Abstract
For several decades, research into the mechanisms of genetic recombination proceeded without a complete understanding of its cellular function or its place in DNA metabolism. Many lines of research recently have coalesced to reveal a thorough integration of most aspects of DNA metabolism, including recombination. In bacteria, the primary function of homologous genetic recombination is the repair of stalled or collapsed replication forks. Recombinational DNA repair of replication forks is a surprisingly common process, even under normal growth conditions. The new results feature multiple pathways for repair and the involvement of many enzymatic systems. The long-recognized integration of replication and recombination in the DNA metabolism of bacteriophage T4 has moved into the spotlight with its clear mechanistic precedents. In eukaryotes, a similar integration of replication and recombination is seen in meiotic recombination as well as in the repair of replication forks and double-strand breaks generated by environmental abuse. Basic mechanisms for replication fork repair can now inform continued research into other aspects of recombination. This overview attempts to trace the history of the search for recombination function in bacteria and their bacteriophages, as well as some of the parallel paths taken in eukaryotic recombination research.
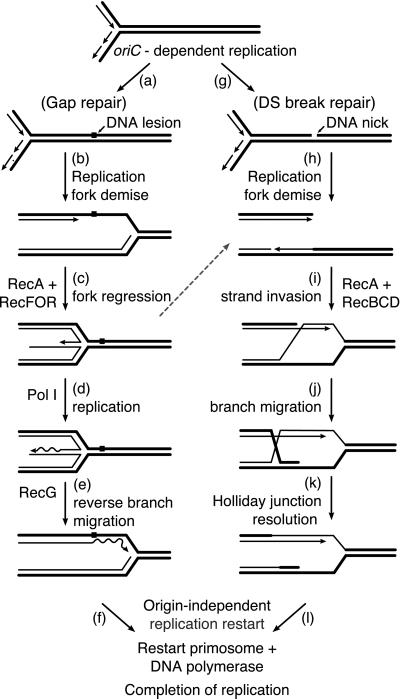
Nonmutagenic replication fork repair can be defined as all of the reactions required to enable a stalled replication fork to restart synthesis, while not compromising genomic integrity (1–4). The progress of a replication fork may be halted by a DNA lesion, a template strand break, a bound protein complex, or other barriers. In many of these instances, repair of the stalled fork requires recombination functions. It recently has become clear that the repair of stalled replication forks is the primary function of bacterial recombination systems. Under normal aerobic growth conditions, most bacterial replication forks undergo recombinational repair at some stage in the replication cycle. The pathways for repair are varied and redundant, reflecting the multitude of DNA structures that might be found at a stalled fork and the importance of nonmutagenic repair. The complete process includes a halt in replication, the action of recombination functions, and the reinitiation of DNA replication (restart) in an origin-independent manner. All of the recombination and replication restart steps are entrusted to enzymatic functions that are both specialized and complex. Some pathways for replication fork repair are concisely illustrated in Fig. 1.
Figure 1.
Pathways for recombinational DNA repair of a stalled replication fork. A pathway involving fork regression is shown for gap repair (a–f), and a double-strand break repair path is shown for the repair of a fork collapsed at the site of a DNA strand break (g–l). The pathways shown are intended to be generic and do not incorporate all current ideas for fork repair. The dashed line represents a pathway in which the Holliday junction is converted to a double-strand break by the action of RuvABC, as observed by Michel and colleagues (180) and others. Arrowheads on DNA strands denote 3′ ends. If a DNA lesion (other than a strand break) is responsible for halting the progress of a replication fork, note that replication fork repair does not entail repair of the lesion itself. Instead, the recombination and replication steps set up the lesion for repair by providing an undamaged complementary DNA strand. The degree to which excision repair and other DNA repair processes are integrated with the recombinational repair pathways is unknown.
This general picture of replication fork repair in bacteria is itself part of a larger convergence in our understanding of recombination and replication in all organisms. Recombination-dependent replication (RDR) is seen in bacterial conjugation or transduction, meiotic recombination in eukaryotes, the repair of replication forks and miscellaneous double-strand breaks in eukaryotes, the life cycle of bacteriophage T4, telomere maintenance in eukaryotic cells lacking telomerase, and many other contexts.
The new awareness of links between replication and recombination was highlighted in a major National Academy of Sciences symposium held Nov. 10–12, 2000 at the Beckman Center in Irvine, California. The breadth of the topic is clear from the talks presented at the conference and the papers in this issue of PNAS (see summary article by Kuzminov, ref. 204). This report is an attempt to outline the historical paths leading to the new focus on the role of recombination at stalled replication forks. I am regularly reminded and readily acknowledge that links between replication and recombination extend well beyond the re-establishment of replication forks. However, nonmutagenic replication fork repair provides a uniquely compelling rationale for the evolution of recombination functions. In a world where frequent nucleic acid damage was prescribed, viable paths for the maintenance of replication forks without sacrificing genome integrity may have been a prerequisite for the evolution of large genomes. Fork repair now also provides a uniquely informative and unifying framework for the discussion of recombination mechanisms in all contexts. Some molecular steps in fork repair pathways, such as DNA strand invasion and RDR, are widespread features of recombination reactions.
Research into recombination mechanisms was originally motivated by the need to understand the genetic information transfers that underpinned so much of the science of genetics. However, it was clear even at the outset that genetic exchange was not the major cellular function of these processes (reviewed in refs. 5–8). Alan Campbell provided a clear definition of the problem in his summary remarks at the 1984 Cold Spring Harbor Symposium on genetic recombination (9):
The function of general recombination has hardly surfaced at this symposium. Perhaps there was nothing constructive to say on the subject. I am left uncertain as to how many investigators in that area consider that the function is too obvious to require discussion, how many think that general recombination serves no useful function, and how many consider the question uninteresting or intractable.
But there is a real question that needs to be answered some time. Our immediate interest in learning that new recombination-deficient mutants have been characterized. (etc.)… derives from our interest in recombinational mechanisms. But we all know that the immediate selective value of those genes and products does not depend on their role in reshuffling genes in natural populations. The typical mutant deficient in general recombination is either sick or dead. Many recombination proteins play essential roles in replication or repair… Even in the case of the highly ordered process of recombination at meiosis, prevention of nondisjunction may be a more direct advantage than recombination itself.
Recent advances in thinking about this topic came from many sources and will be presented as a dozen parallel threads. This number is somewhat arbitrary and could profitably be expanded but for space constraints. Some of the threads were, until recently, independent lines of research. Others were closely related and discussed at the same scientific gatherings, but not put into proper functional context until recently.
Thread 1: The Discovery of Recombination Genes in Bacteria
As the discovery of bacterial conjugation by Lederberg and Tatum in the mid-1940s (10) opened up the field of bacterial genetics, bacterial genetics gave rise to the first insight into the mechanics of the genetic exchanges that accompanied conjugation. The first gene known to be involved in recombination, designated recA, was identified by Clark and Margulies (11, 12). Additional genes soon identified in screens in several laboratories were given the designations recB and recC (13).
In the beginning, sensitivity to DNA damaging agents was not an anticipated property of recombination-deficient mutants (12). However, a comment from Max Delbruck in a seminar and some thought about the uvr mutants that had been isolated by Howard-Flanders led Clark and colleagues (12, 14) to test their first recA mutants for sensitivity to UV irradiation. The dramatic sensitivity of recA mutants to UV light nevertheless came as a surprise (12). Howard-Flanders and Theriot (15) soon demonstrated that recA mutants were also sensitive to x-rays. Thus the links between recombination and DNA repair were established almost immediately after the identification of recombination genes. This eventually contributed to further advances, including hypotheses for postreplication DNA repair (16, 17) and the SOS response (18).
The RecA protein is now known to have three distinct functions important to the continuing discussion. First, RecA protein has a direct role in the recombination process, promoting a class of reactions called DNA strand exchanges. Second, RecA protein promotes the autocatalytic cleavage of certain repressors (particularly LexA protein), a reaction that induces the bacterial SOS response to heavy DNA damage. Finally, RecA protein plays a direct role in the mutagenic translesion synthesis of DNA by DNA polymerase V (19). These different roles have often complicated the interpretations of in vivo experiments using recA mutations.
Thread 2: In Vivo Characterization of Bacterial Recombination Functions
The isolation of recombination genes continues, with dozens of bacterial genes now known to participate (20, 21). Clark (22) went on to contribute the concept of distinct recombination pathways. As more genes were discovered, the link between recombination and DNA repair was strengthened. In most cases, a null mutation in a recombination function results in sensitivity to some or all DNA damaging treatments. Multiple pathways for recombinational repair and functional overlap between the activities of certain recombination proteins initially obscured the importance of some recombination functions. The effects of some single mutant genes (such as recJ, recG, and recQ) on recombination and sensitivity to UV irradiation are modest. However, the effects observed in double mutants are often large and synergistic (23–26). The in vivo study of recombination functions has provided a general appreciation for the complexity of recombination systems in bacteria as well as their importance for survival. Also, some rec− mutants like recF had little effect on conjugation in a wild-type background, but the same recF mutant cell exhibited an enhanced sensitivity to DNA damaging treatments like UV irradiation (27). More recently, genes originally defined as replication functions have been tied to recombination and recombinational DNA repair by these studies (28–31).
Investigation of the in vivo effects of mutations in recombination genes established some intriguing patterns. In general, the results supported the idea that DNA damage was recombinogenic and that recombinational DNA repair was critical for cell survival. Mutations that tended to increase the number of unrepaired DNA strand breaks tend to produce a hyperrec phenotype (32). Cells with such mutations are rendered inviable when they also lack one of the key recombination functions (33–37). DNA strand breaks are thus lethal when the appropriate repair paths are not available. Oxidative damage accounts for the major portion of DNA damage in cells grown aerobically (8, 38–40), and many of these double mutant cells can be grown anaerobically (33, 41).
A better appreciation for the frequency of DNA damage to genomes developed by the early 1990s (42), helping to increase the focus on DNA repair. The gradual shifting of functional emphasis to recombinational DNA repair advanced the discussion of recombination function, but another significant jump was needed to make a link to replication forks.
Thread 3: Early Links Between Replication Forks and Recombinational DNA Repair
Hanawalt (43) was perhaps the first to suggest that replication forks might be halted at the site of a DNA strand break. This theme was elaborated by Skalka (44). Speculating on the apparent replication deficiencies observed in mutants that eliminated the red recombination system of bacteriophage λ, she proposed that a replication fork might collapse upon encounter with a strand break. Going further, she proposed that the resulting double-strand break could be processed by the RecBC enzyme, followed by a DNA strand invasion using mechanisms that had been proposed by Radding just a few years prior (44, 45). In a prescient statement near the end of the article, Skalka also proposed that these general mechanisms might represent a repair pathway to avoid replication disasters in the bacterial genome.
Another proposal for a repair pathway for stalled forks was put forward by Higgins and colleagues 2 years later (46). Working with mammalian cells, those workers found short segments of heavy/heavy DNA shortly after a pulse label of BrdUrd and observed four-pronged replication forks in the electron microscope (the same structures now sometimes called chicken feet; ref. 47). To explain these observations, Higgins et al. (46) proposed that stalled replication forks undergo regression and repair according to a pathway much like the one featured on the left side of Fig. 1.
The chicken foot seems to be a repair intermediate that has been discovered numerous times and has simply awaited wider acknowledgment. Several labs have suggested and/or presented evidence for such structures since the mid-1970s (48–53), and they also have been photographed (46, 49) (Fig. 2). In some cases the apparent regression of replication forks may have been an artifact of sample preparation, but it was clear that this type of reaction could occur.
Figure 2.
A replication intermediate from Drosophila embryos, photographed by Inman (49). One of the apparently regressed forks has a single-stranded tail (arrow). (Reprinted from Biochim. Biophys. Acta, 783, Inman, R. B., “Methodology for the study of the effect of drugs on development and DNA replication in Drosophila melanogaster embryonic tissue,” pp. 205–215, Copyright 1984, with permission from Excerpta Medica, Inc.; ref. 49.)
Some of the early links between recombination and replication were merely circumstantial. For example, both the recR and recF genes are part of operons that encode replication functions rather than classical recombination functions (54–56). Others exhibit a compelling cause and effect. In recBC mutant cells thereby lacking the double-strand break repair pathway, elevated levels of double-strand breaks are seen only if the cells are actively replicating their chromosomes (57). Similarly, recA-dependent repair of DNA gaps is seen only during active chromosome replication (58, 59). Elevated levels of recombination can be observed as a product of DNA damage (17, 60–63), or as a result of replication of the chromosomal region being examined (64).
Additional links between recombination and replication became apparent as a result of the study of bacteria under DNA damaging conditions that induce the SOS response.
Thread 4: Studies of the Bacterial SOS System
DNA damaging treatments, such as exposure to high doses of UV irradiation, lead to a complete cessation of DNA replication. After 30–45 min, replication resumes. The resumption is generally referred to as replication restart (65). The cessation of replication triggers induction of the bacterial SOS system, leading to a staged accumulation of enzymes involved in DNA repair (3, 66–70). The stages reflect a measured response to different levels of damage. The exploration of the mechanisms by which replication is halted, and then restarted, have contributed a wealth of insights to our current understanding of the repair of replication forks.
It is now clear that the cessation of DNA replication during SOS is a result of the collision of replication forks with sites of DNA damage. The subsequent repair is a prerequisite to the restart. Other possibilities have been explored.
Early studies of the effects of UV irradiation on bacterial DNA suggested that damage could be bypassed by reinitiating DNA synthesis downstream, before recombinational DNA repair. Discontinuities detected in the DNA of UV-irradiated uvrA mutant cells were attributed to DNA gaps left behind by replication forks (16, 71). DNA exchanges appeared in UV-irradiated wild-type cells that depended on recA function (17). Models for recombinational DNA repair were built based on the assumption that the replication fork had already continued synthesis downstream (72).
Later work showed that the restart of replication depended on the RecA protein (73, 74), a result broadly inconsistent with a general resumption of replication before recombinational DNA repair. It also became clear that DNA polymerases are halted by many types of DNA damage both in vitro and in vivo (75–78). Some limited bypass may occur in some instances (79). In principle, there is more potential for the bypass of lesions on the lagging strand (where clear mechanisms exist to initiate a new Okazaki fragment upstream) than for bypass on the leading strand. However, in a cell with many genomic lesions, it should not take long for a replication fork to encounter one on the leading strand.
There are two general paths to replication restart (also called induced replisome reactivation or IRR; ref. 73). Nonmutagenic replication fork repair provides one way for a fork to bypass a lesion, and mutagenic translesion DNA synthesis (principally by the specialized DNA polymerase V) provides the other. Induction of the enzymes needed for each of these is organized temporally during SOS so that nonmutagenic repair processes are induced early, followed by mutagenic processes (3, 70). The first enzymes to appear include the components of the excision repair apparatus, DNA polymerase II, and the RuvAB complex involved in the processing of branched DNA intermediates. The RecA and RecN proteins, involved in nonmutagenic repair of replication forks, follow close behind. The induction of DNA polymerase V (umuCD), and its mutagenic translesion DNA synthesis capacity (19, 80), is a late response. If these enzymatic functions are insufficient to repair the DNA damage and restart DNA synthesis, a final induction of colicins occurs to bring about cell lysis.
An analysis of replication restart indicates that the nonmutagenic pathways are more important than the mutagenic translesion synthesis. Replication restart occurs readily in the absence of DNA polymerase V function, but requires RecA as already noted (73, 74). The phenomena of replication restart under SOS conditions thus can now be seen as a ready source of observations relevant to an understanding of the nonmutagenic repair of replication forks in general, with the major caveat that it is not the only process at work. It has the advantage that all replication is halted synchronously, and restart can be readily studied in bacterial populations.
In eukaryotes, the repair of damaged DNA is facilitated by damage checkpoints, which serve to halt DNA replication while repair is underway (81, 82). A similar idea has been put forward for bacteria (83), although there are many observations that argue against a role for a checkpoint system in halting DNA replication (reviewed in ref. 3). More recently, a checkpoint-like phenomenon dependent on uncleaved UmuD protein has been described (84). Although this may not affect the halt in replication brought about by DNA damage, it might play a role in the inhibition of replication restart at some points in the SOS process.
Thread 5: Work on Bacteriophage T4 DNA Metabolism
The connections between replication and recombination have been consistently clear and abundant in the study of DNA metabolism in bacteriophage T4 for over 30 years (85–87). Early rounds of T4 DNA replication are origin-dependent, but then a switch occurs to a new mode of RDR (88, 89). Although several mechanisms have been elucidated (85), they generally involve the invasion of a 3′-ending DNA strand into a homologous duplex DNA to form a D loop (Fig. 1i). The invading end acts as a primer for DNA synthesis, and a replication fork is established. Because the T4 genome is terminally redundant, succeeding rounds of replication lead to the production of longer concatemers. These eventually are cleaved and packaged, with more than one genome-equivalent packaged into each phage head. RDR was first documented in this system (90, 91), and T4 provided the first insights into the mechanism of this process (88, 92, 93). As will become evident in the continuing discussion, this general mechanism of RDR has become generally relevant in bacteria and eukaryotes. Similar replication initiation strategies may even be operative in mitochondria (94). Unfortunately, the RDR seen in bacteriophage T4 was, for over two decades, generally viewed as a unique and unusual process, obscuring its general significance.
Thread 6: Origin-Independent and RDR Restart During the SOS Response
Kogoma and colleagues were among the first to extend the T4 paradigm, demonstrating that RDR also occurs in bacteria. Initiation of replication at the bacterial origin oriC requires ongoing protein synthesis. In an effort to discover why, Kogoma and Lark (95, 96) found that DNA replication could be initiated without protein synthesis under conditions that induced the SOS response. This was called stable DNA replication (SDR). Later, SDR associated with the SOS response was designated induced SDR (iSDR). Initially, the process received little attention. By 1990, it became clear that iSDR depended on recBC function (97). RecA was implicated in 1984 (98), and a clear requirement for the recombinase function of recA was demonstrated in 1993 (99). The expanding list of examples of RDR soon elevated the process to acronym (RDR) status (100).
iSDR was soon seen as a form of RDR. In general, bacterial RDR required the functions of the RecA, RecBCD, RecN, and RecF proteins. Additional recombination functions were implicated under at least some circumstances. Kogoma and colleagues (100) showed that double-strand breaks triggered RDR. The resulting models proposed that DNA ends were processed by RecBCD, and D loops then were formed by the action of RecA protein (100). Following up on observations and proposals by Marians and coworkers (164), Kogoma and colleagues (28, 101, 102) also found that PriA protein and the φX174-type primosome were required for iSDR and bacterial RDR in general.
In a few summarizing reviews, Kogoma (29, 103) suggested a variety of functions for iSDR. These included an alternative mode of replication during SOS that was damage-resistant, error-prone replication, and adaptive mutation. Some of these proposals may have reflected the complexity of events during the SOS response. Indeed, Kogoma (103) noted that iSDR still occurred in umuC mutant cells and was no longer mutagenic in that background. Thus, nonmutagenic replication fork repair is a major component of iSDR, and it is reasonable to link the two. There are a few complexities in making this connection. The apparent reliance of iSDR on novel replication origins (29, 103) does not make sense in the context of replication forks stalled at random sites in the genome. Also, Kogoma (103) noted a few minor differences between the broader process of replication restart during SOS, and iSDR, including some apparent differences in genetic requirements. However, Kogoma and colleagues (29, 100, 103) recognized that whatever its main function, RDR was ideally suited to the repair of stalled or collapsed replication forks and suggested this as early as 1994.
Thread 7: Isolation and Characterization of RecA Protein and Prokaryotic Recombination Proteins
RecA protein is a major player throughout this discussion, and research into its functions and properties has affected the broader development of models for replication fork repair. Purified RecA protein was first described in 1978 as a DNA-dependent ATPase activity (104, 105) that could promote the cleavage of the LexA repressor (104). The DNA strand exchange functions of RecA were soon elucidated in work dominated at first by the Radding and Lehman laboratories (106–111). This effort was joined somewhat later by Howard-Flanders and colleagues (112) and many others. Over 20 years of intensive studies have identified the three distinct functions for RecA protein noted in thread 1, all recently reviewed (80, 113–118).
The list of recombination functions in bacteria is now counted in the dozens, and elucidation of their properties (20, 115, 119–121) also has contributed to model-building efforts. The functional complexity and universal occurrence of RecA and multiprotein recombination systems in bacteria contributed to early arguments that DNA repair was the primary function of bacterial recombination (7, 8, 122).
Thread 8: Recombination Model Development
The decades-long refinement of models for recombination in both prokaryotes and eukaryotes illustrates a gradually increasing awareness of links between recombination and replication. A particularly complete summary of the models advanced up until 1995 can be found in the excellent monograph by Kuzminov (123), and only a few key points need be made here. Replication has been a feature of recombination models at least since the early constructs of Meselson and Weigle (124). Those workers introduced the concept of strand invasion to prime DNA synthesis. Following the cut-and-paste proposal of Holliday (125) which focused attention on crossed strand intermediates, models incorporating replication at some step were elaborated by Hotchkiss (126) and Meselson and Radding (127), both of which envisioned initiation at single-strand breaks.
The focus shifted to double-strand breaks in the constructs of Boon and Zinder (128) and Resnick (129). DNA repair became a dominant theme in the models of Skalka (44) and West et al. (72). The influential double-strand break repair model of Szostak et al. (130), and its more recent iterations (82, 131), now dominate discussion of meiotic recombination, and very similar processes are envisioned for transductional and conjugational recombination in bacteria (132). These models include the T4-like invasion of 3′ ends, which are then used to prime DNA synthesis. The demonstration of break-induced replication in eukaryotes (133–136) provides a rather dramatic manifestation of these processes and extends the list of organism classes where RDR has been observed.
The basic mechanisms of end processing, 3′ end invasion, and the priming of replication, could readily be derived from a system that evolved originally to repair stalled replication forks (137). In eukaryotes, the need to avoid chromosome nondisjunction may have displaced fork repair as a primary function in meiotic recombination. However, fork repair remains a major molecular problem in eukaryotes. Human cells may average about 10 replication-associated double-strand breaks per cell division (137).
Thread 9: Eukaryotic Recombination Proteins
The recombinational DNA repair paradigm appears to be universal. The pantheon of bacteriophage T4 and bacterial recombinational replication fork repair enzymes is paralleled by a functionally equivalent (but sometimes more complex) set of enzymes in eukaryotes (Table 1). The Rad51 protein was identified as a structural and functional homologue of RecA protein early in the 1990s (138–140), followed by the Dmc1 protein (141, 142). Both proteins promote a RecA-like DNA strand exchange reaction. The generally recognized list of needed functions for the repair of stalled replication forks is gradually being filled in for all classes of organisms.
Table 1.
Enzymes involved in the nonmutagenic repair of stalled replication forks
| Bacteriophage T4 | E. coli | Eukaryotes | |
|---|---|---|---|
| Recombinase | UvsX | RecA | Rad51/54 Dmcl |
| Single-strand binding | Gene 32 | SSB | RPA |
| RMP(Rec) | UvsY | RecOR RecBCD | Rad52 Rad55/57 |
| Branch helicase | UvsW | RecG RuvAB | |
| End processing | gp46/47 | SbcCD, RecBCD | Rad50, Mre11, Xrs2/Nbs1 |
| Holliday junction resolution | Endonuclease VII | RuvC/Rus | |
| Replication restart RMP | gp59 | PriA + restart primosome | |
| Replicative helicase | gp41 | DnaB | |
| Polymerase | T4 polymerase | DNA pol II/III | Pol α, δ, ɛ RFC, PCNA, etc. |
The need for recombinational DNA repair in eukaryotes can be seen in the phenotypes of cells mutant in the appropriate repair functions. In eukaryotes, recombination plays a variety of important roles in meiosis, mitosis, and DNA repair. The rad designation for many eukaryotic genes reflects their initial isolation in yeast as mutants that were sensitive to radiation-induced DNA damage. In mammals, homozygous null mutants of Rad51 protein have an embryonic lethal phenotype (143–145). Vertebrate cells lacking Rad51 protein function halt growth at the G2/M phase interface of the cell cycle, and accumulate double-strand breaks before dying (146). Several proteins involved in mammalian carcinogenesis interact with Rad51, including p53 (147), BRCA1 (148), and BRCA2 (145). Recombinational DNA repair is clearly important in all organisms.
Thread 10: The Restart (φX174-Type) Primosome
The restart primosome plays a key role in the restart of replication after the recombination steps of repair, as seen in Fig. 1. This protein complex originally was defined as a factor required for the in vitro initiation of DNA synthesis on φX174 DNA circles (149, 150). The complex consists of seven proteins, with historical names given in parentheses: PriA (protein n′, factor Y), PriB (protein n), PriC (protein n′′), DnaT (protein i), DnaC, the DnaB helicase, and the DnaG primase. Although defined as a system for the replication of a bacterial parasite, all of these proteins were encoded by Escherichia coli. There was every expectation that this complex would function as the lagging strand primosome in the replication of the bacterial chromosome (see, for example, p. 411 in ref. 151). However, when the initiation of replication at the bacterial origin oriC was reconstituted in vitro (152, 153), the DnaB, DnaC, and DnaG proteins were required, but the remaining components of the φX174-type primosome were not (153, 154). A consensus gradually developed that the φX174-type primosome did not play a role in the function of replication forks initiated at oriC (155–157). In coming to this conclusion, Seufert and Messer (156) made the remarkable (for the time) suggestion that this primosome might play a role in the reinitiation of replication when replication had stalled for some reason. Based on a demonstrated requirement for DnaT function in stable DNA replication (158, 159), those workers also suggested that the φX174-type primosome might have a role in Kogoma's iSDR.
Further advance required in vivo analysis of the genes. The genes for the PriA, PriB, and PriC proteins never turned up in a screen for genes defective in chromosomal replication, reinforcing the impression that they lacked a function in oriC-mediated replication. The genes were finally cloned by reverse genetics (160, 161). Despite the apparent lack of a function in oriC-mediated replication, null mutants of priA proved to be only marginally viable, suggesting an important cellular function (162–164). The priA mutants also induced the SOS response under normal growth conditions (163). From here, it was not too hard to connect a few dots. One year earlier, Sassanfar and Roberts (165) had demonstrated that SOS was induced only in cells with active replication forks. Marians and colleagues (163, 164) reasoned that if PriA was not needed at oriC, perhaps these replication forks were stalling at DNA damage. The fork that completed DNA replication thus might not be the same one that initiated it. Picking up on the suggestion of Seufert and Messer (156), Marians and colleagues (163) proposed that PriA and the φX174-type primosome might be necessary to get replication going again. Most important, to explain the marginal viability of the priA mutants, this would have to be happening in a substantial fraction of the cellular replication forks under normal growth conditions (163, 166). This was the first indication that bacterial replication forks underwent a frequent demise, and that restart was a critical cellular function even in the absence of SOS-inducing conditions. This eventually led Sandler and Marians (31) to rename this protein complex the restart primosome. All that remained in the early 1990s was to link this fork repair to recombination, a process already well underway.
Thread 11: Tying the Threads Together, Genetically and Biochemically
In the mid-1990s, multiple lines of research produced results leading to models for the recombinational repair of replication forks. One path arose from an examination of the function of the RecBCD enzyme. RecBCD degrades and unwinds a linear DNA molecule from one end. When this enzyme encounters an asymmetric 8-bp DNA sequence called Chi in the correct orientation, it no longer degrades the 3′ ending strand and produces the 3′ single-strand extensions needed for RecA-mediated strand invasion. Chi sites increase the survival of linear DNAs in bacteria (167, 168). It was clear by the early 1990s that Chi sites were overrepresented in the E. coli genome (169, 170). Furthermore, the Chi sites were not randomly oriented, but overwhelmingly situated so as to modulate the function of a RecBCD enzyme moving through the DNA toward oriC. To explain this pattern, Kuzminov et al. (168) noted that a double-strand break produced by the collision of a replication fork with a single-strand interruption would produce only one DNA end (Fig. 1h). RecBCD would bind to this end and travel back toward oriC. They suggested that the Chi sites had evolved primarily to handle collapsed replication forks, and they presented a model for how the repair of collapsed forks could occur (168). This model was developed further by Kuzminov in the following year (171). A subsequent series of reviews and monographs by Kuzminov and colleagues (3, 123, 172–174) provided a stimulating source of insights and ideas that have been instrumental in focusing this developing field.
Further exploration of the phenotypes of rec− strains led to new advances. Horiuchi and coworkers (175, 176) examined the fate of blocked replication forks in vivo. They observed a growth dependence on RecA and RecBCD function in their strains and independently suggested that stalled forks generate double-strand breaks that must be repaired before replication restart (176). The role of RecBCD was also explored by Michel and colleagues (177, 178), who had earlier noticed the connection between recombination and the pausing of replication forks. They found that double-strand breaks occurred at elevated levels in cells lacking recBCD or recA function (57). Most important, the increase did not occur in the absence of replicating chromosomes, linking the breaks to active replication. Subsequent investigations by this group have begun to elucidate the functions of proteins at stalled forks and have led to increasingly refined models for fork repair (179–181). A similar path has been followed by Lloyd and colleagues (182) in their studies of RecG protein, a helicase that acts at branched recombination intermediates. A connection to replication forks was established with the finding that suppressors of recG- mutations mapped in the priA gene. Subsequent investigations of RecG function have led to new proposals for the molecular situations that can trigger the collapse and repair of a replication fork (183).
Not all of the work has focused on the double-strand break repair pathway. In the context of replication fork repair, the gap repair pathway mediated by RecF and other proteins appears to be as important as the double-strand repair pathway initiated by RecBCD enzyme (39, 184). Potential pathways for replication fork repair without double-strand breaks have been advanced, making use of the fork regression process first postulated by Higgins et al. (46) and McGlynn and Lloyd (183) (Fig. 1 a–f).
Biochemistry increasingly complements the in vivo efforts. The PriA protein binds specifically to branched DNA molecules, establishing another link to intermediates likely to be found during recombinational DNA repair (30, 185, 186). This function is necessary for normal bacterial growth (186). The action of RecBCD and RecA protein in processing a DNA end and forming a D loop has been reconstituted by Kowalczykowski and colleagues (187, 188). Late steps in recombinational processes mediated by the Holliday junction processing enzymes RuvA, RuvB, And RuvC have been reconstituted by West and colleagues (189). The RecA protein promotes a fork regression reaction that may play a role in several pathways for fork repair (190).
When all of these results were put together with the earlier results, it was evident during the last half of the 1990s that the recombinational repair of stalled replication forks was a significant function within bacterial DNA metabolism. To gauge its importance, one now had to determine how often it happened.
Thread 12: XerCD and Frequency Estimates
In terms of frequency of use, the nonmutagenic repair of stalled replication forks is the most important function of the bacterial homologous genetic recombination systems (1, 113, 191, 192). It can thus be argued that this is the primary function of bacterial recombination systems. Under normal growth conditions, replication appears to be the main source of lesions needing recombinational DNA repair, as elaborated above. Conjugation and transduction, which also involve recombination, are normally very rare processes.
Part of this conclusion was derived from an analysis of the XerCD site-specific recombination system, which resolves chromosomal dimers created by the recombinational DNA repair of replication forks (1, 39, 193, 194). This work helped to establish the lower limit (about 15%) for the part of a bacterial cell population that undergoes repair of a stalled fork (1, 192, 195). The higher estimates for recombinational repair of replication forks come from a consideration of the effects of recA and priA mutant cells (1, 192, 195). The priA mutants, as already mentioned, are especially compromised, but not dead. Because all replication fork repair should involve a restart step, this suggests that most, but not all, cells have forks that undergo recombinational DNA repair under normal growth conditions. However, there appear to be some minor and priA-independent pathways for replication fork restart (196). These may permit the survival of priA mutant cells and obscure an even higher frequency of fork stalling and repair.
The End Marks a Beginning
A new understanding of the function of recombination has, more than anything else, revealed how little we really know. The range of events that can occur when a replication fork meets an insurmountable barrier is not known. The structure of stalled forks has been examined in only a very few instances (197, 198). We do not know how long repair takes. Multiple pathways are known only in outline form. We do not have a good understanding of the rates and types of DNA damage as a function of growth conditions. Our view of the roles of the enzymes involved is changing rapidly as this new paradigm becomes established, and new protein components seem likely to emerge (199–202).
Nevertheless, the paradigm promises to help us understand where recombination came from and what we can do with it. Even in eukaryotes, where recombination function is even more complex, one can argue that recombination evolved from a need to repair the molecular train wrecks created by the encounter of replication forks with DNA damage (137). RecA protein arose in bacteria or their predecessors a very long time ago (114, 198, 199) and may have been the precursor of a now-important class of helicases (203). As research continues into the causes of cancer and new pathways for gene therapy, insight into the evolutionarily determinative function of recombination must inevitably brighten the path.
Acknowledgments
I thank Andrei Kuzminov for a careful reading and critique of this manuscript and acknowledge with pleasure the many conversations with colleagues that helped shape this review. Work in my laboratory is supported by National Institutes of Health Grants GM32335 and GM52725.
Abbreviations
- RDR
recombination-dependent replication
- SDR
stable DNA replication
- iSDR
induced SDR
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 3.Kuzminov A. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marians K J. Curr Opin Genet Dev. 2000;10:151–156. doi: 10.1016/s0959-437x(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 5.Michod R E. Eros and Evolution: A Natural Philosophy of Sex. Menlo Park, CA: Addison–Wesley; 1995. [Google Scholar]
- 6.Clark A J, Sandler S J. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 7.Cox M M. Mol Microbiol. 1991;5:1295–1299. doi: 10.1111/j.1365-2958.1991.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 8.Cox M M. BioEssays. 1993;15:617–623. doi: 10.1002/bies.950150908. [DOI] [PubMed] [Google Scholar]
- 9.Campbell A. Cold Spring Harbor Symp Quant Biol. 1984;49:839–844. doi: 10.1101/sqb.1984.049.01.095. [DOI] [PubMed] [Google Scholar]
- 10.Lederberg J, Tatum E L. Cold Spring Harbor Symp Quant Biol. 1946;11:413–441. [Google Scholar]
- 11.Clark A J, Margulies A D. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark A J. BioEssays. 1996;18:767–772. doi: 10.1002/bies.950180912. [DOI] [PubMed] [Google Scholar]
- 13.Emmerson P T. Genetics. 1968;60:19–30. doi: 10.1093/genetics/60.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark A J, Chamberlin M, Boyce R P, Howard-Flanders P. J Mol Biol. 1966;19:442–454. doi: 10.1016/s0022-2836(66)80015-3. [DOI] [PubMed] [Google Scholar]
- 15.Howard-Flanders P, Theriot L. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rupp W D, Howard-Flanders P. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 17.Rupp W D, Wilde C E D, Reno D L, Howard-Flanders P. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 18.Radman M. In: Molecular Mechanisms for Repair of DNA, Part A. Hanawalt P, Setlow R B, editors. New York: Plenum; 1975. pp. 355–367. [Google Scholar]
- 19.Tang M J, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianco P R, Tracy R B, Kowalczykowski S C. Front Biosci. 1998;3:560–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 22.Clark A J. Annu Rev Microbiol. 1971;25:437–464. doi: 10.1146/annurev.mi.25.100171.002253. [DOI] [PubMed] [Google Scholar]
- 23.Lloyd R G. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishioka K, Iwasaki H, Shinagawa H. Genes Genet Syst. 1997;72:91–99. doi: 10.1266/ggs.72.91. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd R G, Buckman C. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd R G, Buckman C. Biochimie. 1991;73:313–320. doi: 10.1016/0300-9084(91)90218-p. [DOI] [PubMed] [Google Scholar]
- 27.Horii Z, Clark A J. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 28.Kogoma T, Cadwell G W, Barnard K G, Asai T. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 30.McGlynn P, Al D A, Liu J, Marians K J, Lloyd R G. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 31.Sandler S J, Marians K J. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konrad E B. J Bacteriol. 1977;130:167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linn S, Imlay J A. J Cell Sci Suppl. 1987;6:289–301. doi: 10.1242/jcs.1984.supplement_6.19. [DOI] [PubMed] [Google Scholar]
- 34.Miguel A G, Tyrrell R M. Biophys J. 1986;49:485–491. doi: 10.1016/S0006-3495(86)83658-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morse L S, Pauling C. Proc Natl Acad Sci USA. 1975;72:4645–4649. doi: 10.1073/pnas.72.11.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T C, Smith K C. J Bacteriol. 1986;165:1023–1025. doi: 10.1128/jb.165.3.1023-1025.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witkin E M, Roegner M V. J Bacteriol. 1992;174:4166–4168. doi: 10.1128/jb.174.12.4166-4168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park E M, Shigenaga M K, Degan P, Korn T S, Kitzler J W, Wehr C M, Kolachana P, Ames B N. Proc Natl Acad Sci USA. 1992;89:3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner W W, Kuempel P L. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morimyo M. J Bacteriol. 1982;152:208–214. doi: 10.1128/jb.152.1.208-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 43.Hanawalt P C. Photochem Photobiol. 1966;5:1–12. [PubMed] [Google Scholar]
- 44.Skalka A. In: Mechanisms in Recombination. Grell R F, editor. New York: Plenum; 1974. pp. 421–432. [Google Scholar]
- 45.Radding C M. Annu Rev Genet. 1973;7:87–111. doi: 10.1146/annurev.ge.07.120173.000511. [DOI] [PubMed] [Google Scholar]
- 46.Higgins N P, Kato K, Strauss B. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 47.Postow L, Ullsperger C, Keller R W, Bustamante C, Vologodskii A V, Cozzarelli N R. J Biol Chem. 2001;267:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 48.Hotchkiss R D. Annu Rev Microbiol. 1974;28:445–468. doi: 10.1146/annurev.mi.28.100174.002305. [DOI] [PubMed] [Google Scholar]
- 49.Inman R B. Biochim Biophys Acta. 1984;783:205–215. doi: 10.1016/0167-4781(84)90030-7. [DOI] [PubMed] [Google Scholar]
- 50.Zannis-Hadjopoulos M, Persico M, Martin R G. Cell. 1981;27:155–163. doi: 10.1016/0092-8674(81)90369-x. [DOI] [PubMed] [Google Scholar]
- 51.Nilsen T, Baglioni C. J Mol Biol. 1979;133:319–338. doi: 10.1016/0022-2836(79)90396-6. [DOI] [PubMed] [Google Scholar]
- 52.Tatsumi K, Strauss B. Nucleic Acids Res. 1978;5:331–347. doi: 10.1093/nar/5.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wanka F, Brouns R M, Aelen J M, Eygensteyn A, Eygensteyn J. Nucleic Acids Res. 1977;4:2083–2097. doi: 10.1093/nar/4.6.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanar M A, Sandler S J, Armengod M E, Ream L W, Clark A J. Proc Natl Acad Sci USA. 1984;81:4622–4626. doi: 10.1073/pnas.81.15.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flower A M, McHenry C S. J Mol Biol. 1991;220:649–658. doi: 10.1016/0022-2836(91)90107-h. [DOI] [PubMed] [Google Scholar]
- 56.Perez-Roge R L, Garcia-Sogo M, Navarro-Avino J, Lopez-Acedo C, Macian F, Armengod M E. Biochimie. 1991;73:329–334. doi: 10.1016/0300-9084(91)90220-u. [DOI] [PubMed] [Google Scholar]
- 57.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith K C, Sharma R C. Mutat Res. 1987;183:1–9. doi: 10.1016/0167-8817(87)90039-3. [DOI] [PubMed] [Google Scholar]
- 59.Smith K C, Wang T V, Sharma R C. J Photochem Photobiol Biol. 1987;1:1–11. doi: 10.1016/1011-1344(87)80002-7. [DOI] [PubMed] [Google Scholar]
- 60.Feng W Y, Hays J B. Genetics. 1995;140:1175–1186. doi: 10.1093/genetics/140.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hays J B, Hays J G. Biopolymers. 1991;31:1565–1579. doi: 10.1002/bip.360311312. [DOI] [PubMed] [Google Scholar]
- 62.Lin P F, Howard-Flanders P. Mol Gen Genet. 1976;146:107–115. doi: 10.1007/BF00268079. [DOI] [PubMed] [Google Scholar]
- 63.Smith K C, Wang T C. BioEssays. 1989;10:12–16. doi: 10.1002/bies.950100104. [DOI] [PubMed] [Google Scholar]
- 64.Morel-Deville F, Ehrlich S D. Mol Microbiol. 1996;19:587–598. doi: 10.1046/j.1365-2958.1996.398936.x. [DOI] [PubMed] [Google Scholar]
- 65.Echols H, Goodman M F. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 66.Smith B T, Walker G C. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shinagawa H. Exs. 1996;77:221–235. doi: 10.1007/978-3-0348-9088-5_14. [DOI] [PubMed] [Google Scholar]
- 68.Sommer S, Boudsocq F, Devoret R, Bailone A. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 69.Walker G C, Smith B T, Sutton M D. In: Bacterial Stress Responses. Storz G, HenggeAronis R, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 131–144. [Google Scholar]
- 70.Defais M, Devoret R. The Encyclopedia of Life Sciences. 2000. www.els.net , Macmillan, www.els.net (on-line only). (on-line only). [Google Scholar]
- 71.Howard-Flanders P, Rupp W D, Wilkins B M, Cole R S. Cold Spring Harbor Symp Quant Biol. 1968;33:195–207. doi: 10.1101/sqb.1968.033.01.023. [DOI] [PubMed] [Google Scholar]
- 72.West S C, Cassuto E, Howard-Flanders P. Nature (London) 1981;294:659–662. doi: 10.1038/294659a0. [DOI] [PubMed] [Google Scholar]
- 73.Khidhir M A, Casaregola S, Holland I B. Mol Gen Genet. 1985;199:133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- 74.Witkin E M, Roegner M V, Sweasy J B, McCall J O. Proc Natl Acad Sci USA. 1987;84:6805–6809. doi: 10.1073/pnas.84.19.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sagher D, Strauss B. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 76.Moore P D, Bose K K, Rabkin S D, Strauss B S. Proc Natl Acad Sci USA. 1981;78:110–114. doi: 10.1073/pnas.78.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banerjee S K, Christensen R B, Lawrence C W, LeClerc J E. Proc Natl Acad Sci USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawrence C W, Borden A, Banerjee S K, LeClerc J E. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livneh Z. Proc Natl Acad Sci USA. 1986;83:4599–4603. doi: 10.1073/pnas.83.13.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goodman M J. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- 81.Kastan M B, Kuerbitz S J. Environ Health Perspect. 1993;101:55–58. doi: 10.1289/ehp.93101s555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bridges B A. BioEssays. 1995;17:63–70. doi: 10.1002/bies.950170112. [DOI] [PubMed] [Google Scholar]
- 84.Opperman T, Murli S, Smith B T, Walker G C. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 86.Karam J D, editor. The Molecular Biology of Bacteriophage T4. Washington, DC: Am. Soc. Microbiol.; 1994. [Google Scholar]
- 87.Kreuzer K N, Morrical S W. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 28–42. [Google Scholar]
- 88.Luder A, Mosig G. Proc Natl Acad Sci USA. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mosig G. Annu Rev Genet. 1987;21:347–371. doi: 10.1146/annurev.ge.21.120187.002023. [DOI] [PubMed] [Google Scholar]
- 90.Yegian C D, Mueller M, Selzer G, Russo V, Stahl F W. Virology. 1971;46:900–919. doi: 10.1016/0042-6822(71)90090-0. [DOI] [PubMed] [Google Scholar]
- 91.Warner H R, Hobbs M D. Virology. 1967;33:376–384. doi: 10.1016/0042-6822(67)90113-4. [DOI] [PubMed] [Google Scholar]
- 92.Dannenberg R, Mosig G. J Virol. 1981;40:890–900. doi: 10.1128/jvi.40.3.890-900.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dannenberg R, Mosig G. J Virol. 1983;45:813–831. doi: 10.1128/jvi.45.2.813-831.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Backert S, Borner T. Curr Genet. 2000;37:304–314. doi: 10.1007/s002940050532. [DOI] [PubMed] [Google Scholar]
- 95.Kogoma T, Lark K G. J Mol Biol. 1970;52:143–164. doi: 10.1016/0022-2836(70)90022-7. [DOI] [PubMed] [Google Scholar]
- 96.Kogoma T, Lark K G. J Mol Biol. 1975;94:243–256. doi: 10.1016/0022-2836(75)90081-9. [DOI] [PubMed] [Google Scholar]
- 97.Magee T R, Kogoma T. J Bacteriol. 1990;172:1834–1839. doi: 10.1128/jb.172.4.1834-1839.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Witkin E M, Kogoma T. Proc Natl Acad Sci USA. 1984;81:7539–7543. doi: 10.1073/pnas.81.23.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Asai T, Sommer S, Bailone A, Kogoma T. EMBO J. 1993;12:3287–3295. doi: 10.1002/j.1460-2075.1993.tb05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Asai T, Bates D B, Kogoma T. Cell. 1994;78:1051–1061. doi: 10.1016/0092-8674(94)90279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Asai T, Kogoma T. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Masai H, Asai T, Kubota Y, Arai K, Kogoma T. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roberts J W, Roberts C W, Craig N L. Proc Natl Acad Sci USA. 1978;75:4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ogawa T, Wabiko H, Tsurimoto T, Horii T, Masukata H, Ogawa H. Cold Spring Harbor Symp Quant Biol. 1979;2:909–915. doi: 10.1101/sqb.1979.043.01.099. [DOI] [PubMed] [Google Scholar]
- 106.Shibata T, Das Gupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Das Gupta C, Shibata T, Cunningham R P, Radding C M. Cell. 1980;22:437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- 108.Shibata T, Das Gupta C, Cunningham R P, Radding C M. Proc Natl Acad Sci USA. 1980;77:2606–2610. doi: 10.1073/pnas.77.5.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weinstock G M, McEntee K, Lehman I R. Proc Natl Acad Sci USA. 1979;76:126–130. doi: 10.1073/pnas.76.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McEntee K, Weinstock G M, Lehman I R. Proc Natl Acad Sci USA. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.West S C, Cassuto E, Mursalim J, Howard-Flanders P. Proc Natl Acad Sci USA. 1980;77:2569–2573. doi: 10.1073/pnas.77.5.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cox M M. Prog Nucleic Acid Res Mol Biol. 1999;63:310–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 114.Kowalczykowski S C, Eggleston A K. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 115.Roca A I, Cox M M. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 116.West S C. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 117.Little J W. Biochimie. 1991;73:411–422. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 118.Shinohara A, Ogawa T. Mutat Res. 1999;435:13–21. doi: 10.1016/s0921-8777(99)00033-6. [DOI] [PubMed] [Google Scholar]
- 119.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 120.Shinagawa H, Iwasaki H. Trends Biochem Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 121.Sharples G J, Ingleston S M, Lloyd R G. J Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roca A I, Cox M M. CRC Crit Rev Biochem Mol Biol. 1990;25:415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- 123.Kuzminov A. Recombinational Repair of DNA Damage. Georgetown, TX: Landes; 1996. [Google Scholar]
- 124.Meselson M, Weigle J J. Proc Natl Acad Sci USA. 1961;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 126.Hotchkiss R D. Adv Genet. 1971;16:325–348. [PubMed] [Google Scholar]
- 127.Meselson M S, Radding C M. Proc Natl Acad Sci USA. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boon T, Zinder N D. Proc Natl Acad Sci USA. 1969;64:573–577. doi: 10.1073/pnas.64.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Resnick M A. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 130.Szostak J W, Orr W T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 131.Hastings P J. BioEssays. 1988;9:61–64. doi: 10.1002/bies.950090206. [DOI] [PubMed] [Google Scholar]
- 132.Smith G R. In: DNA Damage and Repair, Vol. 1: DNA Repair in Prokaryotes and Lower Eukaryotes. Nickoloff J A, Hoekstra M F, editors. Totowa, NJ: Humana; 1998. pp. 135–162. [Google Scholar]
- 133.Bosco G, Haber J E. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morrow D M, Connelly C, Hieter P. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malkova A, Ivanov E L, Haber J E. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Voelkel-Meiman K, Roeder G S. Genetics. 1990;126:851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haber J E. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 138.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 139.Story R M, Bishop D K, Kleckner N, Steitz T A. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- 140.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 141.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 142.Li Z F, Golub E I, Gupta R, Radding C M. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lim D S, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sharan S K, Morimatsu M, Albrecht U, Lim D-S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Nature (London) 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 146.Sonoda E, Sasaki M S, Buerstedde J M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sturzbecher H W, Donzelmann B, Henning W, Knippschild U, Buchhop S. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 148.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 149.Shlomai J, Polder L, Arai K, Kornberg A. J Biol Chem. 1981;256:5233–5238. [PubMed] [Google Scholar]
- 150.Wickner S, Hurwitz J. Proc Natl Acad Sci USA. 1975;72:3342–3346. doi: 10.1073/pnas.72.9.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kornberg A. DNA Replication. San Francisco: Freeman; 1980. [Google Scholar]
- 152.Fuller R S, Kaguni J M, Kornberg A. Proc Natl Acad Sci USA. 1981;78:7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaguni J M, Kornberg A. Cell. 1984;38:183–190. doi: 10.1016/0092-8674(84)90539-7. [DOI] [PubMed] [Google Scholar]
- 154.van der Ende A, Baker T A, Ogawa T, Kornberg A. Proc Natl Acad Sci USA. 1985;82:3954–3958. doi: 10.1073/pnas.82.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Seufert W, Messer W. Cell. 1987;48:73–78. doi: 10.1016/0092-8674(87)90357-6. [DOI] [PubMed] [Google Scholar]
- 156.Seufert W, Messer W. EMBO J. 1986;5:3401–3406. doi: 10.1002/j.1460-2075.1986.tb04656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Funnell B E, Baker T A, Kornberg A. J Biol Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- 158.Masai H, Bond M W, Arai K. Proc Natl Acad Sci USA. 1986;83:1256–1260. doi: 10.1073/pnas.83.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lark K G, Lark C A. Cold Spring Harbor Symp Quant Biol. 1979;43:537–549. doi: 10.1101/sqb.1979.043.01.059. [DOI] [PubMed] [Google Scholar]
- 160.Nurse P, DiGate R J, Zavitz K H, Marians K J. Proc Natl Acad Sci USA. 1990;87:4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zavitz K H, DiGate R J, Marians K J. J Biol Chem. 1991;266:13988–13995. [PubMed] [Google Scholar]
- 162.Lee E H, Kornberg A. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nurse P, Zavitz K H, Marians K J. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zavitz K H, Marians K J. Mol Microbiol. 1991;5:2869–2873. doi: 10.1111/j.1365-2958.1991.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 165.Sassanfar M, Roberts J W. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 166.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 167.Dabert P, Ehrlich S D, Gruss A. Proc Natl Acad Sci USA. 1992;89:12073–12077. doi: 10.1073/pnas.89.24.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kuzminov A, Schabtach E, Stahl F W. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Burland V, Plunkett G D, Daniels D L, Blattner F R. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 170.Medigue C, Viari A, Henaut A, Danchin A. Microbiol Rev. 1993;57:623–654. doi: 10.1128/mr.57.3.623-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kuzminov A. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 172.Kuzminov A. BioEssays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 173.Kuzminov A. BioEssays. 1996;18:757–765. doi: 10.1002/bies.950180911. [DOI] [PubMed] [Google Scholar]
- 174.Kuzminov A, Stahl F W. J Bacteriol. 1997;179:880–888. doi: 10.1128/jb.179.3.880-888.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Horiuchi T, Fujimura Y. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bierne H, Michel B. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 178.Bierne H, Ehrlich S D, Michel B. EMBO J. 1991;10:2699–2705. doi: 10.1002/j.1460-2075.1991.tb07814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Michel B. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 180.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 181.Seigneur M, Ehrlich S D, Michel B. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- 182.Al Deib A, Mahdi A A, Lloyd R G. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 184.Galitski T, Roth J R. Genetics. 1997;146:751–767. doi: 10.1093/genetics/146.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu J, Marians K J. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 186.Liu J I, Xu L W, Sandler S J, Marians K J. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Anderson D G, Kowalczykowski S C. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 188.Churchill J J, Anderson D G, Kowalczykowski S C. Genes Dev. 1999;13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Eggleston A K, Mitchell A H, West S C. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 190.Robu M E, Inman R B, Cox M M. Proc. Natl. Acad. Sci. USA. 2001. 8211–8218. [Google Scholar]
- 191.Cox M M. Proc Natl Acad Sci USA. 1997;94:11764–11766. doi: 10.1073/pnas.94.22.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cox M M. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 193.Michel B, Recchia G D, Penel-Colin M, Ehrlich S D, Sherratt D J. Mol Microbiol. 2000;37:180–191. doi: 10.1046/j.1365-2958.2000.01989.x. [DOI] [PubMed] [Google Scholar]
- 194.Barre F X, Aroyo M, Colloms S D, Helfrich A, Cornet F, Sherratt D J. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cox, M. M. (2001) Annu. Rev. Genet.35, in press. [DOI] [PubMed]
- 196.Sandler S J. Genetics. 2000;155:487–497. doi: 10.1093/genetics/155.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cordeiro-Stone M, Makhov A M, Zaritskaya L S, Griffith J D. J Mol Biol. 1999;289:1207–1218. doi: 10.1006/jmbi.1999.2847. [DOI] [PubMed] [Google Scholar]
- 198.Gruber M, Wellinger R E, Sogo J M. Mol Cell Biol. 2000;20:5777–5787. doi: 10.1128/mcb.20.15.5777-5787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sharples G J, Chan S N, Mahdi A A, Whitby M C, Lloyd R G. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Saveson C J, Lovett S T. Genetics. 1997;146:457–470. doi: 10.1093/genetics/146.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Saveson C J, Lovett S T. Genetics. 1999;152:5–13. doi: 10.1093/genetics/152.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chiu S K, Low K B, Yuan A, Radding C M. Proc Natl Acad Sci USA. 1997;94:6079–6083. doi: 10.1073/pnas.94.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Leipe D D, Aravind L, Grishin N V, Koonin E V. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 204.Kuzminov A. Proc Natl Acad Sci USA. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]