Abstract
Electric fields (EFs) of around 100 mV/mm are present in normal healing wounds and induce the directional migration of epithelial cells. Reepithelialization during wound healing thus may be controlled in part by this electrical signal. In this study, the early transcriptional response of human epidermal keratinocytes to EFs is examined using microarrays. Increased expression of various chemokines, interleukins, and other inflammatory response genes indicates that EFs stimulate keratinocyte activation and immune stimulatory activity. Gene expression activity further suggests that interleukin 1 is either released or activated in EFs. Expression of the chemokine CCL20 steadily increases at 100 mV/mm over time until around 8 h after exposure. This chemokine is also expressed at field strengths of 300 mV/mm—above the level of endogenous wound fields. The early effects of EFs on epithelial gene expression activity identified in these studies suggest the importance of naturally occurring EFs both in repair mechanisms and for the possibility of controlling these responses therapeutically.
Keywords: Electrical stimulation, Wound healing, Keratinocyte, Chemokine, Inflammatory response
Introduction
As part of their role providing barriers to environmental insults and maintaining homeostasis, epidermal cells maintain a potential difference between epidermal and dermal layers through the action of sodium and potassium pumps. Injuries to the skin cause this potential difference to “short circuit”, leading to electric fields (EFs) measured at 40–100 mV/mm [1, 9].
Within hours after injury, wound edge keratinocytes weaken intercellular attachments and begin migrating laterally across the wound bed [5]. As with other cells, keratinocyte activation is influenced by gradients of growth factors and contact with provisional matrix components, such as collagen and fibronectin that comprise the wound microenvironment [12, 30, 50]. Endogenous wound EFs, however, may also contribute to keratinocyte activation during healing. In electric fields of physiological strength, keratinocytes have been shown to migrate toward the negative pole in a process termed galvanotaxis [28]. Galvanotaxis of keratinocytes is promoted by both collagen and fibronectin substrates, but not laminin, an inhibitor of keratinocyte migration [42, 49]. The galvanotactic response in keratinocytes has also been shown to be partially dependent on protein kinase A, tyrosine kinases, and calcium influx; but not on voltage-dependent calcium channels [7, 37, 46]. The absence of growth factors, particularly epidermal growth factor (EGF) and transforming growth factor beta (TGF-β), can diminish the galvanotactic response of epithelial cells [35, 52]. Intensity and directionality of galvanotaxis is also affected by activity of beta-adrenerginic receptors and EGFRs [8, 36, 53].
Although particular growth factors, receptors, extracellular matrix proteins, and soluble factors influence galvanotaxis in epithelial cells, the exact mechanisms underpinning cellular effects of EFs remain unclear. Because studies have shown that multiple growth factors are capable of influencing the response, there may be multiple different pathways activated in EFs. The fact that directed migration still occurs when there are no growth factors present, which suggests that epidermal cells may produce their own growth factors to mediate this response when stimulated with EFs. Another possible mechanism for galvanotaxis and other effects of electrical fields is receptor redistribution. Receptors that are mobile within the membrane, particularly EGFR, have been shown to redistribute toward the negative pole [7], establishing a receptor gradient that may lead to increased signal transduction on the side facing the negative pole [52].
The EF stimulus is present immediately upon wounding and persists until the epidermal layer is reestablished, capable of exerting influence beyond the initial inflammatory phase of healing through the proliferative and migratory events of keratinocytes that occur within hours after injury [6]. However, very little is known about the effects of electric fields on keratinocytes beyond those related to the migratory response. This study examines the changes in gene expression and biochemical pathways activated in keratinocytes upon exposure to physiological strength EFs using microarrays. It is anticipated that EFs trigger regulation of genes associated with reepithelialization and wound repair. The expression of particular genes over time and at varying field strengths is also considered.
Materials and methods
Cell culture
Adult human epidermal keratinocytes (HEKa) (Cascade Biologiecs, Portland, OR, USA) were plated at passage 4 onto 22 mm square coverslips coated with a thin layer of type I bovine collagen (BD Biosciences, Bedford, MA, USA) in a six-well plate in EpiLife® Medium supplemented with Human Keratinocyte Growth Supplement (Cascade Biologics). After reaching approximately 60% confluence, cells were exposed to electric fields in a humidified incubator at 37°C and 5% CO2.
Electric field exposure
Cells on coverslips were placed cell side down into a stimulation apparatus constructed of Delrin® and assembled with vacuum grease, which minimized the cross-sectional area of the media exposed to current to guard against heat production as described in previous studies [16]. Glass tubes containing a gel consisting of 2% agar in saline served as bridges between stimulation chambers and silver/silver chloride electrodes in saline solution, thereby minimizing contamination of media with electrode products. Current through electrodes produced by a power supply was adjusted until the voltage drop across the media wells was measured to produce desired field strengths. Since the commonly reported average field strength occurring in wounds is 100 mV/mm and galvanotaxis response of keratinocytes begins within the first 15 min, microarray experiments were carried out at 100 mV/mm for 1 h [1, 28]. RT–PCR expression measurement of selected transcripts was performed for shorter and longer durations (30 min, 1, 4, 8, and 12 h), and for 1 h at varying field strengths ranging from 50–300 mV/ mm.
RNA isolation
Total RNA was obtained from coverslips using Trizol® (Invitrogen, Carlsbad, CA) and chloroform extraction. A pool of six samples used for each GeneChip®. Total RNA from one sample per experimental condition was used for RT–PCR analysis.
Microarray
RNA was checked for quality and quantity before labeling and hybridization to Affymetrix HG-133 Plus Genechips®. Three microarrays from three experimental replicates were compared to three microarrays for control replicates.
Data analysis
Affymetrix raw data were collected and analyzed using GeneChip® Operating System (GCOS, Affymetrix, Santa Clara, CA, USA). Raw data (.cel and .chp files) were uploaded onto a web-based software program, GeneTraffic™ (Iobion, Inc., La Jolla, CA, USA), for normalization using GC-Robust Multichip Analysis (GC-RMA) with controls as a baseline. p values were obtained using a two-class unpaired unequal variance t test. Transcripts identified as regulated compared to controls were imported into Netaffx™ (Affymetrix), and into Pathway Architect® (Stratagene, La Jolla, CA, USA) software to assist with interpretation of pathway analysis.
RT–PCR
Two transcripts identified as upregulated in microarrays were selected to confirm results using real time RT–PCR primers were designed based on Affymetrix target sequences for upregulated transcripts (Table 1). Total RNA was isolated as described previously; with the exception that experimental material was not pooled and contained total RNA from one experimental condition. First strand cDNA was transcribed from total RNA using Reverse-iT™ MAX first strand synthesis kit (ABGene, Rochester, NY, USA). Real-time PCR using ABsolute™ QPCR SYBR green mix with Fluorescein (ABGene) was performed using an i-Cycler (Biorad, Hercules, CA, USA). Expression levels normalized to an internal control housekeeping gene (GAPDH) were determined using the standard curve method.
Table 1.
Primers
| Gene | Sequences |
|---|---|
| NFKBIZ | |
| Forward | 5′ GATGCTGTCCGCCTGTTGA 3′ |
| Reverse | 5′ CTGGCTGTTCGTTCTCCAAGT 3′ |
| CCL20 | |
| Forward | 5′ CTGGCTGCTTTGATGTCAGTGCT 3′ |
| Reverse | 5′ GCAGTCAAAGTTGCTTGCTGCTTC 3′ |
| GAPDH | |
| Forward | 5′ ATGGGGAAGATAAAGGTCG 3′ |
| Reverse | 5′ TAAAAGCAGCCCTGGTGACC 3′ |
Results
Statistical analysis of multiple microarray experiments identified 161 gene transcripts that were significantly increased over controls, and 245 transcripts significantly decreased after 1 h of exposure to 100 mV/mm EFs. Of these, 17 transcripts were significantly increased and 67 were decreased by more than 1.4-fold (average signal log ratio >0.05 or <−0.05). Above a twofold regulation (average signal log ratio >1 or <−1), 1 transcript was significantly increased (NFKBIZ) and 3 were significantly decreased (ATRX, ZBTB34, and EIF4G3).
Analysis by function
Affymetrix’s NetAffx™ web-based program was used to identify associated biological functions of regulated transcripts. Table 2 lists select genes that are significantly upregulated above 1.4-fold that can be classified according to biological function. Table 3 lists genes identified as downregulated at greater than 1.4-fold and their associated biological functions. Genes that fell into multiple categories of biological functions were classified under functions most relevant to wound healing, and redundant expression results have been omitted. Since relatively few genes were regulated at statistically significant levels, all genes identified as having a greater than twofold average change and their p values are listed in Table 4.
Table 2.
Transcripts significantly increased in HEKa
| Affymetrix Probeset ID | Gene symbol | Gene name | Fold change |
|---|---|---|---|
| Inflammation and immune response | |||
| 223217_s_at | NFKBIZ | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | 2.5 |
| Signal transduction | |||
| 207993_s_at | CHP | Calcium binding protein P22 | 1.7 |
| 1569022_a_at | PIK3C2A | Phosphoinositide-3-kinase, class 2, alpha polypeptide | 1.5 |
| 243452_at | B4GALT6 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 | 1.5 |
| 205868_s_at | PTPN11 | Protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) | 1.4 |
| Lipid metabolism | |||
| 215891_s_at | GM2A | GM2 ganglioside activator | 1.4 |
| Ubiquitin cycle | |||
| 222567_s_at | RKHD2 | Ring finger and KH domain containing 2 | 1.5 |
Transcripts increased above 1.4-fold relative to controls with p < 0.05 were classified using NetAffx™ and listed according to associated biological functions
Table 3.
Transcripts significantly decreased in HEKa
| Affymetrix ID | Gene symbol | Gene name | Fold change |
|---|---|---|---|
| Growth and differentiation | |||
| 241681_at | MBNL1 | Muscleblind-like (Drosophila) | 1.6 |
| 229422_at | NRD1 | Nardilysin (N-arginine dibasic convertase) | 1.4 |
| 225318_at | WHSC1L1 | Wolf-Hirschhorn syndrome candidate 1-like 1 | 1.4 |
| Adhesion and motility | |||
| 236752_at | PKP4 | Plakophilin 4 | 1.4 |
| Apoptosis | |||
| 232565_at | RAB6IP2 | RAB6 interacting protein 2 | 1.7 |
| 216621_at | ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | 1.6 |
| Signal transduction | |||
| 228779_at | LOC440456 | Similar to pleckstrin homology domain containing, family M (with RUN domain) member 1; adapter protein 162 | 1.8 |
| 236007_at | AKAP10 | A kinase (PRKA) anchor protein 10 | 1.7 |
| 240758_at | CENTG2 | Centaurin, gamma 2 | 1.6 |
| 216621_at | ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | 1.6 |
| Transport | |||
| 202940_at | WNK1 | WNK lysine deficient protein kinase 1 | 1.8 |
| 232565_at | RAB6IP2 | RAB6 interacting protein 2 | 1.7 |
| 239274_at | PICALM | Phosphatidylinositol binding clathrin assembly protein | 1.7 |
| 1554557_at | ATP11B | ATPase, Class VI, type 11B | 1.7 |
| 240758_at | CENTG2 | Centaurin, gamma 2 | 1.6 |
| 156920 l_a_at | SEC15L2 | SEC15-like 2 (S. cerevisiae) | 1.6 |
| 240600_at | AP3B1 | Adaptor-related protein complex 3, beta 1 subunit | 1.5 |
| 214244_s_at | ATP6V0E | ATPase, H+ transporting, lysosomal 9 kDa, V0 subunit e | 1.5 |
| 222906_at | FLVCR | Feline leukemia virus subgroup C cellular receptor | 1.5 |
| Cell cycle arrest | |||
| 1553407_at | MACF1 | Microtubule-actin crosslinking factor 1 | 1.5 |
| Transcription | |||
| 236778_at | ATRX | Alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 2.0 |
| 227111_at | ZBTB34 | Zinc finger and BTB domain containing 34 | 2.0 |
| 232716_at | EDG2 | Endothelial differentiation, lysophosphatidic acid G- protein-coupled receptor, 2 | 1.9 |
| 232565_at | RAB6IP2 | RAB6 interacting protein 2 | 1.7 |
| 239238_at | SMARCC1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | 1.7 |
| 238883_at | THRAP2 | Thyroid hormone receptor associated protein 2 | 1.6 |
| 213659_at | ZNF75 | Zinc finger protein 75 (D8C6) | 1.6 |
| 233239_at | ZNF407 | Zinc finger protein 407 | 1.6 |
| 235862_at | RAI1 | Smith-Magenis syndrome chromosome region, candidate 3 | 1.5 |
| Protein modification or biosynthesis | |||
| 243149_at | EIF4G3 | Eukaryotic translation initiation factor 4 gamma, 3 | 2.1 |
| 240240_at | UBE2J2 | Ubiquitin-conjugating enzyme E2, J2 (UBC6 homolog, yeast) | 1.4 |
Transcripts decreased below 1.4-fold relative to controls with p < 0.05 were classified using NetAffx™ and listed according to associated biological functions
Table 4.
Transcripts regulated at a greater than twofold level
| Affymetrix ID | Gene symbol | Gene name | Fold change | p value |
|---|---|---|---|---|
| 211506_s_at | IL8 | Interleukin 8 | 4.3 | 0.12 |
| 205476_at | CCL20 | Chemokine (C–C motif) ligand 20 | 4.1 | 0.08 |
| 220322_at | IL1F9 | Interleukin 1 family, member 9 | 3.5 | 0.15 |
| 209774_x_at | CXCL2 | Chemokine (C–X–C motif) ligand 2 | 3.2 | 0.05 |
| 207850_at | CXCL3 | Chemokine (C–X–C motif) ligand 3 | 3.0 | 0.28 |
| 205207_at | IL6 | Interleukin 6 (interferon, beta 2) | 2.9 | 0.24 |
| 207113_s_at | TNF | Tumor necrosis factor (TNF superfamily, member 2) | 2.8 | 0.26 |
| 210229_s_at | CSF2 | Colony stimulating factor 2 (granulocyte–macrophage) | 2.5 | 0.44 |
| 223217_s_at | NFKBIZ | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | 2.5 | 0.03 |
| 211924_s_at | PLAUR | Plasminogen activator, urokinase receptor /// plasminogen activator, urokinase receptor | 2.3 | 0.36 |
| 208539_x_at | SPRR2B | Small proline-rich protein 2B | 2.3 | 0.14 |
| 1555673_at | KRTAP2-1 | Keratin associated protein 2-1 | 2.2 | 0.10 |
| 205266_at | LIF | Leukemia inhibitory factor (cholinergic differentiation factor) | 2.1 | 0.16 |
| 205220_at | GPR109B | G protein-coupled receptor 109B///G protein-coupled receptor 109B | 2.1 | 0.15 |
| 22407 l_at | IL20 | Interleukin 20 | 2.1 | 0.23 |
| 228964_at | PRDM1 | PR domain containing 1, with ZNF domain | 2.0 | 0.41 |
| 234079_at | RGNEF | Rho-guanine nucleotide exchange factor | −2.0 | 0.08 |
| 244010_at | ABP1 | Amiloride binding protein 1 [amine oxidase (copper-containing)] | −2.2 | 0.12 |
Among genes with increased expression in electric fields, several fell into the category of inflammatory response functions, including interleukins and chemokines. Signal transduction proteins were also expressed at increased levels. Transcripts that were decreased fell into categories of growth and differentiation, adhesion and motility, apoptosis, and transport, among others.
Analysis by pathway
Filtered gene lists imported into Pathway Assist software identified biochemical pathways with multiple genes regulated. Based on the presence of multiple regulated transcripts within certain biological pathways, software suggested that possibly active pathways included chemokine, apoptosis, JAK-STAT, Wnt, and G-protein MAPK activation signaling pathways. Although it is not confirmed that these pathways or receptors were activated, expression of transcripts downstream in these pathways was regulated.
Real-time RT–PCR
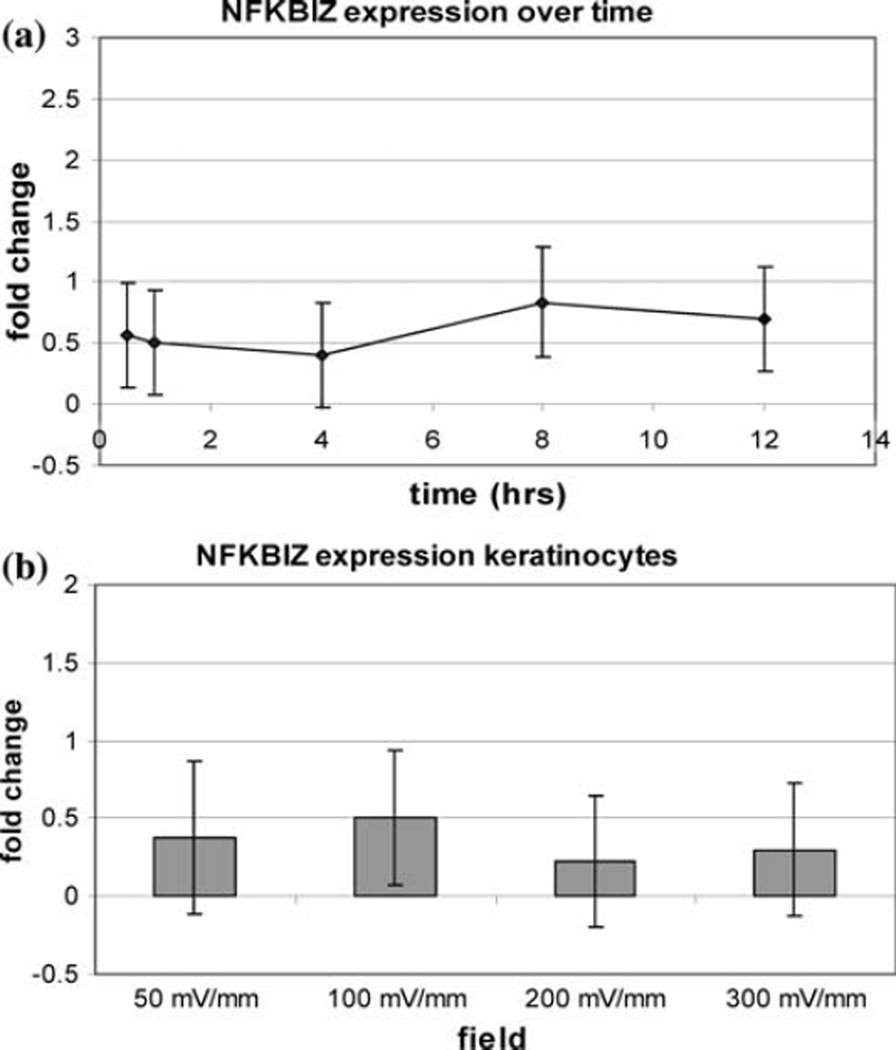
Although nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta (NFKBIZ) was the only gene identified in the microarray study as statistically increased over twofold, increased expression of this transcript was not confirmed by RT–PCR for any of the experimental conditions (Fig. 1).
Fig 1.
NFKBIZ expression in keratinocytes (a) over time at 100 mV/ mm and (b) at different field strengths for 1 h
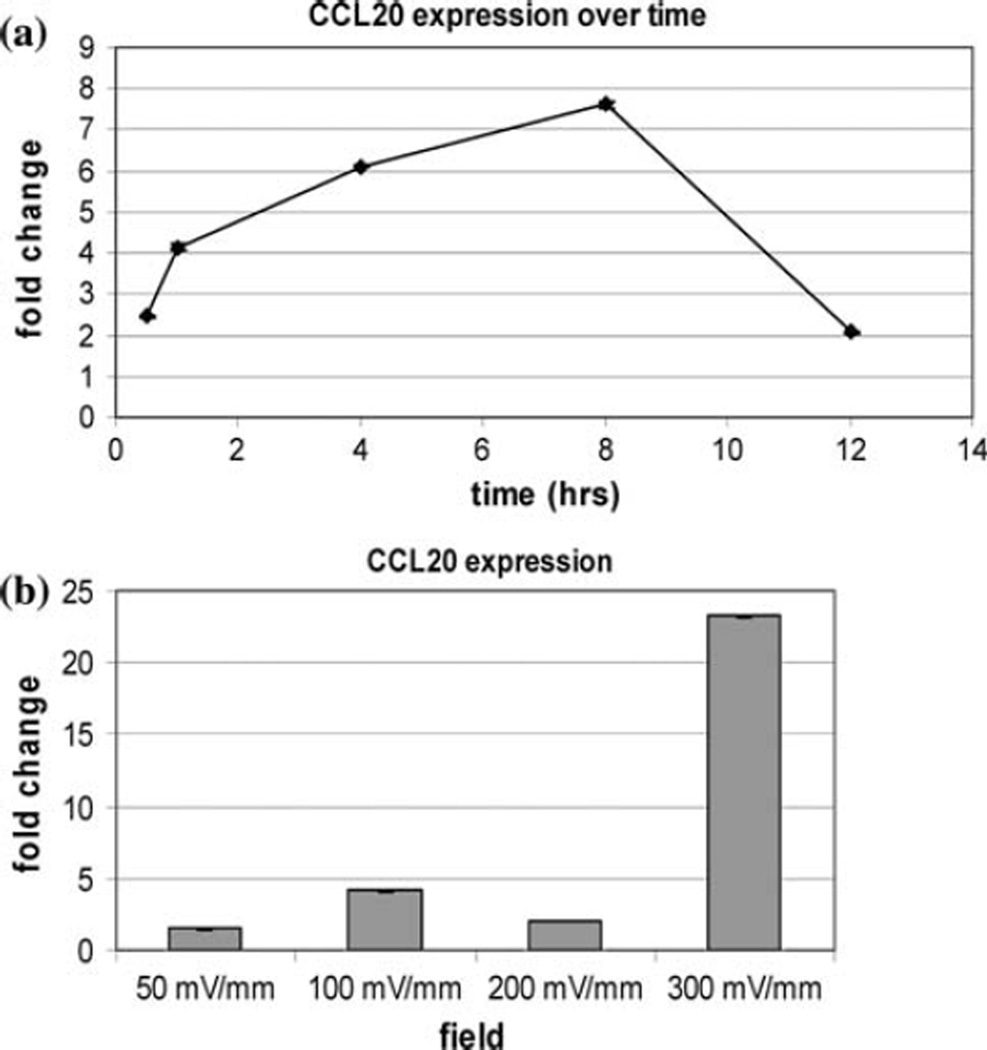
Expression of chemokine (C–C motif) ligand 20 (CCL20) was confirmed to be increased by RT–PCR after 1 h of stimulation with 100 mV/mm. Expression was increased further at 4 and 8 h of continuous stimulation (Fig. 2a). At all field strengths tested, expression of CCL20 was increased over controls and was increased by 23-fold after 1 h of stimulation with 300 mV/mm (Fig. 2b).
Fig 2.
CCL20 expression in keratinocytes (a) over time at 100 mV/ mm and (b) at different field strengths for 1 h
Discussion
EF exposure induces changes in the expression of crucial wound response genes in keratinocytes. Thirty-eight transcripts were identified as statistically different above a 1.4-fold level; and although several transcripts were regulated above a twofold level, only one was detected to be statistically significant in the microarray study, NFKBIZ. This small number of detected gene expression changes contrasts with the pronounced change in the activity of keratinocytes during galvanotaxis. Most galvanotaxis studies, however, report the effects on neonatal or corneal epidermal cells, whereas in this study adult dermal keratinocytes were used. It is possible that responsiveness of cells to electric fields may be dependent on age or tissue location. It has been shown that keratinocytes expressing involucrin, an indicator of differentiation, display diminished galvanotactic responses [31] and subpopulations of neonatal keratinocytes do not respond to EFs or migrate toward the anode [36]. Differentiation of keratinocytes in these culture conditions with low calcium medium is suppressed, and differentiation marker expression has been found to be similar between adult and neonatal keratinocytes in similar culture conditions [25]. The response of adult keratinocytes to electric fields may be reduced compared to neonatal cells or only present in smaller subsets of cells, making the data more variable and decreasing the likelihood of detecting significant differences.
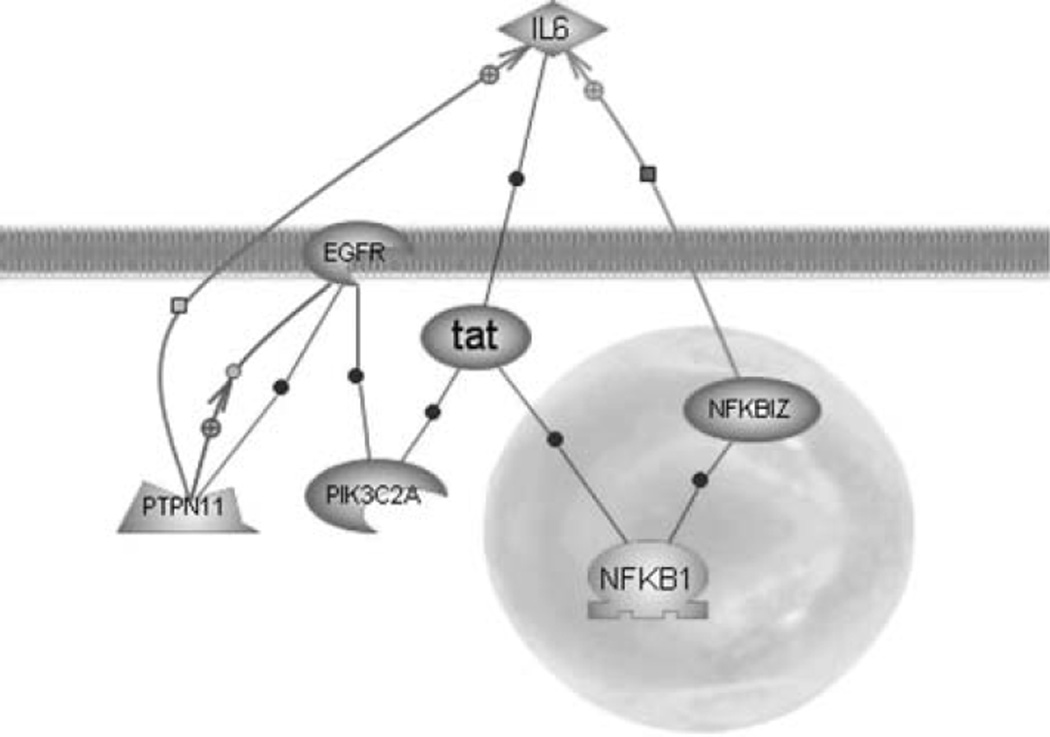
Zhao et al. have linked galvanotaxis to increases in phosphoinositol-3-OH-kinase-γ (PI3K-γ) signaling and decreases in phosphatase and tensin homolog (PTEN) signaling [55]. Microarray analysis identified PTEN expression was decreased significantly by 1.2-fold in keratinocytes exposed to EFs. Although there were no detectable changes in expression of PI3K-γ, another phosphoinositol kinase protein, phosphoinositide-3-kinase, class 2, alpha polypeptide (PIK3C2A) was increased by 1.5-fold. The activity of this newly identified protein in the migratory response remains unknown; however, this protein has a known role in clathrin-mediated membrane trafficking [10]. This protein may be involved in the asymmetric redistribution of membrane proteins through an alternative mechanism to electrophoretic movement [34]. Figure 4 shows the direct interactions between significantly upregulated transcripts—PIK3C2A, protein tyrosine phosphatase, non-receptor type 11 (PTPN11), and NFKBIZ—and EGFRs, which are also active in the galvanotaxis response.
Fig 4.
Statistically significant interactions. Pathway Architect® diagram of relations between the significantly upregulated transcripts NFKBIZ, PIK3C2A, and PTPN11 in EFs and IL-6 whose upregulation was not statistically significant and EGFR which has been shown to be upregulated in other studies of EF effects on epithelial cells
Keratinocyte migration in vivo requires the elimination of intercellular attachments and degradation of surrounding matrix. Expression of plasminogen activated urokinase receptor (PLAUR) was increased over twofold in this study. Binding of urokinase-type plasminogen activator (uPA) to PLAUR converts plasminogen to plasmin, activating growth latent growth factors such as TGF-β1, activating MMPs to digest ECM, and cleaving extracellular components leading to the breakdown of desmosomes [2, 3, 18]. Increased expression of PLAUR in epidermal cells induced by EFs indicates EFs not only increase direct migratory responses of detached keratinocytes but also increase their ability to manipulate intercellular and ECM attachments.
The role of increased chemokine expression in keratinocytes as found in the present study on the galvanotactic response of keratinocytes is unknown but would likely enhance galvanotactic activity. Studies by Pullar et al. indicate that beta 2-adrenergic receptor (B2-AR) antagonists increase galvanotaxis and B2-AR agonists suppress these responses [36]. IL-1 has been shown to attenuate activity within the B-AR adenylyl cyclase system and thus is antagonistic to B2-ARs [17]. Furthermore, inhibition of interleukin and cytokine production by B2-AR agonists may explain their negative regulation of galvanotaxis [15, 47].
The majority of transcripts increased by more than twofold after electric field exposure is associated with inflammatory response after injury and includes chemokines, interleukins, and growth factors. Only NFKBIZ was increased at a statistically significant level, however. Transcription of NFKBIZ, a nuclear protein that interacts with NFkB and the IL-6 promoter, is induced sharply by IL-1, lipopolysaccharide, and toll-like receptor ligands [19, 51]. As it is an inhibitor of the inflammatory protein NFkB, it may be classified as an anti-inflammatory agent, preventing extreme inflammatory reactions during wound healing [26]. Upregulation of NFKBIZ in EFs may be mediated by IL-1 release in keratinocytes, and may be required for the accompanying increase in IL-6 expression. Although the location of receptors that trigger induction of NFKBIZ (toll-like receptors and IL-1 receptors) in EFs are unknown, asymmetric redistribution due to EFs similar to that seen for EGFRs may also enhance this effect [8, 53].
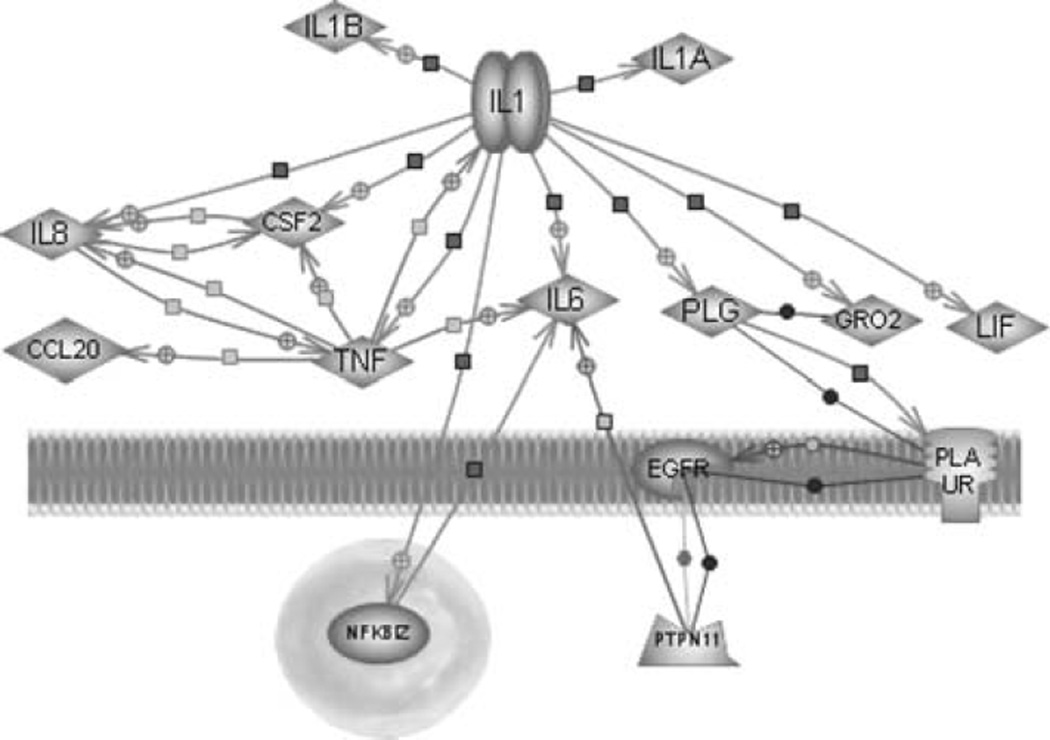
In addition to migrating across the wound bed to recover barrier function, keratinocytes synthesize and secrete factors that signal macrophage and neutrophil migration and promote angiogenesis. Keratinocytes produce and store interleukin 1a (IL-1a), which is quickly released upon exposure to environmental stresses [21–23, 33]. IL-1a secretion signals the activation of endothelial cells, fibroblasts, and other keratinocytes [4, 24, 39]. IL-1 secretion also attracts circulating leukocytes and may induce expression of IL-8, TNF, and other chemokines induced in this study [29, 43]. Results of this study suggest that keratinocytes release IL-1 after exposure to EFs during the process of normal wound healing, which then mediates changes in gene expression during keratinocyte activation (Fig 3.) With increased sample size, these expression results may have reached statistical significance.
Fig 3.
Gene expression in keratinocytes related to IL-1 signaling. Pathway Architect® diagram showing increases in expression of CCL20, CSF2, GRO-beta, IL-6, IL-8, LIF, NFKBIZ, PTPN11, and TNF indicate activity stimulated by IL-1, which is released by keratinocytes upon injury
IL-8 and IL-6, both with increased expression over controls on exposure to EFs, can enhance the healing response by promoting migration, proliferation, and angiogenesis [11, 20, 38, 44]. There were also increases in expression of CXCL2 and CXCL3 (GRO-β and GRO-γ), which are structurally related to IL-8 and share activity in promoting neutrophil infiltration and angiogenesis [40, 45]. TNF-α acts to promote the production of other signaling molecules and their receptors, including EGFR [10, 32]. Upregulation of EGFRs has been observed in corneal epithelial cells exposed to similar EFs, an effect possibly mediated by TNF-α [54]. TNF-α may also be responsible for increased expression of CCL20, which functions to recruit inflammatory cells to tissue and has been found to be expressed in epidermal cells with disrupted barrier function and therefore in the presence of EFs [13, 27, 41]. Increased expression of cytokines and chemokines may also result in the upregulation of leukemia inhibitory factor (LIF), which then may reinforce upregulation of cytokines, particularly interleukins [14, 48].
RT–PCR measurement shows that the expression of the chemokine CCL20 in a 100 mV/mm field increases steadily over time until sometime after 8 h, and then declines. CCL20 expression is also increased dramatically at 300 mV/mm, which is greater than the strength of EFs in normal skin wounds. Although an essential early step in the progression of normal wound healing, expression of chemokines at extremely high levels or over prolonged periods of time interferes with the timely resolution of the inflammatory response during the normal progression of healing. These results suggest that EFs of strengths similar to those present in normal healing wounds induce a moderate expression of chemokines in keratinocytes, which diminishes within hours.
In this study, keratinocytes were stimulated in medium designed to support expansion of keratinocytes and suppress differentiation. This media contained supplements of a defined concentration of epidermal growth factor but an undefined concentration of other growth factors contained within a bovine pituitary extract. EGF is particularly involved in keratinocyte galvanotaxis, but other growth factors such as TGF-β also influence EF effects in epidermal cells [8, 53, 54]. Since the concentration of specific growth factors in bovine pituitary extract is unknown and high concentrations of EGF were supplied in the media, these results may be selective for the effects of EFs that are EGF-dependent. It should be noted that the concentration of ions and growth factors in wound fluid after the collapse of the trans-epithelial potential and cell signaling differs from the medium used in this study. This model system also differs from the in vivo situation in that specific interactions between keratinocytes and matrix as well as between other cell types are absent. Cross talk between keratinocytes and other cells influenced by exposure to EFs, such as fibroblasts and endothelial cells, may play a major role in the coordination of EF-induced effects in vivo.
Statistical analysis of multiple microarray experiments and data mining tools helped to narrow the list of candidate genes for further studies and confirmation of microarray results with RT–PCR. However, expression of NFKBIZ measured by RT–PCR did not agree with microarray data. Additionally, multiple microarray experiments may have revealed induction of different genes at later time points or at different fields. Pathway analysis also identified possible activation of particular cytokine signaling pathways such as JAK-STAT, although specific phosphorylation status or pathway activation was not confirmed in this study. Since gene expression analysis is used to indicate changes in protein synthesis, proteomic analysis of transcripts identified in this study may provide more reliable results. Furthermore, the role that electric field-induced gene expression plays in the processes of migration, cell signaling, and wound healing could be further elucidated by blocking receptors, such as chemokine receptors, or by gene silencing experiments. Ultimately, confirmation of changes in gene expression and cellular activity within an in vivo model is necessary to fully understand the role of EFs in keratinocyte activation. Moreover, the impact of EF-induced effects on keratinocytes in the clinical progression of wound healing deserves further study.
EFs of similar strength to those generated in normal wounds induce gene expression in keratinocytes important to the wound healing response. In addition to promoting migration, increased expression of chemokines induced upon EF exposure may profoundly affect the inflammatory phase of wound healing. Further studies are needed to confirm the production and release of these proteins in EFs and their contribution to galvanotaxis, reepithelialization, and wound healing in general. Visualization of protein location in EFs using labeled antibodies may also identify asymmetries in receptor location or protein secretion capable of controlling the directionality of keratinocyte migration needed for reepithelialization. Overall, this study substantiates the importance of endogenous EFs in reepithelialization and suggests strategies to control keratinocyte response during the inflammatory and proliferative phases of wound healing.
Acknowledgments
This project has been funded in part with US federal funds from the National Cancer Institute, NIH under CA-13148 and from the NCRR, NIH under 1UL1RR025777.
Abbreviations
- CCL20
Chemokine (C–C motif) ligand 20
- EF
Electric field
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- HEKa
Human epidermal keratinocytes
- IL
Interleukin
- LIF
Leukemia inhibitory factor
- NFKBIZ
Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor zeta
- PTEN
Phosphatase and tensin homolog
- PIK3C2A
Phosphoinositide-3-kinase class 2 alpha Polypeptide
- PI3K-γ
Phosphoinositol-3-OH-kinase-γ
- PLAUR
Plasminogen activated urokinase receptor
- TGF-β
Transforming growth factor beta
- TNF
Tumor necrosis factor
- uPA
Urokinase-type plasminogen activator
Contributor Information
Jessica Amber Jennings, Email: jjnnings@memphis.edu, jennings.amber@gmail.com, Department of Biomedical Engineering, University of Alabama at Birmingham, 1075 13th St. South, Birmingham, AL 35294, USA.
Dongquan Chen, Biostatistics and Bioinformatics Unit, Comprehensive Cancer Center, Universtiy of Alabma at Birmingham, 1530 3rd Avenue South, Birmingham, AL 35294, USA; Clinical Translational Science Institute, Health Science Center, West Virginia University, 1 Medical Center Drive, RM 5522, Morgantown, WV 26506, USA.
Dale S. Feldman, Department of Biomedical Engineering, University of Alabama at Birmingham, 1075 13th St. South, Birmingham, AL 35294, USA
References
- 1.Barker A, Jaffe L, Vanable J. The glabrous epidermis of cavies contains a powerful battery. Am J Physiol. 1982;242(3):R358–R366. doi: 10.1152/ajpregu.1982.242.3.R358. [DOI] [PubMed] [Google Scholar]
- 2.Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3(12):932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P, Moons L, Lijnen R, Baes M, Lemaitre V, Tipping P, Drew A, Eeckhout Y, Shapiro S, Lupu F, et al. Urokinase-generated plasmin activates matrix metal-loproteinases during aneurysm formation. Nat Genet. 1997;17(4):439–444. doi: 10.1038/ng1297-439. [DOI] [PubMed] [Google Scholar]
- 4.Chen JD, Lapiere JC, Sauder DN, Peavey C, Woodley DT. Interleukin-1 alpha stimulates keratinocyte migration through an epidermal growth factor/transforming growth factor-alpha-independent pathway. J Invest Dermatol. 1995;104(5):729–733. doi: 10.1111/1523-1747.ep12606970. [DOI] [PubMed] [Google Scholar]
- 5.Clark RA. Cutaneous tissue repair: basic biologic considerations. I. J Am Acad Dermatol. 1985;13(5 Pt 1):701–725. doi: 10.1016/s0190-9622(85)70213-7. [DOI] [PubMed] [Google Scholar]
- 6.Clark RA. Basics of cutaneous wound repair. J Dermatol Surg Oncol. 1993;19(8):693–706. doi: 10.1111/j.1524-4725.1993.tb00413.x. [DOI] [PubMed] [Google Scholar]
- 7.Fang KS, Farboud B, Nuccitelli R, Isseroff RR. Migration of human keratinocytes in electric fields requires growth factors and extracellular calcium. J Invest Dermatol. 1998;111(5):751–756. doi: 10.1046/j.1523-1747.1998.00366.x. [DOI] [PubMed] [Google Scholar]
- 8.Fang KS, Ionides E, Oster G, Nuccitelli R, Isseroff RR. Epidermal growth factor receptor relocalization and kinase activity are necessary for directional migration of keratinocytes in DC electric fields. J Cell Sci. 1999;112(Pt 12):1967–1978. doi: 10.1242/jcs.112.12.1967. [DOI] [PubMed] [Google Scholar]
- 9.Foulds IS, Barker AT. Human skin battery potentials and their possible role in wound healing. Br J Dermatol. 1983;109(5):515–522. doi: 10.1111/j.1365-2133.1983.tb07673.x. [DOI] [PubMed] [Google Scholar]
- 10.Gaidarov I, Smith ME, Domin J, Keen JH. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane traffick-ing. Mol Cell. 2001;7(2):443–449. doi: 10.1016/s1097-2765(01)00191-5. [DOI] [PubMed] [Google Scholar]
- 11.Gallucci RM, Sloan DK, Heck JM, Murray AR, O’Dell SJ. Interleukin 6 indirectly induces keratinocyte migration. J Invest Dermatol. 2004;122(3):764–772. doi: 10.1111/j.0022-202X.2004.22323.x. [DOI] [PubMed] [Google Scholar]
- 12.Grinnell F. Wound repair, keratinocyte activation and integrin modulation. J Cell Sci. 1992;101(Pt 1):1–5. doi: 10.1242/jcs.101.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Harant H, Eldershaw SA, Lindley IJ. Human macrophage inflammatory protein-3alpha/CCL20/LARC/Exodus/SCYA20 is transcriptionally upregulated by tumor necrosis factor-alpha via a non-standard NF-kappaB site. FEBS Lett. 2001;509(3):439–445. doi: 10.1016/s0014-5793(01)03138-6. [DOI] [PubMed] [Google Scholar]
- 14.Hartner A, Sterzel RB, Reindl N, Hocke GM, Fey GH, Goppelt-Struebe M. Cyto-kine-induced expression of leukemia inhibitory factor in renal mesangial cells. Kidney Int. 1994;45(6):1562–1571. doi: 10.1038/ki.1994.206. [DOI] [PubMed] [Google Scholar]
- 15.Hetier E, Ayala J, Bousseau A, Prochiantz A. Modulation of interleukin-1 and tu-mor necrosis factor expression by beta-adrenergic agonists in mouse ameboid micro-glial cells. Exp Brain Res. 1991;86(2):407–413. doi: 10.1007/BF00228965. [DOI] [PubMed] [Google Scholar]
- 16.Jennings J, Chen D, Feldman D. Transcriptional response of dermal fibroblasts in direct current electric fields. Bioelectro-magnetics. 2008;29(5):394–405. doi: 10.1002/bem.20408. [DOI] [PubMed] [Google Scholar]
- 17.Kelsen SG, Anakwe O, Aksoy MO, Reddy PJ, Dhanasekaran N. IL-1 beta alters beta-adrenergic receptor adenylyl cyclase system function in human airway epithelial cells. Am J Physiol. 1997;273(3 Pt 1):L694–L700. doi: 10.1152/ajplung.1997.273.3.L694. [DOI] [PubMed] [Google Scholar]
- 18.Kitajima Y, Aoyama Y, Seishima M. Transmembrane signaling for adhesive regu-lation of desmosomes and hemides-mosomes, and for cell-cell datachment induced by pemphigus IgG in cultured keratinocytes: involvement of protein kinase C. J Investig Dermatol Symp Proc. 1999;4(2):137–144. doi: 10.1038/sj.jidsp.5640197. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura H, Kanehira K, Takahiko S, Morimatsu M, Jung B, Akashi S, Saito M. Bacterial Lipopolysaccharide Induces mRNA Expression of an IκB MAIL through Toll-Like Receptor 4. J Vet Med Sci. 2002;64(5):419–422. doi: 10.1292/jvms.64.419. [DOI] [PubMed] [Google Scholar]
- 20.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, Elner VM, Elner SG, Strie-ter RM. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science. 1992;258(5089):1798–1801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 21.Kondo S, Sauder DN, Kono T, Galley KA, McKenzie RC. Differential modulation of interleukin-1 alpha (IL-1 alpha) and interleukin-1 beta (IL-1 beta) in human epi-dermal keratinocytes by UVB. Exp Dermatol. 1994;3(1):29–39. doi: 10.1111/j.1600-0625.1994.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 22.Kupper TS, Ballard DW, Chua AO, McGuire JS, Flood PM, Horowitz MC, Langdon R, Lightfoot L, Gubler U. Human keratinocytes contain mRNA indistinguishable from monocyte interleukin 1 alpha and beta mRNA. Keratinocyte epidermal cell-derived thymocyte-activating factor is identical to interleukin 1. J Exp Med. 1986;164(6):2095–2100. doi: 10.1084/jem.164.6.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kupper TS, Chua AO, Flood P, McGuire J, Gubler U. Interleukin 1 gene expression in cultured human keratinocytes is augmented by ultraviolet irradiation. J Clin Invest. 1987;80(2):430–436. doi: 10.1172/JCI113090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maas-Szabowski N, Fusenig NE. Interleukin-1-induced growth factor expression in postmitotic and resting fibroblasts. J Invest Dermatol. 1996;107(6):849–855. doi: 10.1111/1523-1747.ep12331158. [DOI] [PubMed] [Google Scholar]
- 25.Michel M, L’Heureux N, Auger FA, Germain L. From newborn to adult: phenotypic and functional properties of skin equivalent and human skin as a function of donor age. J Cell Physiol. 1997;171(2):179–189. doi: 10.1002/(SICI)1097-4652(199705)171:2<179::AID-JCP8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Muta T, Yamazaki S, Eto A, Motoyama M, Takeshige K. IkappaB-zeta, a new anti-inflammatory nuclear protein induced by lipopolysaccharide, is a negative regulator for nuclear factor-kappaB. J Endotoxin Res. 2003;9(3):187–191. doi: 10.1179/096805103125001612. [DOI] [PubMed] [Google Scholar]
- 27.Nelson RT, Boyd J, Gladue RP, Paradis T, Thomas R, Cunningham AC, Lira P, Brissette WH, Hayes L, Hames LM, et al. Genomic organization of the CC chemokine mip-3alpha/ CCL20/larc/exodus/SCYA20, showing gene structure, splice variants, and chromosome localization. Genomics. 2001;73(1):28–37. doi: 10.1006/geno.2001.6482. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura KY, Isseroff RR, Nuccitelli R. Human keratinocytes migrate to the neg-ative pole in direct current electric fields comparable to those measured in mammalian wounds. J Cell Sci. 1996;109(Pt 1):199–207. doi: 10.1242/jcs.109.1.199. [DOI] [PubMed] [Google Scholar]
- 29.Nourshargh S, Larkin SW, Das A, Williams TJ. Interleukin-1-induced leukocyte extravasation across rat mesenteric microvessels is mediated by platelet-activating factor. Blood. 1995;85(9):2553–2558. [PubMed] [Google Scholar]
- 30.O’Keefe EJ, Payne RE, Jr, Russell N, Woodley DT. Spreading and enhanced motility of human keratinocytes on fibronectin. J Invest Dermatol. 1985;85(2):125–130. doi: 10.1111/1523-1747.ep12276531. [DOI] [PubMed] [Google Scholar]
- 31.Obedencio GP, Nuccitelli R, Isseroff RR. Involucrin-positive keratinocytes demonstrate decreased migration speed but sustained directional migration in a DC electric field. J Invest Dermatol. 1999;113(5):851–855. doi: 10.1046/j.1523-1747.1999.00763.x. [DOI] [PubMed] [Google Scholar]
- 32.Palombella VJ, Yamashiro DJ, Maxfield FR, Decker SJ, Vilcek J. Tumor necrosis factor increases the number of epidermal growth factor receptors on human fibrob-lasts. J Biol Chem. 1987;262(5):1950–1954. [PubMed] [Google Scholar]
- 33.Partridge M, Chantry D, Turner M, Feldmann M. Production of interleukin-1 and interleukin-6 by human keratinocytes and squamous cell carcinoma cell lines. J Invest Dermatol. 1991;96(5):771–776. doi: 10.1111/1523-1747.ep12471723. [DOI] [PubMed] [Google Scholar]
- 34.Poo M. In situ electrophoresis of membrane components. Annu Rev Biophys Bioeng. 1981;10:245–276. doi: 10.1146/annurev.bb.10.060181.001333. [DOI] [PubMed] [Google Scholar]
- 35.Pullar CE, Baier BS, Kariya Y, Russell AJ, Horst BA, Marin-kovich MP, Isseroff RR. beta4 integrin and epidermal growth factor coordinately regulate electric field-mediated directional migration via Rac1. Mol Biol Cell. 2006;17(11):4925–4935. doi: 10.1091/mbc.E06-05-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pullar CE, Isseroff RR. Cyclic AMP mediates keratinocyte directional migration in an electric field. J Cell Sci. 2005;118(Pt 9):2023–2034. doi: 10.1242/jcs.02330. [DOI] [PubMed] [Google Scholar]
- 37.Pullar CE, Isseroff RR, Nuccitelli R. Cyclic AMP-depen-dent protein kinase A plays a role in the directed migration of human keratinocytes in a DC electric field. Cell Motil Cyto-skeleton. 2001;50(4):207–217. doi: 10.1002/cm.10009. [DOI] [PubMed] [Google Scholar]
- 38.Rennekampff HO, Hansbrough JF, Kiessig V, Dore C, Sticherling M, Schroder JM. Bioactive interleukin-8 is expressed in wounds and enhances wound healing. J Surg Res. 2000;93(1):41–54. doi: 10.1006/jsre.2000.5892. [DOI] [PubMed] [Google Scholar]
- 39.Romero LI, Zhang DN, Herron GS, Karasek MA. Interleukin-1 induces major phe-notypic changes in human skin microvascular endothelial cells. J Cell Physiol. 1997;173(1):84–92. doi: 10.1002/(SICI)1097-4652(199710)173:1<84::AID-JCP10>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 40.Rosenkilde MM, Schwartz TW. The chemokine system—a major regulator of an-giogenesis in health and disease. Apmis. 2004;112(7–8):481–495. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- 41.Schmuth M, Neyer S, Rainer C, Grassegger A, Fritsch P, Romani N, Heufler C. Expression of the C-C chemokine MIP-3 alpha/CCL20 in human epidermis with im-paired permeability barrier function. Exp Dermatol. 2002;11(2):135–142. doi: 10.1034/j.1600-0625.2002.110205.x. [DOI] [PubMed] [Google Scholar]
- 42.Sheridan DM, Isseroff RR, Nuccitelli R. Imposition of a physiologic DC electric field alters the migratory response of human keratinocytes on extracellular matrix molecules. J Invest Dermatol. 1996;106(4):642–646. doi: 10.1111/1523-1747.ep12345456. [DOI] [PubMed] [Google Scholar]
- 43.Sticherling M, Hetzel F, Schroder JM, Christophers E. Time- and stimulus-dependent secretion of NAP-1/IL-8 by human fibroblasts and endothelial cells. J Invest Dermatol. 1993;101(4):573–576. doi: 10.1111/1523-1747.ep12366023. [DOI] [PubMed] [Google Scholar]
- 44.Strieter RM, Polverini PJ, Kunkel SL, Arenberg DA, Burdick MD, Kasper J, Dzuiba J, Van Damme J, Walz A, Marriott D, et al. The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J Biol Chem. 1995;270(45):27348–27357. doi: 10.1074/jbc.270.45.27348. [DOI] [PubMed] [Google Scholar]
- 45.Thelen M. Dancing to the tune of chemokines. Nat Immunol. 2001;2(2):129–134. doi: 10.1038/84224. [DOI] [PubMed] [Google Scholar]
- 46.Trollinger DR, Isseroff RR, Nuccitelli R. Calcium channel blockers inhibit galva-notaxis in human keratinocytes. J Cell Physiol. 2002;193(1):1–9. doi: 10.1002/jcp.10144. [DOI] [PubMed] [Google Scholar]
- 47.van der Poll T, Jansen J, Endert E, Sauerwein HP, van Deventer SJ. Noradrenaline inhibits lipopolysaccharide-induced tumor necrosis factor and interleukin 6 produc-tion in human whole blood. Infect Immun. 1994;62(5):2046–2050. doi: 10.1128/iai.62.5.2046-2050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villiger PM, Geng Y, Lotz M. Induction of cytokine expression by leukemia inhi-bitory factor. J Clin Invest. 1993;91(4):1575–1581. doi: 10.1172/JCI116363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodley DT, Bachmann PM, O’Keefe EJ. Laminin inhibits human keratinocyte migration. J Cell Physiol. 1988;136(1):140–146. doi: 10.1002/jcp.1041360118. [DOI] [PubMed] [Google Scholar]
- 50.Woodley DT, O’Keefe EJ, Prunieras M. Cutaneous wound healing: a model for cell–matrix interactions. J Am Acad Dermatol. 1985;12(2 Pt 2):420–433. doi: 10.1016/s0190-9622(85)80005-0. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, et al. Regulation of Toll/IL- 1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430(6996):218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 52.Zhao M, Agius-Fernandez A, Forrester JV, McCaig CD. Orientation and directed migration of cultured corneal epithelial cells in small electric fields are serum depen-dent. J Cell Sci. 1996;109(Pt 6):1405–1414. doi: 10.1242/jcs.109.6.1405. [DOI] [PubMed] [Google Scholar]
- 53.Zhao M, Dick A, Forrester JV, McCaig CD. Electric field-directed cell motility involves up-regulated expression and asymmetric redistribution of the epidermal growth factor receptors and is enhanced by fibronectin and laminin. Mol Biol Cell. 1999;10(4):1259–1276. doi: 10.1091/mbc.10.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao M, Pu J, Forrester JV, McCaig CD. Membrane lipids, EGF receptors, and intracellular signals colocalize and are polarized in epithelial cells moving direction-ally in a physiological electric field. Faseb J. 2002;16(8):857–859. doi: 10.1096/fj.01-0811fje. [DOI] [PubMed] [Google Scholar]
- 55.Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442(7101):457–460. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]