Abstract
Escherichia coliO157:H7 is a human pathogen that has emerged from its less pathogenic progenitor, E. coli O55:H7, to form the EHEC 1 clade. In its emergence, E. coli O157:H7 formed three distinct clusters, each of which exists today. Sequencing and SNP analysis of Cluster 1 of this clade demonstrated constrained radiation from the cluster founder. Here we investigated the diversity of Cluster 2 strains by sequencing signature SNPs in six strains collected throughout Washington State. Our results suggest that successful Cluster 2 strains have radiated on only two branches from their founder; one of these two branches leads to Cluster 3. Constrained radiation appears to be a common theme among this pathogenic clade.
Keywords: Escherichia coli O157:H7, SNPs, Evolution
INTRODUCTION
Extant pathogenic Escherichia coli O157:H7 belong to three sequentially emerged clusters (Shaikh and Tarr, 2003; Shaikh, et al., 2007; Leopold, et al., 2009) and are members of the well-characterized EHEC 1 clade. Recently we analyzed backbones of representative E. coli O157:H7 and other members of the EHEC 1 clade (the probable progenitor, E. coli O55:H7, and sorbitol fermenting E. coli O157:H−) to precisely characterize this descent (Leopold, et al., 2009). Our analysis demonstrated that Cluster 1 O157:H7 exhibit constrained and highly non-random radiation from a postulated cluster founder. Cluster 1’s age and many available isolates from around the world particularly enabled us to localize wild-type E. coli O157:H7 to one of only two branches emanating from the Founder with reasonable confidence within this cluster. Cluster 3, which is the most recently emerged E. coli O157:H7 cluster, is also well represented in strain set collections. We sampled intra-cluster (radial) synonymous SNPs in six Cluster 3 E. coli O157:H7, and determined that many of these mutations were shared, suggesting that the concept of constrained radiation applies to that cluster, too. Our study did not address the concept of constrained radiation within Cluster 2 because isolates belonging to this cluster are under-represented among strain sets. However, radiation from a founder was demonstrated for one of its members, strain 86-24, and six of strain 86-24’s radial SNPs were also found in strain 87-23, a non-toxigenic E. coli O157:H7 Cluster 2 isolate (Tarr, et al., 1989; Békássy et al., in press). This result was expected because strains 86-24 and 87-23 were each recovered from patients infected during the same outbreak in the same city (Tarr, et al., 1989). Here, we study a larger collection of Cluster 2 E. coli O157:H7 to determine if there is evidence for constrained radiation in these organisms, too.
MATERIALS AND METHODS
We studied all Cluster 2 E. coli O157:H7 in our collection. Six were from Washington State, collected over a span of nearly two decades (Table 1). To determine if these strains radiated in a constrained pattern from their founder, we used all 14 synonymous and 18 nonsynonymous radial SNPs that are present in the chromosomal backbone of strain 86-24 but not in any other E. coli O157:H7 (strain 86-24 was the only Cluster 2 strain sequenced by us in the initial communication) (Table 2).
Table 1.
Strains used in this study.
| Strain | Cluster | Place | Year | Source | Reference | Role in Study |
|---|---|---|---|---|---|---|
| 86-17 | 2 | Isolated Washington | Isolated 1986 | Human, Apparent Sporadic Isolate | (Tarr, et al., 1989) | 9 of 14 synonymous and 12 of 18 nonsynonymous radial SNPs shared with 86-24 |
| 86-24 | 2 | Walla Walla, Washington | 1986 | Human, Outbreak Isolate | (Griffin, et al., 1988; Tarr, et al., 1989) | Pyrosequenced (GS20) |
| 86-28 | 2 | Washington | 1986 | Human, Apparent Sporadic Isolate | (Tarr, et al., 1989) | 14 of 14 synonymous and 18 of 18 nonsynonymous radial SNPs shared with 86-24 |
| 87-07 | 2 | Washington | 1987 | Human, Apparent Sporadic Isolate | (Tarr, et al., 1989) | 9 of 14 synonymous and 11 of 17 nonsynonymous* radial SNPs shared with 86-24 |
| 87-23 | 2 | Walla Walla, Washington | 1986 | Human, Outbreak Isolate | (Tarr, et al., 1989) | 14 of 14 synonymous and 18 of 18 nonsynonymous radial SNPs shared with 86-24 |
| EK15 | 2 | Seattle, Washington | 1999 | Human, Emergency Dept. Isolate | (Klein, et al., 2002) | 0 of 32 radial SNPs shared with 86-24 10 of 11 linear SNPs shared with O157 Sakai |
| U39 | 2 | Yakima, Washington | 2000 | Child, Region- Wide HUS Study | (Cornick, et al., 2002) | 0 of 32 radial SNPs shared with 86-24 10 of 11 linear SNPs shared with O157 Sakai |
| EK28 | 2 | Seattle, Washington | 2000 | Human, Emergency Dept. Isolate | (Klein, et al., 2002) | 0 of 32 radial SNPs shared with 86-24 7 of 11 linear SNPs shared with O157 Sakai |
| O157 Sakai | 3 | Sakai, Japan | 1996 | Human, Outbreak Isolate | (Hayashi, et al., 2001) | Published Genome |
SNP site 2392826 did not amplify by PCR in this strain. This site was not included.
Table 2. SNP Characteristics and Primers.
SNP locations (based upon O157 Sakai chromosome), amino acid change, and SNP type are listed. The corresponding bases as determined by sequencing are listed for each strain and are color-coded to denote the ancestral (blue) or variant (orange for radial SNPs, green for linear SNPs) designation. An ‘X’ at strain 87-07’s site 2392896 indicates our inability to amplify by PCR at this site. Primers used for PCR amplification of the each SNP are reported.
| Location | Amino Acid Change | SNP Type | 86-17 | 86-24 | 86-28 | 87-07 | 87-23 | U-39 | EK15 | EK28 | O157 Sakai | Forward Primer | Reverse Primer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3798 | Synonymous | Radial | G | T | T | G | T | G | G | G | G | AATGAGCAGGTCAGCTTTGC | GACATCGCTTTCAACATTGG |
| 505838 | Synonymous | Radial | T | T | T | T | T | A | A | A | A | TTCTTCGGTACGTTTGAGCA | TCATCAACCTCCATCAGTGC |
| 795801 | Synonymous | Radial | T | T | T | T | T | C | C | C | C | ATGGTCAACATTCCCTGCAC | GGAGTCTGCGTGTGCAAATA |
| 1098382 | Synonymous | Radial | T | T | T | T | T | C | C | C | C | TTGTCGTGGTTGGTAATCTCACC | TGACGATTACCGCATCAACTGAC |
| 1894260 | Synonymous | Radial | A | A | A | A | A | T | T | T | T | CGCCTGTTCAAGCTGGTATT | GGCGTAAAGATATCCGGTCA |
| 1977261 | Synonymous | Radial | T | T | T | T | T | G | G | G | G | CCATCACTACCAAGGCCATAA | TGAAGAAGTGGTTGATGAATTGC |
| 2388580 | Synonymous | Radial | C | T | T | C | T | C | C | C | C | ATGGTGAAGATAAGCGACTGTTGG | TTGCCGCTGGTCTGATAGATG |
| 2436199 | Synonymous | Radial | A | A | A | A | A | G | G | G | G | GGCGCACAACTGACATTTATC | GGTTAAAGGTCAGCGACAGG |
| 2984974 | Synonymous | Radial | C | A | A | C | A | C | C | C | C | AAAATGGCTCCTTGTTGTGG | TGGTTGAAGCCTTTCGTAGG |
| 4258114 | Synonymous | Radial | G | G | G | G | G | A | A | A | A | CCCTGAAATTTGACCTGCTG | CCATGGAACAACCGTTACAG |
| 4259164 | Synonymous | Radial | G | G | G | G | G | A | A | A | A | GGTTTTACCGTTGGTTTTGC | AATCAGCGTAGCCATTACCG |
| 5153038 | Synonymous | Radial | C | T | T | C | T | C | C | C | C | ATGACGCCTGAACATACCAGC | TACTATACGGAAGCCACAGTCGG |
| 5234150 | Synonymous | Radial | A | A | A | A | A | C | C | C | C | TCAATGCTGAACCACACAGC | GTTTGGCCTGAACCCAGAGT |
| 5391618 | Synonymous | Radial | C | A | A | C | A | C | C | C | C | AAAATGGCTCCTTGTTGTGG | TGGTTGAAGCCTTTCGTAGG |
| 30477 | Nonsynonymous | Radial | T | T | T | T | T | C | C | C | C | ACGCTAAAACTCGACGATGG | ATGCTGATAGCGCGGTCTAC |
| 79307 | Nonsynonymous | Radial | T | A | A | T | A | T | T | T | T | ATATCGCGGGTACGACAGAG | AAAAGGTGAAAGCGATCTGG |
| 571478 | Nonsynonymous | Radial | A | G | G | A | G | A | A | A | A | TGATTCAGGAGCTGCAACAG | CACCGGAATCAGCTGGTAGT |
| 1330848 | Nonsynonymous | Radial | A | A | A | A | A | G | G | G | G | CCGCCAACAATCCACATAAT | CATCCCTCAGGCTAAAGACAA |
| 1480429 | Nonsynonymous | Radial | C | C | C | C | C | T | T | T | T | ATTCGAGGTTCAATGCGTTT | GCGAATTGAAGTCACCATAGC |
| 1492372 | Nonsynonymous | Radial | A | G | G | A | G | A | A | A | A | ATGCAGATCAACCTGAATTCCAGC | AGCAGTTGGGTTTGTTCGTTG |
| 2392896 | Nonsynonymous | Radial | A | A | A | X | A | T | T | T | A | GCAATGTGTGGTTGAGATCG | TGCATCGTGCCTATCTTTCA |
| 2422218 | Nonsynonymous | Radial | A | A | A | A | A | T | T | T | A | CGCAGATTAATGCTGAAGAGG | TTAGTGAAGAATCCGGTAATGG |
| 3006720 | Nonsynonymous | Radial | A | A | A | A | A | C | C | C | A | ATACACAAGCTTTGCGAGTAAC | AAGAGTTGTGTGGCTTCTTGC |
| 3714794 | Nonsynonymous | Radial | A | A | A | A | A | G | G | G | A | TTCAACGCGTTAGAGAACAATC | AATGCAGCGCAAAGAATAGA |
| 4007883 | Nonsynonymous | Radial | T | C | C | T | C | T | T | T | T | TCCTTCTGTCATGATCCGAATCC | ATACCTGGTGCTAGTGCTTCG |
| 4378660 | Nonsynonymous | Radial | T | T | T | T | T | C | C | C | C | TCGCTGATGCTGTAGAGGTG | TCTTTCTGAACTGGGCAACC |
| 4678681 | Nonsynonymous | Radial | G | A | A | G | A | G | G | G | G | GATTATACAGGTTGGCGATAAGC | GACATCAAGCGCATACTCGAC |
| 4912166 | Nonsynonymous | Radial | A | A | A | A | A | G | G | G | A | ATCGAAAGTAGGGGCTCCAG | CAGTGAGAAAAGCACCAGCA |
| 5094375 | Nonsynonymous | Radial | G | G | G | G | G | A | A | A | G | ATTCGAGGTTCAATGCGTTT | GCGAATTGAAGTCACCATAGC |
| 5103324 | Nonsynonymous | Radial | A | A | A | A | A | G | G | G | G | GAACCAGGCGTGATGAGTG | AACTTTCCGTGTCGTTGAGC |
| 5155031 | Nonsynonymous | Radial | A | A | A | A | A | G | G | G | G | TAGCGATTCAGCGTCGAGTA | AACGCTGTGGTGTATCGTTG |
| 5286625 | Nonsynonymous | Radial | G | C | C | G | C | G | G | G | G | TGGTGATGTTGTATGTGAATCC | TGATGACCTGATGGATCACATC |
| 358636 | Nonsynonymous | Linear | G | G | A | G | ACTCACCTTCATAGCGGAAAG | AGACGGAGTGTAGATTAGTCAAC | |||||
| 2030050 | Nonsynonymous | Linear | A | A | G | A | ATTACGACATCATTCTCCGCA | AAGGTTTGTCGTGGACGTG | |||||
| 2373421 | Nonsynonymous | Linear | A | A | A | A | ATATGATGATGGGTGGACTGG | ACTGTGGCGGATAGGATAAGC | |||||
| 4660184 | Nonsynonymous | Linear | G | G | G | G | TCTCTGACTTTGGATGAACGG | TCACTCACATTCATCACGATGG | |||||
| 4757979 | Nonsynonymous | Linear | T | T | T | T | AACCTGAACGACGACGATTAC | ATCGTGTCGGTTTGTTGACAG | |||||
| 421747 | Synonymous | Linear | T | T | C | T | ATAATATCGGTTGCGGAGGTG | ATCCTCTGCATGGTCAGGTC | |||||
| 2100062 | Synonymous | Linear | A | A | A | A | TGTAGAGACTCAGCATTGCTTAG | TACAGATAACCCTGACCAACG | |||||
| 3008913 | Synonymous | Linear | A | A | A | A | TACAGATTTCCTGGTCATCGG | TCTACTCTCCCTGTTGTCTGG | |||||
| 4143190 | Synonymous | Linear | T | T | T | T | GATGACCGTGCAGTTTATCG | GTATGCGGCAGGCCTATAAC | |||||
| 4962486 | Synonymous | Linear | T | T | C | T | TTCTCGAAACCATTACCTGCC | TCTTCACTATCCAGCAGTACG | |||||
| 5303294 | Synonymous | Linear | C | C | C | T | AAGCAATTTAGCGCTCGACAC | TTGGTCATCCAGTGACTGTTG |
Each strain was verified as a Cluster 2 strain according to a set of characteristic Shiga toxin bacteriophage insertion sites as well as a FimH polymorphism (Table 3). PCR amplification was performed using primer pairs as described (Shaikh, et al., 2007).
Table 3. E. coli O157:H7 Cluster Identification.
The status of each of the determinant sites is determined by PCR amplification of key sites. The presence Shiga toxin genes (stx), and the occupation of the yehV site by the stx1 bacteriophage or wrbA by the stx2 bacteriophage are markers in the emergence of these three clusters. A subset of Cluster 1 strains located on a branch that leads to Clusters 2 and 3 have been observed to have sustained an N to K mutation in FimH.
| Determinant | Cluster 1 | Cluster 1* | Cluster 2 | Cluster 3 |
|---|---|---|---|---|
| stx1 | − | − | − | + |
| stx2 | + | + | + | + |
| yehV | Occupied | Occupied | Occupied | Occupied |
| wrbA | Unoccupied | Unoccupied | Occupied | Occupied |
| FimH | Asp | Lys | Lys | Lys |
These isolates have characteristics (FimH allele) typical of Cluster 2
Primer pairs were designed to span each of these 32 SNPs using strain 86-24 as a reference (Table 1). DNA flanking and including each SNP was PCR amplified and Sanger sequenced. Additionally, primer pairs were designed to span all 11 linear SNPs (six synonymous and five nonsynonymous) that were identified lead to Cluster 3 strains, using strain O157 Sakai as a reference (Tables 1 and 2) (Leopold, et al., 2009).
The resulting sequence data at the sites that contained radial or linear SNPs were overlaid on the previously determined Cluster 2 topology.
RESULTS AND DISCUSSION
Strain 86-28, isolated in the same year and state as strains 86-24 and strain 87-23, appears isogenic with these two Cluster 2 strains, in that it possesses the variant nucleotides at all 32 radial sites. Strain 86-17 possess 21 of the 32 radial SNPs. Strain 87-07 possesses 20 of 31 radial SNPs (one nonsynonymous SNP site did not PCR amplify, despite repeated attempts). Both strains had ancestral nucleotides at the remaining five synonymous and five nonsynonymous SNP sites. (Table 2)
Three strains Cluster 2 strains (defined by toxin genotyping, bacteriophage insertion sites, and FimH polymorphisms), EK15, EK28, and U-39, possess the ancestral version (the nucleotide designation shared by all other EHEC 1 strains as determined in a previous study, Leopold, et al., 2009) of the SNPs at all 32 interrogated sites (i.e., they had none of the radial SNPs identified in strain 86-24). To determine if strains EK15, EK28, and U-39 are offshoots of the branch leading from Cluster 2 to the Founder of Cluster 3, we sequenced all 11 “linear” SNPs between the founders of Clusters 2 and 3 (Leopold, et al., 2009). EK29 possesses six of these 11 SNPs, and EK15 and U39 possess 10 of these 11 linear SNPs (Figure 1, Table 2). The SNPs that define the intra-cluster 2 topology are listed in Table 4.
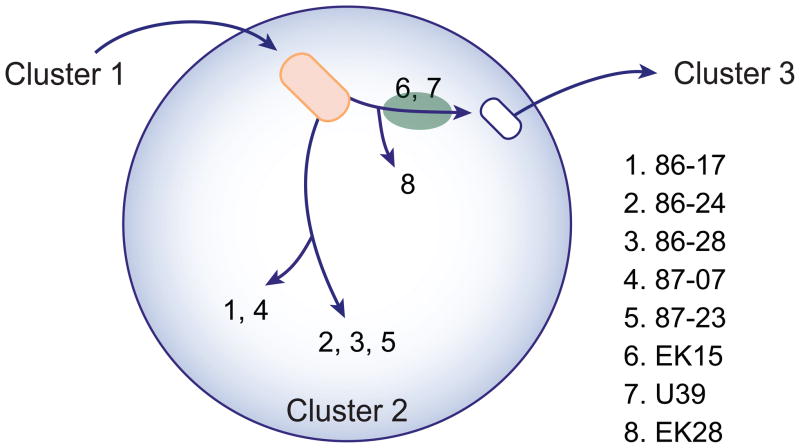
Figure 1. Cluster 2 model of radiation from cluster founder.
Blue circle outlines the boundaries of bacteria belonging to Cluster 2. The red oval depicts the postulated Founder of Cluster 2 and the white oval is a bacterium on the path evolving to Cluster 3. Numeric key refers to each E. coli strain analyzed in this study. The green oval represents the position of strains 6 and 7, which are phylogenetically on, or slightly divergent from, the branch leading from Cluster 2 to Cluster 3. Leopold et al. (2009) Figures 1 and S1 describe an expanded version of the EHEC 1 clade lineage.
Table 4. Phylogenetically Informative SNP Sites.
Key SNP sites at which the topology is assigned for each set of strains are listed. Bases associated with each site are reported within parentheses. Sets 1 and 2 list radial SNPs only. Sets 3 and 4 list key linear SNPs only.
| Sets | Strains | Sites at Which SNPs Assign Topology |
|---|---|---|
| 1 | 86-17, 87-07 | 3798(G), 79307(T), 571478(A), 1492372(A), 1894260(A), 2388580(C), 2984974(C), 4007883(T), 4678681(G), 5153038(C), 5286625(G), 5391618(C) |
| 2 | 86-24, 86-28, 87-23 | 3798(T), 79307(A), 571478(G), 1492372(G), 1894260(A), 2388580(T), 2984974(A), 4007883(C), 4678681(A), 5153038(T), 5286625(C), 5391618(A) |
| 3 | EK28 | 358636(A), 421747(C), 2030050(G), 4962486(C), 5303294(C) |
| 4 | EK15, U39 | 5303294(C) |
Though this study interrogated all members of this group in our collection, the number of Cluster 2 isolates we analyzed is small. For this reason, we cannot state with complete confidence that we have determined the degree of constraint of radiation from a founder within Cluster 2, and anticipate extended analysis as additional Cluster 2 isolates are identified. Nonetheless, the data from these isolates are consistent with a pattern of constrained radiation that was observed previously in Cluster 1, and was suggested in Cluster 3 (Leopold, et al., 2009). Logically, we would have expected a much greater diversity of backbone SNPs among unrelated extant pathogens, but instead we found that all strains could be categorized into two main branches, one containing five of the strains interrogated and another leading to Cluster 3.
Our work has additional implications. The putative isogenicity of strains 86-28, 86-24 and 87-23 is consistent with a deduced statewide outbreak of O157:H7 infections in Washington State in the mid 1980’s caused by an organism resembling strain 86-24 (Ostroff, et al., 1990). The overall paucity of Cluster 2 isolates in our collection is also noteworthy. It is possible that the viability of this pathogenic set of E. coli O157:H7 is limited, and that this subgroup is becoming extinct. Strain 86-24 has been used in many pathogenesis experiments, and in view of the rarity of members of its subgroup in human collections, it is not clear that this isolate is a good representative of pathogenic E. coli O157:H7. It is also interesting to note only two time clusters (“blooms”) of Cluster 2 of E. coli O157 in the State of Washington since our collecting began in 1984: the first consisted of strains 86-17, 86-24, 86-28, 87-07, and 87-23 (which radiated on one branch from the Cluster 2 Founder and which were recovered in the mid 1980’s); the second consisted of strains EK15, EK28, and U39, each of which is on, or an offshoot of, the branch leading to Cluster 3, and which were recovered over a decade later.
In summary, Cluster 2 E. coli O157:H7 portray limited radiation from the Cluster founder. The cluster’s limited SNP repertoire also strengthens our conclusion, and the conclusion of others (Holt, et al., 2008), that bacterial pathogens have small effective population sizes, and that their survival is highly fortuitous. The rarity of Cluster 2 E. coli O157:H7 suggests that they might not be as viable as the more mature and widely disseminated Cluster 1 O157:H7.
Acknowledgments
We are grateful to the late Thomas Whittam for helpful discussions on bacterial evolution that formed the basis for our study. This work was supported by National Institutes of Health Grants AI47499, R56AI063282, and DK52081 (to P.I.T.); 5P30 DK052574 (to Washington University Digestive Diseases Research Core Center Biobank); and 5T32AI007172 (to S.R.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cornick NA, Jelacic S, Ciol MA, Tarr PI. Escherichia coli O157:H7 infections: discordance between filterable fecal Shiga toxin and disease outcome. J Infect Dis. 2002;186:57–63. doi: 10.1086/341295. [DOI] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EJ, Stapp JR, Clausen CR, Boster DR, Wells JG, Qin X, Swerdlow DL, Tarr PI. Shiga toxin-producing Escherichia coli in children with diarrhea: a prospective point-of-care study. J Pediatr. 2002;141:172–177. doi: 10.1067/mpd.2002.125908. [DOI] [PubMed] [Google Scholar]
- Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, Ogura Y, Iguchi A, Hayashi T, Mellmann A, Karch H, Besser TE, Sawyer SA, Whittam TS, Tarr PI. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc Natl Acad Sci U S A. 2009;106:8713–8718. doi: 10.1073/pnas.0812949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff SM, Griffin PM, Tauxe RV, Shipman LD, Greene KD, Wells JG, Lewis JH, Blake PA, Kobayashi JM. A statewide outbreak of Escherichia coli O157:H7 infections in Washington State. Am J Epidemiol. 1990;132:239–247. doi: 10.1093/oxfordjournals.aje.a115653. [DOI] [PubMed] [Google Scholar]
- Shaikh N, Holt NJ, Johnson JR, Tarr PI. Fim operon variation in the emergence of Enterohemorrhagic Escherichia coli: an evolutionary and functional analysis. FEMS Microbiol Lett. 2007;273:58–63. doi: 10.1111/j.1574-6968.2007.00781.x. [DOI] [PubMed] [Google Scholar]
- Shaikh N, Tarr PI. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J Bacteriol. 2003;185:3596–3605. doi: 10.1128/JB.185.12.3596-3605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr PI, Neill MA, Clausen CR, Newland JW, Neill RJ, Moseley SL. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984–1987. J Infect Dis. 1989;159:344–347. doi: 10.1093/infdis/159.2.344. [DOI] [PubMed] [Google Scholar]