Abstract
Biological differences among metazoans, and between cell types in a given organism, arise in large part due to differences in gene expression patterns. The sequencing of multiple metazoan genomes, coupled with recent advances in genome-wide analysis of histone modifications and transcription factor binding, has revealed that among regulatory DNA sequences, gene-distal enhancers appear to exhibit the greatest diversity and cell-type specificity. Moreover, such elements are emerging as important targets for mutations that can give rise to disease and to genetic variability that underlies evolutionary change. Studies of long-range interactions between distal genomic sequences in the nucleus indicate that enhancers are often important determinants of nuclear organization, contributing to a general model for enhancer function that involves direct enhancer-promoter contact. In a number of systems, however, mechanisms for enhancer function are emerging that do not fit solely within such a model, suggesting that enhancers as a class of DNA regulatory element may be functionally and mechanistically diverse.
Introduction
Genomic DNA acts as a carrier of information in two fundamental ways: first, in transcribed genes, by specifying the sequences of mRNAs and functional RNAs that are translated into protein, as well as information encoded in RNA that affects its processing and stability; second, in regulatory sequences, by providing sites for transcription factors to bind and establish the appropriate levels and spatial and temporal expression patterns of those genes. A large proportion of the regulatory information necessary for gene expression is confined to the promoter region immediately upstream of transcription start sites. In single-celled organisms, this information can serve to specify absolute levels of transcription, and in many cases can mediate alternate responses (up- or down-regulation) to external stimuli.
Metazoans present a challenge in this regard. A single genome specifies many morphologically distinct cell types, and also directs the ordered processes of development and differentiation that lead to the varied structures present in an adult multicellular organism. Sequencing of metazoan genomes has not revealed a simple correlation between genome size, as measured by the number of genes, and relative complexity, as measured by number of cells and cell types and by diversity of behavior. The 959 to 1031 cells of the nematode Caenorhabditis elegans and the trillions of cells in a typical human are both specified by ∼20,000-25,000 protein-coding genes. Thus, morphological and developmental complexity is not a function of increased numbers of genes, but of alternative mechanisms. For example, a higher proportion of vertebrate genes are subject to alternative splicing, as compared to invertebrates (Kim et al., 2006). Notably, however, complexity can be generated by diversification of the patterns in which genes are expressed, both spatially and temporally, within an organism.
Such diversification is enabled by a correspondingly more complex set of regulatory information in the genomes of metazoans. In particular, in metazoans transcriptional regulation is often decoupled from the confines of the promoter-proximal region and distributed among distal sequence elements, termed enhancers, which can be located far from the transcription start site. This distribution of regulatory sequences evades the limitations inherent in systems in which transcription is a function solely of the few hundred base pairs immediately upstream of a gene promoter. Recent developments in genomics, coupled with studies of covalent histone modifications, structural features of genomic DNA, functional assays for regulatory elements, methods to investigate nuclear organization, and cross-species sequence comparisons have revealed enhancers, as a class of regulatory element, to be generally and fundamentally
Finding Enhancers: Structure and Sequence
Enhancers were first characterized using, and are still most often functionally defined by, transient reporter gene assays in cultured cell lines. The activity associated with such elements – first for viral sequences (Banerji et al., 1981; Moreau et al. 1981), then subsequently in sequences originating from metazoan gene loci (Banerji et al., 1983; Gillies et al., 1983) – is the activation of transcription regardless of their location or orientation relative to the promoter within a plasmid construct. This flexibility is the defining hallmark of enhancers. They are commonly found within the introns of the genes they regulate (or, in fact, within the introns of neighboring genes), and often at prodigious distances from the promoter. One of the most extreme examples known, for example, is a limb bud enhancer for the mouse sonic hedgehog (Shh) gene, which is located within the intron of another gene more than 1 Mb from the Shh gene promoter (Lettice et al., 2003; Sagai et al., 2005).
This flexibility, however, impedes attempts to comprehensively identify and catalog the full population of enhancers within the genome, or indeed the full complement of enhancers that act upon a single selected gene. Whereas the promoter of a gene can be identified simply by sequencing the 5′ end of its mRNA, no similarly clearcut criterion exists that can pinpoint the location of an enhancer or the target gene for its activity. Enhancer detection therefore relies on a number of imperfect measures of chromatin structure and sequence functionality.
Enhancers are typically found to colocalize with disruptions in chromatin structure revealed by hypersensitivity to digestion by DNaseI. DNaseI hypersensitivity was first discovered at the promoter region of the Drosophila hsp70 gene (Wu, 1980), and is usually thought to result from short (100-300 bp) regions of genomic DNA from which nucleosomes are excluded due to the binding of transcription factors (Elgin, 1988; Gross and Garrard, 1988), although DNA bending by transcription factors has also been implicated (Stamatoyannopoulos et al., 1995; Leach et al., 2001). Enhancers consist of clusters of cognate binding sites for transcription factors that can both exclude nucleosomes and bend DNA, and so are marked by such nuclease hypersensitivity. For several decades, scans of gene loci for putative enhancer elements involved the slow and laborious indirect end-labeling assay, but recent advances have allowed the mapping of nuclease hypersensitivity genome-wide using microarrays or high-throughput sequencing (Crawford et al., 2006).
Another criterion employed for predicting putative enhancer sequences is noncoding sequence conservation. The full utility of this approach has only been realized in the past decade, when multiple fully sequenced genomes have been available for comparison. The principle, however, is well established, resting on the assumption that conservation of DNA sequence across evolution in regions that do not encode proteins implies regulatory function. Nonfunctional sequences, in contrast, will accumulate mutations over evolutionary timescales and eventually diverge. High-level expression of the mammalian β-globin genes in erythroid cells, for example, requires a set of sequence elements termed the locus control region (LCR) that is distributed across a region of 20-30 kb located upstream of the gene cluster (Bender et al., 1990; Moon and Ley, 1990; Grosveld et al., 1987; Hardison et al., 1997). Each of these sequence elements shows significant sequence conservation among all mammalian genomes sequenced thus far. More generalized studies have similarly indicated that evolutionary DNA sequence conservation can predict enhancer activity (Visel et al., 2009; Noonan and McCallion, 2010).
On the other hand, several well-documented examples exist of genes that are expressed in identical patterns in different species, but which require the activities of enhancers that look nothing alike. Both Drosophila and sepsid flies, for example, express orthologous even-skipped genes with identical patterns in the developing embryo, and this expression pattern is governed by multiple enhancers. Sequence comparisons, however, fail to reveal any significant similarities between the DNA sequences of these enhancers (Hare et al., 2008). Similarly, expression of the Pax2 gene in the Drosophila melanogaster eye is regulated in part by a cone-specific enhancer, within which multiple transcription factors bind in a specific pattern that can be disrupted by small deviations in sequence of, or spacing between, binding sites. Surprisingly, however, the cone-specific Pax2 enhancers in other Drosophila species exhibit very little conservation of sequence or binding site spacing, even though they function identically when transferred into D. melanogaster (Swanson et al., 2010). A systematic evaluation of a 40 kb region encompassing the zebrafish phox2b gene showed that a large proportion of sequences with demonstrable regulatory activity were not identified by measures of evolutionary sequence constraint (McGaughey et al., 2008).
In fact, comparisons of genome-wide transcription factor binding patterns across species indicate that a large proportion of enhancers are species-specific. In an analysis of mouse and human hepatocytes, for example, 41-89% of binding sites for four transcription factors were found to be species-specific (Odom et al, 2007). A subsequent study examined genome-wide binding of the liver-specific transcription factors CEBPA and HNF4α in multiple species (Schmidt et al., 2010). Only 10-22% of binding sites were shared between placental mammals (human, mouse or dog), and this number was even lower in comparisons between placental mammals and a nonplacental mammal (opossum) or chicken. Moreover, variation in transcription factor binding patterns appears to have a predominantly genetic origin. In mouse hepatocytes harboring an intact human chromosome 21, binding patterns of the transcription factors HNF1α, HNF4α and HNF6 are nearly identical to those observed on chromosome 21 in human hepatocytes (Wilson et al., 2008). Combined with other indications that a large proportion of functional enhancers is not subject to evolutionary constraint (ENCODE Project Consortium, 2007; McGaughey et al., 2008), such findings indicate that enhancers can “turn over” rapidly over evolutionary timescales, even when associated gene expression patterns are conserved.
Thus, conservation of regulatory function is not always reflected in conservation of DNA sequence. An approach to enhancer prediction that relies on sequence conservation alone will fail to identify a large proportion of enhancers.
Finding Enhancers: Chromatin and Transcription Factor Signatures
Recently, genome-wide studies have suggested that enhancers exhibit a characteristic chromatin “signature”. This signature consists of monomethylation of histone H3 lysine 4 (H3K4Me1) in the absence of significant trimethylation (H3K4Me3) (ENCODE Project Consortium, 2007; Heintzman et al., 2007; Koch et al., 2007). Notably, H3K4Me3 is associated with active gene promoters, which in turn exhibit low levels of H3K4Me1 at the transcription start site. In other studies, however, a sharp divide between H3K4Me3 present at promoters but not enhancers, or H3K4Me1 present at enhancers but not transcription start sites, has not been as obvious (Barski et al., 2007; Wang et al., 2008). The basis for the discrepancy is not clear, although each study utilizes different sources of chromatin (transformed cell lines vs. primary T lymphocytes), different protocols for isolation of chromatin (formaldehyde crosslinking and sonication vs. micrococcoal digestion of native chromatin) and different antibodies. It is nevertheless now possible to catalog enhancers by identifying histone methylation signatures that are not associated with other functional elements.
The H3K4 methylation signature has been correlated with enhancer activity in gain-of-function assays (ENCODE Project Consortium, 2007; Heintzman et al., 2007), but imperfectly. This is perhaps unsurprising, since the transient reporter gene assay is often a poor proxy for the activities of regulatory elements integrated in the genome, which can be active in narrow windows of development and/or cellular differentiation. Enhancer predictions become more significant, however, when the histone methylation signature is combined with other indicators of enhancer activity in a specific cell type, such as transcription cofactor binding – most often, the acetyltransferase p300 (Visel et al., 2009; Blow et al., 2010; Ghisletti et al., 2010).
In addition, studies utilizing embryonic stem (ES) cells and multiple primary cell types suggest that acetylation of histone H3K27 in combination with H3K4Me1 is correlated with enhancers near active genes, while H3K4Me1 in the absence of H3K27 acetylation appears to mark inactive or “poised” enhancers (Creyghton et al., 2010; Rada-Iglesias et al, 2010). Notably, p300 can acetylate H3K27. Moreover, many poised enhancers in ES cells were instead associated with H3K27Me3, and at a subset of these enhancers, the H3K27Me3 mark was replaced with H3K27Ac upon differentiation of ES cells along a neuronal pathway (Rada-Iglesias et al., 2010). The available evidence suggests that H3K4Me1 represents a generalized, although perhaps not all-inclusive, mark for distal enhancers in a given cell type. Additional modifications can then distinguish between enhancers that are active and those that are potentiated for activity in response to growth conditions or cell-fate decisions.
The most accurate predictions for enhancer activity to date are derived from combinatorial analysis of the binding of multiple transcription factors across the genome in Drosophila (Zinzen et al., 2009). In this study, binding of five transcription factors known to be involved in the differentiation of different muscle cell types from mesoderm was mapped genome-wide at different stages of development. A machine learning method was then applied to this data set in order to derive predictions for enhancers active in different cell types. Using this approach, the authors were able to identify 77% of all previously characterized muscle-specific enhancers in five different cell types in Drosophila, and had a roughly equivalent success rate when novel predicted sequences were tested for enhancer activity.
If one accepts the description of a chromatin “signature” for enhancers, current evidence then suggests that mammalian genomes harbor an abundance of such elements, and that they are the major determinant of cell type specificity in gene expression. Examination of genome-wide H3K4 methylation patterns in two cell lines – K562, a human erythroleukemia, and HeLa, a human cervical carcinoma – resulted in an estimate of 24,000-36,000 enhancers in each line (Heintzman et al., 2009). When the locations of these enhancers were compared between the two lines, only 5,000 were found to be present in both. Analysis of histone modification patterns across a region comprising 1% of the human genome, compared among five different cell lines, indicated a frequency of H3K4Me1 distal from promoters that agrees roughly with the genome-wide K562/HeLa study, and also that a much higher number of enhancers defined by this criterion exhibited cell-type specificity, as compared to promoters (Koch et al., 2007).
Genome-wide mapping of nuclease hypersensitive sites (HSs) also indicates that enhancers are the primary determinant of cell-type specificity. A survey of HSs across 1% of the human genome in six cell lines (including HeLa and K562) established a strong correlation between hypersensitive sites that were cell-type specific and enhancer elements as defined by the H3K4Me signature (Xi et al., 2007).
Based on such observations, it has been suggested that the human genome might harbor as many as 1 × 106 enhancers (Heintzman et al, 2009). At present, however, this is little more than the roughest of estimates, since it is not clear how extensive variability in the H3K4Me signature actually is in vivo. For example, as yet no studies have yet addressed the degree of overlap between cells within a specific lineage at different stages of differentiation, or similar cells at different stages of embryonic development. There is also no a priori reason to expect that enhancers as a general class of functional genomic element should necessarily exhibit the same histone methylation pattern. A study of human CD4+ T-cells that investigated 39 distinct chromatin-associated marks identified several different histone modifications that correlated with putative enhancers, but no single modification was completely predictive (Wang et al., 2008). Finally, despite studies such as this, dozens of histone modifications remain untested. Thus, a true enhancer census remains a subject of speculation.
Enhancers, Disease and Evolution
Assuming that each enhancer is marked by a nucleosome-free region that can extend for 200-300 bp, estimates of the abundance of enhancers suggest that they could comprise as much as 10% of the human genome. For comparison, the total extent of protein-coding sequences in the human genome is estimated at 2-3%. Mutations within protein-coding sequences are well established as a basis for human disease, with many thousands of known examples, and so it is unsurprising to find that mutations within enhancers can similarly lead to heritable disorders. For example, some instances of X-linked deafness type 3 are associated with loss of an enhancer region located 900 kb upstream of the POU3F4 gene (de Kok et al., 1996). An enhancer located within the first intron of the RET proto-oncogene occurs in a variant that confers a 20-fold increased risk for Hirschsprung's disease (Emison et al., 2005). Numerous other examples have been presented, indicating that non-coding distal regulatory sequences represent a target for mutations that can lead to disease (Kleinjan and van Heyningen, 2005; Visel et al., 2009; Noonan and McCallion, 2010).
Thus far, however, the proportion of known mutations that localizes to enhancer sequences does not align with the apparent complexity of the enhancer population in the genome. A very small percentage of mutations documented in the Human Gene Mutation Database map to noncoding DNA, and the majority of these correspond to promoter-proximal regions (Noonan and McCallion, 2010). In part, this may stem from the historical difficulty in mapping and characterizing gene-distal enhancers, and thus reflect an artificial bias for protein-coding regions. Another contributing factor, however, may be that enhancers are not as sensitive to single-base pair alterations as protein-coding sequences. While such changes have the potential to fundamentally alter transcription factor association and render an enhancer nonfunctional, they are more likely to alter binding affinities or developmental patterns of association in ways that can change function more subtly without eliminating it. While a mutation in a protein-coding region will affect function in every cell in which the protein is expressed, a mutation in an enhancer may affect only part of the expression pattern. Enhancer mutations are therefore a potentially powerful engine for intraspecies variation, and insofar as such changes affect selectable traits, for evolutionary divergence (Rebeiz et al., 2009; Visel et al., 2009; Levine, 2010; Noonan and McCallion, 2010).
Illustrations of the principle include freshwater stickleback fish populations, which lack pelvic fins that are present in ancestral fish and in modern-day, saltwater stickleback populations. Freshwater sticklebacks have lost an enhancer located 5′ of the gene for the homeobox transcription factor Pitx1. In stickleback fish that have pelvic fins, this enhancer specifically activates Pitx1 gene expression in the pelvic fins during development (Chan et al., 2010; Levine, 2010). Another study defined a series of mutations in an enhancer for the ebony gene in Drosophila (Rebeiz et al., 2009). Populations of Drosophila in Uganda exhibit a correlation between their abdominal pigmentation and the elevation of their primary habitat. Abdominal pigmentation is influenced by the product of the ebony gene, in the absence of which the abdomen exhibits a dark, melanic phenotype. An analysis of dark vs. light-colored lines resulted in the identification of an enhancer located 5′ of the ebony gene, mutations in which give rise to phenotypic variation.
Several studies have systematically attempted to define human-specific mutations in noncoding genomic DNA sequences that are otherwise highly conserved among mammalian species. Such analyses are able to define isolated sequences, termed “human-accelerated conserved noncoding elements” (HACNSs) or “human-accelerated regions” (HARs), that in some cases appear to be enhancers (Pollard et al., 2006; Prabhakar et al., 2006; Noonan, 2009). Notably, one element, HACNS1/HAR2, consists of 81 bp within which 16 human-specific substitutions have accumulated during evolution. When tested in reporter constructs in transgenic mice, the human version acts as an enhancer to drive gene expression in the developing anterior limb and a few other locations (Prabhakar et al., 2008). Versions of this element that lack the human-specific substitutions fail to direct gene expression to the developing limb. Although the biological significance of this expression pattern, including the gene(s) influenced by HACNS1/HAR2, is unknown, such behavior provides a concrete demonstration that changes in enhancer function can alter expression patterns to generate variation.
Mechanisms of Enhancer Function: Enhancer-Promoter Interaction
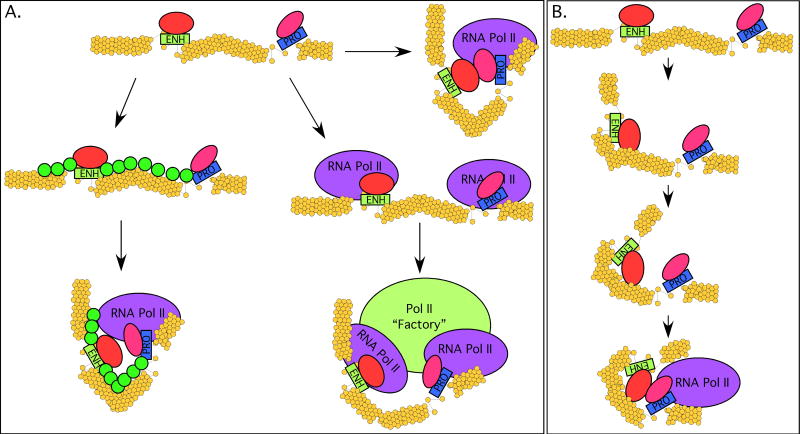
Since the discovery of enhancers, the dominant model for their mechanism of action on promoters has invoked direct interactions (Figure 1A). This model is commonly termed “looping”, since it requires that the intervening DNA be looped out or otherwise organized in order to permit the enhancer-promoter interaction (Bulger and Groudine, 1999; Blackwood and Kadonaga, 1998; de Laat et al., 2008). Alternative models for enhancer function have chiefly differed from the basic “looping” premise only in how the enhancer-promoter interaction is established – whether by free or facilitated diffusion within the nucleus, or by an active “scanning” or “tracking” mechanism (Figure 1B), in which the enhancer diffuses one-dimensionally along the chromatin fiber in search of a promoter (Blackwood and Kadonaga, 1998). In addition, more indirect models have been proposed, including “oozing” or “linking”, in which a complex is nucleated at the enhancer and then polymerizes along the chromatin fiber bidirectionally until it reaches a promoter (Ptashne, 1986; Dorsett, 1999; Bulger and Groudine, 1999). In one variation of this model, RNA polymerase II or other complexes are loaded at the enhancer and then actively move along the DNA until reaching a promoter.
Figure 1. Enhancer Looping and Variants.
(A) Simplified schematic of how enhancers might interact directly with promoters. Nucleosomes are shown as yellow circles, and the default state for chromatin is shown as a 30 nm fiber. Factors are bound to both the enhancer and promoter (red and pink ovals). These factors can potentially interact with each other. The simplest mechanism by which they might do this is via free diffusion in the nucleus (arrow 1). Enhancer-promoter interactions might be facilitated by additional factors (green circles) that bind to the intervening sequences and organize them to bring the enhancer and promoter into proximity (arrow 2). Alternatively, both enhancer and promoter can interact with RNA polymerase II, which then serves to bring the elements into proximity via association with a common RNA polymerase II transcription “factory” (arrow 3). (B) Tracking. The enhancer-bound complex (red oval) actively scans along the chromatin fiber until it encounters the promoter complex.
The “looping” model in particular has received abundant support from studies of nuclear architecture utilizing “chromosome conformation capture” (3C) and its high-throughput derivatives. In this assay, intact nuclei are crosslinked with formaldehyde, digested with restriction endonucleases, and then incubated with DNA ligase (Cullen et al., 1993; Dekker et al., 2002; Miele and Dekker, 2009). Using this procedure, specific genomic restriction fragments, which may be located far from each other on the linear genome, are found to ligate to each other with a greater frequency than other fragments if they are nevertheless colocalized within the three-dimensional space of the nucleus. Such colocalization can be revealed by the amplification of PCR products across the novel junctions produced during the ligation. Moreover, this approach can be adapted to produce genome-wide maps of the three-dimensional associations a specific locus makes among all the other sequences in the genome (“4C”) (Zhao et al., 2006; Simonis et al., 2006), or even associations among multiple sequences located throughout the genome (“5C” and “Hi-C”) (Dostie et al., 2006; Lieberman-Aiden et al., 2010).
3C has been employed to reveal interactions between distal sequence elements – primarily enhancers and promoters – within multiple loci in mammalian genomes (Miele and Dekker, 2008; de Laat et al., 2008). It is now common to find distal enhancers that colocalize with the promoters they regulate, which has uniformly been interpreted to be the result of direct enhancer-promoter interactions that are necessary for gene activation. Moreover, at multiple gene loci strong correlations have been made between active transcription and the ability to reveal such associations. Within the mammalian β-globin locus, for example, transcription factor knockouts that eliminate β-globin gene transcription uniformly result in the loss of colocalization of the gene with the β-globin LCR, as revealed by 3C (Drissen et al., 2004; Vakoc et al., 2005).
Additional support for direct communication between enhancers and promoters is provided by indications that enhancer-promoter interactions can be specific. As a general rule, enhancers are capable of activating transcription from heterologous promoters, and in fact in the majority of gain-of-function assays reporter gene expression follows the pattern governed by the enhancer, not the promoter. Some notable exceptions to this principle have been demonstrated, however. In the Drosophila Antennapedia gene complex, for example, an enhancer specifically mediates activation of the Sex combs reduced (Scr) gene despite the presence of another active gene, fushi tarazu (ftz), located between it and the Scr promoter (Calhoun et al., 2002; Calhoun et al., 2003). This interaction requires a “tethering” element near the Scr promoter, which can direct the enhancer to activate the ftz gene if it is moved to the ftz promoter instead. A similar element within the promoter of the Drosophila yellow gene renders heterologous promoters capable of being activated by the otherwise highly specific yellow gene enhancers (Melnikova et al., 2008). In addition, several studies have indicated that some enhancers can exhibit a preference for specific classes of gene promoters, for example between promoters that harbor a canonical TATA box vs. promoters that contain a DPE (Ohtsuki et al., 1998; Butler and Kadonaga, 2001). Transcription of the ftz gene is mediated by an enhancer bound by the transcription factor Caudal, which specifically activates genes with DPE-containing promoters (Juven-Gershon et al., 2008).
Mechanisms of Enhancer Function: Nuclear Organization
Evidence supporting enhancer-promoter interactions is part of a large and growing body of studies that have suggested that nuclear architecture is a major determinant of gene expression. On a global scale, for example, several generalized properties of genomic organization within the nucleus have been established, and enhancers have been shown to influence many of them.
First, each chromosome occupies a distinct “territory” within the nucleus, although up to 20% of the volume of the nucleus may be comprised of intermingling of neighboring chromosomes (Cremer and Cremer, 2010; Branco and Pombo, 2007). In addition, specific sequences on a given chromosome have been observed to extrude or “loop” out from the main body, and can even be found in other chromosomal territories (CTs). A study of the β-globin locus, for example, found that the region extruded from its CT specifically in erythroid cells (Ragoczy et al., 2003). This extrusion or looping from the CT occurred prior to high-level β-globin gene expression, and was dependent upon the presence of the β-globin LCR. Furthermore, ectopic integration of a β-globin LCR into a gene-dense region of the mouse genome resulted in more frequent extrusion/looping of the region away from its CT (Noordermeer et al., 2008). The limb bud enhancer of the Shh gene is similarly required for extrusion of the gene locus from its CT (Amano et al., 2009). These results suggest that some enhancers mediate a change in nuclear localization for genes in their vicinity, and that this represents a step distinct from transcriptional activation.
Second, in many metazoan cell types, active genes appear to be localized in the interior of the nucleus, while silent genes are found at the periphery. The correlation is far from absolute – the vicinity of the nuclear pore in yeast, for example, has actually been associated with active genes (Taddei, 2007) – and thus localization does not appear to determine expression state, but specific examples indicate that the nuclear periphery is a repressive environment. Outside of numerous correlative studies, some studies have shown that artificial tethering of reporter genes to the nuclear lamina or nuclear periphery results in downregulation of the reporter and of neighboring genes, although not all genes are affected (Andrulis et al., 1998; Finlan et al., 2008; Reddy et al., 2008). A link between this pattern and enhancer function is again provided by the β-globin locus. A study of β-globin locus positioning within the nucleus during erythroid differentiation demonstrated that at early maturational stages, the locus is found near the nuclear periphery (Ragoczy et al., 2006). As differentiation proceeds and β-globin transcription is activated, the locus moves more toward the interior of the nucleus. This process requires the presence of the β-globin LCR, although here it is not clear if relocalization is a function of the LCR directly or if it represents an indirect consequence of LCR-dependent β-globin gene activation. Separate studies of transgenes under the control of an enhancer derived from the β-globin LCR, however, indicated that at ectopic integration sites the enhancer was required for localization of the transgene far from regions of centromeric heterochromatin, and this was in turn associated with a higher, stochastically-determined probability of the gene being active at all (Francastel et al, 1999).
Third, the nucleus harbors a number of self-organized substructures, proximity to which can affect gene expression (Ferrai et al., 2010). In addition to the nuclear lamina, such substructures include nucleoli, Cajal bodies, PML bodies, splicing speckles and other features that represent concentrations of factors that can influence transcription – in the case of splicing speckles, for example, of the splicing machinery and other mRNA processing factors. Such substructures can serve as the basis for colocalization of genes that are otherwise located far apart on a linear chromosome, or even on different chromosomes. For example, erythroid-specific genes have been found to be positioned near common splicing speckles in erythroid cells (Brown et al., 2008), while several muscle-specific genes have been shown to localize to shared speckles in differentiated muscle cells (Moen et al., 2004).
Finally, gene loci can colocalize in the nucleus on the basis of shared associations with specific factors. Perhaps the most notable of these is with RNA polymerase II itself. Visualization using antibodies against RNA polymerase II or labeling of primary mRNA transcripts have suggested that transcription is localized to a limited number of RNA polymerase II “factories” (Iborra et al., 1996; Sutherland and Bickmore, 2009). Transcription factories have been proposed to underlie the observation that active gene loci distributed across single chromosomes, or even located on separate chromosomes, tend to colocalize in the nucleus (Osborne et al., 2004; Simonis et al., 2006). In fact, some studies have suggested that transcription factories occur in different varieties, corresponding to transcription mediated by different factors (Xu and Cook, 2008; Schoenfelder et al., 2010).
In addition to RNA polymerase II, other factors involved in transcriptional regulation appear to organize into discrete foci in the nucleus, and can either directly or indirectly bring distal gene loci into proximity with each other. These include, for example, special AT-rich sequence binding protein 1 (SATB1), a protein expressed in thymocytes and several other cell types that has been implicated in anchoring disparate genomic loci via long-range interactions (Cai et al., 2003; Cai et al., 2006). In thymocytes SATB1 is observed in a “cage-like” distribution around, but not coincident with, concentrations of centromeric heterochromatin. SATB1 in turn binds to promoter-distal regulatory elements in multiple gene loci, and loss of the factor results in disruptions of normal gene expression patterns and locus-wide chromatin structure.
A special case of this category of interaction is presented by CTCF, a zinc finger transcription factor that can have many roles in gene regulation. Notably, CTCF binding to insulator elements can block enhancer-mediated gene activation w hen such elements are located between an enhancer and promoter (Phillips and Corces, 2009). CTCF is capable of self-association (Pant et al., 2004; Yusufzai et al., 2004), and has been implicated in the formation of chromatin loops in vivo by 3C-based approaches (Kurukuti et al., 2006; Splinter et al., 2006; Hou et al., 2010). Interestingly, CTCF associates with cohesins, protein complexes that physically connect sister chromatids during mitosis and meiosis, and this association appears to be necessary for enhancer-blocking activity (Rubio et al., 2008; Wendt et al., 2008). A recent study of enhancers in embryonic stem cells demonstrated that enhancers and promoters were also associated with cohesin (Kagey et al., 2010), and the cohesin-loading factor Nipbl (Nipped-B in Drosophila) was one of two factors that emerged from a genetic screen in Drosophila for factors involved in enhancer-promoter communication (Rollins et al., 1999). Common association with cohesins provides a potential mechanistic link between long-range associations observed between CTCF binding sites and between enhancers and their cognate promoters.
Mechanisms of Enhancer Function: Alternatives and Issues
3C and its variants are inherently descriptive assays, and as with most studies of nuclear organization, they do not provide obvious ways to distinguish between correlation and causation. Thus, while the spatial colocalization of active enhancer and promoter regions revealed by 3C is suggestive of direct interactions that mediate gene activation, it is formally just as likely that it represents a consequence of a distinct activating mechanism. RNA polymerase II transcription factories, for example, provide an alternate mechanism by which enhancer-promoter colocalization might take place (Figure 1A). RNA polymerase II is recruited to many enhancers (Heintzman et al., 2007; Koch et al., 2008; Kim et al., 2010). Although the function of this recruitment, if any, is unknown, the likelihood that RNA polymerase II-bound enhancers and promoters might colocalize by virtue of coincidental association in such transcription factories has rarely elicited much commentary. In some cases – notably, the β-globin locus – it has been speculated that part of the role of the enhancer is to transfer RNA polymerase II to the promoter directly (Zhu et al., 2007; Leach et al., 2001), but this has not been demonstrated, and in the case of the β-globin locus RNA polymerase II still associates with the gene promoter in the absence of the LCR (Sawado et al, 2003).
The function of RNA polymerase II recruitment to enhancers, if it does not involve transfer to the promoter, is not clear, but recent studies have shown that enhancers themselves are transcribed. In one study, a population of ∼12,000 enhancers was defined by the combination of H3K4Me1 and binding of the transcription cofactor CBP (Kim et al., 2010). Roughly 25% of the enhancers defined in this fashion were found to be associated with RNA polymerase II, and this subset was in turn transcribed into RNA. The product RNAs, termed eRNAs, were short, bidirectional and not polyadenylated, and in at least one case, eRNA transcription required the presence of the target promoter. In another study, 70% of extragenic RNA polymerase II binding in primary macrophages was found to map to enhancers, as defined by the H3K4Me1 signature (De Santa et al., 2010).
Such findings complement earlier studies that have shown transcription originating from specific enhancers, for example at HS2 of the β-globin LCR (Kim et al., 2007; Zhu et al., 2007). In a few cases, non-genic transcription originating from enhancers has been shown to be necessary for normal gene activation. For example, at transgenic human growth hormone loci in mice, a large region located upstream of the growth hormone gene is transcribed in an enhancer-dependent fashion (Ho et al., 2006). Insertion of a transcriptional terminator in this region partially eliminates this non-genic transcription pattern and in turn results in a decrease in growth hormone gene expression.
Another study has suggested that some noncoding RNAs (ncRNAs) act as enhancers on neighboring genes (Orom et al., 2010). The authors identified a set of several thousand unique, long ncRNAs, and then focused on a subset that exhibited differential expression during keratinocyte differentiation. RNAi-mediated knockdown of several of these ncRNAs resulted in a decrease in expression of neighboring genes. Further analysis of one ncRNA, located near the gene for the transcription factor Snai1, revealed that it behaved as a classical enhancer in transient reporter gene assays, and furthermore that it was the RNA itself that was required for the effect. The ncRNAs characterized in this study appear to be distinct from eRNAs – the former, for example, are polyadenylated, while the latter are not.
The significance of eRNA transcription is not known, and the mechanism by which ncRNAs might mediate activation of neighboring (but distal) genes is similarly unclear. Such studies, however, reveal an unforeseen complexity to the roles of RNA polymerase II association with enhancers and of the association of enhancers with transcription factories, and suggest functions for noncoding RNA for which conventional models of enhancer-mediated gene activation currently do not account (Figure 2C).
Figure 2. Alternative mechanisms of enhancer function.
(A). “Placeholding”. In ES cells, the enhancer (ENH) is occupied by ES cell-specific transcription factors, but the gene promoter (PRO) is not active. In this representation, nucleosomes are designated by orange circles, and we depict the default state of chromatin as the 30 nm fiber. Upon differentiation along the lineage in which the gene is normally expressed, the ES cell-specific factors are downregulated, but new cell-specific factors occupy the enhancer in their place and mediate gene activation (bottom right). Upon differentiation into another lineage, the ES cell-specific factors are not replaced and the locus is inactivated (top right). (B) Spreading. The enhancer (ENH) is bound by a factor or factors that recruit chromatin-modifying or other activities (blue and yellow ovals), which then spread along the chromatin fiber bidirectionally until they reach a promoter. Histones may be modified throughout the region (nucleosomes shaded green), leading to the formation of a distinct chromatin “domain”. (C) Noncoding RNA. The enhancer (ENH) is bound by RNA polymerase II and transcribed. By as-yet undefined mechanisms, the noncoding RNA then mediates transcriptional activation of a neighboring, but still distal, gene promoter.
Several studies have provided evidence for a mechanism of enhancer function distinct from, or perhaps complementary to, the nuclear colocalization implied by 3C-based studies. Within the β-globin locus, some chromatin modifying enzymes – notably, the histone H3 lysine 4 methyltransferase MLL2 and the histone H3 lysine 9 methyltransferase G9a – have been shown to be recruited by association with the enhancer-binding factor NF-E2, and then to spread locus-wide (Demers et al., 2007; Chaturvedi et al., 2009). In addition, a novel class of enhancer activity is suggested by studies of enhancers within the endogenous mouse β-globin locus, and within a transgenic human growth hormone locus in mice. In both cases, the active genes are embedded within a larger “domain”, extending for 15-30 kb, defined by high levels of histone hyperacetylation. Deletion of binding sites for the transcription factor Pit-1 within the transgenic human growth hormone locus (Ho et al., 2002), or of an evolutionarily conserved enhancer located between the embryonic β-globin genes (G. Fromm and M. Bulger, submitted), results in complete loss or significant decreases in expression, respectively, coupled with complete loss of the hyperacetylated domain. Thus, enhancers can also control chromatin modifications that are distributed continuously over large regions, in a manner analogous to silencers that control heterochromatic domains (Figure 2B; see also below).
Another function for enhancers is emerging from studies of epigenetic marks in pluripotent embryonic stem (ES) cells (Figure 2A). In several cases, “pioneer” transcription factors have been found to associate with distal enhancers in ES cells, although they do not mediate gene activation. These factors are then lost upon ES cell differentiation. If differentiation proceeds along a lineage within which the enhancer is normally active, the “pioneer” factor is replaced with another factor that mediates enhancer-dependent gene activation. If differentiation proceeds along another lineage, loss of the “pioneer” factor results in inactivation of the enhancer and thus the entire gene locus. For example, an enhancer within the liver-specific Alb1 gene locus is bound by the transcription factor FoxD3 in ES cells; binding of FoxD3 prevents DNA methylation at this enhancer. Upon differentiation into endoderm, FoxD3 is replaced by FoxA1, the activating factor, and the Alb1 gene is transcribed (Xu et al., 2009). Differentiation into another lineage is accompanied by DNA methylation at the enhancer. Similar models have been presented for the macrophage/dendritic cell gene IL12b, the thymocyte-specific Ptcra gene (Xu et al., 2009), and for an enhancer within the pre-B cell-specific λ5/VpreB1 gene locus (Liber et al., 2010).
These examples suggest a general model in which developmental and lineage maturational decisions to activate or to silence a given tissue-specific gene locus are postponed until specific timepoints via the binding of “pioneer” transcription factors at enhancer elements. Factor binding to the enhancer thus acts as a “placeholder” for a later step in gene regulation. Although this mechanism for enhancer function does not involve direct activation of gene promoters, it is no less necessary for the proper regulation of transcription, and represents a novel, indirect method by which enhancers can affect gene activation.
Thus, while most studies of enhancer function have focused on models involving direct enhancer-promoter interactions and the assays that can reveal them, other studies indicate that regulation at a distance can involve indirect mechanisms as well. Transcription of enhancers, noncoding RNAs, enhancer “placeholding” and spreading mechanisms are likely to be important for the function of at least some enhancers, and observations of these phenomena raise the larger possibility that simple models of enhancer-promoter looping are not sufficient to account for enhancer activity.
Action at a Distance in Single-Celled Organisms
Such diversity of mechanism is anticipated by long-range regulatory mechanisms in organisms – prokaryotes and single-celled eukaryotes – in which such elements represent the exceptions rather than the rule. In the budding yeast S. cerevisiae, for example, regulatory sequences are usually limited to upstream activation sequences (UASs) that are located within a few hundred base pairs of the promoter. Relocation of a UAS at greater distances results in loss of function (Dobi and Winston, 2007).
Artificial model systems in yeast, however, have shown that bridging of distal sequences can be accomplished. Integration of expression cassettes within yeast telomeres, which naturally fold and can bring distal elements within close proximity of each other, or use of the self-associating Drosophila GAGA factor to mediate looping between two sequences, have demonstrated that gene activation can be accomplished by simple looping interactions (de Bruin et al., 2001; Petrascheck et al., 2005).
In addition, regulatory elements that appear to act like enhancers – activation at a distance and/or from positions downstream of the promoter – have been found at select gene loci in S. cerevisiae. The most notable example in this regard is the gene for the HO endonuclease, an enzyme involved in mating-type switching that is expressed in a brief window during late G1 phase of the cell cycle. The process of HO gene activation requires binding of the transcription factor Swi5 to two sites located more than 1 kb upstream of the promoter (Figure 3A). Swi5 in turn is required for recruitment of chromatin remodeling factors, and results in histone acetylation across the entire region between the binding sites and the promoter. Interestingly, binding of Swi5 is a transient event, and HO gene activation occurs well after Swi5 is no longer present, suggesting that these distal regulatory elements initiate an activation mechanism that is maintained by chromatin remodeling at the promoter in an indirect fashion (Cosma et al., 1999; Krebs et al., 1999).
Figure 3. Mechanisms of distal regulatory element function in yeast and bacteria.
(A) HO gene activation in yeast. The locus is diagrammed in the top panel, with the gene represented by the green box (“HO”), nucleosomes by orange circles, and binding sites for Swi5p, located >1 kb upstream of the gene, by open boxes. Swi5p enters the nucleus and binds its cognate sites in late anaphase of the cell cycle (second panel), and is required for recruitment of the SWI/SNF chromatin remodeling complex. SWI/SNF recruitment in turn leads to later recruitment, in mid- to late telophase, of the acetyltransferase-containing SAGA complex, which acetylates nucleosomes throughout the region (yellow circles), and in turn recruits additional factors (Swi4/Swi6) required for gene activation. Swi5p, however, is present only transiently and does not participate directly in these subsequent steps. (B) Mating-type silencing in S. cerevisiae. In this simplified representation, a discrete silencer element is bound by several factors (ORC, Abf1p, Rap1p) which in turn recruit a complex of SIR proteins that includes a histone deacetylase. Deacetylated nucleosomes (orange circles) are in turn recognized by the SIR complex, which can then deacetylate additional nucleosomes, resulting in a progressive spread of histone deacetylation and associated SIR complexes. Genes within the region are silenced. (C) Enhancer-binding protein interactions with σ54-dependent promoters in bacteria. An enhancer sequence (ENH) is recognized by enhancer-binding protein (EBP), and in turn the promoter (PRO) is recognized by a σ54 holoenzyme that includes RNA polymerase. This closed complex is only activated upon direct interaction between the EBP and σ54 (bottom), which can be facilitated by DNA bending mediated by integration host factor (IHF) binding between the enhancer and promoter. (D) Activation of phage T4 late genes by a tracking complex. A sliding clamp, consisting of a trimer of gp45 polypeptides, is loaded at a promoter-distal site by the gp44-gp62 complex. The gp45 trimer then tracks or slides along the DNA until it reaches the promoter, where it mediates gene activation. In this representation, RNA polymerase and additional factors (gp33 and gp55) are present at the promoter in a closed complex prior to the arrival of gp45, but this is not known; in other models the polymerase can track with gp45, for example.
Studies in yeast also provide a cautionary note that may be applied to enhancer studies in metazoans. For example, early investigations of the S. cerevisiase rRNA genes identified an apparent transcriptional enhancer located downstream of the 35S rRNA gene. In transgene constructs, this enhancer region mediated 10- to 30-fold increases in 35S rRNA transcription, and was also selective, having no effect on the 5S rRNA gene located between the enhancer region and the 35S rRNA promoter (Neigeborn and Warner, 1990; Morrow et al., 1993). The results suggested a simple model in which the enhancer interacted with the 35S rRNA gene promoter by a looping mechanism, with the intervening 5S gene isolated within the loop. Subsequent studies, however, demonstrated that the enhancer was completely unnecessary for normal 35S rRNA expression (Wai et al., 2001), and instead functioned in ectopically integrated reporter constructs to recruit RNA polymerase I, which is otherwise primarily localized within the nucleolus. Such findings sound a cautionary note for any study in which the potential function of a distal enhancer is investigated outside of its normal context.
Finally, yeast heterochromatin provides a well-established model for action at a distance in single-celled eukaryotes. Notably, the simplest version of this model, exemplified by silencing at the yeast mating-type gene loci, is entirely indirect: silencer elements within the locus serve to nucleate a complex of Sir proteins (Rusche et al., 2003). The complex includes the histone deacetylase Sir2, which deacetylates the histone tails of nearby nucleosomes. Other components of the complex bind with high affinity to deacetylated histone tails, and then serve to recruit more of the Sir2 deacetylase, resulting in a progressive spreading of histone deacetylation and silencing factors along the chromatin fiber (Figure 3B). In this way DNA sequences that in themselves do not encode information sufficient for silent regulation are packaged in a repressive chromatin structure. It has been shown that once established, silencing is maintained in yeast cells even after the silencer itself is excised by an in vivo recombination strategy, although the silencer is still required for re-establishment of the silent state after cell division (Holmes and Broach, 1996).
Silent domains in yeast, however, can be established and maintained discontinuously. For example, silent domains at yeast telomeres or within the HMR locus can encompass artificially inserted active genes. Such behavior involves the activity of “proto-silencer” elements located elsewhere in the domain that have no activity by themselves, but can augment the activity of canonical silencers (Talbert and Henikoff, 2006). Recently, it has been demonstrated that silencers within the HMR locus in yeast colocalize, as determined by 3C (Valenzuela et al., 2008). Thus far, however, it is not clear if discontinuous silencing requires such direct interactions, or if it occurs indirectly by localization to nuclear subcompartments enriched in silencing factors. Such issues directly parallel corresponding aspects of enhancer function, as described above.
Studies of enhancer elements in bacteria have provided elegant demonstrations of both the looping and tracking models (reviewed by Xu and Hoover, 2001). A number of bacterial genes regulated by the σ54-holoenzyme are also controlled by factors that bind to enhancer sequences that can be located up to 3 kb upstream and 1.5 kb downstream of the promoter. These enhancer binding proteins then interact directly with the σ54-holoenzyme via DNA looping, and notably, in some cases the looping mechanism is facilitated by additional factors that bind between the enhancer and promoter and bend DNA (Figure 3C). In contrast, a distal enhancer that is required for activation of phage T4 late genes functions by the loading of a ring-shaped trimer of a factor termed gp45, followed by one-dimensional diffusion of the trimeric protein along the DNA until it reaches the promoter (Figure 3D).
In summary, studies in organisms that typically lack enhancer activity indicate that distal sequences can affect gene expression by widely different mechanisms. In considering such mechanisms, there is no reason to exclude the possibility that any given enhancer might work by a combination of them. Thus, such studies argue against simple, unified models for enhancer function in metazoans.
Conclusion
Enhancers were discovered nearly 30 years ago, and for most of the intervening period they existed in the experimental literature largely as an odd feature of certain tissue-specific loci, with a peculiar ability to activate transcription from promoters over large genomic distances. It is only recently that advances in technology and understanding have provided a fuller delineation of the role distal enhancers play in gene regulation in metazoans. Their function is not only likely to constitute a primary basis for differential gene expression that underlies cell-type specificity, but also to have crucial roles in stem cell pluripotency, human disease and metazoan evolution. Mechanistically, enhancers lie at the nexus of transcription, nuclear organization, chromatin structure, epigenetics and noncoding RNA. In accordance with such a complex spectrum of biological functions, it seems unlikely that enhancers constitute a monolithic class of regulatory element that works via a single, unified mechanism.
Acknowledgments
We are grateful to M. Bender, M. Conerly, R. Kamakaka, J. Palis, T. Ragoczy, H. Rincon-Arano. J. Ritlund, and D. Strongin for critical reading of the manuscript and helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Chromosome organization: new facts, new models. Curr Biol. 2007;17:127–134. doi: 10.1016/j.tcb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for longdistance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Calhoun VC, Levine M. Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc Natl Acad Sci USA. 2003;100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci USA. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jónsson B, Schluter D, Bell MA, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi CP, Hosey AM, Palii C, Perez-Iratxeta C, Nakatani Y, Ranish JA, Dilworth FJ, Brand M. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci USA. 106:18303–18308. doi: 10.1073/pnas.0906769106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor Perspect Bio. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- de Bruin D, Zaman Z, Liberatore RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- de Kok YJ, Vossenaar ER, Cremers CW, Dahl N, Laporte J, Hu LJ, Lacombe D, Fischel-Ghodsian N, Friedman RA, Parnes LS, Thorpe P, Bitner-Glindzicz M, Pander HJ, Heilbronner H, Graveline J, den Dunnen JT, Brunner HG, Ropers HH, Cremers FP. Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet. 1996;5:1229–1235. doi: 10.1093/hmg/5.9.1229. [DOI] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. Three-dimensional organization of gene expression in erythroid cells. Curr, Top, Dev, Biol. 2008;82:117–139. doi: 10.1016/S0070-2153(07)00005-1. [DOI] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC. The formation and function of DNaseI hypersensitive sites in the process of gene activation. J Biol Chem. 1988;263:19259–19262. [PubMed] [Google Scholar]
- Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. Gene positioning. Cold Spring Harbor Perspect Biol. 2:a000588. doi: 10.1101/cshperspect.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C, Walters MC, Groudine M, Martin DI. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;29:259–969. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global celltype- specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Cooke N, Liebhaber S. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Liebhaber SA, Cooke NE. Locus control region transcription plays an active role in long-range gene activation. Mol Cell. 23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Holmes SG, Broach JR. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon R, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Zhao H, Ifrim I, Dean A. Beta-globin intergenic transcription and histone acetylation dependent on an enhancer. Mol Cell Biol. 2007;27:2980–2986. doi: 10.1128/MCB.02337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucl Acids Res. 2006;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Jourquin F, Ferrier P, Andrau JC. Genome-wide RNA polymerase II: not genes only! Trends Biochem Sci. 2008;33:265–273. doi: 10.1016/j.tibs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach KM, Nightingale K, Igarashi K, Levings PP, Engel JD, Becker PB, Bungert J. Reconstitution of human beta-globin locus control region hypersensitive sites in the absence of chromatin assembly. Mol Cell Biol. 2001;21:2629–2640. doi: 10.1128/MCB.21.8.2629-2640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff EA. long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7:114–126. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. 10. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromosoma. 2008 Apr;117(2):137–45. doi: 10.1007/s00412-007-0132-6. Epub 2007 Nov. [DOI] [PubMed] [Google Scholar]
- Melnikova L, Kostuchenko M, Silicheva M, Georgiev P. Drosophila gypsy insulator and yellow enhancers regulate activity of yellow promoter through the same regulatory element. Chromosoma. 2008;117:137–145. doi: 10.1007/s00412-007-0132-6. [DOI] [PubMed] [Google Scholar]
- Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Dekker J. Mapping cis- and trans- chromatin interaction networks using chromosome conformation capture (3C) Methods Mol Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Ley TJ. Conservation of the primary structure, organization, and function of the human and mouse beta-globin locus-activating regions. Proc Natl Acad Sci U S A. 1990;87:7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, McCallion AS. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen PT, Johnson CV, Byron M, Shopland LS, de la Serna IL, Imbalzano AN, Lawrence JB. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Cell Biol. 2004;15:197–206. doi: 10.1091/mbc.E03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow BE, Johnson SP, Warner JR. The rRNA enhancer regulates rRNA transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1283–1289. doi: 10.1128/mcb.13.2.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Warner JR. Expression of yeast 5S RNA is independent of the rDNA enhancer region. Nucl Acids Res. 1990;18:4179–4184. doi: 10.1093/nar/18.14.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP. Regulatory DNAs and the evolution of human development. Curr Opin Genet Dev. 2009;19:557–564. doi: 10.1016/j.gde.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Noonan JP, McCallion AS. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet. 2008;4:e1000016. doi: 10.1371/journal.pgen.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long Noncoding RNAs with Enhancer-like Function in Human Cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucl Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin E. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, Pennacchio LA, Rubin EM, Noonan JP. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010 doi: 10.1038/nature09692. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosphila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M. The beta -globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between coregulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Goodwin A, Joyce T, Lowrey CH. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nature Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A. Active genes at the nuclear pore complex. Curr Opin Cell Biol. 2007;19:305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nature Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT. Long-range communication between the silencers of HMR. Mol Cell Biol. 2008;28:1924–1935. doi: 10.1128/MCB.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai H, Johzuka K, Vu L, Eliason K, Kobayashi T, Horiuchi T, Nomura M. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol Cell Biol. 2001;21:5541–5553. doi: 10.1128/MCB.21.16.5541-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VLJ, Fisher EMC, Tavare S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNaseI. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Hoover TR. Transcriptional regulation at a distance in bacteria. Curr Opin Microbiol. 2001;4:138–144. doi: 10.1016/s1369-5274(00)00179-x. [DOI] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucl Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]