Abstract
Although somatic cell reprogramming to generate inducible pluripotent stem cells (iPSCs) is associated with profound epigenetic changes, the roles and mechanisms of epigenetic factors in this process remain poorly understood. Here we identify Jmjd3 as a potent negative regulator of reprogramming. Jmjd3-deficient MEFs produced significantly more iPSC colonies than did wild-type cells, while ectopic expression of Jmjd3 markedly inhibited reprogramming. We show that the inhibitory effects of Jmjd3 are produced through both histone demethylase-dependent and -independent pathways. The latter pathway is entirely novel and involves Jmjd3 targeting of PHF20 for ubiquitination and degradation via recruitment of an E3 ligase, Trim26. Importantly, PHF20-deficient MEFs could not be converted to fully reprogrammed iPSCs, even with knockdown of Jmjd3, Ink4a or p21, indicating that this protein exerts predominant effects on reprogramming. Our findings demonstrate a previously unrecognized role of Jmjd3 in cellular reprogramming and provide molecular insight into the mechanisms by which the Jmjd3-PHF20 axis controls this process.
Introduction
Both human and mouse somatic cells can be reprogrammed to a pluripotent embryonic stem cell (ESC)-like state, giving rise to induced pluripotent stem cells (iPSCs), by the use of four key transcription factors: Oct4, Sox2, Klf4 and c-Myc (Takahashi et al., 2007b; Takahashi and Yamanaka, 2006; Yu et al., 2007). Because of their similarity to ESCs in terms of gene expression profile, epigenetics/genetic marks, and their ability to self-renew and differentiate into many different cell types, iPSCs hold great promise for human disease modeling, drug screening and perhaps therapeutic applications (Robinton and Daley, 2012; Stadtfeld and Hochedlinger, 2010). Although somatic cell reprogramming can be achieved by several strategies (Hanna et al., 2009; Robinton and Daley, 2012; Stadtfeld and Hochedlinger, 2010), its efficiency and the kinetics of iPSC generation are still suboptimal, suggesting the existence of substantial genetic and epigenetic barriers during reprogramming.
Many factors, including cell proliferation and cycling, mesenchymal-to-epithelial transitions and epigenetic regulation of histone modification and DNA methylation, influence reprogramming efficiency (Plath and Lowry, 2011; Stadtfeld and Hochedlinger, 2010). Transiently enforced expression of reprogramming factors leads to separable events, beginning with mesenchymal-to-epithelial transitions associated with loss of the somatic marker THY1, followed by the activation of embryonic markers such as alkaline phosphatase (AP) and stage-specific embryonic antigen 1 (SSEA1) (Li et al., 2010; Plath and Lowry, 2011). Induction and maintenance of endogenous pluripotency genes such as Nanog and Oct4 require further epigenetic reprogramming changes at the DNA methylation and histone modification levels (Stadtfeld and Hochedlinger, 2010). Failure to achieve these epigenetic changes in a timely manner can lead to partially reprogrammed iPSCs.
Global analysis of euchromatin dynamics during the reprogramming process has revealed orchestrated epigenetic changes at the histone modification level (Gaspar-Maia et al., 2011; Hemberger et al., 2009; Koche et al., 2011). Both ESCs and iPSCs contain “bivalent domains”, where nucleosomes are marked with trimethylation at histone3-lysine27 (H3K27me3) and histone3-lysine4 (H3K4me3) (Gaspar-Maia et al., 2011). While the Polycomb group (PcG) complex mediates H3K27 methylation and inhibits gene repression (Margueron and Reinberg, 2011), Jmjd3 and Utx mediate H3K27 demethylation (Agger et al., 2007; Lan et al., 2007). Thus, given the importance of epigenetic factors in defining cell lineages, it is reasonable to suggest that some of these factors are required for efficient somatic reprogramming, while others may function as negative regulators. Removal of such roadblocks to successful reprogramming will require increased insight into the molecular mechanisms by which epigenetic factors control cell lineage and hence the dynamic process of reprogramming. Here we report identification of Jmjd3 as a potent negative regulator of somatic cell reprogramming in screening studies of a panel of histone-modifying proteins. Knockdown or ablation of Jmjd3 enhanced the efficiency and kinetics of reprogramming, apparently by dual mechanisms: 1) Jmjd3 partially inhibits iPSC reprogramming by promoting cell senescence through upregulation of p21 and Ink4a, and 2) Jmjd3 targets PHF20 (plant homeodomain finger protein 20) for ubiquitination and proteasomal degradation via the E3 ubiquintin ligase Trim26 in a demethylase activity-independent manner. Knockdown or ablation of PHF20 blocks the reactivation of endogenous Oct4 expression, thus leading to partially programmed cells. Our results implicate the Jmjd3-PHF20 axis as a key pathway in somatic cell reprogramming, and provide novel insights into the molecular mechanisms used by Jmjd3 to impede efficient reprogramming.
Results
Identification of Jmjd3 as an Inhibitor of Reprogramming
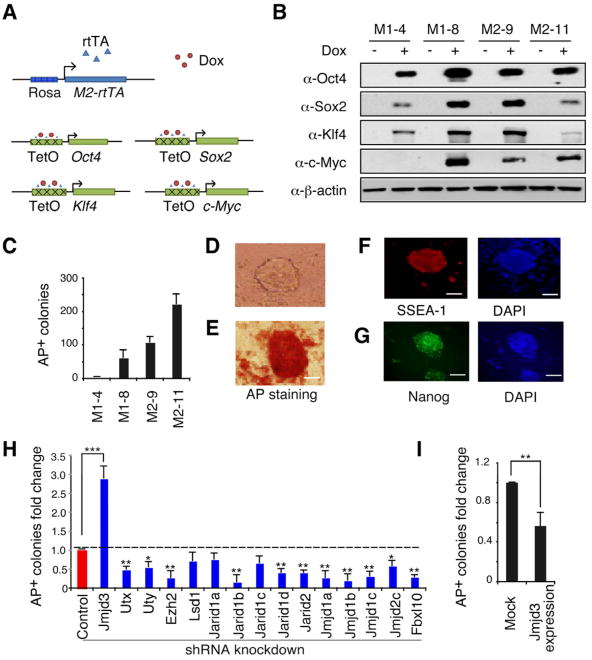
To establish a simpler and inducible 4F-based method to generate iPSCs, we created transgenic mice expressing tetracycline (Tet)-O-inducible Oct4, Sox2, Klf4 and Myc transgenic mice carrying rtTA-M2 reverse tetracycline transactivator (Figure 1A). Mouse embryonic fibroblasts (MEFs) were generated from intercrossing transgenic mice (Figure S1A). As shown in Figure 1B, Oct4, Sox2, Klf4, and Myc proteins were readily detected by immunoblot analysis after treatment with Dox for 24 h. These 4F-expressing MEFs (Tet-O-4F MEFs) could be efficiently reprogrammed to generate iPSCs in the presence of Dox (Figure 1C). Withdrawal of Dox before or at day 8 markedly reduced AP+ colony formation, but withdrawn at day 10 or later showed little or no effect on AP+ colony number using three different types of MEFs (WT, Tet-O-4F and Oct4-GFP) (Figure S1B-D). The fully programmed iPSCs stained positively for AP, SSEA-1 and Nanog (Figures 1D-G), suggesting that Tet-O-4F MEF-based reprogramming would provide a reliable system to screen for epigenetic factors that either enhance or reduce the efficiency of reprogramming.
Figure 1. Identification of Jmjd3 and Other Key Epigenetic Factors that Regulate Reprogramming.
(A) Outline of generation of transgenic mice expressing rtTA, together with Oct4 (O), Sox2 (S), Klf4 (K) and Myc (M) (OSKM, 4F) under control of a tetracycline-on promoter (Tet-O). (B) Western blot analysis of 4F expression in Tet-O MEFs treated with or without Dox. (C) Alkaline phosphatase (AP)-positive colonies were counted at day12 after Dox treatment. (D) Bright field images of an iPSC colony derived from Tet-O 4F MEFs. (E-G) Staining of representative iPSC colonies with antibodies against AP, stage-specific embryonic antigen 1 (SSEA1) and Nanog. Scale bars in panels D, E, F and G, 50μm (H) Fold changes in number of AP-positive colonies generated from Tet-O 4F MEFs transduced with specific shRNA, compared with control shRNA. AP-positive colonies were counted on day14 after Dox treatment. (I) Fold changes in number of AP-positive colonies generated from Tet-O 4F MEFs transduced with Jmjd3 expression or empty vector. Ectopic expression of Jmjd3 inhibits reprogramming. The data in panels H and I are reported as the means ± SD with indicated significance (*p <0.05, **p < 0.01 ***p < 0.001 by Student's t test). See also Figure S1.
We predicted that epigenetic factors play critical roles in reactivating the expression of stem cell-enriched genes, while shutting down the expression of cell lineage-specific differentiation genes, thus greatly increasing the efficiency of 4F-mediated reprogramming. To test this notion, we selected a panel of shRNAs with high knockdown efficiency (>70%) against a subset of genes encoding histone methyltransferases or demethylases based on PCR or western blot analysis (Figures S1E-S1F and Tables S1-S2). After three rounds of screening, we found that knockdown of the H3K27 methyltransferase Ezh2 and many histone demethylase genes, including Fbxl10, Jarid1b, Jarid1d, Jarid2, Jmjd1a, Jmjd2c and Utx, markedly decreased reprogramming efficiency (Figure 1H), consistent with recent findings for Utx and Fbxl10 (Mansour et al., 2012; Wang et al., 2011). By contrast, knockdown of Jmjd3 markedly increased the efficiency of 4F-mediated reprogramming, while its ectopic expression resulted in decreased reprogramming efficiency (Figure 1I), suggesting that Jmjd3 functions as a barrier to somatic reprogramming. This unique feature of Jmjd3 led to its selection for further study.
Jmjd3 Ablation Enhances the Efficiency and Kinetics of Reprogramming
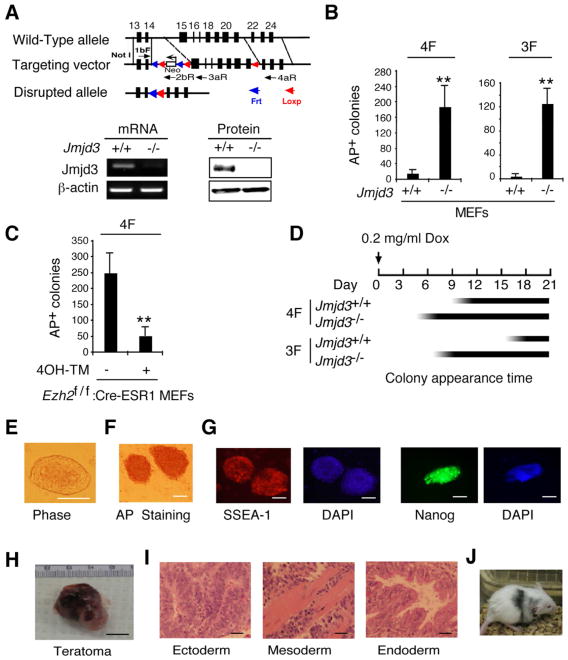
To further define the role of Jmjd3 in reprogramming, we generated Jmjd3 knockout mice by targeted deletion of exon 15-21 using a Cre-LoxP system (Figure 2A). Global deletion of Jmjd3 died shortly after birth due to lung dysfunction (to be reported elsewhere). Expression of Jmjd3 was abrogated in Jmjd3-deficient MEFs (Figure 2A). Consistent with results obtained by Jmjd3 knockdown, 4F-reprogramming of Jmjd3-deficienct MEFs produced significantly more iPSC colonies than did WT MEFs (Figure 2B). Robust reprogramming was also achieved with Jmjd3-deficient 3F-transduced MEFs (Oct4, Sox2 and Klf4), compared with WT MEFs (Figure 2B). By contrast, Ezh2-deficient MEFs, which were generated by treating Ezh2flox/flox:Cre-ESR MEFs with tamoxifen, strikingly inhibited the efficiency of 4F-mediated reprogramming of MEFs (Figure 2C), further confirming that Ezh2 is necessary for reprogramming. More importantly, we found that AP+ iPSC colonies appeared much earlier in Jmjd3-deficient MEFs than in WT MEFs (Figure 2D). The iPSCs generated from Jmjd3-deficient MEFs showed characteristic ESC morphology and markers such as AP, SSEA-1 and Nanog (Figures 2E-2G). They also formed teratomas comprising all three embryonic germ layers (ectoderm, mesoderm and endoderm) (Figures 2H-2I), and contributed to chimeras after injection into BALB/C host blastocysts (Figure 2J). Taken together, these results suggest that loss of Jmjd3 markedly enhances the efficiency and kinetics of iPSC reprogramming.
Figure 2. Jmjd3 Ablation Enhances the Efficiency and Kinetics of Reprogramming.
(A) Establishment of Jmjd3 knockout mice and Jmjd3-deficient MEFs. PCR and immunoblot (IB) analyses confirmed loss of Jmjd3 expression in Jmjd3-deficient MEFs. (B) AP-positive colonies were counted on day 14 of 4F-mediated or on day 21 of 3F-mediated reprogramming of WT and Jmjd3-deficient MEF cells. (C) AP-positive colonies were counted on day 14 of 4F-mediated reprogramming after tamoxifen treatment of Ezh2f/f :Cre-ESR1 MEFs. (D) Schematic representation of the time of AP+ colony appearance during reprogramming. (E) Bright-field image of a representative iPSC colony generated from Jmjd3-deficient MEFs. (F and G) Staining of representative iPSC colonies with antibodies against AP, SSEA1 and Nanog. Scale bars, 50 μm. (H and I) Photomicrograph and HE (hematoxylin and eosin) staining of teratomas generated from immune-deficient mice harboring a Jmjd3-deficient iPSC clone. Three layers of teratoma (ectoderm, mesoderm and endoderm) are presented. Scale bars, 100 μm. (J) Jmjd3−/− MEF-derived iPSCs were pluripotent and contributed to chimeras after injection into BALB/C host blastocysts as indicated by coat color. The data in panels B and C are reported as means ± SD with indicated significance (*p < 0.05, **p < 0.01 by Student's t test).
Jmjd3 Negatively Regulates Reprogramming by Modulating Expression of the Ink4a/Arf Locus
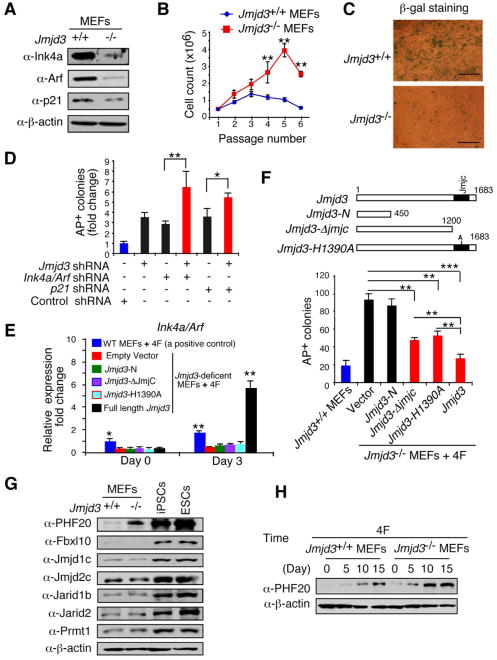
We next asked how Jmjd3 ablation enhances reprogramming. Jmjd3 expression is thought to increases the expression of Ink4a/Arf in MEFs by modifying H3K27 methylation in the promoter region of the Ink4a/Arf locus (Agger et al., 2009; Barradas et al., 2009). Knockdown or deletion of Ink4a/Arf and p21 reduces cell senescence and markedly increases the efficiency of reprogramming (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009). To assess the expression level of these molecules in Jmjd3-deficient MEFs, we showed that Jmjd3 deletion markedly reduced the expression of Ink4a/Arf mRNA and protein (Figure 3A and Figure S2A). The p21 protein, but not its mRNA, was also reduced in Jmjd3-deficient MEFs (Figure 3A and Figure S2A). Consistent with this, we found that Jmjd3-deficient MEF cells grew faster than WT cells and reduced cellular senescence based on β-galactosidase (β-gal) staining, compared with WT MEFs (Figures 3B-3C). Because the Jmjd3-deficient MEFs eventually underwent a senescence crisis after 5-7 passages, it is likely that transient effects of Jmjd3 deficiency on cell proliferation and senescence may have contributed to the improved efficiency and kinetics of reprogramming.
Figure 3. Identification of Jmjd3 Targets Responsible for Enhanced Reprogramming.
(A) Western blot analysis of Ink4a, Arf and p21 in WT and Jmjd3-deficient MEFs at passage 4. (B) Growth of WT versus Jmjd3-deficient MEFs. (C) β-gal staining of WT versus Jmjd3-deficient MEFs at passage 4. (D) Number of AP-positive iPSC colonies counted on day 14 of 4F-mediated reprogramming of WT MEFs transduced with Jmjd3-, Ink4a/Arf-, p21-specific or control shRNAs. (E) Jmjd3-deficient MEFs transduced with lentiviruses expressing Jmjd3 or its mutants together with 4F for 3 days. Expression of Ink4a/Arf was determined by real-time PCR analysis and was normalized by internal control beta-actin. (F) Number of AP-positive iPSC colonies counted on day 11 of 4F-mediated reprogramming of Jmjd3-deficient MEFs transduced with lentiviruses expressing Jmjd3 or its mutants. (G) Western blot analysis of 7 epigenetic proteins in WT and Jmjd3-deficient MEFs, iPSCs and ES cells. (H) Western blot analysis of PHF20 expression in WT and Jmjd3-deficient MEFs during 4F-mediated reprogramming. The data in panels B, D-F are reported as means ± SD with indicated significance (*p < 0.05, **p < 0.01, ***p<0.001). See also Figures S2 and S3.
To determine the relative contribution of downregulation of Ink4a/Arf and p21 to increased efficiency of reprogramming in Jmjd3-deficient MEFs, we knocked down the expression of these genes with specific shRNAs (Figure S2B). Although knockdown of Jmjd3, Ink4a/Arf or p21 alone by shRNAs increased reprogramming efficiency, compared to that in MEFs transduced with a control shRNA, the efficiency nearly doubled with simultaneous knockdown of Jmjd3 and Ink4a/Arf or p21 (Figure 3D), suggesting that Jmjd3 might have additional effects on reprogramming. To test this possibility, we generated the Jmjd3-N (containing the N-terminal 450 aa), Jmjd3-ΔJmjC (containing a deletion in the catalytic Jumonji domain) and Jmjd3-H1390A (containing a point mutation in the catalytic domain) constructs, all of which lack the H3K27me3 demethylase activity of H3K27 trimethylation (Figure S2C). To determine whether Jmjd3-mediated inhibition of reprogramming depends upon expression of Ink4a/Arf and p21, we showed that ectopic expression of full-length Jmjd3, but not Jmjd3-N, Jmjd3-ΔJmjC or Jmjd3-H1390A, in Jmjd3-deficient MEFs restored the expression of Ink4a/Arf (Figure 3E) and markedly inhibited reprogramming (Figure 3F). Surprisingly, two Jmjd3 mutants (Jmjd3-ΔJmjC and Jmjd3-H1390A) that lacked H3K27 demethylase activity and failed to upregulate Ink4a/Arf expression were still capable of inhibiting reprogramming in Jmjd3-deficient MEFs (Figure 3F), suggesting that Jmjd3 can modulate reprogramming through a previously unknown, demethylase activity-independent pathway.
PHF20 is a Key Target of the Jmjd3 Protein
To search for the targets of Jmjd3, we performed a comparative analysis of miRNA and mRNA gene expression between WT and Jmjd3-deficient MEFs, but failed to identify any gene that could be responsible for the increased reprogramming efficiency in Jmjd3-deficient MEF cells (data not shown). Hence, we turned our attention to the expression levels of epigenetic factors, because they are critical in reprogramming somatic cells to an ESC-like state (Plath and Lowry, 2011; Stadtfeld and Hochedlinger, 2010). By comparing the expression of 59 genes that encode for histone modification proteins, we identified 18 genes that were markedly upregulated at the RNA level in iPSCs/ESCs, versus MEFs, and 11 genes that were upregulated between iPScs/ESCs versus human fibroblasts (Figure S2D and S2E). Among them, seven genes were highly expressed in both mouse and human iPSCs/ESCs (Figure S2F). Of these, only PHF20 (also called GLEA2) showed a marked increase of expression in Jmjd3-deficient MEFs, iPSCs and ESCs, versus WT MEFs (Figure 3G). However, no appreciable differences in PHF20 mRNA and H3K27 trimethylation were observed between WT and Jmjd3-deficient MEFs (Figure S3A-S3B). PHF20 was strongly expressed in testis, ovary and ES cells, but weakly in other tissues (Figures S3C and S3D). Interestingly, PHF20 expression was gradually increased in WT MEFs during reprogramming, which was accelerated in Jmjd3-deficient MEFs (Figure 3H). These results suggest that the PHF20 protein is an important target of Jmjd3, and thus may play a role in ESCs and iPSCs.
Requirement for PHF20 in the Maintenance and Reprogramming of ESCs and iPSCs
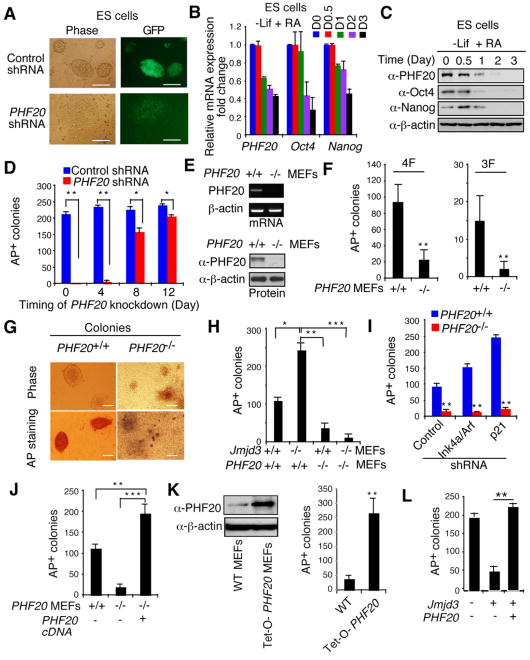
Because the PHF20 protein is abundantly expressed in both ESCs and iPSCs, we next sought to determine its importance in the maintenance of these cell types. After knocking down PHF20 in ESCs with specific shRNAs containing a GFP cassette (Figure S4A), ESCs underwent differentiation, while ESCs transduced with control shRNA remained undifferentiated (Figure 4A). Furthermore, RT-PCR and western blot analyses revealed that PHF20 expression in ESCs, like that of Oct4 and Nanog, was markedly reduced after withdrawal of leukemia-inhibiting factor (LIF) and addition of retinoic acid (RA) in the culture medium (Figure 4B and 4C). Similar results were obtained with iPSCs (Figures S4B and S4C). To determine whether stable ESC lines could be derived from WT and PHF20 knockout mice, we found that ESC lines could be readily generated from WT mice, but not from PHF20 knockout mice (Figure S4D-S4E). WT ESCs expressed AP, Nanog and Oct4 proteins, whereas cells from PHF20 knockout blastocysts did not (Figure S4E) and differentiated rapidly into trophectoderm, based on downregulation of Oct4 and upregulation of Cdx2 (Figure S4F). These results suggest that PHF20 is required for the generation and maintenance of both ESCs and iPSCs.
Figure 4. PHF20 Is Essential for Maintenance and Reprogramming of iPSCs.
(A) Bright-field and GFP images of ESCs transduced with PHF20-specific or control shRNA. Scale bars, 50 μm. (B) Real-time PCR analysis of PHF20, Oct4 and Nanog expression in ESCs treated with 1 mM RA and LIF withdrawal. (C) Western blot analysis of PHF20, Oct4 and Nanog expression after treatment of ESCs with 1 μM RA and LIF withdrawal. (D) Colony formation after PHF20 knockdown by PHF20-specific or a control lentivirus-based constitutive shRNA at different time points (i.e., day 0, 4, 8 or 12) during reprogramming of Tet-O-4F MEFs. AP-positive colonies were counted on day 14. (E) PCR and Western blot analyses of PHF20 expression in WT and PHF20-deficient MEFs. (F) AP-positive colony numbers were counted on day 14 of 3F- or 4F-mediated reprogramming of WT and PHF20-deficient MEFs. (G) Bright-field images and AP staining of iPSC-like colonies from WT and PHF20-deficient MEFs. Scale bars, 50 μm. (H) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT and PHF20 single, or Jmjd3 and PHF20-double KO MEFs. (I) Number of AP-positive colonies on day 14 of 4F-mediated reprogramming of WT and PHF20 KO MEFs transduced with Ink4a/Arf- or p21-specific or control shRNA. (J) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT and PHF20 knockout MEFs with or without PHF20 cDNA expression. (K) Western analysis of PHF20 expression in WT and Tet-O-PHF20 transgenic MEFs expressing rtTA in the presence of Dox. AP-positive colonies were counted on day 14 of 4F-mediated reprogramming of rtTA-expressing Tet-O-PHF20 transgenic MEFs. (L) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT MEFs transduced with Tet-O-Jmjd3 or Tet-O-PHF20 in the presence of Dox until day 10. The data in panels D, F and H-L are plotted as means ± SD with indicated significance (*p < 0.05, **p < 0.01). See also Figure S4.
To further define the role of PHF20 in iPSC generation, we first knocked down the protein in Tet-O-4F MEFs at different time points and tested its ability to form iPSC colonies. Knockdown of PHF20 in the early stages of reprogramming (i.e., at day 0 or 4) markedly blocked iPSC generation, whereas in the intermediate or later stages (day 10 or 12) it led to a decreased (but still significant) inhibitory effect on the numbers of iPSC colonies (Figure 4D). Using PHF20 knockout MEFs (Figure 4E), we showed that reprogramming to iPSCs with either 3F or 4F was significantly inhibited in PHF20-deficient MEFs (Figure 4F), and the few iPSC colonies that were generated from PHF20-deficient MEFs showed only partially reprogrammed iPSCs (Figure 4G), suggesting that PHF20 is required for the efficient generation of fully reprogrammed iPSCs.
Like the results obtained with Jmjd3-deficient MEFs, we found that Jmjd3 knockdown in human fibroblasts enhanced reprogramming, while PHF20 knockdown blocked this process (Figure S4G and S4H). To clarify how Jmjd3 and PHF20 reciprocally regulate reprogramming, we generated Jmjd3/PHF20 single- or double-knockout MEFs and tested their ability to regulate reprogramming. Both Jmjd3-deficient and Jmjd3/PHF20 double-knockout MEF cells grew faster than WT and PHF20-deficient cells, but no appreciable difference was observed in the growth between WT and PHF20-deficient cells (Figure S4I). As expected, Jmjd3 deletion enhanced reprogramming, but PHF20 ablation inhibited this process (Figure 4H). Remarkably, Jmjd3 deletion failed to improve reprogramming in Jmjd3 and PHF20 double-knockout MEFs (Figure 4H), suggesting that the proliferative advantage of Jmjd3-deficient MEFs cannot overcome the failure of reprogramming in PHF20-deficient MEFs. Similar results were obtained when either Ink4a or p21 was knocked down in PHF20-deficient MEFs: that is, loss of each of these regulators increased reprogramming in WT MEFs, but failed to rescue defective reprogramming in PHF20-deficient MEFs (Figure 4I). Ectopic expression of PHF20, by contrast, restored reprogramming in PHF20-deficient MEFs (Figure 4J), suggesting a requirement for expression of this gene in both WT and Jmjd3-deficient MEFs.
To further examine the ability of PHF20 expression to facilitate reprogramming, we generated Tet-O-PHF20 MEFs from rtTA:Tet-O-PHF20 transgenic mice and treated them with Dox, resulting in increased expression of PHF20, compared with findings in Dox-treated rtTA-expressing WT MEFs (Figure 4K). More importantly we observed that Dox-induced expression of PHF20 in these cells led to a marked increase in the efficiency of 4F-mediated reprogramming, compared with Dox-treated rtTA-expressing WT MEFs (Figure 4K). Furthermore, overexpression of PHF20 could reverse the Jmjd3-mediated inhibition of reprogramming (Figure 4L). The increased reprogramming efficiency in Tet-O-PHF20 MEFs was not due to cellular proliferative activity, because there was no appreciable difference in cell growth between WT and Tet-O-PHF20 MEFs, with or without Dox treatment (Figures S4J). Instead, Dox-induced expression of PHF20 markedly blocked downregulation of Oct4, Sox2 and Nanog in iPSCs and thus their differentiation after LIF withdrawal (Figures S4K and S4L). Nonetheless, PHF20 overexpression could not substitute for any of the 4F (Figure S4M). These results indicate as essential requirement for PHF20 in somatic cell reprogramming.
Jmjd3 Interacts with PHF20 and Mediates its Proteasomal Degradation
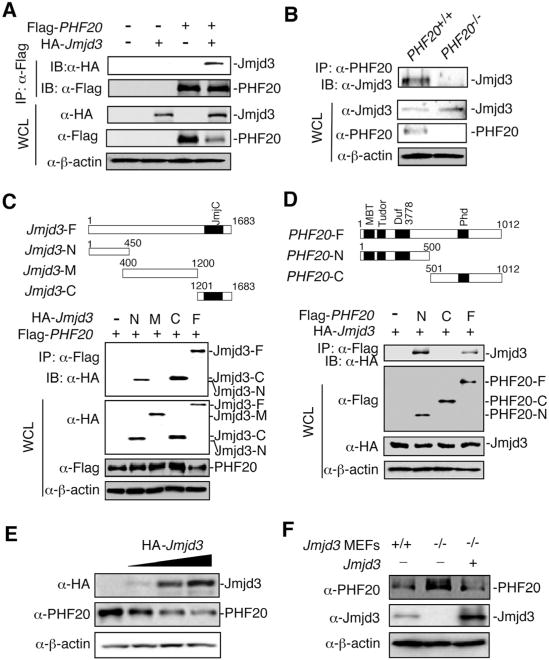
To dissect the molecular mechanisms by which Jmjd3 and PHF20 reciprocally control reprogramming, we first showed their localization in the nucleus by immunofluorescent staining and fractionation experiment of ESCs and iPSCs (Figures S5A-S5B). Coimmunoprecipitation (co-IP) and western blot analyses of 293T cells transfected with Flag-PHF20 and HA-Jmjd3 revealed that Jmjd3 interacted with PHF20 (Figure 5A). Similar results were obtained with WT MEFs but not with PHF20-deficient MEFs (Figure 5B), suggesting that Jmjd3 interacts with PHF20 under physiological conditions. We then performed domain-mapping experiments with Jmjd3-N (1-450 aa), Jmjd3-M (400-1200 aa) and Jmjd3-C (1201-1683 aa), showing that the Jmjd3-N and Jmjd3-C constructs, but not Jmjd3-M, interacted with PHF20 (Figure 5C). Similarly, the N-terminal region (1-332 aa containing a DNA binding domain), but not the C-terminal region, of PHF20 interacted with Jmjd3 (Figure 5D and Figure S5C). Further experiments showed that Jmjd3, but not Utx or Uty, interacted with PHF20 (Figure S5D), suggesting that Jmjd3 specifically interacts with PHF20 via their functional domains.
Figure 5. Jmjd3 Interacts with PHF20 and Causes its Degradation.
(A) 293T cells were transfected with Flag-PHF20 and HA-Jmjd3. Cell extracts were immunoprecipitated with anti-Flag beads, followed by immunoblot (IB) with an anti-HA antibody. (B) Detection of interaction between Jmjd3 and PHF20 in WT and PHF20 knockout MEFs by immunoprecipitation (IP) with a PHF20 antibody, followed by IB with an anti-Jmjd3 antibody. (C and D) 293T cells were transfected with different HA-Jmjd3 domain constructs (C) and different Flag-PHF20 constructs (D). Cell extracts were immunoprecipitated with anti-Flag beads, followed by IB with an anti-HA antibody. (E) 293T cells were transfected with increasing amounts of HA-Jmjd3 cDNA, followed by IB with anti-PHF20 and anti-HA antibodies. (F) Jmjd3 WT and KO MEF cells were infected with or without Flag-tagged Jmjd3 retrovirus for24 h, and cell extracts were analyzed by IB with anti-PHF20 and anti-Jmjd3 antibodies. See also Figure S5.
What are the functional consequences of the Jmjd3-PHF20 interaction? To address this question, we transfected 293T cells with a fixed amount of Flag-PHF20 together with increasing amounts of HA-Jmjd3, and found that the amounts of PHF20 protein decreased with increasing expression of Jmjd3 protein (Figure S5E). Similarly, the amounts of endogenous PHF20 protein were decreased in 293T cells transfected with increasing amounts of Jmjd3 cDNA (Figure 5E). Furthermore, the amount of endogenous PHF20 protein in Jmjd3-deficient MEFs was much higher than in WT MEFs, while ectopic expression of Jmjd3 cDNA in Jmjd3-deficient MEFs reduced the amount of PHF20 protein to a level similar to that in WT MEFs (Figure 5F). It, therefore, appears that Jmjd3 negatively regulates PHF20 protein by targeting it for degradation.
Trim26 is an E3 Ubiquitin Ligase Required for PHF20 Ubiquitination and Degradation
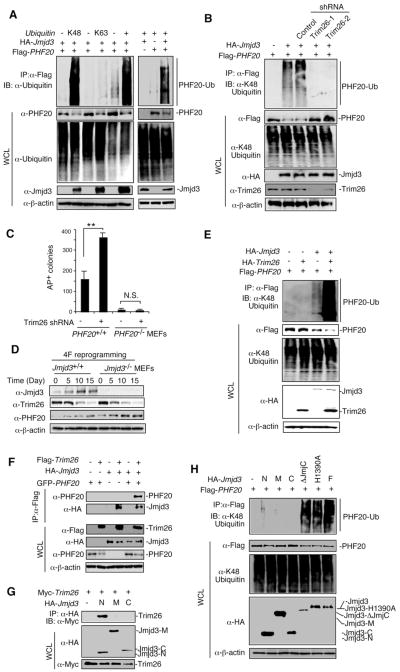
To determine how Jmjd3 causes the degradation of PHF20, we first tested whether this protein is ubiquitinated in 293T cells expressing WT, K48 or K63 ubiquitin. PHF20 strongly underwent K48-linked ubiquitination, with little or no K63-linked ubiquitination, and such an ubiquitination was observed only when Jmjd3 and PHF20 were coexpressed (Figure 6A), suggesting that Jmjd3 specifically targets PHF20 for K48-linked polyubiquitination.
Figure 6. Jmjd3 Targets PHF20 for Ubiquitination by Recruiting an E3 Ligase Trim26.
(A) 293T cells transfected with HA-tagged ubiquitin, Flag-tagged PHF20 and HA-tagged Jmjd3. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with an anti-ubiquitin antibody. (B) 293T cells transfected with Flag-tagged PHF20, HA-tagged Jmjd3 and different Trim26 shRNA constructs as indicated. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed with an anti-K48 ubiquitin. (C) Number of AP-positive colonies counted on day 14 of 4F-mediated reprogramming of WT and PHF20-deficient MEFs transduced with Trim26-specific or control shRNAs. Asterisks indicate significant differences between groups (*p < 0.05, **p < 0.01, N.S. p > 0.05). (D) Western blot analysis of Jmjd3, Trim26 and PHF20 expression in WT and Jmjd3-deficient MEFs during cellular reprogramming. (E) 293T cells transfected with HA-Trim26, Flag-PHF20 and HA-Jmjd3 as indicated. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with an anti-K48 ubiquitin antibody. (F) 293T cells transfected with Flag-Trim26, GFP-PHF20 and HA-Jmjd3 as indicated. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with PHF20 and HA antibodies. (G) 293T cells transfected with Myc-Trim26 and HA-Jmjd3 constructs. Cell lysates were immunoprecipitated with anti-HA beads, followed by IB with an anti-Myc antibody. (H) 293T cells transfected with Flag-PHF20 and HA-Jmjd3 constructs. Cell lysates were immunoprecipitated with anti-Flag beads, followed by IB with an anti-K48 ubiquitin antibody. See also Figure S6.
Since Jmjd3 is not an E3 ubiquitin ligase, we reasoned that Jmjd3 might function as an adaptor to recruit an E3 ubiquitin ligase to PHF20 for ubiquitination. To test this prediction, we designed a screen using 293T cells transfected with Jmjd3 expression vector and a lentivirus-based shRNA sublibrary against human E3 ubiquitin ligases, as previously described (Cui et al., 2012). In an initial screening of about 600 shRNAs, we identified an E3 ubiqtuitin ligase (Trim26)-specific shRNA that was associated with increased PHF20 protein amounts, relative to results with control shRNA (Figures S6A and S6B). To substantiate this finding, we selected two shRNAs for human Trim26 and three for murine Trim26, with 60-90% knockdown efficiency (Figures S6C and S6D). Knockdown of endogenous Trim26 by shRNAs markedly abrogated Jmjd3-mediated ubiquitination of PHF20 in 293T cells (Figure 6B), with similar results obtained when either Jmjd3 or Trim26 was knocked down in PHF20+/+ MEFs (Figure S6E). Consistent with these results, we found that knockdown of Trim26 increased reprogramming efficiency in PHF20 WT MEFs, but not in PHF20-deficient MEFs (Figure 6C). Furthermore, knockdown of Trim26 reversed Jmjd3-mediated inhibition of reprogramming (Figures S6F), while overexpression of Trim26 inhibited reprogramming efficiency enhanced by Jmjd3 knockdown (Figure S6G).
Because Trim26 and Jmjd3 could act in concert to modulate reprogramming through targeting PHF20 for ubiquitination and degradation, we next determined their expression patterns during reprogramming, and found that Trim26 was decreased while Jmjd3 was increased (Figure 6D). As expected, PHF20 expression gradually increased, but PHF20 protein in Jmjd3−/− MEFs was significantly higher than that in WT cells during reprogramming (Figures 6D and S6H). Although treatment with the protease inhibitor MG132 blocked protein degradation even when both Trim26 and Jmjd3 were overexpressed (Figure S6I), it was non-specific and caused cell death. Thus, we did not observe any iPSC colony formation after MG132 treatment (Figure S6J).
We then asked whether ectopic expression of Trim26 would promote PHF20 ubiquitination and degradation. Indeed, coexpression of Trim26 and Jmjd3 led to a remarkable increase in K48-linked ubiquitination and degradation of PHF20, compared with Trim26 or Jmjd3 alone (Figure 6E). To determine whether Trim26 interacts with Jmjd3 or PHF20, we performed immunoprecipitation experiments using cells that expressed Jmjd3, Trim26 alone or PHF20 and Trim26 together. Although Trim26 interacted with Jmjd3 but not PHF20 (Figure 6F), we detected both Jmjd3 and PHF20 in the anti-Flag-Trim26 immunoprecipitants of the cells that expressed Jmjd3, PHF20 and Trim26 (Figure 6F), suggesting that Jmjd3 is an adaptor protein that recruits Trim26 to PHF20. To determine which domain of Jmjd3 is responsible for recruiting Trim26 to PHF20, we transfected 293T cells with Jmjd3-N, Jmjd3-M or Jmjd3-C together with Trim26. Immunoprecipitation and western blot experiments revealed that the N-terminus of Jmjd3 (Jmjd3-N), but not Jmjd3-M and Jmjd3-C, was capable of binding to Trim26 (Figure 6G). To identify the domain of Jmjd3 that is required for Trim26-mediated ubiquitination of PHF20, we transfected 293T cells with Flag-PHF20 together with HA-tagged Jmjd3-N, Jmjd3-M, Jmjd3-C, Jmjd3-ΔJmjC, Jmjd3-H1390A, or full-length Jmjd3. After immunoprecipitation with anti-Flag, we assessed K48-linked ubiquitination of PHF20, observing that none of the Jmjd3-N, Jmjd3-M and Jmjd3-C constructs were sufficient to cause PHF20 ubiquitination (Figure 6H). By contrast, like full-length Jmjd3, Jmjd3-ΔJmjC and Jmjd3-H1390A were able to mediate PHF20 ubiquitination (Figure 6H), consistent with results showing that Jmjd3-ΔJmjC and Jmjd3-H1390A could still inhibit iPSC reprogramming in Jmjd3-deficient MEFs (Figure 3F). These results suggest that the N-terminus of Jmjd3 (Jmjd3-N) can interact with Trim26, but is not sufficient to cause PHF20 ubiquitination. Jmjd3 containing the first 1200 aa or a point mutation (Jmjd3-ΔJmjC or Jmjd3-H1390A) is necessary and sufficient to target PHF20 for ubiquitination by recruiting the E3 ligase Trim26.
PHF20 Is Required for Endogenous Oct4 Expression and Interacts with Wdr5 During Reprogramming
Since PHF20 is essential for reprogramming in both WT and Jmjd3-deficient MEFs, we reasoned that it might be required for the reactivation of endogenous key genes such as Oct4 and other markers of ESCs. To test this prediction, we examined the effects of PHF20 loss on the activation of 11 ESC markers during Tet-O-4F reprogramming, using WT and PHF20−/− MEFs in the presence of Dox for the first 10 days (and then withdrawal). Real-time PCR analysis on day 14 revealed that expression of Oct4, Sox2, Nanog, Dnmt3l, Esg1, Eras, Rex1, and Cripto could not be activated or substantially reduced in PHF20-deficient MEFs, but were highly activated in WT MEFs. By contrast, Stat3, Grb2 and β-catenin were activated normally in both WT and PHF20-deficient cells (Figure 7A). Notably, Sox2 and Nanog could be reactivated when Dox was retained during reprogramming (Figure S7A). Overexpression of Oct4 or even 4F could not rescue the incompletely reprogrammed phenotype of PHF20-deficient MEFs after reprogramming (Figure S7B), suggesting that PHF20 is an upstream factor that controls many key reprogramming and pluripotency factors.
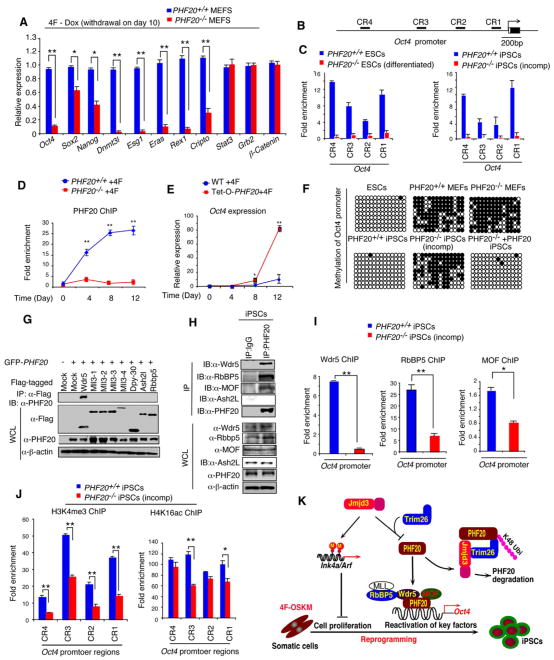
Figure 7. PHF20 is Required forOct4Expression during Reprogramming by Interacting with Wdr5.
(A) WT and PHF20 knockout MEFs were reprogrammed by 4F in the presence of Dox for 10 days, followed by Dox withdrawal. Real-time PCR analysis of endogenous Oct4, Sox2, Nanog and other ES genes was performed on day 14. (B) Schematic diagram of Oct4 promoter regions. (C) Determination of PHF20 binding to the CR regions of Oct4 promoter in iPSCs and ESCs by ChIP-qPCR analysis with PHF20-specific antibody. Values are reported as fold enrichment relative to input DNA. (D) ChIP-PCR analysis of timing of PHF20 binding to the Oct4 promoter region during 4F-mediated reprogramming using a PHF20-specific antibody. (E) Real-time PCR analysis of Oct4 expression in rtTA-expressing WT and Tet-O-PHF20 transgenic MEFs during reprogramming. (F) Bisulfite sequencing of the Oct4 promoter region in ESCs, PHF20+/+ MEFs, PHF20−/− MEFs,iPSCs from PHF20+/+ MEFs, iPS-like (incompletely reprogrammed) cells from PHF20−/− MEFs,and iPSCs from PHF20-expressing PHF20−/− MEFs. Open circles (○) stand for unmethylated CpG dinucleotides, while closed circles (•) for methylated CpGs. (G) 293T cells were transfected with GFP-PHF20, Flag-tagged Wdr5, MLL3, Dpy-30, Ash2L orRbbp5. Cell extracts were immunoprecipiated with anti-Flag beads, followed by IB with an anti-PHF20 antibody. (H) Detection of endogenous interaction between PHF20 and Wdr5 complex in iPSCs by IP with a PHF20 antibody, followed by IB with Wdr5, RbBP5, MOF and Ash2L antibodies. (I) ChIP-qPCR analysis of Wdr5, RbBP5 and MOF binding to Oct4 promoter region during 4F-mediated reprogramming of WT and PHF20-deficient MEFs. (J) ChIP-qPCR analysis of H3K4me3 and H4K16ac mark of the Oct4 promoter in WT MEF-derived iPSCs and PHF20-deficient derived iPSC-like (incompletely reprogrammed) cells. (K) Proposed a working model by which Jmjd3 regulates somatic cell reprogramming. The data in panels A, C-E, I and J are plotted as means ± SD with indicated significance (*p < 0.05, **p < 0.01, by Student's t test). See also Figure S7.
Because reactivation of endogenous Oct4 is essential for the generation of completely reprogrammed iPSCs (Ang et al., 2011), we next determined whether PHF20 could directly bind to the Oct4 promoter in vivo. ChIP-PCR assay revealed that PHF20 was strongly bound to this promoter in WT ESCs and iPSCs, but not in PHF20-deficient (differentiated) ESCs and (incompletely reprogrammed) iPSCs (Figures 7B and 7C). PHF20 was unable to bind to the promoter regions of Cripto, Dnmt3l, Esg1, Eras, Nonog, Rex1 or Sox2 (Figure S7C). Furthermore, the binding of PHF20 to the Oct4 promoter increased gradually over the course of reprogramming (Figure 7D). To further determine whether overexpression of PHF20 could promote expression of endogenous Oct4, we treated both WT and Tet-O-PHF20 MEFs expressing rtTA with Dox during 4F-mediated reprogramming. The expression level of Oct4 was markedly increased in Dox-treated Tet-O-PHF20 MEFs, compared with Dox-treated rtTA-expressing WT MEFs (Figure 7E), suggesting that PHF20 promotes endogenous Oct4 gene expression during reprogramming.
Because the DNA methylation status of the Oct4 promoter serves as an important marker of fully reprogrammed iPSCs (Stadtfeld and Hochedlinger, 2010), we undertook bisulfate sequencing analysis of ESCs and iPSCs generated from WT MEFs, which showed robust DNA demethylation in the Oct4 promoter regions. By contrast, incompletely reprogrammed iPSC colonies from PHF20-deficient MEFs retained their DNA methylation pattern (Figure 7F). More importantly, we showed that ectopic expression of PHF20 could rescue the incompletely reprogrammed state of PHF20-deficient iPSCs and the status of the Oct4 promoter demethylation, similar to results for WT iPSCs (Figure 7F).
PHF20 is a component of mixed-lineage leukemia (MLL) H3K34 methyltransferase complexes with the core components MLL, ASH2L, WDR5 and RBBP5, as well as a component of the H4K16 acetyltransferase MOF (male-absent on the first, also called MYST1, KAT8) complex (Cai et al., 2010; Dou et al., 2005; Wysocka et al., 2005). Importantly, Wdr5 is also a key component shared by MLL H3K4 methyltransferase and the H4K16 acetyltransferase MOF (Cai et al., 2010; Dou et al., 2005; Wysocka et al., 2005). However, it is not known whether PHF20 interacts with Wdr5 or other components of these two complexes. Because PHF20 is upregulated and binds to the Oct4 promoter during reprogramming, we predicted that PHF20 might interact with Wdr5 to promote endogenous Oct4 expression during reprogramming. To test this possibility, we transfected 293T cells with PHF20 together with Wdr5, MLL3, Dpy-30, Ash2l or RbBP5, all core components of the H3K4 methyltransferase complex (Trievel and Shilatifard, 2009). PHF20 interacted with Wdr5, but not with other proteins tested (Figures 7G and S7D). Endogenous interactions between PHF20 and Wdr5 or RbBP5 (but not Ash2L) were observed in iPSCs (Figure 7H). ChIP-seq analysis of ESCs and iPSCs confirmed that both PHF20 and Wdr5 were bound to the Oct4 promoter (Figure S7E). Among 6209 genes bound by PHF20 and 7774 genes by Wdr5, approximately 2348 genes were co-occupied by PHF20 and Wdr5 in ESCs (Figure S7F and Table S3). To determine the PHF20 and Wdr5 binding distribution relative to gene structure (i.e. 5′ distal, 5′ proximal, 5′ UTR, Coding, intron, 3′ UTR, 3′ proximal and 3′ distal regions), we found that the majority of PHF20 and Wdr5 binding sites were mapped to gene body (coding and intron regions) and 5′ proximal region in ESCs (Figure S7G). The distribution of PHF20 was more pronounced toward 5′ end of genes with 9.7% peaks at 5′ proximal region, compared to 2.8% at 3′ proximal region (Figure S7G). Consistent with this, we found that both PHF20 and Wdr5 binding peaks centered on TSS within a 7 kb region (from -2 to +5 kb) (Figure S7H). To determine functional relevance of target genes bound by PHF20 or by PHF20 and Wdr5, we performed gene ontology (GO) analysis and found that binding targets were enriched for genes involved in cell and organ developmental process, embryonic development, and cell differentiation (Figure S7I).
To determine whether PHF20 interacts with MOF, we performed co-immunoprecipitation with anti-PHF20, and found that PHF20 interacted with endogenous MOF in iPSCs (Figure 7H), consistent with the results of a recent report showing that H3K4 methylation is closely associated with H4K16 acetylation (Wang et al., 2009). Thus, PHF20 interacts with Wdr5 and MOF to bring the H3K4 methyltransferase complex and H4K16 aceyltransferase MOF complex together. To understand how the loss of PHF20 results in failure to reactivate endogenous Oct4 expression, we tested the possibility that PHF20 might affect the ability of Wdr5, RbBP5 and MOF to bind to the Oct4 promoter region. In ChIP-PCR experiments with WT and PHF20-deficient cells, Wdr5 failed to bind to the Oct4 promoter in PHF20-deficient cells, but bound strongly to the Oct4 promoter in WT cells (Figure 7I). Similarly, the ability of RbBP5 and MOF to bind to the Oct4 promoter was markedly reduced in PHF20-deficient cells. Consistent with these results, ChIP-qPCR experiments revealed a sharp reduction in H3K4 trimethylation in the Oct4 promoter, while H4K16 acetylation was also affected but to a lesser extent (Figure 7J). Taken together, these results suggest that binding of PHF20 to the Oct4 promoter may be required for recruiting H3K4 methyltransferase complex and perhaps H4K16 acetlytransferase complex to bind to the same promoter through the interaction with Wdr5 and MOF, leading to reactivation of endogenous Oct4 expression during reprogramming.
Discussion
Using a shRNA knockdown screen in Tet-O-4F MEFs, we identified a number of histone-modifying proteins that are required for the reprogramming of somatic cells to iPSCs, but only one, Jmjd3, functions as a negative regulator of this process. Given the sequential multistep nature of iPSC generation (Plath and Lowry, 2011), it is not surprising that negative regulators of reprogramming exist. Indeed, DOT1L was recently shown to negatively control Nanog and LIN28 expression, posing an unexpected barrier to efficient reprogramming (Onder et al., 2012). Until the full spectrum of such regulators is delineated and their mechanisms of action deciphered, it will be difficult to revise current approaches to iPSC generation with any degree of confidence.
Jmjd3 plays a critical role in the upregulation of Ink4a/Arf by modulating the levels of H3K27 trimethylation in the promoter (Agger et al., 2009; Barradas et al., 2009). These effects on the expression of Ink4a/Arf and p21, in turn, induce senescence and inhibit reprogramming (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009), consistent with our demonstration that Jmjd3 ablation reduces cell senescence and promotes reprogramming of Jmjd3-deficient MEFs. However, we provide several lines of evidence indicating that Jmjd3 can regulate reprogramming through a previously unrecognized, histone demethylase activity-independent pathway. First, the combined knockdown of Jmjd3 with Ink4a or p21 resulted in significantly more iPSC colonies than did knockdown of any single gene alone. Second, although ectopic expression of full-length Jmjd3 in Jmjd3-deficient MEFs restored Ink4a/Arf expression and strongly inhibited the efficiency of reprogramming, the Jmjd3 mutants Jmjd3-ΔJmjC and Jmjd3-H1390A, defined by their lack of H3K27me3 demethylase activity and inability to upregulate Ink4a/Arf expression, could still inhibit reprogramming in Jmjd3-deficient MEFs. We therefore propose that Jmjd3 exploits both demethylase activity-dependent and -independent mechanisms to regulate somatic cell reprogramming, with the latter having a predominant role.
An extensive search for target molecules that might be involved in a Jmjd3-mediated but demethylase activity-independent pathway led to the identification of PHF20. We provide evidence of a critical role for PHF20 expression in the maintenance of the pluripotent state. PHF20 was first identified as an antibody-reactive protein that is highly expressed in several types of cancer (Fischer et al., 2001; Wang et al., 2002). PHF20 has since been identified as a histone code reader that specifically recognizes the dimethylation of H3K4, H3K9, H4K20, and H4K79 (Adams-Cioaba et al., 2012; Kim et al., 2006). Recent studies show that it also regulates p53 protein at both the transcriptional and posttranscriptional levels in response to DNA damage (Li et al., 2012b; Park et al., 2012). Mice with PHF20 ablation die shortly after birth (Badeaux et al., 2012). Consistently, we failed to generate ESC lines from PHF20 knockout mice. PHF20 deficiency almost completely inhibits reprogramming of PHF20-deficient MEFs, suggesting an absolute requirement for this factor in iPSC reprogramming and generation of ESCs.
Jmjd3 has been shown to play an important role in T cell-lineage commitment by interacting with T-bet and Brg-1 in a demethylase activity-independent manner (Miller et al., 2010). Here we show that Jmjd3 interacts with PHF20, targeting it for ubiquitination and proteasomal degradation. Although both the N- and C-terminal regions of Jmjd3 protein can bind to the N-terminus of PHF20, Jmjd3 itself cannot ubiquitinate PHF20 in a K48-linkage. Instead, it recruits an E3 ligase, Trim26, to PHF20 for K48-linked polyubiquitination. Indeed, knockdown of Trim26 abolishes PHF20 ubiquitination and degradation, thus enhancing iPSC reprogramming. Further experiments demonstrated that like full-length Jmjd3, certain Jmjd3 mutants (Jmjd3-ΔJmjC and Jmjd3-H1390A), but not Jmjd3-N, Jmjd3-M, or Jmjd3-C, target PHF20 for ubiquitination. These results emphasize the importance of Jmjd3-Trim26 mediated ubiquitination in the negative regulation of reprogramming.
Fully reprogrammed iPSCs are accompanied by changes in distinct DNA methylation patterns associated with reactivation of endogenous Oct4 and several other ESC marker genes (Plath and Lowry, 2011; Stadtfeld and Hochedlinger, 2010). How, then, does PHF20 deficiency lead to failure to reactivate these endogenous marker genes, thus imposing a major barrier to successful reprogramming? A recent study shows that exogenous Oct4 together with other key reprogramming factors first induce Wdr5 expression in MEFs, which in turn forms a Wdr5-Oct4 complex that binds to the Oct4 promoter, leading to reactivation of endogenous Oct4 expression (Ang et al., 2011). To directly link PHF20 to Oct4 expression, we show that PHF20 not only binds to the Oct4 promoter region, but also specifically interacts with Wdr5 and MOF. A recent study shows that MOF is required for ESC self-renewal and function, and regulates Nanog expression (Li et al., 2012a). Deletion of PHF20 reduces the ability of Wdr5 and MOF to bind to the Oct4 promoter, suggesting a critical requirement for this protein in reactivation of endogenous Oct4 expression and hence for successful generation of fully reprogrammed iPSCs. Consistent with this notion, our results further demonstrated that PHF20 deficiency results in failure to reactivate expression of many endogenous ESC maker genes during reprogramming, suggesting that PHF20 affects expression of many key ESC genes directly or indirectly. ChIP-PCR and ChIP-seq analyses show that PHF20 and Wdr5 bind to the Oct4 promoter, but not to the promoter regions of Sox2, Nanog, Dnmt3l, Esg1, Eras, Rex1 and Cripto. In addition, ChIP-seq analysis revealed that both PHF20 and Wdr5 bind to several key epigenetic factor genes, including Baf155, Brg-1 and Sall4. Hence, these findings explain why the incompletely reprogrammed phenotype of PHF20-deficient MEFs cannot be rescued by overexpression of Oct4 or 4F (OSKM), and further suggest that PHF20 functions as a upstream regulator that control Oct4 and many other critical ESC marker genes, thus providing a mechanistic link between Jmjd3-mediated PHF20 degradation and deficient reprogramming.
On the basis of these findings, we propose a working model to explain how the Jmjd3-PHF20 axis regulates iPSC reprogramming. Increased expression of Jmjd3 due to 4F-mediated reprogramming in WT MEFs initiates at least two distinct pathways (Figure 7K): 1) Jmjd3upregulates Ink4a/Arf and p21 by modulating H3K4 and H3K27 methylation through itsH3K27me2/3 demethylase activity. Increased amounts of Ink4a, Arf and p21 induce cell senescence or apoptosis and reduce cell proliferation, thus decreasing the efficiency and kinetics of reprogramming; 2) Jmjd3 protein also targets PHF20 for ubiquitination and degradation by recruiting an E3 ligase, Trim26, in a H3K27 demethylase activity-independent manner. The resultant decrease in PHF20 protein leads to the loss or negligible expression of endogenousOct4, thereby greatly reducing reprogramming efficiency. We conclude that the demethylase activity-dependent and -independent pathways used by Jmjd3 act in concert to potently restrain the kinetics and efficiency of reprogramming. The observations that Jmjd3 loss reduces cells senescence and apoptosis and increases cell proliferation, and that increased amounts of PHF20lead to reactivation of endogenous Oct4 expression, suggest means to enhance the outcome of somatic cell reprogramming overall.
Experimental Procedures
Mice
Tet-O-Sox2 and -Klf4 transgenic mice were generated and crossed with Rosa-rtTA, Tet-O-Oct4 (from the Jackson Laboratories) and Tet-O-Myc transgenic mice. Tet-O-4F MEFs that express rtTA and Tet-O-Oct4, Sox2, Klf4 and Myc were established and used for reprogramming. Tet-O-PHF20 mice were generated as Tet-O-Sox2 and -Klf4 transgenic mice. Jmjd3 was targeted deletion of exon 15-21 using a Cre-LoxP system. Jmjd3 was globally deleted by crossing Jmjd3f/f mice with Hprt-Cre mice (Jackson Laboratories).
Generation of iPSCs from MEFs and Tet-O-4F MEFs
Mouse iPSCs were generated from MEFs, as previously described (Takahashi et al., 2007a) with some minor modifications. Tet-O-4F MEFs were used to generate iPSCs by treating MEFs with Dox in mESC medium. The efficiency of iPSC formation as calculated based on the number of AP+ iPSC colonies and the initial cell number of seeded MEFs. Human iPSCs were generated as previously described (Park et al., 2008). Lentiviral particles were generated and concentrated, as previously described (Peng et al., 2005).
Screening for Epigenetic Factors that Modulate Reprogramming Efficiency
Tet-O-4F transgenic MEF cells (M2-11) were transduced with lentiviras-based shRNAs specific for 15 epigenetic factors and then reseeded on irradiated feeder cells at the desired density. The next day, mESC medium containing 2 μg/ml Dox was added and replenished every day. The colonies were stained for AP activity on Days 12-14.
Co-IP, ChIP-PCR and ChIP seq Analysis
The cells were lysed in RIPA buffer, and analyzed, as previously described (Cui et al., 2010). ChIP assay was performed with Imprint Ultra Chromatin Immunoprecipitation kit (Sigma). ChIP-Seq libraries were prepared, sequenced and analyzed. See the Extended Experimental Procedures for details.
Accession Numbers
The ChIP-seq data are available at the Gene Expression Omnibus under the accession number GSE43247.
Supplementary Material
Acknowledgments
We would like to thank Drs Yi Li at Baylor College of Medicine for providing Tet-O-Myc transgenic mice, Mark Bedford at M.D. Anderson Cancer Center for PHF20 knockout mice, Dan Liu at Baylor College of Medicine for assistance for shRNA screening, and Paul Webb for assistance in ChIP-seq experiments. We would also like to thank John Gilbert and Adebusola A. Ajibade for their critical reading of this manuscript. This work was in part supported by grants from National Cancer Institute, NIH (P01CA094237, R01CA09327, R01CA101795, R01CA116408, R01CA121191 and R01 DA030338) and Cancer Prevention and Research Institute of Texas (RP121048) to R.F.W and The Methodist Hospital Research Institute.
Footnotes
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, seven supplemental Figures and three Tables and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams-Cioaba MA, Li Z, Tempel W, Guo Y, Bian C, Li Y, Lam R, Min J. Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins. FEBS Lett. 2012;586:859–865. doi: 10.1016/j.febslet.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeaux AI, Yang Y, Cardenas K, Vemulapalli V, Chen K, Kusewitt D, Richie E, Li W, Bedford MT. Loss of the methyl lysine effector protein PHF20 impacts the expression of genes regulated by the lysine acetyltransferase MOF. J Biol Chem. 2012;287:429–437. doi: 10.1074/jbc.M111.271163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Fischer U, Struss AK, Hemmer D, Pallasch CP, Steudel WI, Meese E. Glioma-expressed antigen 2 (GLEA2): a novel protein that can elicit immune responses in glioblastoma patients and some controls. Clin Exp Immunol. 2001;126:206–213. doi: 10.1046/j.1365-2249.2001.01635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012a;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Park J, Piao L, Kong G, Kim Y, Park KA, Zhang T, Hong J, Hur GM, Seok JH, et al. PKB-mediated PHF20 phosphorylation on Ser291 is required for p53 function in DNA damage. Cell Signal. 2012b;25:74–84. doi: 10.1016/j.cellsig.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Park S, Kim D, Dan HC, Chen H, Testa JR, Cheng JQ. Identification of Akt Interaction Protein PHF20/TZP That Transcriptionally Regulates p53. J Biol Chem. 2012;287:11151–11163. doi: 10.1074/jbc.M111.333922. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng G, Guo Z, Kiniwa Y, Voo KS, Peng W, Fu T, Wang DY, Li Y, Wang HY, Wang RF. Toll-like receptor 8 mediated-reversal of CD4+ regulatory T cell function. Science. 2005;309:1380–1384. doi: 10.1126/science.1113401. [DOI] [PubMed] [Google Scholar]
- Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007a;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007b;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, Peng JR, Zhao HT, Rui JA, Leng XS, et al. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol. 2002;169:1102–1109. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.