Abstract
CHRNA4, the gene that encodes the nicotinic acetylcholine receptor α4 subunit, is a potential candidate gene for nicotine dependence (ND). However, studies of the association of CHNRA4 with smoking behavior have shown inconsistent results. Our meta-analysis of linkage studies of smoking behavior identified a genome-wide significant linkage of the phenotype maximum number of cigarettes smoked in a 24-hour period to a region (20q13.12-q13.32) harboring CHRNA4. This motivated us to examine the association of CHRNA4 with smoking behavior in two independent samples. In this study, we examined five single nucleotide polymorphisms (SNPs) within CHRNA4 and three smoking-related behaviors: one quantitative trait [cigarettes smoked per day (CPD)], and two binary traits [DSM-IV diagnosis of ND and dichotomized Fagerstrom test of ND (FTND)], in 1,249 unrelated European-Americans (EAs) and 1,790 unrelated African-Americans (AAs). Using the combined sample with sex, age and race as covariates, the synonymous SNP rs1044394 was significantly associated with ND (P = 0.001) and FTND (P = 0.01). Rs2236196, which has a low correlation with rs1044394, was also significantly associated with CPD (P = 0.003). The pattern of association for these SNPs was similar in AAs and EAs. After correction for multiple testing, the association between rs1044394 and ND in the combined sample remained significant (P = 0.033). In summary, our study supports association between CHRNA4 common variation and ND in AA and EA samples. Additional studies will be necessary to evaluate the role of rare variants at CHRNA4 for ND.
Keywords: smoking behavior, nicotine dependence, FTND, SNP, association
Introduction
Cigarette smoking is the most preventable cause of mortality in the world. Despite increasing awareness of the risks associated with smoking, the World Health Organization (2002) estimated that around 1.2 billion people worldwide were active smokers. Nicotine contained in cigarettes is a rewarding drug and the primary substance responsible for continued smoking and addiction (Benowitz 2008). Thus, an understanding of the various factors that influence the risk of nicotine dependence (ND) is critical to the prevention and cessation of smoking. Although the etiology of ND is complex, a genetic contribution to ND has been well established from twin studies. The heritability was estimated to be 0.56 for all smokers on average in a meta-analysis of 17 twin studies (Li et al., 2003). Numerous ND candidate loci or genes have been identified by genome-wide linkage studies (for review see Li et al., 2008), candidate gene association studies (Gelernter et al., 2006; Saccone et al., 2007; Conti et al., 2008; Weiss et al., 2008; Bergen et al., 2009; Saccone et al., 2009a,b) and recent genome-wide association studies (GWAS) (Berrettini et al., 2008; Thorgeirsson et al., 2008; Caporaso et al., 2009; Liu et al., 2009; Vink et al., 2009; Tobacco and Genetics Consortium 2010; Liu et al., 2010; Thorgeirsson 2010).
The gene CHRNA4, which encodes the nicotinic acetylcholine receptor α4 subunit, is a candidate gene for ND risk. The gene maps to 20q13.3 and contains 6 exons spanning ~17kb of genomic DNA. CHRNA4 is highly expressed in the central nervous system, and its protein product is part of high affinity receptors (α4β2) for nicotine. It plays a major role in tolerance, reward, and the modulation of mesolimbic dopamine function, all of which are critical to the development of ND (Tapper et al., 2004). Although some genetic association studies support the role of CHRNA4 in smoking-related behaviors (Feng et al., 2004; Li et al., 2005; Hutchison et al., 2007; Breitling et al., 2009), there have been conflicting results (Ehringer et al., 2007; Weiss et al., 2008; Etter et al., 2009). GWAS studies on smoking behavior have not indentified CHRNA4 gene as a risk locus (Berrettini et al., 2008; Thorgeirsson et al., 2008; Caporaso et al., 2009; Liu et al., 2009; Vink et al., 2009; Tobacco and Genetics Consortium 2010; Liu et al., 2010; Thorgeirsson 2010). Our meta-analysis of linkage scans of smoking behavior identified a genome-wide significant linkage for the maximum number of cigarettes smoked in a 24-hour period in a region (20q13.12-q13.32) harboring CHRNA4 (Han et al., 2010). This motivated us to evaluate the association of CHRNA4 with smoking behavior in two independent samples.
Materials and Methods
Subjects and smoking phenotypes
Subjects were recruited for participation in studies of the genetics of cocaine, opioid, and alcohol dependence from the communities around five sites in the United States: the University of Connecticut Health Center (UConn), Yale University School of Medicine (Yale), the Medical University of South Carolina (MUSC), the University of Pennsylvania (UPENN) and McLean Hospital (McLean). Subjects were interviewed using an electronic version of the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) instrument (Gelernter et al., 2005; Pierucci-Lagha et al., 2005). Most controls were recruited at the same recruitment sites (excluding McLean) and were screened to exclude those with a diagnosis of DSM-IV substance abuse or dependence, and a major Axis I psychiatric disorders. The study protocol was approved by the institutional review board at each site. A complete description of the study was provided to all subjects, who then gave written informed consent. Subjects were paid for their study participation.
A total of 1,249 unrelated European-Americans (EAs) and 1,790 unrelated African-Americans (AAs) were genotyped and included in analysis. The number of included subjects across the five recruiting sites are 1233, 1226, 310, 249 and 21 for UConn, Yale, MUSC, UPENN and McLean, respectively. Subjects were considered “affected” based on smoking behavior as defined by a DSM-IV diagnosis of ND and the Fagerstrom Test of ND (FTND). Lifetime, as opposed to current, measures were considered. Cigarettes smoked per day (CPD) was also tested for genetic association as a quantitative trait. Never-smokers are included in the current study. The diagnosis of DSM-IV ND was made using a computerized algorithm that applied the DSM-IV diagnostic criteria for the disorder (American Psychiatric Association, 1994). The FTND score (Heatherton et al., 1991) (range 0–10) was derived from the SSADDA, in which the FTND questions were embedded. Based on prior work (Saccone et al., 2009b), we dichotomized FTND scores as follows: and FTND score ≥4 was defined as a “case”, an FTND score ≤1 was defined as a “control,” and subjects with scores in-between those values were considered to have an “unknown” phenotype. Most ND/FTND cases were diagnosed with alcohol dependence (AD), cocaine dependence (CD), and/or opioid dependence (OD) using the SSADDA instrument. The basic demographic information for participants in this study for the analysis of each of the smoking behaviors is summarized in Table 1.
Table 1.
Basic demographic information of the three smoking phenotypes by study population
| African-American | European-American | |
|---|---|---|
| Number of genotyped subjects | 1790 | 1249 |
| Males (%) | 1003 (56.0) | 721 (57.7) |
| Age (years±SD) | 41.2±9.7 | 38.6±11.9 |
| Household gross income (dollars/year±SD) | 16889±20768 | 30541±34971 |
|
| ||
| Cigarettes smoked per day (CSPD) | 1385 | 915 |
| CSPD (Means±SD) | 13.5±9.1 | 18.8±9.6 |
| Males (%) | 834 (60.2) | 549 (60.0) |
| Age (years±SD) | 42.2±8.8 | 38.4±11.3 |
|
| ||
| DSM-IV diagnosis of ND | ||
| Cases | 990 | 716 |
| Males (%) | 585 (59.0) | 434 (60.6) |
| Age (years±SD) | 41.9±8.5 | 38.1±10.9 |
| Controls | 237 | 223 |
| Males (%) | 59 (24.9) | 92 (41.3) |
| Age (years±SD) | 36.6±12.8 | 38.8±14.7 |
|
| ||
| Dichotomized FTND | ||
| FTND (Means±SD) | 3.3±2.7 | 3.7±3.1 |
| Cases | 897 | 705 |
| Males (%) | 540 (60.2) | 434 (61.6) |
| Age (years±SD) | 42.1±8.6 | 38.8±10.8 |
| Controls | 229 | 214 |
| Males (%) | 53 (23.6) | 86 (41.3) |
| Age (years±SD) | 36.2±12.9 | 38.5±14.9 |
SNP selection and genotyping
Five SNPs (rs2236196, rs3787138, rs1044396, rs1044394 and rs6010918) across CHRNA4 were selected for genotyping. This included the 2 SNPs (rs2236196, rs1044396) that were associated with smoking behavior in previous studies (Feng et al., 2004; Li et al., 2005; Hutchison et al., 2007; Breitling et al., 2009). The proportions of HapMap Phase II common SNPs (MAF > 0.05) of CHRNA4 tagged using an r2 of 0.6 by the five SNPs are 0.83 and 0.80 for CEU and YRI, respectively. SNPs were genotyped with a fluorogenic 5’ nuclease assay method, i.e., the TaqMan technique, using the ABI PRISM 7900 Sequence Detection System (ABI, Foster City, CA, USA). For genotyping quality control, 8% of samples were re-genotyped with 100% concordance.
Statistical analysis
Single-marker analysis
A Pearson Chi-square test was used to test departures from Hardy–Weinberg equilibrium (HWE) expectations in controls defined by the absence of a DSM-IV diagnosis of ND and an FTND score ≤1 and in cases of ND or FTND. Only the SNPs in HWE in controls (P > 0.01, Bonferroni corrected) were included in subsequent analyses. The CPD phenotype was log transformed to achieve approximate normality and the association between log (CPD) and each SNP was tested using linear regression. The association between each SNP and the two binary traits was estimated in a logistic regression framework. We tested the genetic effects under a log-additive model with age, sex and race as covariates in the combined sample, and age and sex were included as covariates in each population-specific subgroup analysis. All of above analysis was performed in the R package “SNPassoc” (González et al., 2007).
Haplotype and conditional analysis
LD plots were drawn using the R package “snp.plotter” (Luna et al., 2007) for the unrelated controls. We used r2 as the measure of LD strength. The haplotype association test was performed by WHAP (Purcell et al., 2007). WHAP calculates likelihood estimates, likelihood ratios and P-values, and takes into account the loss of information due to haplotype phase uncertainty and missing genotypes. To disentangle the correlation structure in the highly correlated regions, conditional haplotype analysis was performed using WHAP. To test the hypothesis that all significant associations surrounding a particular assumed “causal” SNP reflected the high correlation between each candidate SNP and the “causal” SNP, we performed a two-SNP haplotype analysis pairing the “causal” SNP with every other SNP one at a time and conditional on that SNP. Disappearance of the global haplotype association was interpreted to mean that the conditional SNP explained the entire association.
Controlling population stratification
To control for possible confounding due to population stratification, we estimated individual ancestry proportions for each subject, which were then used as covariates in the regression model. A panel of 41 ancestry informative markers (AIMs), including 36 highly ancestry-informative short tandem repeat markers and 5 SNPs (rs1540771, rs2814778, rs1805007, rs1426654 and rs12896399) were genotyped in a majority of subjects (93.4%), among which 1592 AAs and 1093 EAs with genotyping rate > 50% were further included to estimate the individual admixture proportion. The exact sample size for each phenotype analyzed with admixture proportion as a covariate is provided in Supplementary table 1. Detailed genotyping methods for 37 of the 41 AIMs (including an FY SNP) have been described in detail previously (Yang et al., 2005). The remaining four SNPs were genotyped by the same TaqMan technique as was used for the CHRNA4 SNPs. We used the STRUCTURE program (Pritchard et al., 2000) to estimate the admixture proportion for each individual based on the AIMs. The log-likelihood of each analysis at a different number of population groups (k) was estimated from the average of 3 independent runs (20,000 burn in and 30,000 iterations) and, as expected, the result favored a two-ancestry population model. We included the admixture proportion of each individual as a covariate in the combined analysis and in each population specific subgroup analysis.
Correction for multiple testing
We tested the genetic associations between five SNPs and three smoking-related behaviors in three groups (the combined sample, AAs and EAs), requiring adjustment of the α level to avoid inflation of the type 1 error rate. Because traditional Bonferroni correction is too conservative when different tests are correlated, we used a permutation-based correction procedure. Briefly, we randomly reshuffled the phenotypes and genotypes while keeping the phenotype and genotype correlation structure unchanged. In each randomly permuted sample, we ran the association test in exactly the same way as in the real dataset and a minimum P-value was recorded. This process was repeated 10,000 times and 10,000 minimum P-values were obtained to get the empirical distribution of the minimum P-values. For each point-wise association test P-value, the empirical P-value corrected for multiple testing was estimated by counting the proportion of the minimum P-values less than or equal to the observed one across the 10,000 randomly permuted samples.
Power evaluation
We used QUANTO software (Gauderman 2002) to calculate the power of current association studies for each investigated phenotype at the 0.05 level of significance with a two-sided test. In the AA sample, for the quantitative log transformed CPD trait with a mean 2.38 and standard deviation 0.70, under a log-additive genetic model with MAF = 0.1 (0.3), the CPD sample (N=1385) had 62% (93%) power to detect a main effect of 0.1. For the two binary traits in AAs, under a log-additive genetic model with MAF = 0.1 (0.3), our study had 71% (96%) and 68% (95%) power to detect an effect with a relative risk 1.5 for ND (990 cases/237 controls) and FTND (897 cases/225 controls), respectively. In the EA sample, for the quantitative log transformed CPD trait with a mean 2.78 and standard deviation 0.62, under a log-additive genetic model with MAF = 0.1 (0.3), the CPD sample (N = 915) had 54% (89%) power to detect a main effect of 0.1. For the two binary traits in EAs, under a log-additive genetic model with MAF = 0.1 (0.3), our study had 66% (94%) and 63% (93%) power to detect an effect with a relative risk of 1.5 for ND (716 cases/223 controls) and FTND (705 cases/208 controls), respectively.
Results
Marker information and related summary statistics are presented in Table 2. For each SNP, the genotyping rate was >95% and the genotype distribution in the control sample was consistent with HWE. However, three SNPs (rs2236196, rs1044394, rs6010918) in ND cases and four SNPs (rs2236196, rs3787138, rs1044394, rs6010918) in FTND cases had genotype distributions that were not in HWE (0.004 < P < 0.05). Considering all the SNPs are in HWE in control samples, we included all the SNPs for association tests in subsequent analysis. In addition, there were large differences in allele frequencies between EAs and AAs for each SNP tested. Therefore, to avoid the spurious associations that can arise from stratification in the analysis of the combined sample, race was included as a categorical covariate in the association test.
Table 2.
SNP information and quality criteria in African-American and European-American samples.
| SNP | Positiona | Function | African-Americans | European-Americans | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Alleles (Major/Minor) | Call rate (%) | MAFb (%) | MAFc (%) | HWE | Alleles (Major/Minor) | Call rate (%) | MAFb (%) | MAFd (%) | HWE | |||
| rs2236196 | 61448000 | 3’-UTR | G/A | 97.1 | 30.9 | 34.1 | 0.64 | A/G | 98.5 | 27.6 | 29.2 | 1 |
| rs3787138 | 61449668 | Intron 5 | A/G | 96.9 | 49.8 | 49.2 | 1 | A/G | 97 | 14.4 | 9.2 | 0.62 |
| rs1044396 | 61451578 | Exon 5 (Synonymous) | G/A | 98 | 7.7 | 2.5 | 0.37 | A/G | 97.8 | 47.7 | 40.5 | 0.94 |
| rs1044394 | 61452529 | Exon 5 (Synonymous) | G/A | 97.7 | 41.3 | 33.9 | 0.42 | G/A | 97.1 | 8.3 | 6.9 | 1 |
| rs6010918 | 61459945 | Intron 2 | G/A | 95.8 | 39.1 | 35.0 | 0.33 | G/A | 97 | 6.6 | 5.8 | 1 |
Abbreviation: UTR, untranslated region; MAF, minor allele frequency; HWE, p-value for Hardy-Weinberg equilibrium. MAFb and HWE were calculated from the controls without a DSM-IV diagnosis of ND and dichotomized FTND cases.
coordinate based on Genome Build 36.3;
MAF from our study samples.
MAF from Hapmap YRI samples (except rs2236196 for which the MAF was from African American sample in dbSNP);
MAF from Hapmap CEU samples (except rs2236196 for which the MAF was from pilot 1 CEU sample in dbSNP).
We note that our observed allele frequency for rs1044396 differs from the HapMap frequency by about 7%; our observation however is very close to that reported in an article with a much larger sample (Breitling, 2009)
Table 3 illustrates the association results in detail. In the combined analysis, the strongest signal came from the association of a synonymous SNP (rs1044394) with ND; the minor allele “A” at this locus was associated with a lower risk of ND (P = 0.001, OR = 0.73, 95% CI = 0.61–0.89). Subgroup analysis demonstrated a similar pattern of association in AAs (P = 0.009, OR = 0.74, 95% CI = 0.60–0.93) and EAs (P = 0.05, OR = 0.66, 95% CI = 0.43–1.00). Rs1044394 was also significantly associated with FTND (P = 0.01), and showed non-significant evidence for association with CPD (P = 0.083) in the combined samples. Another SNP (rs2236196), which has a low correlation with rs1044394 (r2 = 0.26 and 0.22 for AAs and EAs, respectively), was significantly associated with CPD (P = 0.003) in the combined sample, as well as in AAs (P = 0.022) and EAs (P = 0.049) separately. Note that the coefficient for rs2236196 is opposite in sign for CPD in AAs and EAs; this is attributable to the fact that the minor allele is opposite for AAs and EAs. That is, the allele effects are congruent. However, only the association between rs1044394 and ND in the combined sample survived the correction for multiple testing (P = 0.033).
Table 3.
Associations of CHRNA4 polymorphisms with three smoking-related behaviors in African-Americans, European-Americans, and the full sample.
| SNP/Phenotype | African-Americans | European-Americans | Combined Sample | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Smoking Quantity (CPD) | Minor allele | β (95%CI) | P | P* | Minor allele | β (95%CI) | P | P* | Minor allele | β (95%CI) | P | P* |
| rs2236196 | A | -0.063 (-0.12--0.009) | 0.022 | 0.34 | G | 0.063 (0.0004-0.13) | 0.049 | 0.58 | A | -0.060 (-0.10--0.020) | 0.0034 | 0.075 |
| rs3787138 | G | 0.031 (-0.020-0.083) | 0.23 | 0.98 | G | 0.004 (-0.079-0.087) | 0.92 | 1 | G | 0.023 (-0.019-0.067) | 0.29 | 0.99 |
| rs1044396 | A | -0.031 (-0.11-0.05) | 0.45 | 0.99 | G | 0.038 (-0.018-0.09) | 0.19 | 0.96 | A | -0.035 (-0.083-0.013) | 0.15 | 0.92 |
| rs1044394 | A | 0.049 (-0.005-0.1) | 0.075 | 0.72 | A | 0.015 (-0.1-0.13) | 0.81 | 1 | A | 0.042 (-0.006-0.09) | 0.083 | 0.76 |
| rs6010918 | A | 0.027 (-0.028-0.082) | 0.34 | 0.99 | A | -0.005 (-0.14-0.13) | 0.95 | 1 | A | 0.019 (-0.029-0.069) | 0.43 | 0.99 |
|
| ||||||||||||
| DSM-IV ND | Minor allele | OR (95%CI) | P | Minor allele | OR (95%CI) | P | Minor allele | OR (95%CI) | P | |||
|
| ||||||||||||
| rs2236196 | A | 1.07 (0.86-1.34) | 0.54 | 1 | G | 0.95 (0.75-1.22) | 0.70 | 1 | G | 0.95 (0.80-1.11) | 0.50 | 1 |
| rs3787138 | G | 0.91 (0.73-1.12) | 0.37 | 0.99 | G | 0.89 (0.64-1.22) | 0.46 | 0.99 | G | 0.89 (0.75-1.06) | 0.21 | 0.97 |
| rs1044396 | A | 1.41 (0.95-2.08) | 0.081 | 0.75 | G | 1.01 (0.81-1.26) | 0.93 | 1 | A | 1.07 (0.89-1.30) | 0.46 | 0.99 |
| rs1044394 | A | 0.74 (0.60-0.93) | 0.0085 | 0.16 | A | 0.66 (0.43-1.00) | 0.052 | 0.60 | A | 0.73 (0.61-0.89) | 0.0013 | 0.033 |
| rs6010918 | A | 0.76 (0.61-0.95) | 0.015 | 0.26 | A | 0.70 (0.44-1.10) | 0.13 | 0.89 | A | 0.76 (0.62-0.92) | 0.0056 | 0.11 |
|
| ||||||||||||
| FTND | Minor allele | OR (95%CI) | P | Minor allele | OR (95%CI) | P | Minor allele | OR (95%CI) | P | |||
|
| ||||||||||||
| rs2236196 | A | 1.03 (0.81-1.3) | 0.81 | 1 | G | 1.02 (0.79-1.31) | 0.89 | 1 | G | 0.98 (0.83-1.16) | 0.84 | 1 |
| rs3787138 | G | 0.97 (0.78-1.21) | 0.77 | 1 | G | 0.90 (0.65-1.25) | 0.54 | 1 | G | 0.93 (0.77-1.11) | 0.41 | 0.99 |
| rs1044396 | A | 1.41 (0.93-2.13) | 0.094 | 0.79 | G | 1.02 (0.82-1.27) | 0.87 | 1 | A | 1.07 (0.88-1.31) | 0.47 | 0.99 |
| rs1044394 | A | 0.82 (0.65-1.02) | 0.081 | 0.75 | A | 0.65 (0.43-0.98) | 0.046 | 0.56 | A | 0.78 (0.64-0.94) | 0.011 | 0.20 |
| rs6010918 | A | 0.80 (0.64-1.01) | 0.059 | 0.64 | A | 0.67 (0.42-1.05) | 0.089 | 0.78 | A | 0.78 (0.64-0.95) | 0.014 | 0.24 |
SNP, single nucleotide polymorphism; CPD, cigarettes per day; DSM-IV ND, Nicotine dependence according to the Diagnostic and Statistical Manual of Mental Disorders, 4th ed., FTND, Fagerstrom Test for Nicotine Dependence dichotomized as cases and controls.
Nominal significant P-values and P-values remaining significant after multiple testing correction are shown in bold.
multiple testing corrected P-values.
The LD map and each SNP-associated P-value are depicted in Figure 1. Rs1044394 and rs6010918 are in strong LD in both AAs and EAs (r2 = 0.91 and 0.81 for AAs and EAs, respectively), which explained the evidence for association of both SNPs with ND (FTND) in the combined sample and in each population group (Table 3). To exclude the possibility that multiple observed effects were caused by LD with a single true risk allele, we performed conditional haplotype analysis on the combined sample with race, age and sex as covariates. As expected, both SNPs showed a significant global association with ND (P = 0.0046). The global association disappeared when conditioned on rs1044394 (P = 0.24), but remained significant when conditioned on rs6010918 (P = 0.047). This analysis suggests that the significant association of rs6010918 with ND arises from its high correlation with rs1044394. When a similar analysis was used for the FTND binary trait, the effects of rs1044394 and rs6010918 could not be disentangled.
Figure 1.
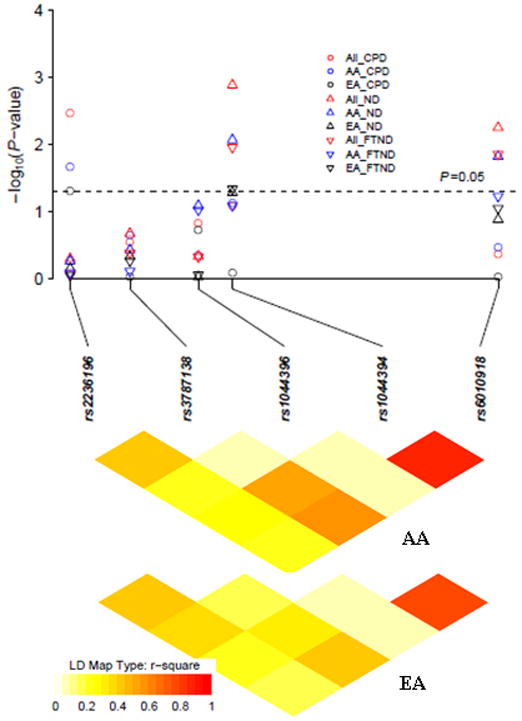
Linkage disequilibrium (LD) structure and association P values (multiple testing uncorrected P values) in African-Americans (AA), European-Americans (EA) and the full samples (All) for the three smoking-related behaviors. In each population-specific analysis, P values were adjusted by age and sex. In the combined analysis, P values were adjusted by age, sex and race. LD was calculated from the genotype data in AA and EA controls.
Additional analyses were conducted to define more clearly the association of the CHRNA4 polymorphisms and the three smoking-related behaviors. First, an association test was performed in the subset of individuals with AIMs data available. The admixture proportion was estimated for each individual and included as a covariate with age and sex in the regression model. The association results remained similar, arguing against stratification as a confounder (Supplementary Table 2). Second, we examined whether recruitment site was a significant confounder by including site as a categorical covariate, and the association results did not change substantially (Supplementary Table 3). Third, haplotype analysis was performed using different SNP combinations but did not yield more significant results (data not shown).
Discussion
The biological link between CHRNA4 and nicotine addiction phenotypes in animal models (Ross et al., 2000; Labarca et al., 2001) and humans (Breese et al., 1997; Fenster et al., 1999; Buisson et al., 2001) has made this gene a notable candidate in genetic association studies of smoking behavior. However, prior association studies in small samples have shown inconsistent results, and recent GWAS studies as reflected by three meta-analyses of GWAS studies on smoking behavior, also did not identify CHRNA4 as a susceptibility gene for nicotine dependence. There are many possible explanations for this lack of association, including genetic heterogeneity, phenotypic heterogeneity, and the limited power of GWAS studies arising from the necessity to correct for multiple comparisons.
In the first report, Feng et al. (2004) identified two synonymous SNPs in exon 5 (rs1044396, rs1044397), which were significantly associated with reduced risk for ND in Chinese men. Li et al. (2005) replicated and extended the association of CHRNA4 with smoking behavior in EAs and AAs, but the association results were not entirely consistent with those published by Feng. For example, rs1044396 showed a significant association with ND in the Chinese male sample but showed no association in the EA or AA samples and rs2236196 showed significant association in AA females but not in Chinese males. More recently, an association study in 5500 Germans showed that rs2236196 was significantly associated with FTND and the “G” allele was associated with a higher risk of nicotine addiction and higher odds of being a smoker (Breitling et al., 2009). The accumulating evidence from genetic association studies plus the additional findings on clinical effects (Hutchison et al., 2007) suggest that rs2236196 could be one of the most promising risk variants for smoking-related behaviors.
Consistent with this prior work, we detected a significant association of rs2236196 with a quantitative trait (CPD) in the combined sample, as well as in AAs and EAs. Specifically, we found that the rs2236196 “G” allele was associated with higher number of CPD, a finding consistent in magnitude and direction with the findings from previous studies (Hutchison et al., 2007; Saccone et al., 2007; Breitling et al., 2009). Although the association between rs2236196 and CPD in the combined sample did not survive multiple testing (Pcorrected = 0.075), given the prior findings of association between rs2236196 and smoking behavior, a nominally significant p-value could be considered of interest, which would provide evidence of replication of the finding that rs2236196 plays a role in smoking behavior. However, no significant evidence was observed for association between rs2236196 and the two binary traits of ND or dichotomized FTND. Because the SSADDA does not include an assessment of the maximum number of cigarettes smoked in a 24-hour period (the trait that we identified through the linkage signal with CHRNA4 in the meta-analysis), we cannot directly test the association of CHRNA4 variants with this phenotype.
In addition, we found that rs1044394, another synonymous SNP in exon 5, was nominally significantly associated with ND and FTND in the combined sample, as well as in AAs and in EAs separately. However, only the association of rs1044394 with ND in the combined sample remained significant after correction for multiple testing (Pcorrected = 0.033). We are aware of only one other study that investigated the role of rs1044394 in smoking-related behaviors; that yielded a negative result (Etter et al., 2009). Interestingly, the correlation between rs2236196 and rs1044394 was low in both AAs and EAs, suggesting that multiple independent variants within this gene could be associated with different dimensions of smoking behavior. However, all of the published studies investigated a very limited number of variants in this gene, thus we would speculate that other independent variants associated with smoking behavior may have been missed.
An exploration of the correlation among the three investigated phenotypes could be helpful to interpret the consistency (or lack of consistency) among the association results. The correlation between CPD and ND was low for both AAs (Spearman correlation coefficient ρ = 0.14) and EAs (ρ = 0.19). A low correlation between CPD and FTND was also observed in AAs (ρ = 0.16) and EAs (ρ = 0.26). The low correlation between CPD and ND/FTND could explain the inconsistent results for rs2236196, which demonstrated association for CPD but not for ND or FTND. There is a strong correlation between the ND and FTND phenotype (ρ = 0.83 and 0.88 in AAs and EAs, respectively), which could explain the similar patter of association for ND and FTND (Table 3).
Since the ND or FTND cases included in our samples are affected with other substance dependence disorders, including cocaine dependence (CD), alcohol dependence (AD) and/or opioid dependence (OD), we also investigated whether the signals associated with smoking behavior could be driven by other substance dependence disorders. To control the effect of other substance dependence disorders, we reanalyzed the data by including CD, AD and OD status as covariates in the regression model. As a result, rs2236196 remained significant for CPD (P = 0.0078) and was less significant for ND (P = 0.042). However, the signal disappeared for rs1044394 for both ND and FTND. We speculate that this loss of a signal for rs1044394 is attributable to either random fluctuation or that other substance dependence disorders drive the association between rs1044394 and ND (or FTND). It will be necessary to examine the role of CHRNA4 in other substance dependence disorders to determine the basis for this finding.
This study also has limitations. The positive linkage finding by genome scan meta-analysis in our previous study is consistent with a role for multiple rare (or less common) variants mapped to this region for ND; however, these were not investigated in present study. Further, there is evidence that the rare missense variants at CHRNA4 are associated with sporadic amyotrophic lateral sclerosis (Sabatelli et al., 2009). The role of rare variants at this locus for ND and related behaviors thus needs to be evaluated. With increasing evidence for a role of rare variants in psychiatric disorders (Carroll et al., 2010; Knight et al., 2009), we believe that sequencing the whole gene will be necessary to discover all of the causal variants in CHNRA4, especially rare variants, that are associated with smoking behavior. In addition, the AA and EA samples differed significantly on MAF for the five SNPs. There is a possibility that adjustment for self-reported race in the combined analysis may not be sufficient to address this confounding by race. However, in a subgroup of samples with AIMs available, the association results remained similar when using admixture proportion as a covariate, arguing against stratification as a confounder. Finally, the control samples used in the binary traits analysis include many “never-smokers” which may have become dependent if they had been exposed to nicotine, and this could arguably have reduced the power.
In summary, our study confirms the previously reported genetic association between CHRNA4 polymorphisms and smoking behavior in independent samples of AAs and EAs. Although rs2236196 could be one of the causal variants for smoking behavior, other independently associated functional variants, especially rare variants, may have been missed due to the limitations of the current study design. To identify the full range of CHRNA4 variants and to evaluate their role in smoking behavior, complete sequencing of the gene is necessary.
Supplementary Material
Acknowledgments
The authors are grateful to the volunteer families and individuals who participated in this research study. We are also grateful to Ann Marie Lacobelle and Greg Kay for their excellent technical assistance and to the many SSADDA interviewers who obtained the phenotypic data used in this study. Kathleen Brady, M.D., Ph.D. oversaw recruitment of some of the subjects from the Medical University of South Carolina. Roger Weiss, M.D. of McLean Hospital and Harvard Medical School oversaw study recruitment at that site. This work was supported by the U.S. National Institutes of Health (R01 AA11330, R01 AA017535, R01 DA12849, R01 DA12690, R01 DA018432, K01 DA024758, and M01 RR06192) and by the US Department of Veterans Affairs (VA CT REAP and New England MIRECC Center; VA CT Alcohol Research Center).
Footnotes
Financial Disclosures:
Drs. Yang, Han, and Gelernter report no competing interests. Dr. Kranzler reports consulting arrangements with Gilead Sciences and Alkermes, Inc. and research support from Merck & Co.. Dr. Kranzler also reports a current association with the following pharmaceutical companies: Eli Lilly, Janssen, Schering Plough, Lundbeck, Alkermes, GlaxoSmithKline, Abbott, and Johnson & Johnson, as these companies provide support to the Alcohol Clinical Trials Initiative (ACTIVE) and Dr. Kranzler receives support from ACTIVE.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. American Psychiatric Press; Washington, D.C: 1994. [Google Scholar]
- Benowitz NL. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin Pharmacol Ther. 2008;83:531–541. doi: 10.1038/clpt.2008.3. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Conti DV, Van Den Berg D, Lee W, Liu J, Li D, Guo N, Mi H, Thomas PD, Lessov-Schlaggar CN, Krasnow R, He Y, Nishita D, Jiang R, McClure JB, Tildesley E, Hops H, Tyndale RF, Benowitz NL, Lerman C, Swan GE. Dopamine genes and nicotine dependence in treatment-seeking and community smokers. Neuropsychopharmacology. 2009;34:2252–2264. doi: 10.1038/npp.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese CR, Marks MJ, Logel J, Adams CE, Sullivan B, Collins AC, Leonard S. Effect of smoking history on [3H]nicotine binding in human postmortem brain. J Pharmacol Exp Ther. 1997;282:7–13. [PubMed] [Google Scholar]
- Breitling LP, Dahmen N, Mittelstrass K, Rujescu D, Gallinat J, Fehr C, Giegling I, Lamina C, Illig T, Müller H, Raum E, Rothenbacher D, Wichmann HE, Brenner H, Winterer G, et al. Association of nicotinic acetylcholine receptor subunit alpha 4 polymorphisms with nicotine dependence in 5500 Germans. Pharmacogenomics J. 2009;9:219–224. doi: 10.1038/tpj.2009.6. [DOI] [PubMed] [Google Scholar]
- Buisson B, Bertrand D. Chronic exposure to nicotine upregulates the human a4b2 nicotinic acetylcholine receptor function. J Neurosci. 2001;21:1819–1829. doi: 10.1523/JNEUROSCI.21-06-01819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, Ziegler RG, Hunter DJ, Chanock S, Hankinson S, Kraft P, Bergen AW. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll LS, Williams NM, Moskvina V, Russell E, Norton N, Williams HJ, Peirce T, Georgieva L, Dwyer S, Grozeva D, Greene E, Farmer A, McGuffin P, Morris DW, Corvin A, Gill M, Rujescu D, Sham P, Holmans P, Jones I, Kirov G, Craddock N, O’Donovan MC, Owen MJ. Evidence for rare and common genetic risk variants for schizophrenia at protein kinase C, alpha. Mol Psychiatry. 2010;15:1101–1111. doi: 10.1038/mp.2009.96. [DOI] [PubMed] [Google Scholar]
- Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, Bergen AW, Swan GE, Tyndale RF, Benowitz NL, Lerman C. Pharmacogenetics of Nicotine Addiction and Treatment Consortium. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, Schlaepfer I, Smolen A, Stallings MC, Young SE, Zeiger JS. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet (Neuropsychiatr Genet) 2007;144:596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Etter JF, Hoda JC, Perroud N, Munafò M, Buresi C, Duret C, Neidhart E, Malafosse A, Bertrand D. Association of genes coding for the alpha-4, alpha-5, beta-2 and beta-3 subunits of nicotinic receptors with cigarette smoking and nicotine dependence. Addict Behav. 2009;34:772–775. doi: 10.1016/j.addbeh.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Feng Y, Niu T, Xing H, Xu X, Chen C, Peng S, Wang L, Laird N, Xu X. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. Am J Hum Genet. 2004;75:112–121. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CP, Hicks JH, Beckman ML, Covernton PJ, Quick MW, Lester RA. Desensitization of nicotinic receptors in the central nervous system. Ann N Y Acad Sci. 1999;868:620–623. doi: 10.1111/j.1749-6632.1999.tb11335.x. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet (Neuropsychiatr Genet) 2005;136:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Kranzler HR, Farrer L. Haplotype Spanning TTC12 and ANKK1, Flanked by the DRD2 and NCAM1 Loci, is Strongly Associated to Nicotine Dependence in Two Distinct American Populations. Human Molecular Genetics. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- González JR, Armengol L, Solé X, Guinó E, Mercader JM, Estivill X, Moreno V. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- Han S, Gelernter J, Luo X, Yang BZ. Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biol Psychiatry. 2010;67:12–19. doi: 10.1016/j.biopsych.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, Stitzel J, Bryan A, McGeary J, Haughey HM. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch Gen Psychiatry. 2007;64:1078–1086. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- Knight HM, Pickard BS, Maclean A, Malloy MP, Soares DC, McRae AF, Condie A, White A, Hawkins W, McGhee K, van Beck M, MacIntyre DJ, Starr JM, Deary IJ, Visscher PM, Porteous DJ, Cannon RE, St Clair D, Muir WJ, Blackwood DH. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am J Hum Genet. 2009;85:833–846. doi: 10.1016/j.ajhg.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C, Schwarz J, Deshpande P, Schwarz S, Nowak MW, Fonck C, Nashmi R, Kofuji P, Dang H, Shi W, Fidan M, Khakh BS, Chen Z, Bowers BJ, Boulter J, Wehner JM, Lester HA. Point mutant mice with hypersensitive a4 nicotinic receptors show dopaminergic deficits and increased anxiety. Proc Natl Acad Sci USA. 2001;98:2786–2791. doi: 10.1073/pnas.041582598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, Duenes AS, Crews KM, Elston RC. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schäfer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Völzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A, Mooser V, Francks C, Marchini J Wellcome Trust Case Control Consort. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YZ, Pei YF, Guo YF, Wang L, Liu XG, Yan H, Xiong DH, Zhang YP, Levy S, Li J, Haddock CK, Papasian CJ, Xu Q, Ma JZ, Payne TJ, Recker RR, Li MD, Deng HW. genome-wide association analyses suggested a novel mechanism for smoking behavior regulated by IL15. Mol Psychiatry. 2009;14:668–680. doi: 10.1038/mp.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23:774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- Ross SA, Wong JY, Clifford JJ, Kinsella A, Massalas JS, Horne MK, Scheffer IE, Kola I, Waddington JL, Berkovic SF, Drago J. Phenotypic characterization of an a4 neuronal nicotinic acetylcholine receptor subunit knock-out mouse. J Neurosci. 2000;20:6431–6441. doi: 10.1523/JNEUROSCI.20-17-06431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Eusebi F, Al-Chalabi A, Conte A, Madia F, Luigetti M, Mancuso I, Limatola C, Trettel F, Sobrero F, Di Angelantonio S, Grassi F, Di Castro A, Moriconi C, Fucile S, Lattante S, Marangi G, Murdolo M, Orteschi D, Del Grande A, Tonali P, Neri G, Zollino M. Rare missense variants of neuronal nicotinic acetylcholine receptor altering receptor function are associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:3997–4006. doi: 10.1093/hmg/ddp339. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D, Johnson EO, Madden PA, Swan GE, Wang JC, Goate AM, Rice JP, Bierut LJ. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet (Neuropsychiatr Genet) 2009a;150:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, Culverhouse RC, Fox L, Hinrichs AL, Steinbach JH, Wu M, Rice JP, Goate AM, Bierut LJ. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009b;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapper AR, McKinney SL, Nashmi R, Schwarz J, Deshpande P, Labarca C, Whiteaker P, Marks MJ, Collins AC, Lester HA. Nicotine activation of alpha4* receptors: Sufficient for reward, tolerance, and sensitization. Science. 2004;306:1029–1032. doi: 10.1126/science.1099420. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsäter A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Mägi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tönjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Döring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Järvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K ENGAGE Consortium. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, Smit JH, Hoogendijk WJ, Zitman FG, Peltonen L, Kaprio J, Pedersen NL, Magnusson PK, Spector TD, Kyvik KO, Morley KI, Heath AC, Martin NG, Westendorp RG, Slagboom PE, Tiemeier H, Hofman A, Uitterlinden AG, Aulchenko YS, Amin N, van Duijn C, Penninx BW, Boomsma DI. Genome-wide association study of smoking initiation and current smoking. Am J Hum Genet. 2009;84:367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, Piper ME, Fiore MC, Scholand MB, Connett JE, Kanner RE, Gahring LC, Rogers SW, Hoidal JR, Leppert MF. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e100012–5. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. The World Health Report 2002. World Health Organization; 2002. [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


