structural process beginning with the adhesion of the two grafted partners, followed by callus formation and the establishment of a functional vascular system. At the molecular level, the sequence of events underlying graft union formation remains largely uncharacterized. The present study investigates the transcriptome of grapevine rootstock and graft interface tissues sampled 3 d and 28 d after grafting of over-wintering stems in the spring. Many genes were differentially expressed over time, from 3 d to 28 d after grafting, which could be related to the activation of stem growth and metabolic activity in the spring. This hypothesis is supported by the up-regulation of many genes associated with cell wall synthesis, and phloem and xylem development. Generally, there was an up-regulation of gene expression in the graft interface tissue compared with the rootstock, particularly genes involved in cell wall synthesis, secondary metabolism, and signalling. Although there was overlap between the genes differentially expressed over time (from 3 d to 28 d after grafting) with the gene differentially expressed between the rootstock and the graft interface, numerous graft interface-specific genes were identified.
Key words: Gene expression, grafting, grapevine, perennial plant, spring, stem.
Introduction
Grafting is widely used in horticulture (e.g. in the cultivation of grapevine, apples, Prunus spp., and vegetables) to aid plant adaptation to potentially deleterious biotic and abiotic soil conditions and/or to control the vigour of the shoot (e.g. Lee et al., 2010). In viticulture, grafting is primarily used in Europe to facilitate grapevine cultivation in soils infected with the phylloxera, a soil-dwelling insect pest introduced to Europe from America at the end of the 19th century.
Successful grafting is a complex biochemical and structural process that begins with the adhesion of the two grafted partners, followed by callus formation and the establishment of a functional vascular system (as reviewed by Pina and Errea, 2005). At the molecular level, graft union formation presumably requires considerable re-programming of gene expression, protein translation, and metabolism, but the sequence of events underlying graft union formation remains largely uncharacterized. Gene expression changes during the process of graft union formation have been analysed in hickory (Carya cathayensis) with cDNA amplified fragment length polymorphism (AFLP) 3, 7, and 14 d after grafting, and this work revealed that some genes related to indole-3-acetic acid (IAA), cell cycle, metabolism, and signal transduction are differentially expressed (Zheng et al., 2009).
Graft union development in Arabidopsis has been recently studied at the histological and transcriptional level ~24h after grafting, and graft union development was shown to involve wound and hormone signalling as well as the clearing of cellular debris (Yin et al., 2012). However, grafting in horticulture is often performed on dormant over-wintering stems in which the development of the graft union and the reactivation of growth in the spring occur in parallel.
During the autumn, perennial plants enter a phase of dormancy in which metabolic activity is minimized and growth is apparently absent. Environmental signals (light and temperature) play a central role in regulating this activity–dormancy cycle in perennial plants. Hormone signalling has been shown to be involved, such as changes in concentrations of and/or sensitivity to abscisic acid (ABA), auxin, and gibberellic acid (Wareing, 1958; Lachaud, 1989; Baba et al., 2011). The transition between active growth and dormancy involves changes at the cellular, physiological, anatomical, and metabolic levels; initially cell growth ceases, then cold hardiness develops and storage compounds accumulate. Temperature is the main factor triggering spring reactivation of growth, and this growth is fuelled by the catabolism of the accumulated storage compounds (e.g. Druart et al., 2007).
The present work analyses the genes differentially expressed in the rootstock and the graft interface tissues 3 d and 28 d after grafting in grapevine in autografted plants; the objective of this work was to study the genes differentially expressed in the graft interface zone during graft union formation. Because grafting was performed on over-wintering lignified stems, many genes were also differentially expressed over time (i.e. from 3 d to 28 d after grafting) in both the rootstock and graft interface tissue. The differential expression of genes over time in both the graft interface and rootstock tissues is presumably related to the reactivation of stem growth in the spring. Overlap between the genes differentially expressed between the graft interface and rootstock, and regulation of gene expression over time suggests that some similar processes may be involved. However, many interesting genes specifically differentially expressed in the graft interface tissue were also identified.
Materials and methods
Two independent grafting experiments were done in 2011 and 2012; the samples from 2011 were used for microarray and quantitative real-time (q) PCR analysis, whereas the samples from 2012 were only used for qPCR analysis.
Plant material
Vitis vinifera cv. ‘Cabernet-Sauvignon N’ hardwood was collected in January from a vineyard in Bordeaux (France) and stored in a cold room (4 °C) until grafting in March. Dormant stems were taken out of the cold room 2 d before grafting and soaked in water at room temperature in order to rehydrate. One-bud cuttings were made for scions and two-node de-budded cuttings were made for rootstocks shortly before grafting. Mechanical omega grafting was performed on scion/rootstock pairs of approximately the same diameter. The Cabernet Sauvignon autografts (Cabernet Sauvignon grafted with Cabernet Sauvignon) were briefly dipped into melted wax and placed into humid sawdust-filled boxes for callusing at 28 °C.
RNA extraction
Three pools of 15 graft interface zones (the omega shape shown in Fig. 1A and B, ~5mm in length that included both scion and rootstock wood tissue) and sections of rootstock wood (~5mm in length and >1.5cm below the graft interface) were harvested 3 d and 28 d after grafting and immediately snap-frozen in liquid nitrogen. Total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich) with some modifications to the first step of the extraction: 20mg ml–1 (w/v) poly(vinylpolypyrrolidone) was added to the lysis buffer and the resulting solution was extracted with chloroform:isoamyl alcohol (24:1, v/v) once; the supernatant was used for the subsequent steps in the protocol according to the manufacturer’s instructions. In some cases, RNA yields were low in the samples harvested 3 d after grafting, therefore requiring the pooling of more than one extraction.
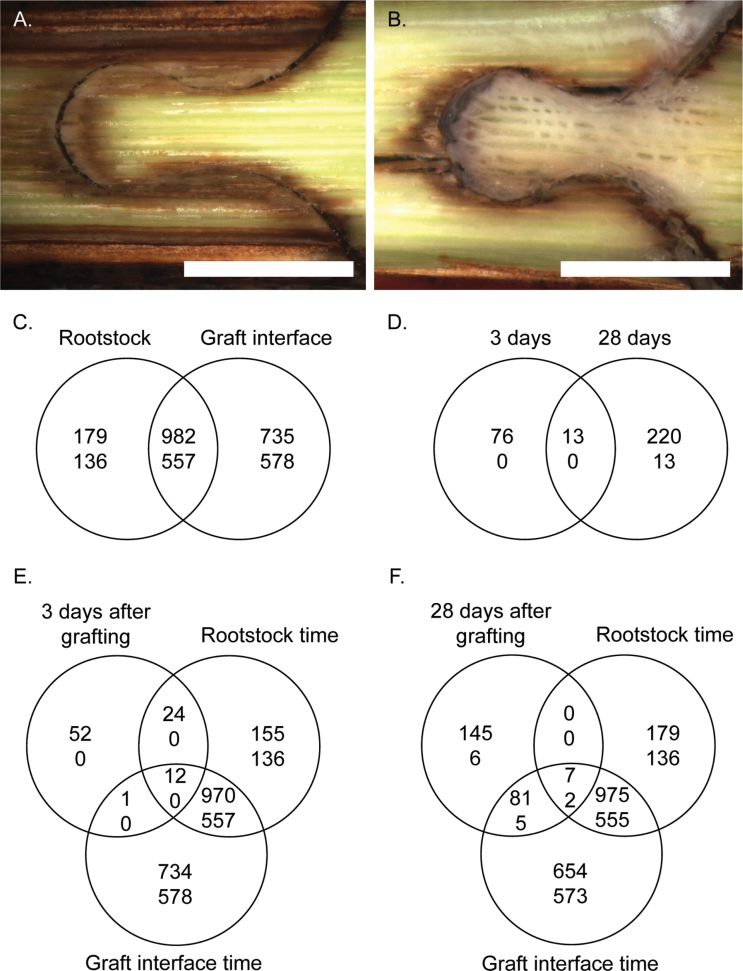
Fig. 1.
Photographs of the graft interface of Vitis vinifera cv. Cabernet Sauvignon grafted with Cabernet Sauvignon (A) 8 d and (B) 29 d after grafting; scale bar=4mm. Venn diagrams showing the genes differentially expressed (C) from 3 d to 28 d after grafting in the rootstock wood tissue and the callus and (D) in the graft interface callus zone compared with the rootstock wood tissue 3 d and 28 d after grafting, and Venn diagrams showing the overlap between the differential expression over time in the rootstock and graft interface tissues from 3 d to 28 d after grafting compared with the genes differentially expressed (E) 3 d and (F) 28 d after grafting. Text thickness indicates the direction of gene expression change, the number of genes up- and down-regulated are represented by bold and normal text, respectively (genes with significant differences were selected with a log fold change >1 and P-value <0.05, adjusted with Holm).
Microarray analysis
The microarrays used were the grape whole-genome microarrays from Nimblegen, Roche (Design name 090918 Vitus exp HX12). The microarray probe design is based on the 12× genome assembly (Jaillon et al., 2007) using the grapevine V1 gene model prediction from Centro Ricerche Interdipartimentale Biotecnologie Innovative, Italy (http://genomes.cribi.unipd.it/). The probe design is available online at the following address: http://ddlab.sci.univr.it/FunctionalGenomics/. The microarray contains 118 015 probes with an average of four probes per gene; the expression of 29 549 genes can be quantified with the microarray. The correspondence between probe identifiers and gene identifiers was obtained from http://genomes.cribi.unipd.it/. The microarray hybridizations were done for the 10 samples (two tissues harvested on two dates with three and two biological replicates for the graft interface and rootstock tissues, respectively) by the Plateforme Biopuces, Institut National des Sciences Appliquées, Toulouse, France according to the manufacturer’s instructions.
The microarray data were analysed using the statistical package R, version 2.14.0 (R Development Core Team, 2005) with various Bioconductor packages (http://www.bioconductor.org, Gentleman et al., 2004). Microarray quality controls were performed using the arrayQualityMetrics package (Kauffmann et al., 2009). Expression intensities were background corrected, quantile-normalized, and summarized using the rma function of the oligo package (Carvalho and Irizarry, 2010). The raw and normalized microarray data are available on http://www.ebi.ac.uk/arrayexpress; the accession number is E-MEXP-3803. Differentially expressed genes were identified using the limma package (Smyth, 2005); genes with absolute log2 fold changes >1 and Holm (Holm, 1979) corrected P-values <0.05 were considered significant.
Differences in gene expression were visualized using MapMan (Thimm et al., 2004; Usadel et al., 2005) and PageMan (Usadel et al., 2006). The MapMan mapping file was obtained from http://www.gomapman.org/; 27 212 of the 29 549 genes on the microarray are present in the mapping file. Enrichments of functional categories of the MapMan annotation in the significantly differentially expressed genes were tested for significance by applying Fisher tests with a Bonferroni correction for multiple tests using Mefisto Version 0.23beta (http://www.usadellab.org). Enrichment of Gene Ontology (GO) terms in significantly differentially expressed genes was evaluated using analysis tool from AgriGO (http://bioinfo.cau.edu.cn/agriGO/, Du et al., 2010) with Fisher tests and Bonferroni multiple testing correction (P < 0.05).
qPCR analysis
For qPCR experiments, genomic DNA contamination was removed from the RNA with the Turbo DNA-free kit from Ambion (according to the manufacturer’s instructions) and reverse transcription was done using the Superscript III kit from Invitrogen [using oligo(dT) primers, 1.5 µg of RNA, and according to the manufacturer’s instructions). Gene expression was analysed on a Biorad CFX96 machine using iQ Sybr Green Supermix (according to the manufacturer’s instructions). The quality (and quantity) of cDNA synthesized was tested using two sets of primers that amplified the 3′ and 5′ regions of the same reference gene (a SAND protein, VIT_06s0004g02820) and genomic DNA contamination was checked by qPCR using intron-specific genes (Supplementary Table S1 available at JXB online). The expression of genes of interest was normalized with VIT_06s0004g02820, and two additional reference genes were used to confirm the stability of expression of VIT_06s0004g02820 (Supplementary Table S1). qPCR expression data are presented as 40–∆Ct with the reference gene VIT_06s0004g02820. PCR efficiency for each primer pair was calculated using LinRegPCR (Ramakers et al., 2003).
Results
The objective of this work was to determine the transcriptional changes associated with graft union formation in over-wintering stems of grapevine 3 d and 28 d after grafting in autografted plants. The callus tissue was absent during the first week after grafting (Fig. 1A), but by 28 d after grafting considerable callus tissue had developed (Fig. 1B). Ten times more genes were differentially expressed over time (from 3 d to 28 d after grafting) than were differentially expressed between the graft interface and rootstock tissues (Fig. 1C, D). This Results section begins with the description of the genes commonly differentially expressed over time in both the rootstock and graft interface tissues, then the genes differentially expressed over time specifically in the graft interface are described, and finally the genes differentially expressed between the graft interface and rootstock tissue are presented. Some of the genes differentially expressed in the microarray data were confirmed by qPCR in the same samples (from 2011) and in an independent experiment in 2012 (Supplementary Fig. S1 at JXB online). The direction of gene expression changes could generally be repeated in the independent experiment; however, the magnitude of change was frequently reduced.
Up-regulation of genes from 3 d to 28 d after grafting
From the third to the 28th day after grafting, the expression of 1161 and 1717 genes was up-regulated in rootstock wood and graft interface tissues, respectively (Fig. 1C); 982 of these genes were up-regulated in both tissues. MapMan functional category and GO term enrichment analysis of the 982 common up-regulated genes identified the coordinated regulation of genes from various functional categories (Supplementary Table S2A, B at JXB online). The following MapMan functional categories were significantly enriched (Supplementary Table S2A): cell, cell wall, cell organization, lipoxygenase genes associated with jasmonate signalling, miscellaneous genes from the functional categories beta 1,3 glucan hydrolases, plastocyanin-like, and gluco-, galacto-, and mannosidases, as well as protein degradation by subtilases, receptor kinases [particularly from the functional category leucine rich repeat (LRR) III], and pathogenesis-related (PR) proteins. The GO terms enriched in the up-regulated genes were the functional categories motor and hydrolase activity and the compartments extracellular and membrane (Supplementary Table S2B).
Examination of the 982 genes up-regulated between 3 d and 28 d after grafting in both the rootstock and graft interface tissues (Supplementary Table S3A at JXB online) shows that some of the most strongly up-regulated genes (~250-fold) were associated with phloem development; for example, the F4P13 genes, which code for proteins required for phloem filament protein formation (e.g. VIT_14s0066g000060, VIT_14s0066g000100, VIT_14s0066g000120, and VIT_14s0066g00070), and VIT_19s0177g00200, the plastocyanin-like grapevine orthologue of At3g20570, which is an Arabidopsis sieve element-specific EARLY NODULIN-LIKE PROTEIN9 (ENOD9) (Khan et al., 2007). Genes associated with the MapMan functional category cell wall were also up-regulated, such as a cell wall–plasma membrane linker protein (VIT_11s0016g05830), a cell wall LRR (VIT_18s0001g10040), and two genes (VIT_08s0058g00480 and VIT_08s0105g00500) similar to the Arabidopsis gene TRICHOME BIREFRINGENCE-LIKE 28, involved in cellulose biosynthesis.
Genes associated with protein degradation such as subtilases (F6N15, VIT_15s0048g01210, and four cucumisin precursor genes, VIT_13s0019g02530, VIT_15s0048g01120, VIT_15s0048g01180, and VIT_13s0019g02490) were also strongly up-regulated. In addition, a serine protease (a carboxypeptidase type III, VIT_06s0061g00890) and two xylem cysteine protease 1 precursors (VIT_18s0122g00790 and VIT_03s0038g04000) were in some of the most highly up-regulated genes.
Genes specifically up-regulated in the graft interface zone from 3 d to 28 d after grafting
A large number of genes (735) were specifically up-regulated in the graft interface tissue from 3 d to 28 d after grafting. The MapMan functional categories cell wall, miscellaneous, and non-assigned were enriched in this gene list (Supplementary Table S4A at JXB online) along with the GO term oxidoreductase (Supplementary Table S4B). One of the most highly up-regulated genes was a SENESCENCE ASSOCIATED GENE 101, SAG101 (VIT_14s0066g01840). Other highly up-regulated genes included transcription factors (e.g. APETALA2_Ethylene-responsive element binding protein family transcription factors VIT_11s0016g03350 and VIT_03s0063g00460), a number of cytochrome P450 genes, genes involved in cell wall modification, one arachidonic acid-induced DEA1 (VIT_02s0154g00280), and four ankyrin repeat family protein-like genes (Supplementary Table S5A). The up-regulation of a DEA1 gene could suggest that phospholipid signalling processes occur at the graft interface from 3 d to 28 d after grafting.
Down-regulation of genes from 3 d to 28 d after grafting
From the third to the 28th day after grafting, the expression of 693 and 1135 genes was down-regulated in the rootstock wood and graft interface tissues, respectively (Fig. 1C); 557 genes were down-regulated in both tissues. The following MapMan functional categories were enriched in the 557 common down-regulated genes (Supplementary Table S2C at JXB online): photosynthesis and jasmonate signalling [namely 12-oxo phytodienoic acid (PDA) reductase]. Examination of the genes down-regulated in the graft interface and rootstock from 3 d to 28 d after grafting (Supplementary Table S3B) shows that the most strongly down-regulated genes (250-fold) were EXPANSIN-LIKE PROTEIN B genes (VIT_00s0309g00090 and VIT_00s1355g00020). Many genes from the functional category non-assigned were down-regulated, as well as genes involved in calcium signalling.
Genes specifically down-regulated in the graft interface tissue from 3 d to 28 d after grafting.
Many genes (578) were specifically down-regulated in the graft interface tissue from 3 d to 28 d after grafting. The MapMan functional categories glycolysis [particularly phosphofructokinase (PPFK)], photosynthesis (particularly light reaction), redox (heme), and wounding were particularly enriched in the down-regulated genes (Supplementary Table S4C at JXB online) in addition to the GO term extracellular region (Supplementary Table S4D). The most highly down-regulated gene was an amino acid transporter (VIT_14s0108g00690); this was followed by a PPFK gene (VIT_02s0109g00080), a peroxidase anionic precursor (VIT_01s0010g02010), and a cytochrome P450 (VIT_14s0006g01670) (Supplementary Table S5B).
Co-ordinated transcriptional re-programming of various functional categories from 3 d to 28 d after grafting
The gene expression changes during the reactivation of metabolic activity and growth over time in both the roostock and graft interface tissues required the differential expression of numerous genes involved in transcriptional regulation (Supplementary Fig. S2 at JXB online), jasmonate synthesis and degradation (Fig. 2A, B), receptor kinases, in particular LRR III receptor kinases (Supplementary Fig. S3), and metabolism (Supplementary Fig. S4).
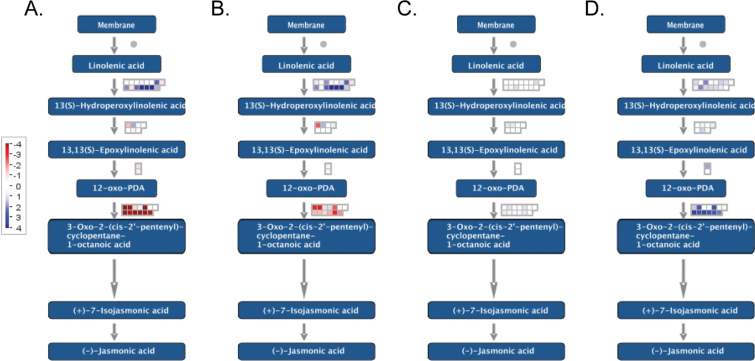
Fig. 2.
MapMan visualization of genes assigned to the functional category jasmonic acid and differentially expressed from 3 d to 28 d after grafting in the (A) rootstock and (B) callus, and between the callus and rootstock (C) 3 d and (D) 28 d after grafting.
Global up-regulation of gene expression in the graft interface compared with the rootstock tissue
The analysis of the genes differentially expressed between the rootstock and the graft interface shows that globally there was an up-regulation of gene expression in the graft interface zone. Eighty-nine and 233 genes were up-regulated 3 d and 28 d after grafting, respectively, whereas no genes were down-regulated 3 d after grafting and only 13 genes were down-regulated 28 d after grafting in the graft interface compared with the rootstock (Fig. 1D). The general up-regulation of gene expression can also been seen in the MapMan images of the functional categories metabolism (Fig. 3), regulation (Supplementary Fig. S5 at JXB online), jasmonate biosynthesis (Fig. 2C, D), and transcription (Supplementary Fig. S6).
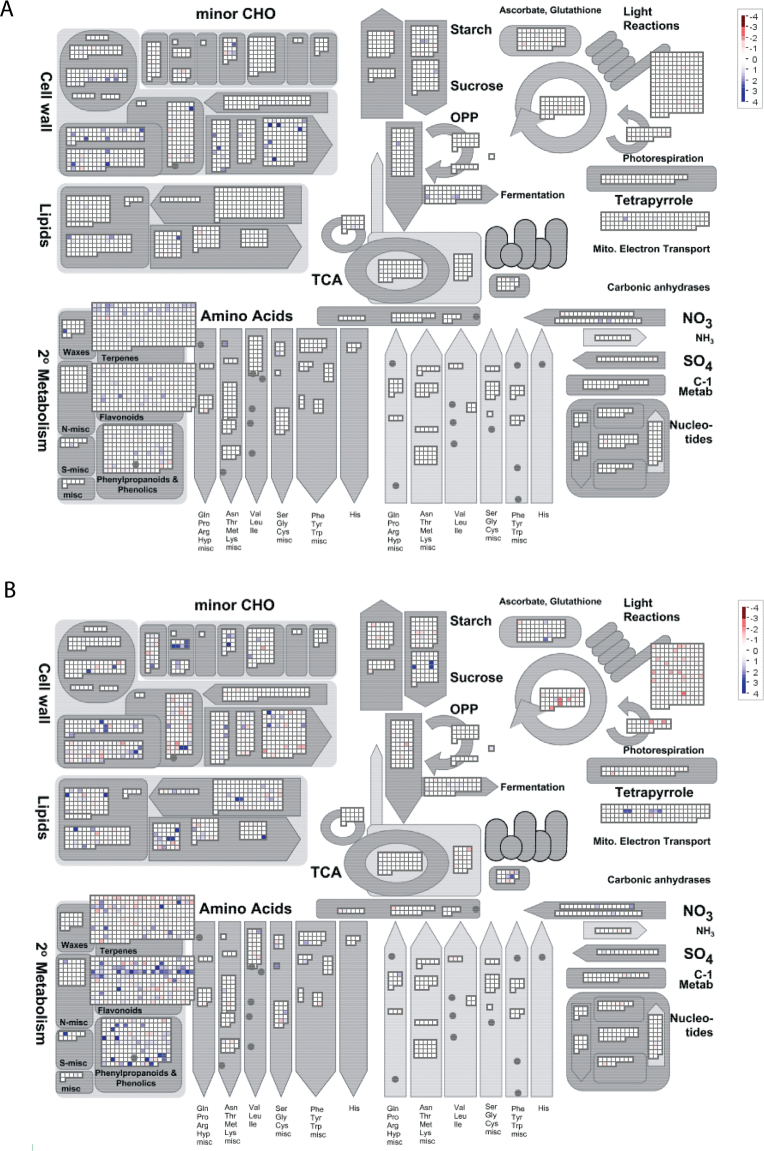
Fig. 3.
MapMan visualization of genes assigned to the functional category metabolism differentially expressed between the graft interface and rootstock (A) 3 d and (B) 28 d after grafting. Squares represent the expression of individual genes from each subcategory; the degree of up- and down-regulated gene expression is shown in shades of blue and red, respectively.
Thirteen genes were up-regulated in the graft interface compared with the rootstock both 3 d and 28 d after grafting; these genes were from a variety of functional groups (Supplementary Table S6 at JXB online). One of the common up-regulated genes was a MYB transcription factor (VIT_19s0014g03820), an orthologue of the Arabidopsis gene MYB-like 102, which is involved in wound and osmotic stress signalling (Denekamp and Smeekens, 2003). The up-regulated late embryogenesis abundant (LEA) protein (LEA 14-A, VIT_15s0046g02110) is the orthologue of the Arabidopsis gene At1g01470, which has a structural similarity to fibronectin type III domains (Singh et al., 2005). Interestingly, in animals, fibronectin domains are involved in cell adhesion, motility, wound healing, and maintenance of cell shape (Singh et al., 2005). However, LEA proteins are generally associated with the cellular dehydration tolerance (Hundertmark and Hincha, 2008).
Genes up-regulated in the graft interface compared with rootstock tissue 3 d after grafting
Three days after grafting, 89 genes were up-regulated in the graft interface zone. The functional categories enriched in this list were cell wall and miscellaneous genes (Supplementary Table S7A at JXB online). The GO terms enriched in the up-regulated genes were associated with the extracellular compartment, namely the extracellular region, external encapsulating structure, and extracellular space (Supplementary Table S7B).
The most strongly up-regulated genes 3 d after grafting were associated with the cell wall (Supplementary Table S8 at JXB online); two pectate lyase genes (VIT_17s0000g09800 and VIT_17s0000g09810) were up-regulated 16-fold, three glycosyltransferases were up-regulated ~10-fold, and one expansin (VIT_06s0004g04860) was up-regulated 8-fold. Germin-like proteins and peroxidises were also among some of the most highly up-regulated genes. In addition, genes coding for an auxin influx carrier protein (VIT_03s0038g02140) and LATERAL ORGAN BOUNDARIES DOMAIN PROTEIN 4 (LBD4; VIT_06s0004g03330) were up-regulated.
Genes up-regulated in the graft interface compared with rootstock tissue 28 d after grafting
In grapevine 28 d after grafting, the callus tissue is normally well developed (Fig. 1B) and the shoot and root have started to grow (data not shown). The functional categories hormone metabolism (particularly related to jasmonate and cytokinin signalling), carbohydrate metabolism, miscellaneous, mitochondrial electron transport, and secondary metabolism were enriched in genes up-regulated 28 d after grafting as well as genes associated with stress, signalling, and the transport of nitrate (Supplementary Table S7C at JXB online). The GO process response to stimulus and the functions receptor, oxidoreductase, molecular transducer, signal transducer, and transferase activity, as well as the extracellular compartment were also enriched (Supplementary Table S7D).
The most highly up-regulated gene in the graft interface compared with the rootstock tissue 28 d after grafting was a cytochrome P450 (VIT_08s0007g04060), followed by a serine protease gene (VIT_08s0040g01130) similar to At1g28110 (SERINE CARBOXYPEPTIDASE-LIKE 45 which has been identified in the cell wall), and the third most up-regulated gene has an unknown function, but contains an LRR domain (VIT_18s0041g02440) (Supplementary Table S9 at JXB online). The up-regulated genes related to cytokinin metabolism were associated with its degradation (VIT_00s2191g00010, VIT_00s2520g00010, and VIT_07s0005g05960), and eight genes coding for Lithospermum erythrorhizon dark-inducible proteins (LeDI-5c) from the functional category jasmonate metabolism (specifically 12-oxo PDA reductase) on chromosome 18 were up-regulated (Fig. 3D; Supplementary Table S7). Many receptor kinases from a variety of classes were also up-regulated: S-locus glycoprotein-like, LRR XI and XII, domain of unknown function 26, receptor-like protein kinase 10-like, cytoplasmic receptor-like kinase, and miscellaneous kinases. In addition, a SAG101 (VIT_14s0066g01830) was up-regulated along with eight glutathione S-transferases and three glycosyltransferases.
Overlap between the functional categories enriched in the genes differentially expressed over time and between the graft interface and rootstock tissues
Globally the expression changes of genes from a number of functional categories were similar during from 3 d to 28 d after grafting and between the graft interface and rootstock, such as the up-regulation of the categories cell wall (particularly 3 d after grafting, Supplementary Table S10A at JXB online), secondary metabolism, jasmonate signalling, and some miscellaneous categories (particularly 28 d after grafting; Supplementary Table S10E) (Fig. 4). However, some functional categories were differently regulated, such as various subcategories of secondary metabolism, RNA, and regulation of transcription (Fig. 4).
Fig. 4.
Pageman visualization of MapMan functional categories enriched in the genes differentially expressed between the callus and rootstock tissue (A) 3 d and (B) 28 d after grafting, and in the (C) rootstock and (D) callus tissue from 3 d to 28 d after grafting. The degree of enrichment of functional categories in up- and down-regulated genes is given by shades of blue and red, respectively.
At the individual gene level, of the 89 genes up-regulated in the graft interface tissue 3 d after grafting, 37 genes were also up-regulated over time (in either or both the graft interface and rootstock tissues from 3 d to 28 d after grafting) and 52 genes were specifically up-regulated in the graft interface tissues (Fig. 1E). The 37 genes up-regulated both over time from 3 d to 28 d after grafting and at the graft interface 3 d after grafting included many genes involved in cell wall synthesis, degradation, and modification, an AUX1-like auxin influx carrier protein (VIT_03s0038g02140) and two legumin-like proteins (VIT_12s0034g01950 and VIT_12s0034g01900). The 52 genes specifically up-regulated in the graft interface tissue 3 d after grafting included a number of peroxidases and berberine bridge enzymes, LEA14-A (VIT_15s0046g02110) and LBD4 (VIT_06s0004g03330) discussed above and genes involved in isoprenoid metabolism (VIT_00s2280g00010 and VIT_00s1350g00030) (Supplementary Table S8 at JXB online).
Of the 233 genes up-regulated in the graft interface tissue 28 d after grafting, 88 were also up-regulated over time and 145 genes were specifically up-regulated in the graft interface tissue (Fig. 1F). The 88 genes up-regulated both in the graft interface 28 d after grafting and over time included many genes involved in carbohydrate metabolism (particularly cell wall invertases), miscellaneous genes, nitrate transporters, a SAG101 (VIT_14s0066g01830), and a number of transcription factors (Supplementary Table S9 at JXB online). The 145 genes specifically up-regulated in the graft interface included many genes associated with hormone signaling, for example cytokinin and jasmonate (particularly 12-oxo PDA reductase) signalling, secondary metabolism (particularly flavonoids and lignin biolsynthesis), abiotic stress, and receptor kinase signalling (Supplementary Table S9).
Thirteen genes were down-regulated in the graft interface 28 d after grafting; seven of these genes were also down-regulated in the graft interface over time and six were specifically down-regulated in the graft interface (Fig. 1F; Supplementary Table S9 at JXB online).
Discussion
The differential expression of genes over time is often the reversal of gene expression changes during the onset of dormancy, suggesting that these changes could be related to the reactivation of growth in the spring
In the autumn, the tissues of woody perennials enter a phase of dormancy in which active growth is (visibly) absent and metabolic activity decreases considerably (as reviewed by van der Schoot and Rinne, 2011). In grapevine, grafting is traditionally (and in this study) performed on re-hydrated over-wintering stems in the early spring, therefore on physiologically dormant material. As such, a large proportion of genes differentially regulated during the grafting processes are presumably related to the reactivation of metabolic activity and growth for the new season of growth. Although there have been many studies of the transition from active growth to dormancy at the transcriptional level (e.g. Schrader et al., 2004; Druart et al., 2007; Galindo Gonzalez et al., 2012), there have been surprisingly few studies of the transcriptional changes that occur in woody tissue during the reactivation of growth that takes place in the first few weeks in the spring. Therefore, there are few studies with which the gene expression changes observed in this work can be compared. However, many of the genes that are up- or down-regulated during the onset of dormancy (e.g. Schrader et al., 2004) show reverse responses from 3 d to 28 d after grafting, such as genes annotated as cytochrome P450, chitinases, LRR-containing proteins, and APETALA2 transcription factors.
Transcriptional re-programming of genes involved with cell wall, phloem, and xylem development during the reactivation of growth in the spring
It appears that many genes involved in cell wall production, cell organization, metabolism, and xylem and phloem development are differentially expressed from 3 d to 28 d after grafting in both the graft interface and rootstock tissues; this could be interpreted as the up-regulation of genes associated with the reactivation of cell division and differentiation in the spring. Surprisingly, only four genes from the MapMan functional category cell cycle were up-regulated from 3 d to 28 d after grafting, suggesting that the increase in cell division in the spring is not strongly regulated at the transcriptional level. This is in agreement with the large absence of differential expression of cell cycle genes during the onset of dormancy in poplar (Schrader et al., 2004) and the absence of correlation between cell division and cell cycle gene expression in the spring in aspen trees (Druart et al., 2007).
The up-regulation of beta 1,3 glucan hydrolases could be associated with the removal of callose from the sieve elements, which allows the phloem to re-establish functional conduits in the spring (as reviewed by van der Schoot and Rinne, 2010). Additionally, the up-regulation of genes encoding phloem filament-associated proteins suggests that new phloem cells were being produced. The up-regulation of subtilases, as well as other classes of proteases, could be related to vascular tissue differentiation and development during the beginning of the spring growth cycle (Petzold et al., 2012). The up-regulation of proteases has been widely reported in transcriptomic studies of xylem differentiation (e.g. Moreau et al., 2005; Friedmann et al., 2007). However, subtilases are known to have wider roles in protein turnover and different aspects of developmental regulation, as well as pathogen and environmental responses (as reviewed by Schaller et al., 2012).
Transcriptional reprogramming of hormone signalling during the reactivation of growth in the spring
The global gene expression changes in the cambium during the onset of dormancy suggest that ethylene, ABA, and light signalling pathways are involved (van der Schoot and Rinne, 2012). However, from 3 d to 28 d after grafting, the jasmonate signalling categories lipoxygenase and 12-oxo-PDA reductase were the only hormone-related categories enriched in the responding genes. The up-regulation of lipoxygenase suggests that the first steps of jasmonate biosynthesis were up-regulated, but the down-regulation of 12-oxo PDA reductase suggests that 12-oxo PDA was the signal molecule synthesized. 12-Oxo PDA has been implicated in wound-induced gene expression (Taki et al., 2005) and in seed germination (Dave et al., 2011) in Arabidopsis. Potentially 12-oxo PDA could have similar functions in seedling germination and the reactivation of growth in stems of grapevine in the spring.
Genes specifically differentially expressed in the graft interface over time suggest that wounding and defence responses are specifically regulated in the graft interface zone from 3 d to 28 d after grafting
Of the genes specifically differentially expressed in the graft interface tissue over time, some merit further attention, for example SAG101 (VIT_14s0066g01840) is specifically up-regulated in the graft interface over time. SAG101 is known to have an important role in programmed cell death and plant innate immunity responses in Arabidopsis (Feys et al., 2005). In addition, the MapMan functional category wounding is enriched in the genes down-regulated specifically in the graft interface from 3 d to 28 d after grafting. This could suggest that by 28 d after grafting the graft interface has partially recovered from the damage caused by the grafting process.
Graft union formation triggered the differential expression of genes involved in wound responses, hormone signalling, and callus maintenance in the graft interface compared with the rootstock tissue
Autografting of over-wintering stems of grapevine resulted in the up-regulation of many genes in the zone of the graft interface compared with the rootstock, whereas autografting in Arabidopsis triggered particularly the down-regulation of gene expression (Yin et al., 2012). Generally wounding is associated with the local up-regulation of gene expression (e.g. Reymond et al., 2000), which is consistent with the up-regulation of gene expression at the graft interface of grapevine. Many of the genes up-regulated in the graft interface compared with the rootstock were associated with defence and wound responses, such as MYB-like 102, germin-like proteins, peroxidases, and chitinases.
Many regulatory genes were up-regulated in the graft interface compared with the rootstock tissue both 3 d and 28 d after grafting, particularly genes involved in calcium signalling, receptor kinase, protein modification and degradation, and hormone signalling. Twenty-eight days after grafting, many genes associated with jasmonate signalling were up-regulated, particularly 12-oxo PDA reductase, potentially suggesting that although 12-oxo PDA may be involved in the reactivation of growth in the spring, jasmonate or methyl jasmonate was involved in graft union formation. Many transcription factors were also up-regulated, suggesting that callus formation requires considerable regulation at the transcriptional level. The up-regulation of an LBD4 gene 3 d after grafting could be related to the formation and maintenance of non-differentiated callus cells as four LBD proteins have been implicated in callus maintenance in Arabidopsis (Fan et al., 2012). The down-regulation of this gene 28 d after grafting is also consistent with this hypothesis because 28 d after grafting the shoot and roots have started to develop and the graft union is functional. The up-regulation of an LEA protein could be related to a local cellular dehydration at the graft interface. However, the transcriptionally responding LEA, LEA 14-A, has structural similarity to animal fibronectin domain proteins, which are involved in cell adhesion, motility, wound healing, and maintenance of cell shape, potentially suggesting a role in graft union formation. Both the LBD and LEA14-A genes were specifically up-regulated in the graft interface tissue and not up-regulated over time (from 3 d to 28 d after grafting) suggesting that their expression is specific to graft union formation. These genes are accompanied by graft interface-specific genes such as genes involved in secondary metabolism, hormone signalling, peroxidases, and abiotic stress responses.
The comparison between gene expression in the graft interface of a woody perennial and a summer annual
In Arabidopsis the biosynthesis of ethylene and jasmonate was identified as important in graft union formation 24h after grafting (Yin et al., 2012), whereas in grapevine, genes associated with cytokinin and jasmonate biosynthesis were only enriched in the up-regulated genes 28 d after grafting. Examination of the genes up-regulated at the graft interface 24h after grafting in Arabidopsis (Yin et al., 2012) and 3 d after grafting in grapevine identifies a number of similar genes in the responding gene lists. Yin et al. (2012) highlighted the role of auxin in graft union formation, and in this work one gene encoding an auxin influx carrier was up-regulated 3 d after grafting in the graft interface zone of grapevine. Furthermore, the Arabidopsis gene MYB-like 102 (At4g21440) was up-regulated 24h after grafting (Yin et al., 2012) and the grapevine homologue (VIT_19s0014g03820) was also up-regulated both 3 d and 28 d after grafting. This could suggest that MYB-like 102 is a key regulator of wound responses (and potentially graft union formation) in a wide range of plant species.
Conclusion
The genes differentially expressed from 3 d to 28 d after grafting in both the rootstock and graft interface tissues could be related to the reactivation of stem growth in the spring; this hypothesis is supported by the up-regulation of many genes involved in cell wall synthesis, and phloem and xylem development. On the other hand, in the graft interface compared with the rootstock, many genes associated with cell wall modification, hormone signalling, secondary metabolism, and wound responses were up-regulated. Traditionally, grafting in perennial crops is done using over-wintering stems in the spring when the reactivation of stem growth and recovery from the grafting process occur in parallel. Several genes are similarly up-regulated at the graft interface and from 3 d to 28 d after grafting, suggesting that some similar mechanisms are involved in graft union development and spring reactivation of growth. However, the majority of genes differentially regulated in the graft interface are specific to graft union formation.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Validation of microarray data (open bars, samples from 2011) by qPCR (filled bars, samples from 2011 and 2012) in the graft interface or rootstock wood tissue 3 d or 28 d after grafting for (A) VIT_08s0007g04060, (B) VIT_11s0016g05830, (C) VIT_17s0000g09810, (D) VIT_19s0177g00200, (E) VIT_08s0040g01130, (F) VIT_14s0128g00540, and (G) VIT_18s0001g06120.
Figure S2. MapMan visualization of the expression of genes from the functional category transcription from 3 d to 28 d after grafting in (A) the rootstock and (B) the graft interface tissues. Squares represent the expression of individual genes from each subcategory; the degree of up- and down- regulated gene expression is shown in shades of blue and red, respectively.
Figure S3. MapMan visualization of genes assigned to the functional category receptor kinases and differentially expressed in (A) the rootstock and (B) the graft interface from 3 d to 28 d after grafting. Squares represent the expression of individual genes from each subcategory; the degree of up- and down- regulated gene expression is shown in shades of blue and red, respectively.
Figure S4. MapMan visualization of genes assigned to the functional category metabolism differentially expressed in (A) the rootstock and (B) the callus from 3 d to 28 d after grafting. Squares represent the expression of individual genes from each subcategory; the degree of up- and down- regulated gene expression is shown in shades of blue and red, respectively.
Figure S5. MapMan visualization of genes assigned to the functional category regulation differentially expressed between the graft interface and rootstock (A) 3 d and (B) 28 d after grafting. Squares represent the expression of individual genes from each subcategory; the degree of up- and down-regulated gene expression is shown in shades of blue and red, respectively.
Figure S6. MapMan visualization of genes assigned to the functional category transcriptional regulation differentially expressed between the graft interface and rootstock (A) 3 d and (B) 28 d after grafting. Squares represent the expression of individual genes from each subcategory; the degree of up- and down-regulated gene expression is shown in shades of blue and red, respectively.
Table S1. Sequence and mean PCR efficiency of primers used for qPCR analysis.
Table S2. Enrichment of MapMan functional categories and Gene Ontology (GO) terms in genes differentially expressed from 3 d to 28 d after grafting in both the graft interface and rootstock tissues: (A) MapMan functional categories in up-regulated genes, (B) GO terms in up-regulated genes, (C) MapMan functional categories in down-regulated genes, and (D) GO terms in down-regulated genes.
Table S3. Genes (A) up- and (B) down-regulated in the graft interface and rootstock tissues from 3 d to 28 d after grafting.
Table S4. Enrichment of MapMan functional categories and Gene Ontology (GO) terms in genes differentially expressed from 3 d to 28 d after grafting only in the graft interface tissue: (A) MapMan functional categories in up-regulated genes, (B) GO terms in up-regulated genes, (C) MapMan functional categories in down-regulated genes, and (D) GO terms in down-regulated genes.
Table S5. Genes (A) up- and (B) down-regulated in the graft interface tissue from 3 d to 28 d after grafting.
Table S6. Genes differentially expressed in the graft interface tissue at both 3 d and 28 d after grafting.
Table S7. MapMan and Gene Ontology (GO) term enrichment in genes up-regulated in the graft interface compared with the rootstock tissue. (A) MapMan enrichment of genes up-regulated 3 d after grafting and (B) GO enrichment of genes up-regulated 3 d after grafting. (C) MapMan enrichment of genes up-regulated 28 d after grafting and (D) GO enrichment of genes up-regulated 28 d after grafting.
Table S8. Genes up-regulated in the graft interface compared with the rootstock 3 d after grafting
Table S9. Genes up-regulated in the graft interface compared with the rootstock 28 d after grafting
Table S10. Enrichment of MapMan functional categories and GO terms in genes differentially expressed between the graft interface and rootstock. (A) MapMan functional categories enriched in genes up-regulated 3 d after grafting and over time (from 3 d to 28 d after grafting). (B) GO terms enriched in genes up-regulated 3 d after grafting and over time. (C) MapMan functional categories enriched in genes up-regulated 3 d after grafting but not up-regulated over time. (D) GO terms enriched in genes up-regulated 3 d after grafting but not up-regulated over time. (E) MapMan functional categories enriched in genes up-regulated 28 d after grafting and up-regulated over time. (F) GO terms enriched in genes up-regulated 28 d after grafting and up-regulated over time. (G) MapMan functional categories enriched in genes up-regulated 28 d after grafting but not up-regulated over time. (H) GO terms enriched in genes up-regulated 28 d after grafting but not up-regulated over time. No MapMan Bins or GO terms were enriched in the genes down-regulated in the graft interface and down-regulated or non-responding over time.
Acknowledgements
The authors thank Bernard Douens, Guillaume Pacreau, Jean-Pierre Petit, Jean-Paul Robert, and the Bordeaux Imaging Centre (Pôle Végétal) for their technical help. This work was supported by a grant from the Institut National de la Recherche Agronomique department Génétique et Amélioration des Plantes.
References
- Baba K, Karlberg A, Schmidt J, Schrader J, Hvidsten TR, Bako L, Bhalerao RP. 2011. Activity–dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proceedings of the National Academy of Sciences, USA 108, 3418–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. 2010. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Hernandez ML, He ZS, Andriotis VME, Vaistij FE, Larson TR, Graham IA. 2011. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. The Plant Cell 23, 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denekamp M, Smeekens SC. 2003. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiology 132, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart N, Johansson A, Baba K, Schrader J, Sjodin A, Bhalerao RR, Resman L, Trygg J, Moritz T, Bhalerao RP. 2007. Environmental and hormonal regulation of the activity–dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. The Plant Journal 50, 557–573. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan MZ, Xu CY, Xu K, Hu YX. 2012. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Research 22, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. 2005. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. The Plant Cell 17, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann M, Ralph SG, Aeschliman D, Zhuang J, Ritland K, Ellis BE, Bohlmann J, Douglas CJ. 2007. Microarray gene expression profiling of developmental transitions in Sitka spruce (Picea sitchensis) apical shoots. Journal of Experimental Botany 58, 593–614. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology 5, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo Gonzalez LM, El Kayal W, Ju CJT, Allen CCG, King-Jones S, Cooke JEK. 2012. Integrated transcriptomic and proteomic profiling of white spruce stems during the transition from active growth to dormancy. Plant, Cell and Environment 35, 682–701. [DOI] [PubMed] [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics 6, 65–70. [Google Scholar]
- Hundertmark M, Hincha DK. 2008. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana . BMC Genomics 9, 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R C, Huber W. 2009. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25, 415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JA, Wang Q, Sjolund RD, Schulz A, Thompson GA. 2007. An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis. Plant Physiology 143, 1576–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaud S. 1989. Participation of auxin and abscisic acid in the regulation of seasonal variations in cambial activity and xylogenesis. Trees-Structure and Function 3, 125–137. [Google Scholar]
- Lee JM, Kubota C, Tsao SJ, Bie Z, Echevarria PH, Morra L, Oda M. 2010. Current status of vegetable grafting: diffusion, grafting techniques, automation. Scientia Horticulturae 127, 93–105. [Google Scholar]
- Moreau C, Aksenov N, Lorenzo MG, Segerman B, Funk C, Nilsson P, Jansson S, Tuominen H. 2005. A genomic approach to investigate developmental cell death in woody tissues of Populus trees. Genome Biology 6, R34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold HE, Zhao MZ, Beers EP. 2012. Expression and functions of proteases in vascular tissues. Physiologia Plantarum 145, 121–129. [DOI] [PubMed] [Google Scholar]
- Pina A, Errea P. 2005. A review of new advances in mechanism of graft compatibility–incompatibility. Scientia Horticulturae 106, 1–11. [Google Scholar]
- R Development Core Team. 2005. R: A language and environment for statistical computing, reference index version 2.8.1. http://www.R-project.org Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. 2000. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. The Plant Cell 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Stintzi A, Graff L. 2012. Subtilases—versatile tools for protein turnover, plant development, and interactions with the environment. Physiologia Plantarum 145, 52–66. [DOI] [PubMed] [Google Scholar]
- Schrader J, Moyle R, Bhalerao R, Hertzberg M, Lundeberg J, Nilsson P, Bhalerao RP. 2004. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. The Plant Journal 40, 173–187. [DOI] [PubMed] [Google Scholar]
- Singh S, Cornilescu CC, Tyler RC, Cornilescu G, Tonelli M, Lee MS, Markley JL. 2005. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Science 14, 2601–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S R, Irizarry WH, eds. Bioinformatics and computational biology solutions using R and Bioconductor New York: Springer; 397–420. [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 2005. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology 139, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, et al. 2006. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7, 535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, et al. 2005. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiology 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schoot C, Rinne PLH. 2011. Dormancy cycling at the shoot apical meristem: transitioning between self-organization and self-arrest. Plant Science 180, 120–131. [DOI] [PubMed] [Google Scholar]
- Wareing P. 1958. Interaction between indole-acetic acid and gibberellic acid in cambial activity. Nature Biotechnology 181, 1744–1745. [DOI] [PubMed] [Google Scholar]
- Yin H, Yan B, Sun J, Jia PF, Zhang ZJ, Yan XS, Chai J, Ren ZZ, Zheng GC, Liu H. 2012. Graft-union development: a delicate process that involves cell–cell communication between scion and stock for local auxin accumulation. Journal of Experimental Botany 63, 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng BS, Chu HL, Jin SH, Huang YJ, Wang ZJ, Chen M, Huang JQ. 2010. cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiology 30, 297–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.