Abstract
Identifying neuronal molecular markers with restricted patterns of expression is a crucial step in dissecting the numerous pathways and functions of the brain. While the dorsomedial nucleus of the hypothalamus (DMH) has been implicated in a host of physiological processes, current functional studies have been limited by the lack of molecular markers specific for DMH. Identification of such markers would facilitate the development of mouse models with DMH-specific genetic manipulations. Here we used a combination of laser-capture microdissection (LCM) and gene expression profiling to identify genes that are highly expressed within the DMH relative to adjacent hypothalamic regions. Six of the most highly expressed of these genes, Gpr50, 4930511J11Rik, Pcsk5, Grp, Sulf1, and Rorβ, were further characterized by real-time polymerase chain reaction (PCR) analysis and in situ hybridization histochemistry. The genes identified in this article will provide the basis for future gene-targeted approaches for studying DMH function.
INDEXING TERMS: DMH, LCM, hypothalamus
The dorsomedial nucleus of the hypothalamus (DMH) has been implicated in a wide range of physiological processes, ranging from reproduction, thermogenesis, stress response, pancreatic nerve activity, and plasma glucose levels (Elmquist et al., 1998b; Thompson and Swanson, 1998; Zhang et al., 2011). Recently, there has been renewed interest in the potential role of the DMH in regulating food intake and circadian rhythms (Chou et al., 2003; Gooley et al., 2006). The DMH is positioned between key circadian and metabolic centers, and thus is ideally situated to integrate signals from these two systems. It receives projections from circadian nuclei, including the superchiasmatic nucleus (SCN) and subparaventricular zone (SPZ) (Watts, 1991; Kalsbeek et al., 1996; Huang et al., 2011). In turn, the DMH sends abundant projections to the paraventricular hypothalamic nucleus (PVH), preoptic area, arcuate nucleus (ARC), and lateral hypothalamus, important centers for energy balance, sleep, and wakefulness (Elmquist et al., 1998a; Chou et al., 2003; Gautron et al., 2010).
Functional and molecular studies also support the role of DMH in metabolism. DMH expresses a number of neuropeptides known to be important in energy homeostasis, including leptin receptor (LepR), neuropeptide Y, (Npy), orexin, cholecystokinin (Cck), several opioids, melanocortin 4 receptor (Mc4r), cocaine and amphetamine-regulated transcript (Cart), melanin concentrating hormone (Mch), and agouti-related peptide (Agrp) (Bellinger and Bernardis, 2002). Importantly, DMH neurons also respond to changes in metabolic stimuli. For instance, mice fed a high-fat diet (HFD) show increases in Npy expression and c-fos immunoreactivity within the DMH (Guan et al., 1998; Lin and Huang, 1999). Furthermore, intravenous injections of the adipocyte-derived hormone leptin, a key metabolic hormone, strongly activate neurons within the DMH (Elmquist et al., 1998b; Elias et al., 2000).
The role of DMH in circadian rhythmicity, however, is not as clear. While the circadian genes, period homolog 1 (Per1) and period homolog 2 (Per2), have been shown to oscillate in the compact DMH in a food entrainable manner (Mieda et al., 2006), DMH ablation experiments have produced variable effects on food intake (Bellinger and Bernardis, 2002), food anticipatory behavior, and other circadian rhythms (Chou et al., 2003; Gooley et al., 2006; Landry et al., 2006; Landry et al., 2007; Moriya et al., 2009). While these discrepancies are most likely due to variations in lesioning methods and experimental designs, neuron-specific, genetic manipulations are ultimately needed to resolve these issues.
The genetic and chemical makeup of DMH is poorly understood. While other hypothalamic nuclei can be defined by molecular markers, such as Agrp in the arcuate (ARC), or steroidogenic factor 1 (Sf-1) in the ventromedial hypothalamus (VMH), no such marker genes have been identified in the DMH. Identifying DMH-specific genes would be invaluable for studying cell lineages, cell biology, and the creation of a set of molecular tools for genetic DMH deletion studies.
With recent advances in whole genome sequencing and high-throughput gene expression assays, it is now possible to conduct large-scale efforts to identify and map the expression of all genes within the rodent brain (Gray et al., 2004; Lein et al., 2007). While these approaches have produced comprehensive expression databases, their high-throughput natures have necessitated the use of broad neuroanatomical classifications, and have preferentially selected for highly expressed genes. However, genes that are expressed at low levels, or in small, discrete regions of the brain, such as the DMH, are often overlooked.
To address these limitations, we set out to use a neuroanatomically guided approach to identify genes that are highly expressed in the ventral DMH relative to other hypothalamic nuclei. Laser-capture microdissection (LCM) was used to isolate the DMH and three adjacent nuclei: the PVH, ARC, and the dorsal medial portion of the ventromedial hypothalamus (dmVMH). Gene expression arrays were used to create preliminary expression profiles of each nucleus, and to identify genes that were expressed preferentially in the DMH relative to the other nuclei. RNA in situ hybridization histochemistry and real-time quantitative polymerase chain reaction (qPCR) were performed on the putative DMH genes to confirm their expression patterns within the hypothalamus, and throughout the entire brain. Using this approach, we have compiled the first comprehensive description of genes from the adult DMH.
MATERIALS AND METHODS
Animals and tissue collection
Animal protocols were approved by the Beth Israel Deaconess Medical Center and the University of Texas Southwestern Medical Center Animal Care and Use Committees. Adult male C57Bl/6 mice were housed with ad libitum access to both food and water in a light (12/12-hour on/off, 7 am to 7 pm) and temperature (21.5–22.5°C)-controlled environment.
For the microarray expression analysis, C57Bl/6 male mice (6–7 weeks old, n = 2, Taconic, Farms, German-town, NY) were fed standard chow ad libitum and sacrificed via intraperitoneal injections of chloral hydrate between 3–4 pm. Brains were rapidly isolated, imbedded in OCT compound (Tissue-Tek, Sakura Finetek, Torrance, CA), frozen on dry ice, and stored at −80°C.
For diet-induced obesity studies, 6-week-old C57Bl/6 males (Jackson laboratories, Bar Harbor, ME) were fed either a standard maintenance chow (n = 24; Harlan Teklad TD.7912, Madison, WI) or an HFD with 42% of calories derived from fat (n = 12; Harlan Teklad TD.88137). Mice were maintained on their respective diets for 12–13 weeks. Half of the mice on standard chow (n = 12) were fasted for 24 hours prior to being sacrificed. Six-week-old ob/ob males (n = 24, Jackson laboratories) were fed standard maintenance chow for 12–13 weeks. Fed mice were injected with either 100 µg of recombinant mouse leptin (n = 12; A.F. Parlow, National Hormone and Peptide Program) or saline (n = 12) and sacrificed 45 minutes later. All mice were sacrificed between 7–9 am and brains were isolated as described above.
For circadian studies, 10-week-old C57Bl/6 males (n = 6; Jackson Laboratories) were fed maintenance chow (TD.2916 Teklad Global Diet, Harlan Teklad) and sacrificed at lights on (7 am) or lights off (7 pm). Animals sacrificed during the dark phase were decapitated in complete darkness to reduce light-induced gene expression changes. Brains were isolated as described above.
Laser-capture microdissection
Brains were cryosectioned at a thickness of 14–30 µm and thaw-mounted onto either silane-coated glass slides (Labscientific, Livingston, NJ) or silane-coated PEN membrane glass slides (Applied Biosystems, Foster City, CA) and stored at −80°C. Slides were lightly fixed in 75% ethanol immediately prior to thionin staining. Slides were then dehydrated in a graded ethanol series followed by 5 minutes in xylenes. The Arcturus Autopix (Applied Biosystems) and Arcturus Veritas Microdissection System (Applied Biosystems) were used to isolate the superchiasmatic nucleus (SCN) (−0.34 mm to −0.92 mm from Bregma), retrochiasmatic nucleus (RCN) (−0.94 mm to − 1.06 mm from Bregma), PVH (−0.70 mm to − 1.22 mm from Bregma), dmVMH (−1.34 mm to −1.70 mm from Bregma), vlVMH (− 1.34 mm to − 1.70 mm from Bregma), ARC (− 1.46 mm to − 1.70 mm from Bregma), and ventral DMH (−1.94 mm from Bregma), as defined by Paxinos and Franklin (2001). RNA was extracted using a PicoPure RNA Isolation Kit (Applied Biosystems) with an on-column DNAseI treatment to remove genomic contamination (Qiagen, Valencia, CA) and stored at −80°C. RNA was visualized on the Experion Automated Electrophoresis system (Bio-Rad, Hercules, CA). The anatomic specificity of the LCM dissections was confirmed by qPCR for the expression of known marker genes in each nucleus (see below).
Amplification and microarray hybridization
One ng of total RNA underwent two rounds of linear amplification using the Arcturus RiboAmp HS RNA Amplification Kit (Molecular Devices, Palo Alto, CA). RNA transcript was labeled with the ENZO BioArray High Yield RNA Transcript Labeling Kit (Enzo Biochem, New York, NY). The resulting labeled cRNA was cleaned up via the Affymetrix GeneChip Cleanup Module (Affymetrix, Santa Clara, CA). Samples were quantified using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). Samples were hybridized to the GeneChip 430 2.0 Array (Affymetrix).
Microarray analysis
Microarray data were analyzed using the GeneSpring GX 7.3 Gene Expression Analysis software (Agilent). Briefly, linear signals were generated using the GCRMA summarization algorithm (Wu et al., 2004). To normalize the values, measurements less than 0.01 were set to 0.01. Each chip was then normalized to the 50th percentile of all measurements in that sample, and each gene was then normalized to its median value across all samples. Logarithms of the expression ratios were used as the basis for statistical analysis and the Cross-Gene Error Model (CGEM) was applied because of the small sample numbers. Genes were filtered based on the control strength as calculated by CGEM. Genes were considered ‘‘DMH-enriched’’ if they were upregulated at least 2-fold in the DMH compared to the PVH, ARC, and VMH. Twelve genes showing the highest hybridization signals in the DMH relative to the other nuclei were selected for further analysis by RNA in situ hybridization and qPCR expression analysis.
Generation of in situ hybridization histochemistry (ISHH) probes
Probes for RNA in situ hybridization were derived from PCR fragments amplified with iTaq DNA polymerase (Bio-Rad) from cDNA generated with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) from total mouse hypothalamic RNA (BD Biosciences, Palo Alto, CA). The PCR products were cloned with the TOPO TA Cloning Kit for Sequencing (Invitrogen). The probe positions and reference GenBank sequences are as follows: the Gpr50 probe includes positions 1218–1737 of GenBank accession number NM_010340. The 4930511J11Rik probe corresponds to nucleotide positions 784–1354 of GenBank accession number BC027071. The Pcsk5 probe spans nucleotides 2314–2743 of GenBank accession number BC013068. The Grp probe contains nucleotides 17–456 of GenBank accession number NM_175012. The Sulf1 probe is designed from positions 3932–4547 of GenBank accession number NM_172294. The Rorβ probe spans 877–1381 of Gen-Bank accession number NM_146095.1. Antisense and sense 35S-labeled probes were generated with MAXI-script In Vitro Transcription Kits (Ambion, Austin, TX).
ISHH
ISHH of coronal sections through the entire mouse brain were performed as previously described (Marcus et al., 2001; Kishi et al., 2003; Zigman et al., 2006). Briefly, 6–8-week-old male C57Bl/6 mice (n = 3, Taconic Farms) were intracardially perfused with 10% formalin between 1–3 PM and brains were sectioned, then mounted onto SuperFrost slides (Fisher Scientific, Pittsburgh, PA). Sections were hybridized with either sense or antisense 35S-labeled probes. ISHH signal was visualized first on autoradiographic film, then on slides dipped in Kodak NTB Autoradiography Emulsion (Carestream Health, Rochester, NY) and exposed for 1–3 weeks, developed in Kodak Dektol Black & White Paper Developer (Eastman Kodak, Rochester, NY), and counterstained with thionin. The hybridization signal was estimated subjectively. Control procedures included hybridization with sense probes and tissue pretreatment with RNase A (200 µg/ml). No specific hybridization was observed following either procedure.
Dual-label ISHH/immunohistochemistry (IHC)
Nine-week-old C57Bl/6 males (n = 3, Taconic Farms) were fasted overnight and then intraperitoneally injected with 100 µg of recombinant mouse leptin (A.F. Parlow, National Hormone and Peptide Program) or phosphate-buffered saline (PBS) and perfused with 10% formalin 45 minutes later. Free-floating brain sections were processed sequentially by ISHH for the DMH-enriched genes and IHC for phosphorylated STAT3 (p-STAT3) as previously described (Liu et al., 2003; Zigman et al., 2006). Briefly, after ISHH tissues were incubated overnight at 4°C in primary rabbit p-STAT3 antiserum (Cell Signaling Technology, Danvers, MA). Next, tissues were incubated with a biotinylated donkey antirabbit secondary (1:1,000; Jackson ImmunoResearch Laboratories, West Grove PA) for 1 hour at room temperature, followed by a 1-hour incubation in avidin-biotin complex from the Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA, 1:500). The sections were then incubated in a solution of 0.04% diaminobenzidine tetrahydrochloride (DAB; Sigma-Aldrich, St. Louis, MO). Sections were mounted onto SuperFrost slides, visualized with x-ray film, followed by photographic emulsion, as described above.
Antibody characterization
The p-STAT3 antisera used in this study is commercially available and has been tested and reported for use in IHC (Scott et al., 2009). The key features are summarized in Table 1. The antiserum recognizes a single band with the predicted molecular weight on western blots of cell extracts from SK-N-MC neuroblastoma cells (manufacturer’s datasheet). The antibody specificity was tested by enzyme-linked immunosorbent assay (ELISA), as well as by reabsorption of the antiserum with a synthetic phospho-STAT3 peptide (Scott et al., 2009). Furthermore, in these experiments p-STAT3 immunoreactivity was only detected in the brains of animals treated with leptin, and not in saline controls (data not shown).
TABLE 1.
Antibody Used
| Antigen | Immunogen | Manufacturer | Dilution used |
|---|---|---|---|
| p-STAT3 | Synthetic peptide, from mouse ADPGSAAPyLKTKFIC |
Cell Signaling Technologies (Danvers, MA) cat. no. 9131L lot 9 |
1: 4,000 |
Production of photomicrographs
Images were obtained with a Carl Zeiss Axioskop 2 microscope and a Zeiss Stemi 2000-C dissecting microscope using both brightfield and darkfield optics. Images were captured using a Zeiss digital camera and the Axiovision 3.1 software. Adobe PhotoShop CS2 (San Jose, CA) was used to combine the images into plates, make minor adjustments to contrast and brightness, and remove any obvious dust from the darkfield images.
Quantitative PCR (qPCR)
All gene expression levels were measured with an Applied Biosystems 7900HT Sequence Detection System as previously described (Bookout et al., 2006a). Real-time qPCR gene expression analysis was performed using inventoried TaqMan Gene Expression Assays (Applied Biosystems) and previously published gene assays (Fu et al., 2005; Bookout et al., 2006b).
Anatomic specificity of the LCM-isolated nuclei was confirmed by testing for the expression of the following marker genes in each nuclei by qPCR, SCN: vasoactive intestinal peptide receptor 2 (Vipr2; ABI, Cat. no. Mm00437316_m1), leptin receptor (Lepr; ABI, Cat. no. Mm00440181_m1); RCN: Lepr; ARC: Neuropeptide Y (Npy; ABI, Cat. no. Mm00445771_m1), proopiomelanocortin (Pomc; ABI, Cat. no. Mm00435874_m1), Lepr; PVH: single-minded (Sim1; ABI, Cat. no. Mm00441390_m1), arginine vasopressin (Avp; ABI, Cat. no. Mm00437761_g1), Lepr; dmVMH and vlVMH: steroidogenic factor 1 (Sf-1) (Fu et al., 2005; Bookout et al., 2006b), estrogen receptor α (Esr1) (Fu et al., 2005), estrogen receptor β (Esr2) (Fu et al., 2005), Lepr; DMH: cocaine- and amphetamine-regulated transcript (Cart; ABI, Cat. no. Mm00489086_m1), neurotensin (Nts; ABI, Cat. no. Mm00481140_m1), Lepr. The expression of marker genes were assayed in the individual nuclei samples and samples that did not show the expected patterns of marker gene expression were excluded from further analysis.
Because of the low levels of starting material, 0.5 ng of RNA from each isolated hypothalamic nuclei was converted to cDNA using the High Capacity Reverse Transcription Kit (Applied Biosystems) and subjected to 14 rounds of preamplification using the TaqMan Preamp mastermix (Applied Biosystems) using a cocktail of the marker genes listed above, as well as DMH-specific genes identified by microarray analysis: Gpr50 (ABI, Cat. no. Mm00439147_m1), 4930511J11Rik (ABI, Cat. no. Mm00512483_m1), Pcsk5 (ABI, Cat. no. Mm01206138_m1), Grp (ABI, Cat. no. Mm00612977_m1), Sulf1 (ABI, Cat. no. Mm00552283_m1), Rorβ (Fu et al., 2005). Primers for the normalizer gene 18S were not included in the preamplification assay mix. Prior to this study, the amplicons were tested to confirm unbiased, uniform amplification. Preamplified products were diluted 1/20 and PCR-amplified for 50 cycles with TaqMan Gene Expression Master Mix (Applied Biosystems) with a final concentration of 900 mM of Taq-Man Gene Expression Assays (Applied Biosystems). Analysis of gene expression was performed using the TaqManbased efficiency-corrected ΔCt assay. mRNAs with cycle times ≥30 cycles were considered below detection.
qPCR data were analyzed using ABI instrument software SDS2.1. Baseline values of amplification plots were set automatically and threshold values were kept constant to obtain normalized cycle times and linear regression data. For each sample, normalized mRNA levels were expressed as arbitrary units and were obtained by dividing the averaged, efficiency-corrected values for each gene by that for 18S RNA. The resulting values were multiplied by 105 for graphical representation and plotted ± standard deviation from triplicate sample wells. Fold changes were calculated by dividing normalized signal intensities for fasted and HFD conditions by those obtained under standard chow. Fold changes of less than 0.8 or greater than 1.2 were considered physiologically significant.
RESULTS
LCM and microarray analysis
The DMH can be divided into two anatomically and functionally distinct parts, the compact DMH, and the leptin receptor-rich dorsal and ventral portions (Elmquist et al., 1997, 1998a; Elias et al., 2000). To compare gene expression profiles between the leptin receptor expressing portion of the DMH and the adjacent metabolically important regions of the hypothalamus, LCM was used to isolate RNA from the PVH, dmVMH, ARC, and ventral DMH (Fig. 1). Hypothalamic nuclei were collected from two male C57Bl/6 mice fed on standard chow ad libitum. Unpooled RNA from each nuclei was subjected to two rounds of amplification, biotin-labeled, and hybridized to the Affymetrix GeneChip 430 2.0 Array, consisting of over 39,000 transcripts.
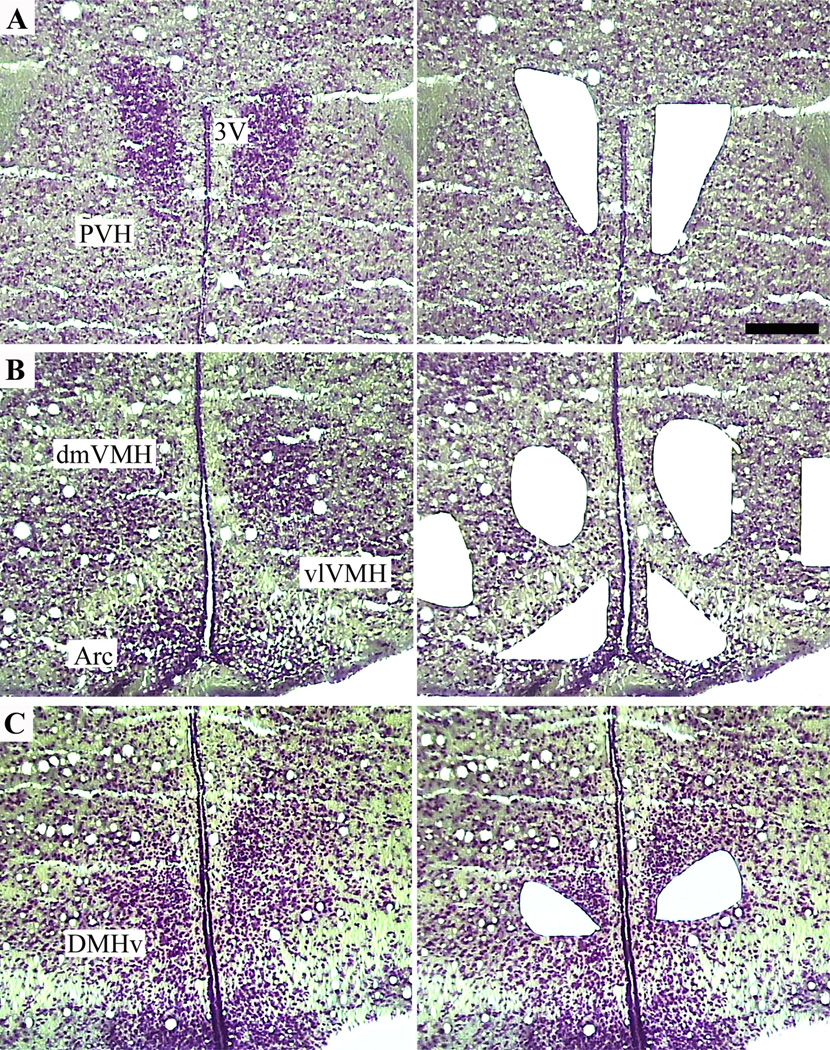
Figure 1.
Isolation of specific hypothalamic nuclei from mouse brain by laser-capture microdissection. Left: Hypothalamic sections stained with thionin allow visualization of the neuroanatomical subdivisions. Right: The remainder of the hypothalamus after selective capture and removal of the cells of interest. 3V, third ventricle; Arc, arcuate nucleus; dmVMH, dorsomedial ventromedial hypothalamic nucleus; PVH, paraventricular nucleus; vlVMH, ventrolateral ventromedial hypothalamic nucleus. Scale bar 200= µm.
To confirm the specificity of the dissections and the fidelity of the RNA amplification, we first screened the data generated from the microarrays for genes known to be specifically or preferentially expressed in each of the dissected nuclei. We confirmed the exclusive and robust expression of Npy, Pomc, and Agrp in the ARC (Elmquist et al., 1998c); Sf-1 and Cbln1 in the dmVMH (Segal et al., 2005); Sim1 in the PVH (Michaud et al., 1998); and the preferential expression of orexin (Ox) (Chou et al., 2001) and neurotensin (Nts) (Watts et al., 1999) in the DMH as compared to the other nuclei examined (data not shown).
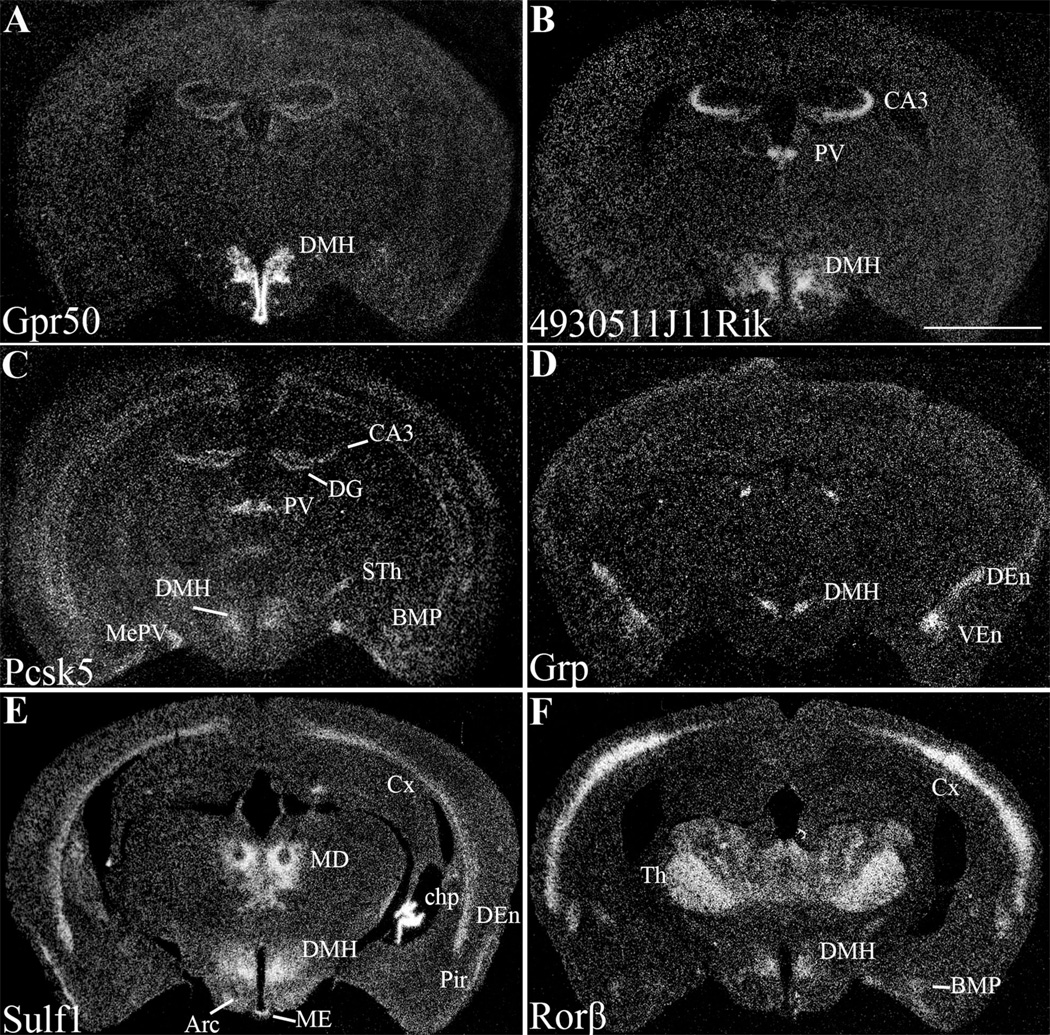
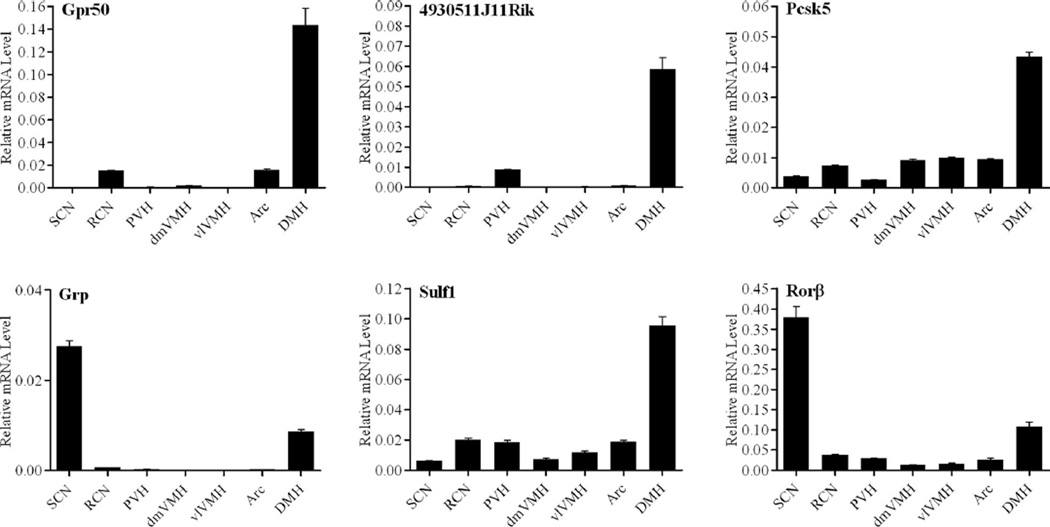
After normalization, 67 genes were found to be expressed at least 2-fold above background levels in the DMH. Many of the 67 genes were also coexpressed at high levels in the ARC. To restrict the population to DMH-specific genes, we selected genes that were expressed at least 2-fold higher in the DMH as compared to the PVH, dmVMH, and ARC. We found that 36 genes met this initial criteria for DMH enrichment. Next, we removed genes that were previously described to be strongly expressed in regions adjacent to the DMH, but which were restricted to only a few cells within the ventral DMH; these included melanin-concentrating hormone (Mch) (Nahon et al., 1989) and Ox (de Lecea et al., 1998; Chou et al., 2001), which are predominantly expressed in the lateral hypothalamic area (LHA), and histidine decarboxylase (Hdc) (Castren and Panula, 1990), which is expressed in the medial tuberal nucleus (MTu). These exclusions produced a list of 33 genes preferentially expressed in the DMH and 14 of these were selected for further analysis by ISHH and/or qPCR (Table 2). We then eliminated those genes whose expression could not be confirmed by both in situ hybridization and qPCR, or which were found to have high levels of expression outside of the DMH. Using these criteria, we selected six genes: G-protein-coupled receptor 50 (Gpr50), clone 4930511J11Rik, proprotein convertase subtilisin/kexin type 5 (Pcsk5), gastrin releasing peptide (Grp), sulfatase 1 (Sulf1), and retinoid-related orphan nuclear receptor β (Rorb) for further expression analysis (Fig. 2). RNA in situ hybridization showed the Gpr50 and 4930511J11Rik were most strongly expressed in the DMH, with limited expression in other brain sites. Pcsk5, Grp, Sulf1, and Rorβ; were moderately expressed in the DMH and also showed restricted expression in other sites. The gene distribution patterns within the hypothalamus were confirmed and quantified using real-time qPCR on additional LCM samples taken from broader sampling of hypothalamic nuclei, including the RCN, SCN, PVH, dmVMH, ventrolateral portion of the ventromedial hypothalamus (vlVMH), ARC, and DMH (Fig. 3). qPCR analysis allowed us to determine the relative expression levels of these genes across the hypothalamus and confirmed the robust expression of all six genes within the DMH. Furthermore, our qPCR findings also confirmed the previously described expression of Grp (Aida et al., 2002) and Rorβ (Schaeren-Wiemers et al., 1997; Andre et al., 1998; Sumi et al., 2002) in the SCN.
TABLE 2.
DMH-Enriched Genes
| Gene symbol | Gene name | Accession no. | Normalized intensity |
|---|---|---|---|
| Gpr50*# | G-protein-coupled receptor 50 | NM_010340 | 28.17 |
| Sulf1*# | sulfatase 1 | NM_172294 | 8.10 |
| 4930511J11Rik*# | RIKEN cDNA 4930511J11 gene | BC027071 | 7.29 |
| Grp*# | gastrin releasing peptide | NM_175012 | 5.52 |
| 4930547N16Rik# | RIKEN cDNA 4930547N16 gene | XM_125861 | 4.67 |
| Lypd6* | LY6/PLAUR domain containing 6 | NM_177139 | 4.55 |
| Gpr64# | G protein-coupled receptor 64 | NM_178712 | 4.29 |
| Pcsk5*# | proprotein convertase subtilisin/kexin type 5 | BC013068 | 3.58 |
| Nrp1# | Neuropilin 1 | NM_008737 | 3.24 |
| Synpr* | synaptoporin | NM_028052 | 3.00 |
| Trhr* | thyrotropin releasing hormone receptor | NM_013696 | 2.81 |
| Rorb*# | RAR-related orphan receptor beta | NM_146095 | 2.80 |
| 2310010M24Rik | RIKEN cDNA 2310010M24 gene | NM_027990 | 2.76 |
| Cbln2 | cerebellin 2 precursor protein | NM_172633 | 2.68 |
| Met | met proto-oncogene | NM_008591 | 2.68 |
| Erbb4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 | NM_010154 | 2.55 |
| Rgs16* | regulator of G-protein signaling 16 | NM_011267 | 2.54 |
| Ccnd2 | cyclin D2 | NM_009829 | 2.47 |
| Zmiz1/Rai17# | zinc finger, MIZ-type containing 1 | NM_183208 | 2.34 |
| 4921525O09Rik | RIKEN cDNA 4921525O09 gene | AV312506 | 2.31 |
| 3110001A13Rik | RIKEN cDNA 3110001A13 gene | NM_025626 | 2.31 |
| Transcribed locus | AV133559 | 2.27 | |
| Dlx6os2 | Dlx6 opposite strand transcript 2 | BB023120 | 2.25 |
| C130034I18Rik | RIKEN cDNA C130034I18 gene | NM_177233 | 2.23 |
| Cachd1 | cache domain containing 1 | NM_198037 | 2.22 |
| Galr1 | galanin receptor 1 | NM_008082 | 2.21 |
| Nts | neurotensin | NM_024435 | 2.21 |
| Lbxcor1 | ladybird homeobox 1 homolog corepressor 1 | NM_172446 | 2.15 |
| Kirrel3# | kin of IRRE like 3 | NM_026324 | 2.13 |
| EG226654 | predicted gene, EG226654 | XM_129558 | 2.10 |
| Herc4# | hect domain and RLD 4 | NM_026101 | 2.09 |
| Ankrd38 | ankyrin repeat domain 38 | NM_172872 | 2.08 |
| Vwf | Von Willebrand factor homolog | NM_011708 | 2.08 |
Figure 2.
A series of low-power darkfield photomicrographs summarizing the distribution of the candidate genes in the mouse brain. Arc, arcuate nucleus; BMP, posterior basomedial amygdaloid nucleus; CA3, field CA3 of hippocampus; chp, choroid plexus; Cx, cortex; DEn, dorsal endopiriform nucleus; DG, dentate gyrus; DMH, dorsal medial hypothalamus; MD, mediodorsal thalamus; ME, median eminence; MePV, posteroventral medial amygdaloid nucleus; Pir, piriform cortex; PV, paraventricular thalamic nuclei; STh, subthalamic nucleus; Th, thalamus; VEn, ventral endopiriform nucleus. Scale bar = 2 mm.
Figure 3.
Relative distribution of candidate genes across the hypothalamus. Graphs are representative of duplicate experiments. Values are plotted as the mean of triplicate measurements ± standard deviation error bars. Arc, Arcuate nucleus; DMH, dorsomedial hypothalamus; dmVMH, dorsomedial ventromedial hypothalamic nucleus; PVH, paraventricular nucleus; RCN, retrochiasmatic nucleus; SCN, supra-chiasmatic nucleus; vlVMH, ventrolateral ventromedial hypothalamic nucleus.
Characterization of DMH-enriched genes
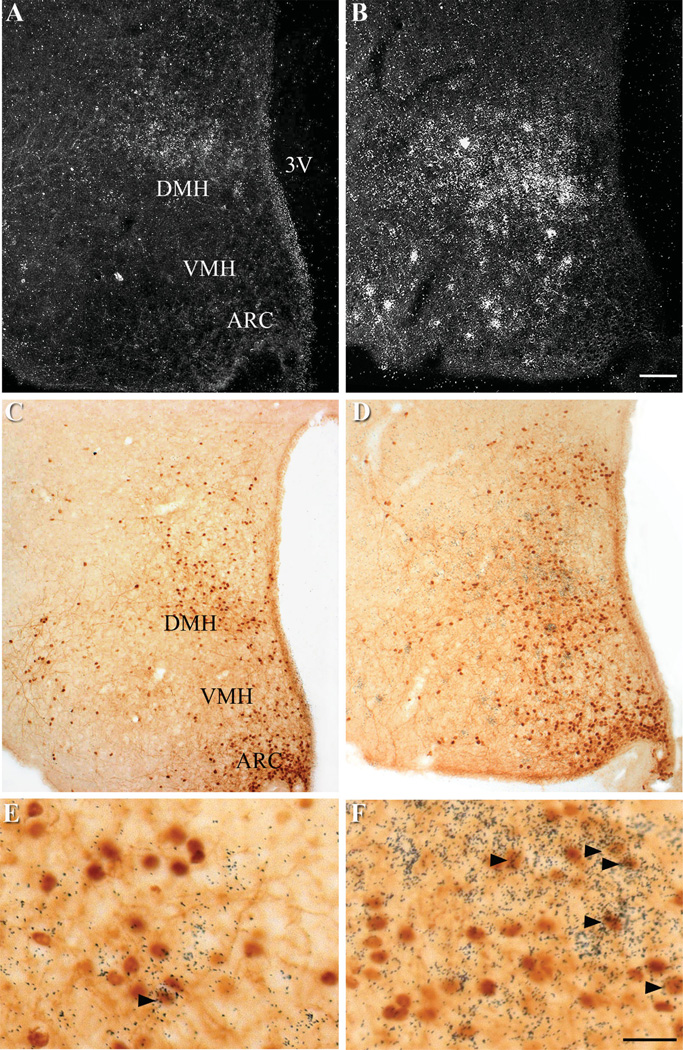
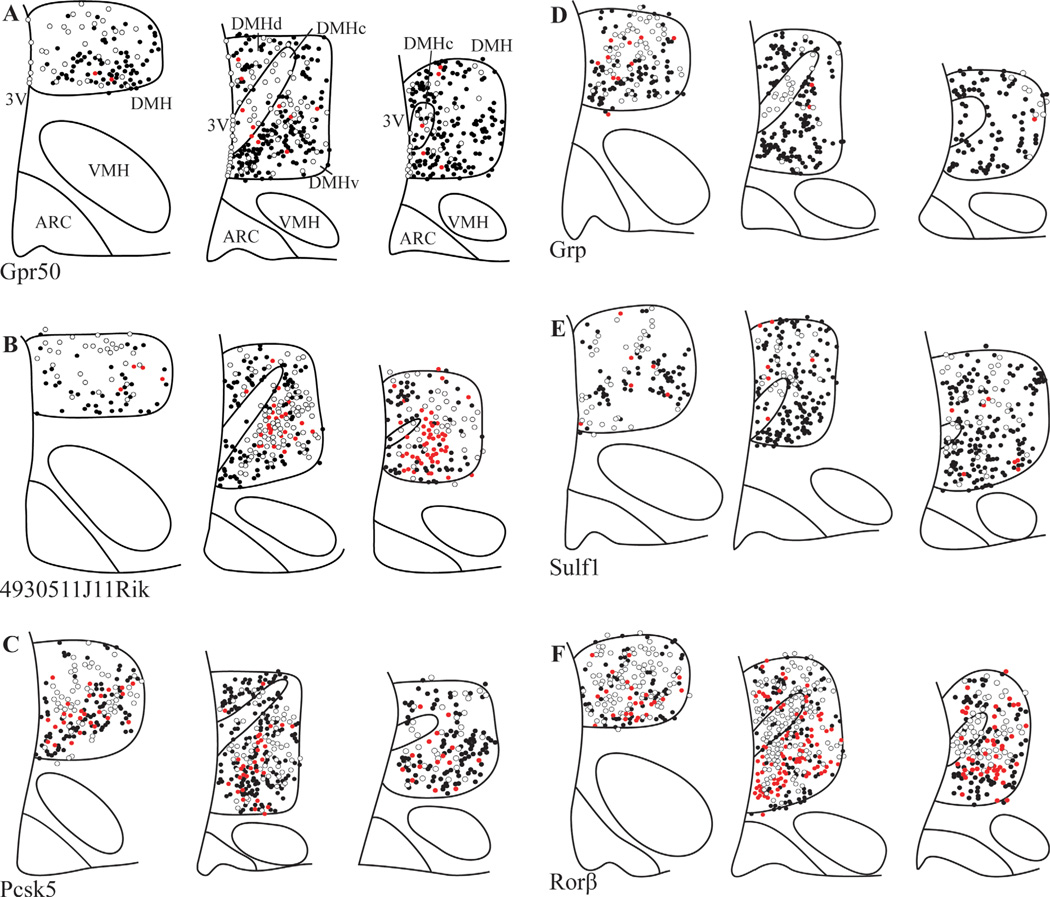
To further map the distribution of these genes within the DMH, we subdivided the DMH into rostral, central, and caudal levels, and within each level, into dorsal, ventral, and compact regions based on previously described distributions of leptin-responsive cells (Elias et al., 2000). To visualize the distribution of the six DMH-enriched genes relative to leptin responsive cells we performed dual-label ISHH for the genes of interest with IHC for p-STAT3, a protein downstream of leptin receptor that is commonly used as a marker of leptin signaling (Figs. 4, 5) (Vaisse et al., 1996). All of the genes examined were expressed throughout the rostral, central, and caudal levels of the DMH. Interestingly, Gpr50, 4930511J11Rik, Pcsk5, and Sulf1 were distributed in a pattern similar to those seen for leptin responsive cells, with greatest expression in the dorsal and ventral portions of the DMH, and limited and scattered expression within the compact DMH. Despite this general similarity, there were only limited occurrences of colocalization between the RNA in situ silver grains and cells immunopositive for p-STAT3 (Figs. 4, 5). In contrast, while silver grains representing Grp mRNA were observed throughout the DMH, they were concentrated in the rostral and compact DMH, in regions that were devoid of leptin responsive cells. While Rorβ had the most extensive distribution of silver grains throughout the brain, with especially robust signal within the DMH, there was only moderate colocalization with neurons immunopositive for p-STAT3. Genes expressed primarily in nonneuronal cells would also show limited colocalization with p-STAT3 immunostaining. However, careful examination of dual-label ISHH/IHC sections showed silver grain deposition patterns consistent with that seen for neurons, with no histological evidence of gene expression in glial cells (Fig. 4E,F).
Figure 4.
Distribution of p-STAT3 immunoreactivity and Grp50 and 4930511J11Rik mRNA in the DMH. A,C,E: Gpr50 ISHH with p-STAT3 IHCC. B,D,F: 4930511J11Rik ISHH with p-STAT3 IHCC. mRNA hybridization is represented by silver grains, while p-STAT3 immunoreactivity is represented by brown staining. A,B: Darkfield image. C–F: Brightfield images. The DMH regions of C,D are magnified in E,F. Black arrows indicate examples of neurons doubly labeled with either Gpr50 or 4930511J11Rik riboprobe and anti-p-STAT3 antisera. Scale bars = 100 µm in B; 25 µm in F.
Figure 5.
Camera lucida drawings of dual-label ISHH/IHC showing the relative distribution of p-STAT3 and the DMH-enriched genes. Drawings were made from representative sections from three rostral-caudal levels of the DMH, and only include signals detected within the DMH. Sections are organized from rostral (left) to caudal (right). Each circle represents one neuron. White circles, neurons with silver grains (ISHH); black circles, neurons with DAB (p-STAT3 IHC), red circles, double stained. Arc, Arcuate nucleus; 3V, third ventricle; ARC, arcuate nucleus; DMHc, compact subdivision of the DMH, DMHd, dorsal subdivision of the DMH, DMHv, ventral subdivision of the DMH; VMH, ventromedial nucleus.
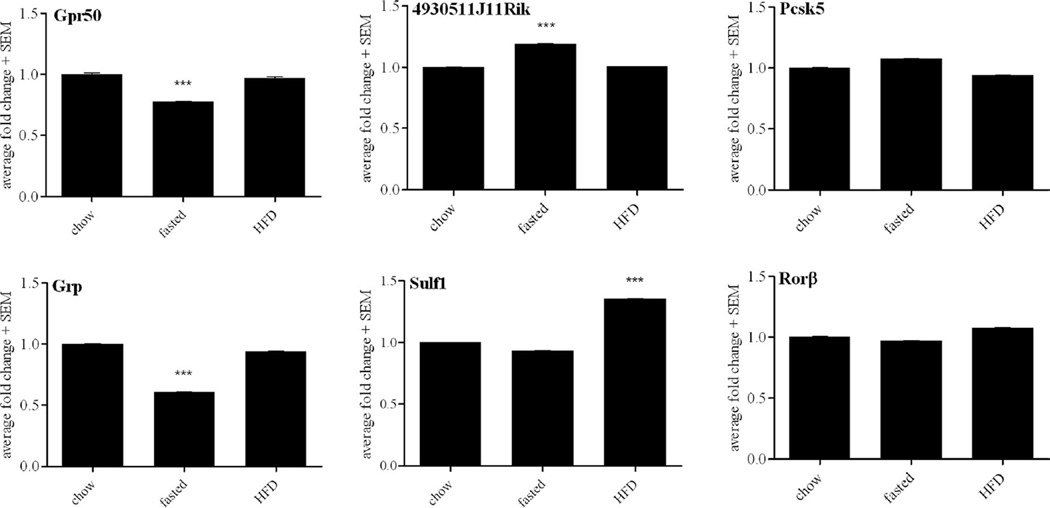
The DMH has been shown to innervate the ARC and PVH (Gautron et al., 2010), sites critical for the control of energy balance, glucose homeostasis, and food intake (Balthasar et al., 2005; Coppari et al., 2005). To assess the potential role of the DMH-enriched genes on metabolism we performed real-time qPCR assays from LCM dissected samples to assess gene expression levels in mice fed a standard maintenance chow, a high-fat and high-cholesterol diet (HFD), or mice that underwent an extended fast for 24 hours (Fig. 6). Of the genes assayed, moderate and statistically significant changes in response to fasting were seen in the expression of Gpr50 (0.8-fold), 4930511J11Rik (1.2-fold), and Grp (0.6-fold). Sulf1 showed moderate upregulation (1.4-fold) in response to HFD. LCM dissected samples obtained from leptin-deficient, ob/ob mice did not show significant changes in gene expression between those that were intraperitoneally injected with leptin and those treated with saline (data not shown).
Figure 6.
Metabolic regulation of candidate genes. Values are plotted as the mean of triplicate measurements ± standard deviation error bars. Fold change is compared to standard chow, expressed as 1. Only values less than 0.8 or greater than 1.2 fold change are reported as significant. ***P < 0.001.
A number of the newly identified genes have been shown to be expressed in a circadian manner in other tissues. For instance, Gpr50 expression oscillates in tanycytes lining the third ventricle (Kamphuis et al., 2005), Rorβ in retina and peripheral white adipose tissue (Kamphuis et al., 2005; Yang et al., 2006), and Grp in the SCN (Zoeller et al., 1992). Despite these reports and the innervations between the DMH and the SCN and SPZ, we did not detect any day–night expression differences in the six DMH-enriched genes (data not shown).
DISCUSSION
Historically, hypothalamic gene expression studies were performed on samples from homogenized whole hypothalamus or from tissue punches of the approximate region of interest. These methods were imprecise and limited to hypothalamic regions that could be grossly identified. Using LCM, we are able to obtain pure, nuclei specific samples with minimal cross-contamination from adjacent tissues. This is particularly important because of the spatial and functional organization within the hypothalamus and the sensitivity of the microarray and qPCR expression assays that we used.
We identified six genes that are strongly enriched in the ventral portion of the DMH relative to other areas of the hypothalamus. Five of these genes (Gpr50, Pcsk5, Grp, Sulf1, and Rorβ) can be seen to be expressed in the DMH in the Allen Mouse Brain Atlas (Lein et al., 2007) when sections at the level of the DMH are individually analyzed, but the overall expression levels were too restricted to be included on their annotated list of hypothalamic genes.
Other groups have also used microarray analysis to investigate the molecular makeup of the DMH. Draper et al. (2010) examined differential gene expression between NPY neurons in the DMH and the ARC in juvenile mice. While they identified 41 genes that were preferentially expressed in the DMH, there was no overlap with the genes from our assay. These differences are most likely due to differences between juvenile and adult mice, and between isolated NPY neurons and the heterogeneous population of cells found in the ventral DMH.
While we initially predicted that genes highly expressed in the DMH would be strongly regulated in response to dietary manipulations, we detected modest changes in gene expression in only four of the genes. It is possible that the technical limitations of profiling gene expression from minute cell populations, which requires cDNA amplification, may mask or suppress differences in gene expression. However, our initial tests suggest that the expression levels of most genes are faithfully and repeatedly assayed after amplification. It is also possible that these genes are important for DMH-associated physiological processes other than metabolism, such as reproduction, body temperature, or corticosterone secretion.
While p-STAT3 immunoreactivity is commonly used to identify leptin-responsive neurons, it has recently been shown that the assessment of p-STAT3 immunoreactivity shortly after leptin administration may underrepresent the total number of leptin-responsive cells in certain areas of the brain, including the DMH (Scott et al., 2009). Thus, while we found a limited degree of colocalization between our DMH-specific genes and p-STAT3 immunoreactivity, a more detailed study following prolonged leptin treatment, or using one of the newly described LepRb reporter mice may be necessary (Scott et al., 2009). Alternatively, it is possible that the DMH-enriched genes respond to HFD and fasting through leptin-independent pathways. It has recently been shown that NPY neurons of the DMH are not leptin-responsive (Draper et al., 2010), suggesting that there is indeed a leptin-insensitive population of DMH neurons that responds to metabolic cues.
Food entrainment or food anticipatory activity describes the range of physiological behaviors that are associated with expected mealtimes (Bolles and Stokes, 1965; Krieger, 1974; Herzog and Muglia, 2006). In rodents, this includes increased wakefulness, activity such as running, eating, licking, body temperature, hormone secretion, digestive activity, and hepatic gene transcription. Furthermore, these anticipatory behaviors persist during prolonged fasts (Boulos et al., 1980). The existence of a food entrainable oscillator, similar to the light entrainable oscillator located in the SCN, has been postulated since the late 1970s (Krieger et al., 1977). It is unclear whether this oscillator exists as a unique cluster of cells within the DMH, is distributed among various hypothalamic nuclei and/or the brainstem (Mistlberger, 2011), or if it requires a functional interaction between the SCN and DMH (Acosta-Galvan et al., 2011). It is intriguing to note that Grp and Rorβ are also expressed in the SCN, while Grp, Gpr50, and Rorβ have been reported to have oscillating patterns of expression in other tissues (Zoeller et al., 1992; Kamphuis et al., 2005; Yang et al., 2006). While our preliminary analyses did not reveal circadian expression in the DMH, these genes remain intriguing candidates for mediators of circadian and metabolic interactions.
Candidate genes
Gpr50 was initially cloned from the pituitary but has since been shown to be expressed in the DMH and tanycytes lining the third ventricle (Reppert et al., 1996; Drew et al., 1998, 2001; Sidibe et al., 2010). Despite high sequence similarity to the melatonin receptor family, Gpr50 does not bind to radiolabeled melatonin (Reppert et al., 1996); however, recent work has shown that it does form heterodimers with the melatonin receptor MT1 to reduce the activity of MT1 itself (Levoye et al., 2006). Furthermore, Gpr50 expression is decreased in short-day photoperiods (Barrett et al., 2006). Interestingly, sequence variants in humans have been associated with circulating triglyceride levels, high-density lipoprotein (HDL) levels, bipolar affective disorder, and autism (Thomson et al., 2005; Bhattacharya et al., 2006). Most recently, a genetically modified mouse lacking Gpr50 was shown to have decreased body weight accompanied by increased activity and basal metabolic rate (Ivanova et al., 2008). The authors examined the regulation of Gpr50 expression levels using radioactive RNA in situ hybridization, and found that Gpr50 was strongly decreased by a 36-hour fast, or after a 5-week exposure to a high-calorie diet. In our experiments, we found Gpr50 expression in the DMH to be only moderately regulated by fasting, but did not show regulation in response to HFD. These differences could be due to diet formulation, sample collection, or the assay methodology. It should also be noted that we also detected very low levels of Gpr50 expression in the RCN and ARC, two metabolically significant regions and thus potentially important sites of action for Gpr50.
Clone 4930511J11Rik has recently been identified by sequence homology as claudin 26, a member of the claudin protein family that is important for tight junction formation (Mineta et al., 2011). Claudin 26 was shown to be expressed in mouse embryo and in adult brain and intestine. In the brain, claudins are known to be important for the maintenance of the blood–brain barrier, and more recent work suggests that they may have broader functions, such as stress response and embryonic morphogenesis (Cardoso et al., 2010). The expression of 4930511J11Rik/claudin 26 in the DMH is puzzling, but may reflect a role for these proteins outside of tight junctions. Indeed, the expression of several claudins have recently detected in the oligodendrocytes, neurons, and astrocytes of the cortex in humans (Romanitan et al., 2010).
Pcsk5 is a proprotein convertase involved in the processing of secretory proteins (Lissitzky et al., 2000). Pcsk5 has been previously shown to be expressed in the rodent brain, where it is speculated to be involved in processing of the neuropeptides, including cholecystokinin and neurotensin (Dong et al., 1995; Villeneuve et al., 1999; Villeneuve et al., 2000; Cain et al., 2003). Pcsk5 is also essential for development, and deletion of the catalytic subunit is embryonic lethal at the implantation stage (Essalmani et al., 2006, 2008). Intriguingly, variants have been associated with HDL cholesterol levels in humans (Iatan et al., 2009), although this is thought to be most likely due to an effect on endothelial lipase activity rather than a central effect of Pcsk5.
Grp is a bombesin-like peptide known for stimulating the release of gastrin and inhibiting feeding. It is expressed throughout the brain and periphery, especially in the gastrointestinal tract (Aida et al., 2002; Ohki-Hamazaki et al., 2005; Gonzalez et al., 2008). Its receptor, Grp-R, is expressed peripherally, as well as in the brain, with strong expression in the hypothalamus. Grp and its receptor are involved in numerous physiological processes, including, smooth-muscle contraction in the gastrointestinal/urogenital tract, immune function, insulin secretion, cancer, regulation of circadian rhythms, thermoregulation, behavior, and satiety (reviewed in Gonzalez et al., 2008). In fact, Grp can directly excite NPY neurons in electrophysiological slice preparations (van den Pol et al., 2009). While a Grp mouse has not been yet reported, several groups have produced Grpr-deficient mice. The loss of the receptor caused behavioral changes, including increased locomotor activity and social responses, and eliminated the effects of bombesin administration on feeding suppression (Wada et al., 1997; Hampton et al., 1998). Grp also plays a crucial role in the photic signaling cascade in the SCN (Piggins et al., 1995; Albers et al., 1995; Aida et al., 2002; Karatsoreos et al., 2004). Grp cells within the SCN receive direct retinal input via the retinohypothalamic tract, and microinjections of exogenous Grp into the SCN can cause a phase shift of the circadian rhythms.
Rorβ is a member of the nuclear receptor family of transcription factors (Carlberg et al., 1994). It is highly expressed in the brain, especially in the regions involved in sensory processing, such as the cortex, thalamus; and circadian timing, such as the pineal gland, retina, and the SCN, where it oscillates with a diurnal rhythm (Schaeren-Wiemers et al., 1997; Andre et al., 1998; Sumi et al., 2002). Furthermore, Rorβ-deficient mice are ataxic, with decreased fertility and retinal degeneration (Andre et al., 1998; Masana et al., 2007). Despite the retinal degeneration, these mice still respond and entrain to a light-dark cycle, but show an altered free-running period during the dark cycle.
Sulf1 is a heparin sulfate 6-O-endosulfatase and modifies heparin sulfate proteoglycans (HSPGs) at the cell surface to regulate signaling molecules, such as fibroblast growth factor, bone morphogenic protein (BMP), wingless (WNT), Decapentaplegic (Dpp), and hedgehog (Hh) (Perrimon and Bernfield, 2000; Morimoto-Tomita et al., 2002; Ai et al., 2003; Hacker et al., 2005; Lamanna et al., 2007). Sulf1-deficient mice have defects in neurite outgrowth, behavioral problems, and reduced spine density in the hippocampus (Kalus et al., 2009). The loss of either Sulf1 or the closely related Sulf2 disrupts cartilage homeostasis through the BMP and fibroblast growth factor (FGF) pathways (Otsuki et al., 2010). Mice that are missing both Sulf1 and Sulf2 show increased neonatal lethality, accompanied by skeletal and renal defects (Holst et al., 2007). Sulf1 and Sulf2 also appear to function together for proper esophageal innervation (Ai et al., 2007).
CONCLUSION
We described six genes that are highly expressed within the DMH. Of these genes, Gpr50, 4930511J11Rik, Grp, and Sulf1 respond to basic metabolic manipulations, but none of them exhibit diurnal fluctuations. Future experiments that include gene targeting will be required to define the role of these genes in the DMH. In addition to extending our knowledge about the molecular makeup of the DMH, the identification of these and other DMH-specific genes will undoubtedly add to the molecular tools available for studying this elusive part of the hypothalamus, including the development of reporter constructs and recombinase drivers.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant numbers: P01 DK088761, PL1 DK081182, and UL1 RR024923; Grant numbers: R01DK53301 and RL1DK081185 (to J.K.E.).
LITERATURE CITED
- Acosta-Galvan G, Yi CX, van der Vliet J, Jhamandas JH, Panula P, Angeles-Castellanos M, Del Carmen Basualdo M, Escobar C, Buijs RM. Interaction between hypothalamic dorsomedial nucleus and the suprachiasmatic nucleus determines intensity of food anticipatory behavior. Proc Natl Acad Sci U S A. 2011;108:5813–5818. doi: 10.1073/pnas.1015551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134:3327–3338. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- Aida R, Moriya T, Araki M, Akiyama M, Wada K, Wada E, Shibata S. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- Albers HE, Gillespie CF, Babagbemi TO, Huhman KL. Analysis of the phase shifting effects of gastrin releasing peptide when microinjected into the suprachiasmatic region. Neurosci Lett. 1995;191:63–66. doi: 10.1016/0304-3940(95)11559-1. [DOI] [PubMed] [Google Scholar]
- Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M. Disruption of retinoid-related orphan receptor beta changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 1998;17:3867–3877. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T, Ferreira M, Tang V, McGovern RA, Kenny CD, Christiansen LM, Edelstein E, Choi B, Boss O, Aschkenasi C, Zhang CY, Mountjoy K, Kishi T, Elmquist JK, Lowell BB. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Barrett P, Ivanova E, Graham ES, Ross AW, Wilson D, Ple H, Mercer JG, Ebling FJ, Schuhler S, Dupre SM, et al. Photoperiodic regulation of cellular retinoic acid-binding protein 1, GPR50 and nestin in tanycytes of the third ventricle ependymal layer of the Siberian hamster. J Endocrinol. 2006;191:687–698. doi: 10.1677/joe.1.06929. [DOI] [PubMed] [Google Scholar]
- Bellinger LL, Bernardis LL. The dorsomedial hypothalamic nucleus and its role in ingestive behavior and body weight regulation: lessons learned from lesioning studies. Physiol Behav. 2002;76:431–442. doi: 10.1016/s0031-9384(02)00756-4. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Luan J, Challis B, Keogh J, Montague C, Brennand J, Morten J, Lowenbeim S, Jenkins S, Farooqi IS, et al. Sequence variants in the melatonin-related receptor gene (GPR50) associate with circulating triglyceride and HDL levels. J Lipid Res. 2006;47:761–766. doi: 10.1194/jlr.M500338-JLR200. [DOI] [PubMed] [Google Scholar]
- Bolles RC, Stokes LW. Rat’s anticipation of diurnal and a-diurnal feeding. J Comp Physiol Psychol. 1965;60:290–294. doi: 10.1037/h0022308. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol. 2006a doi: 10.1002/0471142727.mb1508s73. Chapter 15, Unit 15.8. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006b;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulos Z, Rosenwasser AM, Terman M. Feeding schedules and the circadian organization of behavior in the rat. Behav Brain Res. 1980;1:39–65. doi: 10.1016/0166-4328(80)90045-5. [DOI] [PubMed] [Google Scholar]
- Cain BM, Connolly K, Blum A, Vishnuvardhan D, Marchand JE, Beinfeld MC. Distribution and colocalization of cholecystokinin with the prohormone convertase enzymes PC1, PC2, and PC5 in rat brain. J Comp Neurol. 2003;467:307–325. doi: 10.1002/cne.10924. [DOI] [PubMed] [Google Scholar]
- Cardoso FL, Brites D, Brito MA. Looking at the blood-brain barrier: molecular anatomy and possible investigation approaches. Brain Res Rev. 2010;64:328–363. doi: 10.1016/j.brainresrev.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-Andre M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- Castren E, Panula P. The distribution of histidine decarboxylase mRNA in the rat brain: an in situ hybridization study using synthetic oligonucleotide probes. Neurosci Lett. 1990;120:113–116. doi: 10.1016/0304-3940(90)90181-8. [DOI] [PubMed] [Google Scholar]
- Chou TC, Lee CE, Lu J, Elmquist JK, Hara J, Willie JT, Beuckmann CT, Chemelli RM, Sakurai T, Yanagisawa M, et al. Orexin (hypocretin) neurons contain dynorphin. J Neurosci. 2001;21:RC168. doi: 10.1523/JNEUROSCI.21-19-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Scammell TE, Gooley JJ, Gaus SE, Saper CB, Lu J. Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms. J Neurosci. 2003;23:10691–10702. doi: 10.1523/JNEUROSCI.23-33-10691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppari R, Ichinose M, Lee CE, Pullen AE, Kenny CD, McGovern RA, Tang V, Liu SM, Ludwig T, Chua SC, Jr, et al. The hypothalamic arcuate nucleus: a key site for mediating leptin’s effects on glucose homeostasis and locomotor activity. Cell Metab. 2005;1:63–72. doi: 10.1016/j.cmet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, Fukuhara C, Battenberg EL, Gautvik VT, Bartlett FS, 2nd, et al. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci U S A. 1998;95:322–327. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Marcinkiewicz M, Vieau D, Chretien M, Seidah NG, Day R. Distinct mRNA expression of the highly homologous convertases PC5 and PACE4 in the rat brain and pituitary. J Neurosci. 1995;15:1778–1796. doi: 10.1523/JNEUROSCI.15-03-01778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper S, Kirigiti M, Glavas M, Grayson B, Chong CN, Jiang B, Smith MS, Zeltser LM, Grove KL. Differential gene expression between neuropeptide Y expressing neurons of the dorsomedial nucleus of the hypothalamus and the arcuate nucleus: microarray analysis study. Brain Res. 2010;1350:139–150. doi: 10.1016/j.brainres.2010.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew JE, Barrett P, Williams LM, Conway S, Morgan PJ. The ovine melatonin-related receptor: cloning and preliminary distribution and binding studies. J Neuroendocrinol. 1998;10:651–661. doi: 10.1046/j.1365-2826.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- Drew JE, Barrett P, Mercer JG, Moar KM, Canet E, Delagrange P, Morgan PJ. Localization of the melatonin-related receptor in the rodent brain and peripheral tissues. J Neuroendocrinol. 2001;13:453–458. doi: 10.1046/j.1365-2826.2001.00651.x. [DOI] [PubMed] [Google Scholar]
- Elias CF, Kelly JF, Lee CE, Ahima RS, Drucker DJ, Saper CB, Elmquist JK. Chemical characterization of leptin-activated neurons in the rat brain. J Comp Neurol. 2000;423:261–281. [PubMed] [Google Scholar]
- Elmquist JK, Ahima RS, Maratos-Flier E, Flier JS, Saper CB. Leptin activates neurons in ventrobasal hypothalamus and brainstem. Endocrinology. 1997;138:839–842. doi: 10.1210/endo.138.2.5033. [DOI] [PubMed] [Google Scholar]
- Elmquist JK, Ahima RS, Elias CF, Flier JS, Saper CB. Leptin activates distinct projections from the dorsomedial and ventromedial hypothalamic nuclei. Proc Natl Acad Sci U S A. 1998a;95:741–746. doi: 10.1073/pnas.95.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmquist JK, Bjorbaek C, Ahima RS, Flier JS, Saper CB. Distributions of leptin receptor mRNA isoforms in the rat brain. J Comp Neurol. 1998b;395:535–547. [PubMed] [Google Scholar]
- Elmquist JK, Maratos-Flier E, Saper CB, Flier JS. Unraveling the central nervous system pathways underlying responses to leptin. Nat Neurosci. 1998c;1:445–450. doi: 10.1038/2164. [DOI] [PubMed] [Google Scholar]
- Essalmani R, Hamelin J, Marcinkiewicz J, Chamberland A, Mbikay M, Chretien M, Seidah NG, Prat A. Deletion of the gene encoding proprotein convertase 5/6 causes early embryonic lethality in the mouse. Mol Cell Biol. 2006;26:354–361. doi: 10.1128/MCB.26.1.354-361.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essalmani R, Zaid A, Marcinkiewicz J, Chamberland A, Pasquato A, Seidah NG, Prat A. In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc Natl Acad Sci U S A. 2008;105:5750–5755. doi: 10.1073/pnas.0709428105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Sun T, Bookout AL, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. A Nuclear Receptor Atlas: 3T3-L1 adipogenesis. Mol Endocrinol. 2005;19:2437–2450. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- Gautron L, Lazarus M, Scott MM, Saper CB, Elmquist JK. Identifying the efferent projections of leptin-responsive neurons in the dorsomedial hypothalamus using a novel conditional tracing approach. J Comp Neurol. 2010;518:2090–2108. doi: 10.1002/cne.22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Moody TW, Igarashi H, Ito T, Jensen RT. Bombesin-related peptides and their receptors: recent advances in their role in physiology and disease states. Curr Opin Endocrinol Diabetes Obes. 2008;15:58–64. doi: 10.1097/MED.0b013e3282f3709b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley JJ, Schomer A, Saper CB. The dorsomedial hypothalamic nucleus is critical for the expression of food-entrainable circadian rhythms. Nat Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Guan XM, Yu H, Trumbauer M, Frazier E, Van der Ploeg LH, Chen H. Induction of neuropeptide Y expression in dorsomedial hypothalamus of diet-induced obese mice. Neuroreport. 1998;9:3415–3419. doi: 10.1097/00001756-199810260-00015. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Hampton LL, Ladenheim EE, Akeson M, Way JM, Weber HC, Sutliff VE, Jensen RT, Wine LJ, Arnheiter H, Battey JF. Loss of bombesin-induced feeding suppression in gastrin-releasing peptide receptor-deficient mice. Proc Natl Acad Sci U S A. 1998;95:3188–3192. doi: 10.1073/pnas.95.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Muglia LJ. You are when you eat. Nat Neurosci. 2006;9:300–302. doi: 10.1038/nn0306-300. [DOI] [PubMed] [Google Scholar]
- Holst CR, Bou-Reslan H, Gore BB, Wong K, Grant D, Chalasani S, Carano RA, Frantz GD, Tessier-Lavigne M, Bolon B, et al. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE. 2007;2:e575. doi: 10.1371/journal.pone.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatan I, Dastani Z, Do R, Weissglas-Volkov D, Ruel I, Lee JC, Huertas-Vazquez A, Taskinen MR, Prat A, Seidah NG, et al. Genetic variation at the proprotein convertase subtilisin/kexin type 5 gene modulates high-density lipoprotein cholesterol levels. Circ Cardiovasc Genet. 2009;2:467–475. doi: 10.1161/CIRCGENETICS.109.877811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EA, Bechtold D, Dupre S, Brennand J, Barrett P, Luckman S, Loudon AS. Altered metabolism in the melatonin-related receptor (GPR50) knock out mouse. Am J Physiol Endocrinol Metab. 2008;294:E176–E182. doi: 10.1152/ajpendo.00199.2007. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Drijfhout WJ, Westerink BH, van Heerikhuize JJ, van der Woude TP, van der Vliet J, Buijs RM. GABA receptors in the region of the dorsomedial hypothalamus of rats are implicated in the control of melatonin and corticosterone release. Neuroendocrinology. 1996;63:69–78. doi: 10.1159/000126937. [DOI] [PubMed] [Google Scholar]
- Kalus I, Salmen B, Viebahn C, von Figura K, Schmitz D, D’Hooge R, Dierks T. Differential involvement of the extracellular 6-O-endosulfatases Sulf1 and Sulf2 in brain development and neuronal and behavioural plasticity. J Cell Mol Med. 2009;13:4505–4521. doi: 10.1111/j.1582-4934.2008.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem Biophys Res Commun. 2005;330:18–26. doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Aschkenasi CJ, Lee CE, Mountjoy KG, Saper CB, Elmquist JK. Expression of melanocortin 4 receptor mRNA in the central nervous system of the rat. J Comp Neurol. 2003;457:213–235. doi: 10.1002/cne.10454. [DOI] [PubMed] [Google Scholar]
- Krieger DT. Food and water restriction shifts corticosterone, temperature, activity and brain amine periodicity. Endocrinology. 1974;95:1195–1201. doi: 10.1210/endo-95-5-1195. [DOI] [PubMed] [Google Scholar]
- Krieger DT, Hauser H, Krey LC. Suprachiasmatic nuclear lesions do not abolish food-shifted circadian adrenal and temperature rhythmicity. Science. 1977;197:398–399. doi: 10.1126/science.877566. [DOI] [PubMed] [Google Scholar]
- Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. The heparanome—the enigma of encoding and decoding heparan sulfate sulfation. J Biotechnol. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Landry GJ, Simon MM, Webb IC, Mistlberger RE. Persistence of a behavioral food-anticipatory circadian rhythm following dorsomedial hypothalamic ablation in rats. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1527–R1534. doi: 10.1152/ajpregu.00874.2005. [DOI] [PubMed] [Google Scholar]
- Landry GJ, Yamakawa GR, Webb IC, Mear RJ, Mistlberger RE. The dorsomedial hypothalamic nucleus is not necessary for the expression of circadian food-anticipatory activity in rats. J Biol Rhythms. 2007;22:467–478. doi: 10.1177/0748730407307804. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Levoye A, Dam J, Ayoub MA, Guillaume JL, Couturier C, Delagrange P, Jockers R. The orphan GPR50 receptor specifically inhibits MT1 melatonin receptor function through heterodimerization. EMBO J. 2006;25:3012–3023. doi: 10.1038/sj.emboj.7601193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Huang XF. Altered hypothalamic c-Fos-like immunoreactivity in diet-induced obese mice. Brain Res Bull. 1999;49:215–219. doi: 10.1016/s0361-9230(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Lissitzky JC, Luis J, Munzer JS, Benjannet S, Parat F, Chretien M, Marvaldi J, Seidah NG. Endoproteolytic processing of integrin pro-alpha subunits involves the redundant function of furin and proprotein convertase (PC) 5A, but not paired basic amino acid converting enzyme (PACE) 4, PC5B or PC7. Biochem J. 2000;346(Pt 1):133–138. [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kishi T, Roseberry AG, Cai X, Lee CE, Montez JM, Friedman JM, Elmquist JK. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003;23:7143–7154. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JN, Aschkenasi CJ, Lee CE, Chemelli RM, Saper CB, Yanagisawa M, Elmquist JK. Differential expression of orexin receptors 1 and 2 in the rat brain. J Comp Neurol. 2001;435:6–25. doi: 10.1002/cne.1190. [DOI] [PubMed] [Google Scholar]
- Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORbeta knockout. Am J Physiol Regul Integr Comp Physiol. 2007;292:R2357–R2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Williams SC, Richardson JA, Tanaka K, Yanagisawa M. The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc Natl Acad Sci U S A. 2006;103:12150–12155. doi: 10.1073/pnas.0604189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–612. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol Behav. 2011 doi: 10.1016/j.physbeh.2011.04.015. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya T, Aida R, Kudo T, Akiyama M, Doi M, Hayasaka N, Nakahata N, Mistlberger R, Okamura H, Shibata S. The dorsomedial hypothalamic nucleus is not necessary for food-anticipatory circadian rhythms of behavior, temperature or clock gene expression in mice. Eur J Neurosci. 2009;29:1447–1460. doi: 10.1111/j.1460-9568.2009.06697.x. [DOI] [PubMed] [Google Scholar]
- Nahon JL, Presse F, Bittencourt JC, Sawchenko PE, Vale W. The rat melanin-concentrating hormone messenger ribonucleic acid encodes multiple putative neuropeptides coexpressed in the dorsolateral hypothalamus. Endocrinology. 1989;125:2056–2065. doi: 10.1210/endo-125-4-2056. [DOI] [PubMed] [Google Scholar]
- Ohki-Hamazaki H, Iwabuchi M, Maekawa F. Development and function of bombesin-like peptides and their receptors. Int J Dev Biol. 2005;49:293–300. doi: 10.1387/ijdb.041954ho. [DOI] [PubMed] [Google Scholar]
- Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, Wong CH, Lotz MK. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc Natl Acad Sci U S A. 2010;107:10202–10207. doi: 10.1073/pnas.0913897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. New York: Academic Press; 2001. [Google Scholar]
- Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR, Ebisawa T, Mahle CD, Kolakowski LF., Jr Cloning of a melatonin-related receptor from human pituitary. FEBS Lett. 1996;386:219–224. doi: 10.1016/0014-5793(96)00437-1. [DOI] [PubMed] [Google Scholar]
- Romanitan MO, Popescu BO, Spulber S, Bajenaru O, Popescu LM, Winblad B, Bogdanovic N. Altered expression of claudin family proteins in Alzheimer’s disease and vascular dementia brains. J Cell Mol Med. 2010;14:1088–1100. doi: 10.1111/j.1582-4934.2009.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Andre E, Kapfhammer JP, Becker-Andre M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Scott MM, Lachey JL, Sternson SM, Lee CE, Elias CF, Friedman JM, Elmquist JK. Leptin targets in the mouse brain. J Comp Neurol. 2009;514:518–532. doi: 10.1002/cne.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal JP, Stallings NR, Lee CE, Zhao L, Socci N, Viale A, Harris TM, Soares MB, Childs G, Elmquist JK, et al. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci. 2005;25:4181–4188. doi: 10.1523/JNEUROSCI.0158-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidibe A, Mullier A, Chen P, Baroncini M, Boutin JA, Delagrange P, Prevot V, Jockers R. Expression of the orphan GPR50 protein in rodent and human dorsomedial hypothalamus, tanycytes and median eminence. J Pineal Res. 2010;48:263–269. doi: 10.1111/j.1600-079X.2010.00750.x. [DOI] [PubMed] [Google Scholar]
- Sumi Y, Yagita K, Yamaguchi S, Ishida Y, Kuroda Y, Okamura H. Rhythmic expression of ROR beta mRNA in the mice suprachiasmatic nucleus. Neurosci Lett. 2002;320:13–16. doi: 10.1016/s0304-3940(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Thompson RH, Swanson LW. Organization of inputs to the dorsomedial nucleus of the hypothalamus: a reexamination with Fluorogold and PHAL in the rat. Brain Res Brain Res Rev. 1998;27:89–118. doi: 10.1016/s0165-0173(98)00010-1. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Thomson AM, Dunbar DR, Grassie MA, Condie A, Walker MT, Smith DJ, Pulford DJ, Muir W, et al. Sex-specific association between bipolar affective disorder in women and GPR50, an X-linked orphan G protein-coupled receptor. Mol Psychiatry. 2005;10:470–478. doi: 10.1038/sj.mp.4001593. [DOI] [PubMed] [Google Scholar]
- Vaisse C, Halaas JL, Horvath CM, Darnell JE, Jr, Stoffel M, Friedman JM. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- van den Pol AN, Yao Y, Fu LY, Foo K, Huang H, Coppari R, Lowell BB, Broberger C. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J Neurosci. 2009;29:4622–4639. doi: 10.1523/JNEUROSCI.3249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve P, Seidah NG, Beaudet A. Immunohistochemical distribution of the prohormone convertase PC5-A in rat brain. Neuroscience. 1999;92:641–654. doi: 10.1016/s0306-4522(98)00763-5. [DOI] [PubMed] [Google Scholar]
- Villeneuve P, Lafortune L, Seidah NG, Kitabgi P, Beaudet A. Immunohistochemical evidence for the involvement of protein convertases 5A and 2 in the processing of pro-neurotensin in rat brain. J Comp Neurol. 2000;424:461–475. doi: 10.1002/1096-9861(20000828)424:3<461::aid-cne5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Wada E, Watase K, Yamada K, Ogura H, Yamano M, Inomata Y, Eguchi J, Yamamoto K, Sunday ME, Maeno H, et al. Generation and characterization of mice lacking gastrin-releasing peptide receptor. Biochem Biophys Res Commun. 1997;239:28–33. doi: 10.1006/bbrc.1997.7418. [DOI] [PubMed] [Google Scholar]
- Watts AG. The efferent projections of the suprachiasmatic nucleus: anatomical insights into the control of circadian rhythms. In: Klein D, Moore RY, Reppert SM, editors. The suprachiasmatic nucleus: the mind’s clock. London: Oxford University Press; 1991. pp. 75–104. [Google Scholar]
- Watts AG, Sanchez-Watts G, Kelly AB. Distinct patterns of neuropeptide gene expression in the lateral hypothalamic area and arcuate nucleus are associated with dehydration-induced anorexia. J Neurosci. 1999;19:6111–6121. doi: 10.1523/JNEUROSCI.19-14-06111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Irizarry RA, Gentleman R, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. Johns Hopkins University; 2004. Department of Biostatistics Working Papers, Working Paper 1. [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kerman IA, Laque A, Nguyen P, Faouzi M, Louis GW, Jones JC, Rhodes C, Munzberg H. Leptin-receptor-expressing neurons in the dorsomedial hypothalamus and median preoptic area regulate sympathetic brown adipose tissue circuits. J Neurosci. 2011;31:1873–1884. doi: 10.1523/JNEUROSCI.3223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494:528–548. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller RT, Broyles B, Earley J, Anderson ER, Alberst HE. Cellular levels of messenger ribonucleic acids encoding vasoactive intestinal peptide and gastrin-releasing peptide in neurons of the suprachiasmatic nucleus exhibit distinct 24-hour rhythms. J Neuroendocrinol. 1992;4:119–124. doi: 10.1111/j.1365-2826.1992.tb00354.x. [DOI] [PubMed] [Google Scholar]