Abstract
Transmissible spongiform encephalopathies (TSEs) or prion diseases are characterized by the accumulation of an aggregated isoform of the prion protein (PrP). This pathological isoform, termed PrPSc, appears to be the primary component of the TSE infectious agent or prion. However, it is not clear to what extent other protein co-factors may be involved in TSE pathogenesis or whether there are PrPSc-associated proteins which help to determine TSE strain-specific disease phenotypes. We enriched PrPSc from the brains of mice infected with either 22L or Chandler TSE strains and examined the protein content of these samples using nanospray liquid chromatography-tandem mass spectrometry. These samples were compared to “mock” PrPSc preparations from uninfected brains. Prion protein was the major component of the infected samples and ferritin was the most abundant impurity. By contrast, mock enrichments contained no detectable prion protein but did contain a significant amount of ferritin. Of the total proteins identified, 32% were found in both mock and infected samples. The similarities between PrPSc samples from 22L and Chandler TSE strains suggest that the non-PrPSc protein components found in standard enrichment protocols are not strain-specific.
Keywords: Transmissible spongiform encephalopathies, TSE, Prion protein, prion strains, Chandler, LC-MS, MS, 22L, proteomics
1 Introduction
Transmissible spongiform encephalopathies (TSEs) or prion diseases are a group of neurodegenerative disorders affecting humans and other mammals [1,2]. A central event in the prion disease process is the conversion of the normal host-encoded prion protein (PrPC) into a pathological form, termed PrPSc [1]. Relative to PrPC, the PrPSc isoform is aggregated, more protease-resistant, and significantly higher in β-sheet structure [3,4]. TSEs are typically characterized post-mortem by intense vacuolation of brain tissue, gliosis, and the deposition of PrPSc aggregates, occasionally in the form of amyloid plaque [2]. Similar plaque deposition is seen in other protein misfolding disorders such as Alzheimer’s and Parkinson’s disease [5], but only TSEs are transmissible between mammals [1].
Another distinguishing feature of TSEs is the existence of unique strains [6]. Strains in TSE disease are defined by distinct clinical features, including disease incubation time and neuropathology, as well as by biochemical features such as PrPSc-specific conformations and glycosylation patterns [7]. Multiple TSE strains have been maintained in mice with a single PrP genotype [6]. Although a definitive mechanism for TSE pathogenesis and strains has not yet emerged, the widely accepted prion hypothesis provides a key constraint by arguing that PrPSc is the sole infectious agent and propagates by binding to PrPC in order to form new PrPSc [8]. Recent evidence from in vitro studies [9], including reports describing the de novo generation of TSE infectivity [10,11], seem to support the prion hypothesis. However, it remains unclear to what extent other molecules are required for the conversion of PrPC to PrPSc and whether such molecules are TSE strain specific.
One strategy would be to examine which proteins co-purify with PrPSc derived from different TSE strains. Researchers have developed a variety of methods over the last three decades that significantly enrich PrPSc from infected animal tissue [12–24]. Such enriched PrPSc mixtures have been reported to contain a wide variety of additional components, such as nucleic acids [17,23], polysaccharides [25], fatty acids [26] and ferritin [17,24]. Most recently, protein identification approaches using tandem mass spectrometry have found multiple proteins that co-enriched with PrPSc [27,28]. It is not yet clear whether any of these non-PrP proteins are specific to the enrichment protocol itself, whether they contribute to pathogenesis, or whether they are TSE-strain specific. Nevertheless, enriched PrPSc mixtures are highly infectious [17,18,21,29]. Thus, if there are molecules other than PrPSc required for TSE transmission, they should be present in these highly enriched samples.
We have isolated PrPSc from 22L and RML/Chandler infected mouse brains and compared these to mock samples enriched from age-matched uninfected brains using nanospray liquid chromatography tandem mass spectrometry (LC-MS/MS). PrPSc was consistently found in every infected sample but not in any of the uninfected samples. A significant portion of the total protein identifications in this study were common to both the infected and non-infected samples. Proteins other than PrPSc that were found exclusively in the infected preparations were variable and no protein was uniquely associated with every sample from one TSE strain but not the other. Our data suggest that TSE strain-specific phenotypes are not determined by non-PrP proteins.
2 Materials and Methods
2.1 Reagents and Supplies
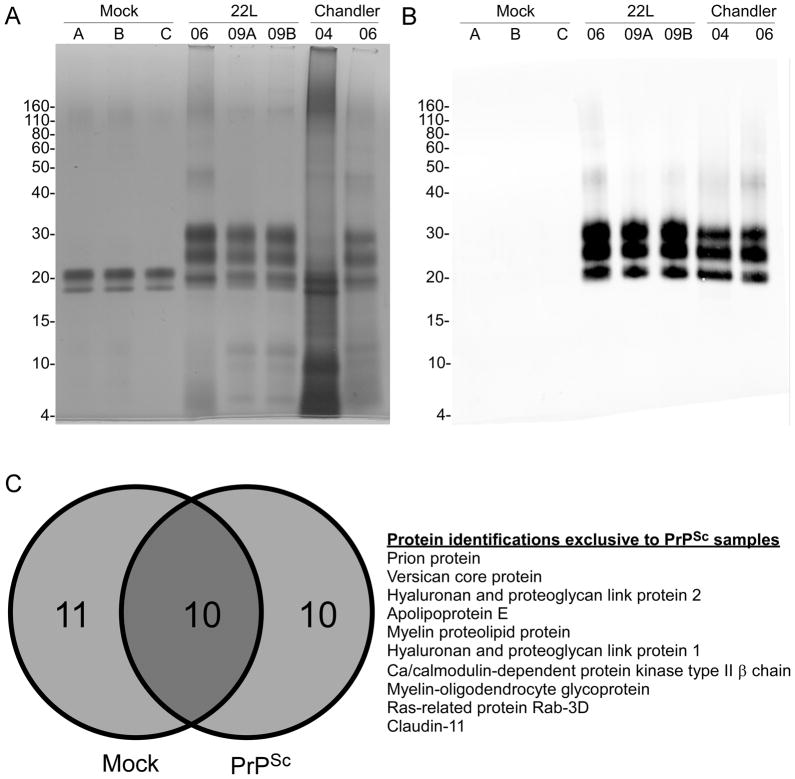
Dithiothreitol, iodoacetamide, EDTA and NaCl solutions, detergent SB3-14, tributylphosphine and membrane solubilization buffer were purchased from Sigma (St. Louis, MO, USA). Trypsin (#V5111) was purchased from Promega (Madison, WI, USA). Protease inhibitors were from Roche Diagnostics (Indianapolis, IN, USA). Burdick & Jackson brand water and acetonitrile were purchased from VWR (Pittsburg, PA, USA). Formic acid (FA) ampules and Imperial Coomassie blue stain were purchased from Thermo-Fisher Scientific (Pittsburg, PA, USA). SDS-PAGE was run using reagents and gels from Life Technologies-Invitrogen Corporation (Carlsbad, CA, USA), with NuPAGE Bis-Tris gels. The PlusOne Silver Stain kit (GE Healthcare) was used for staining total protein (Figure 1A).
Figure 1.
Comparison of mock, 22L or Chandler-derived PrPSc samples. A) Approximately 3.5 μg of total protein from mock, 22L and Chandler samples were loaded for SDS-PAGE and silver stained. B) For immunoblot, 10-fold less protein was loaded and probed with monoclonal antibody 6D11 as described in the methods. C) Venn diagram of protein distribution in mock and PrPSc samples (left panel). A total of 21 (68%) of the 31 total proteins could be identified in the absence of PrPSc. The 10 proteins that were uniquely associated with PrPSc and not identified in the mock samples are listed in the right panel in order of relative apparent abundance as estimated by the number of assigned spectra for each protein.
2.2 Preparation of enriched PrPSc
Enriched PrPSc samples were prepared from TSE-infected brain tissue by the method of Bolton [18] with minor modifications [19]. All of the PrPSc was derived from mice that displayed clear signs of TSE disease at the time of euthanasia. Samples of 22L and RML/Chandler PrPSc prepared in 2006 (22L-06, CH-06) and 2004 (CH-04) were generous gifts from Dr. Byron Caughey. Non-infected 7 – 9 month old age-matched C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Brain tissues from these age-matched control mice were subjected to the same protocol as the infected samples. Three non-infected mock preparations were done in parallel and termed A, B, and C. The final enriched pellets were kept suspended in 0.5% SB3-14/PBS at a concentration of 0.5 – 1 mg/mL total protein at 4 °C. Protein concentrations of enriched PrPSc samples were estimated by the BCA method (Thermo-Fisher Scientific).
2.3 SDS-PAGE and in-gel digestion
Each sample was diluted 4-fold with membrane solubilization buffer (7M urea, 2M thiourea, 1% C7BzO; Sigma #C0356) and incubated at 37 °C for 60min. The sample was reduced with 15mM DTT and 3mM tributylphosphine, and then alkylated with 74mM iodoacetamide for 30 min in the dark. Remaining iodoacetamide was quenched by the addition of DTT to a final concentration of 90mM. Invitrogen LDS sample buffer (4X) was added, resulting in a final dilution of 7-fold from the original sample. Each sample was subjected to centrifugation (22,000 g) for 5 min prior to electrophoresis using NuPAGE 10% Bis-Tris 1.5mm gels with MES running buffer using 175V constant voltage. Gels used for analysis by LC-MS/MS were stained with Coomassie blue Imperial stain (Thermo-Fisher Scientific) and destained with water prior to choosing bands. Stained bands from each of the 3 mock, 22L (09-A, 09-B, −06), and Chandler (−06 and −04) were selected for in-gel digestion from a single lane of SDS-PAGE (gel images shown in Supplementary Figure 1). Final digestion of each band was conducted overnight in a mixture of 10% trifluoroethanol in 50mM ammonium bicarbonate, pH 8.0 at 37°C for 16 hrs. The digests were quenched with 8% FA, then transferred to another vial and dried in a centrifugal vacuum concentrator. Each digest was then dissolved in 14 μL of LC buffer A (water/3% ACN/0.1% FA), subjected to centrifugation at 22,000 g and the upper 12 μL transferred to an autosampler vial for nanospray LC-MS/MS analysis. This process was repeated at least 3 times per sample and the identifications in Table 1 are a cumulative representation of more than 340 LC/MS-MS runs.
Table 1.
SEQUEST-derived protein identifications from mock and PrPSc samples. All proteins listed had at least two unique peptides derived from MS/MS spectra passing discriminate score cutoffs as described in the methods section. Keratin and trypsin sequences were filtered from the list. The total number of assigned spectra and the number of unique peptides (e.g, the number of peptides with distinct m/z values) were cumulatively tabulated from all of the samples examined. Proteins are listed in order of relative apparent abundance based upon spectral counts for the each of the proteins listed.
| Protein (Swiss-prot) | Spectral Count | Unique Peptides | 22L-06 | 22L-09A | 22L-09B | CH-04 | CH-06 | Mock |
|---|---|---|---|---|---|---|---|---|
| Major Prion protein (P04925) | 1639 | 61 | √ | √ | √ | √ | √ | |
|
|
|
|
||||||
| Ferritin light chain 1 (P29391) | 946 | 30 | √ | √ | √ | √ | √ | √ |
|
|
|
|
||||||
| Ferritin heavy chain (P09528) | 867 | 35 | √ | √ | √ | √ | √ | √ |
|
|
|
|
||||||
| Versican core protein (Q62059) | 94 | 14 | √ | √ | √ | |||
|
|
|
|
||||||
| Myelin-associated oligodendrocyte basic protein (Q9D2P8) | 90 | 6 | √ | √ | √ | √ | ||
|
|
|
|
||||||
| Hyaluronan and proteoglycan link protein 2 (Q9ESM3) | 80 | 17 | √ | √ | ||||
|
|
|
|
||||||
| Proteasome subunit beta type-5 (O55234) | 74 | 11 | √ | |||||
|
|
|
|
||||||
| Apolipoprotein E (P08226) | 63 | 7 | √ | √ | √ | √ | ||
|
|
|
|
||||||
| Collagen alpha-1(I) chain (P11087) | 56 | 2 | √ | √ | √ | √ | ||
|
|
|
|
||||||
| Proteasome subunit beta type-2 (Q9R1P3) | 43 | 7 | √ | |||||
|
|
|
|
||||||
| Ca/calmodulin-dependent protein kinase type II α chain (P11798) | 42 | 7 | √ | √ | √ | √ | √ | |
|
|
|
|
||||||
| Proteasome subunit beta type-4 (P99026) | 38 | 5 | √ | |||||
|
|
|
|
||||||
| Proteasome subunit beta type-1 (O09061) | 33 | 7 | √ | |||||
|
|
|
|
||||||
| Proteasome subunit alpha type-7 (Q9Z2U0) | 29 | 7 | √ | √ | ||||
|
|
|
|
||||||
| Myelin proteolipid protein (P60202) | 26 | 3 | √ | √ | √ | |||
|
|
|
|
||||||
| Proteasome subunit beta type-3 (Q9R1P1) | 22 | 5 | √ | |||||
|
|
|
|
||||||
| Hyaluronan and proteoglycan link protein 1 (Q9QUP5) | 21 | 8 | √ | |||||
|
|
|
|
||||||
| 60S ribosomal protein L35 (Q6ZWV7) | 19 | 3 | √ | √ | √ | √ | √ | √ |
|
|
|
|
||||||
| Ca/calmodulin-dependent protein kinase type II β chain (P28652) | 15 | 4 | √ | √ | √ | √ | ||
|
|
|
|
||||||
| Proteasome subunit alpha type-6 (Q9QUM9) | 13 | 5 | √ | |||||
|
|
|
|
||||||
| Proteasome subunit alpha type-4 (Q9R1P0) | 11 | 6 | √ | |||||
|
|
|
|
||||||
| Proteasome subunit alpha type-2 (P49722) | 10 | 5 | √ | |||||
|
|
|
|
||||||
| Proteasome subunit beta type-6 (Q60692) | 10 | 3 | √ | |||||
|
|
|
|
||||||
| Myelin-oligodendrocyte glycoprotein (Q61885) | 8 | 2 | √ | √ | ||||
|
|
|
|
||||||
| Junction plakoglobin (Q02257) | 8 | 4 | √ | √ | √ | √ | ||
|
|
|
|
||||||
| 60S ribosomal protein L13a (P19253) | 6 | 2 | √ | √ | √ | √ | ||
|
|
|
|
||||||
| Actin, cytoplasmic 1 (P60710) | 6 | 5 | √ | √ | ||||
|
|
|
|
||||||
| Beta-actin-like protein 2 (Q8BFZ3) | 5 | 4 | √ | |||||
|
|
|
|
||||||
| Ras-related protein Rab-3D (P35276) | 4 | 2 | √ | |||||
|
|
|
|
||||||
| 60S ribosomal protein L3 (P27659) | 3 | 2 | √ | |||||
|
|
|
|
||||||
| Claudin-11 (Q60771) | 3 | 3 | √ | √ | ||||
2.4 Western blotting
Proteins were separated by SDS-PAGE as described above and transferred to PVDF membrane by wet transfer using Towbin’s buffer. Samples were probed with mouse monoclonal antibody 6D11 (Covance, Emeryville, CA, USA) at a dilution of 1:15,000. 6D11 recognizes residues 93–109 of mouse PrP. The secondary antibody was IRDye 800CW Goat Anti-Mouse (Li-Cor Biosciences, Lincoln, NE, USA) used at a 1:15,000 dilution in TBST buffer. Imaging was performed with an Odyssey imaging system (Li-Cor) with PrPSc visualized fluorescently in the 800 channel. The proteasome and ferritin immunoblots were done using the ECL Advance kit (GE) according to the directions of the manufacturer. Proteins were transferred to PVDF at 14 Volts/cm for one hour and the membrane was probed for proteasomal subunits using primary antibody MCP196 at 1:1000 and a 1:200,000 dilution of horseradish peroxidase (HRP) labeled anti-mouse IgG secondary antibody. For the detection of ferritin, samples that had not been urea denatured and reduced/alkylated were boiled in LDS sample buffer (Invitrogen) for 5min and then loaded onto 4–12% gradient NuPAGE gels for electrophoresis and transferred to PVDF membrane (Millipore). Anti-ferritin heavy chain antibody (Abcam #71562) was used at 1:5000, HRP-labeled anti-rabbit IgG secondary antibody at 1:400,000, and Streptactin-HRP (BIO-RAD) at 1:400,000. The Streptactin allows visualization of tagged markers in the molecular weight standard (Western C, BIO-RAD). After one hour of post secondary antibody wash in TBST buffer, the blot was developed on film with ECL Advance. The PVDF membrane used for ferritin western blot was then washed in TBST, and re-probed with anti-PrP monoclonal antibody 6D11 at 1:10,000, anti-mouse IgG at 1:200,000, and Streptactin-HRP (BIO-RAD) at 1:400,000. Western C molecular weight standards were used to align scanned images of the ferritin and PrP stains.
2.5 HPLC chip-based nanospray LC-MS/MS
Proteins from the tryptic digests were identified by LC-MS/MS using an Agilent 1200 connected to an XCT Ultra Ion Trap via a microfluidic HPLC chip interface and a nanospray source. The tryptic digests were loaded onto the chip (Agilent #G4240-62001, Zorbax 300SB-C18, 5μm, 75μm × 43mm) with an autosampler and washed with Buffer A (3% acetonitrile/H2O and 0.1% formic acid) prior to elution at 300nl/min by reverse-phase chromatography. The gradient was 6 – 25% Buffer B (90% acetonitrile and 0.1% formic acid) over 18 min, 25% B for an additional 4 min, 25 – 50% B in 8 min, 50 – 80% B in 2 min, 80 – 97% B in 4min at 300nL/min, followed by post-run re-equilibration at 3% B, with a total run time of 38 min. A blank run consisting of an injection of buffer A preceded each analysis in order to rule out contamination from previous runs.
The ion trap was calibrated externally using a tuning mix provided by Agilent specifically for this instrument. Data-dependent MS acquisition was performed with dry gas (nitrogen/air) set to 6L/min at 350 °C, MS capillary voltage 1800V, maximum accumulation time 150ms, with a maximum target of 100,000. The MS scan range was set to 300 – 1400 m/z in the Ultrascan mode. Four parent ions were selected for each MS/MS cycle with a fragmentation amplitude of 1.0V and the SmartFrag setting on. Spectra were actively excluded for fragmentation after 2 spectra in order to facilitate detection of less abundant ions. Separate HPLC chips were used for infected and non-infected samples in order to minimize the possibility of contamination.
2.6 Data analysis and database searching
Post-run data were deconvoluted and peak lists were exported as MGF files using an automated algorithm that was part of the Ion Trap data analysis software, version 3.4, Rev 6.1. These data were searched against the Swiss-prot database in independent searches using both the SEQUEST [30] and MASCOT [31] search engines. For the SEQUEST-based analysis, a mouse species subset of the Sprot protein database (v57.4; 16,140 proteins) was prepared and analysis software [32] was used to calculate Peptide Prophet-like [33] discriminant scores and filter out incorrect peptide identifications using sequence-reversed matches from a decoy database to estimate false discovery rates, which ranged from 1.4 to 3.4% (Supplementary Table 2). No reversed proteins are reported and keratin sequences were excluded. A parent ion mass tolerance of 2.5 Da was used with a 1.0 monoisotopic fragment ion mass tolerance in no enzyme specificity searches. Cysteine was assigned a static modification mass of +57 Da and methionine had a variable modification mass of +16 Da. Proteins were considered present in a given sample if they had two or more peptides with distinct sequences, having a unique count greater than or equal to one in the respective sample. Peptides had to be consistent with fully- or semi-tryptic cleavage since protein fragments produced from previous PK treatment may have resulted in N- or C-terminal peptides having only a single tryptic cleavage site. Total protein spectral counts were corrected by partitioning counts of peptides shared between proteins based upon relative unique-peptide-per-protein evidence. Corrected counts were also normalized so that the total numbers of identified peptides in each sample were the same.
Confirmatory MASCOT searches were run with the MGF files described above using Mascot Daemon with parameters set to semi-trypsin enzyme specificity, allowing for one missed cleavage site with cysteine carbamidomethylation as a fixed modification and methionine oxidation as a variable modification and the taxonomy restricted to mouse proteins. The peptide and MS/MS tolerances were set to 2.4 and 1.4 Da, respectively, and all searches were concomitantly run against a decoy database. The Mascot search results were visualized using Scaffold software version 2_03_01 (Proteome Software, Portland, OR, USA). Protein identifications were accepted only if they could be established at greater than 99% probability from Scaffold’s implementation of the Protein Prophet algorithm [34] and contained two or more unique peptides.
3 Results
3.1 Preparation of PrPSc and mock samples
Two mouse-adapted TSE agents with distinct pathologies were selected for this study. RML/Chandler (Chandler) causes vacuolation in various regions of the cerebrum while 22L presents with severe vacuolation in the cerebellum and cerebellar cortex [35,36]. Freshly enriched 22L PrPSc (22L-09A, 22L-09B) was compared to PrPSc samples that had been prepared earlier (22L-06, CH-06, CH-04) by the same protocol [19] in order to identify TSE-associated protein signatures. The key steps in this procedure include a low speed centrifugation in which bulk contaminants are pelleted, followed by higher speed centrifugations in which PrPSc itself pellets and soluble contaminants do not. Subsequent treatment with Proteinase K (PK) removes a majority of the remaining proteins, leaving behind highly concentrated PrPSc, which numerous studies have shown to be infectious [17,18,21,29,37]. Uninfected brains from age-matched C57BL/6 mice were subjected to the same procedure in order to identify proteins that were specific to the enrichment protocol.
Each reduced and alkylated sample was subjected to SDS-PAGE as described in the methods. The mock preparations clearly displayed two silver-stained protein bands of approximately 20–21 kDa (Figure 1A). The infected samples revealed the characteristic di, mono, and non-glycosylated banding patterns commonly observed with PrPSc (Figure 1A). Western blot analysis of the same samples confirmed the presence of PrPSc in the infected samples (Figure 1B). Even CH-04 clearly displayed the characteristic PrPSc banding pattern by western blot, despite its apparent lack of homogeneity by silver stain (Figure 1B). The 20–21 kDa bands in the mock preparations are not PrPSc since they were not detected by immunoblot using the PrP-specific antibody 6D11 (Figure 1B).
3.2 Protein identifications
Approximately 40 micrograms of total protein from each sample shown in Figure 1 was subjected to SDS-PAGE. In-gel digests were performed on bands excised from the Coomassie blue stained regions of each gel (Supplementary Figure 1). Each replicate sample set consisted of 10–12 in-gel digests that were then analyzed by LC-MS/MS as described. Searches of the Sprot database were performed using the SEQUEST and MASCOT search engines as described in methods. The two independently implemented searches gave very similar protein identifications, with the SEQUEST-generated results generally yielding slightly higher spectral counts (i.e., the total number of assigned tandem MS spectra for each protein listed) for most of the proteins identified. A complete list of the identified peptides, including their rankings, XCorr scores and charge states is given in Supplementary Table 1. The false discovery rates ranged between 1.4 and 3.4% and are given in Supplementary Table 2.
The proteins identified in this study are summarized in Table 1 and listed in descending order of spectral counts tallied cumulatively from all of the samples that were examined. A total of 31 different proteins from the combined mock and PrPSc enrichments met the criteria for identification as described in the experimental section. The most consistently identified protein in all of the 22L and Chandler-derived samples was PrPSc (Table 1), consistent with the silver stain and western blot results (Figure 1). The same PrPSc-derived peptides were consistently found for both the 22L and Chandler samples (see list in Supplementary Table 1). No PrPSc was found in any of the mock samples by nanospray LC-MS/MS (Table 1).
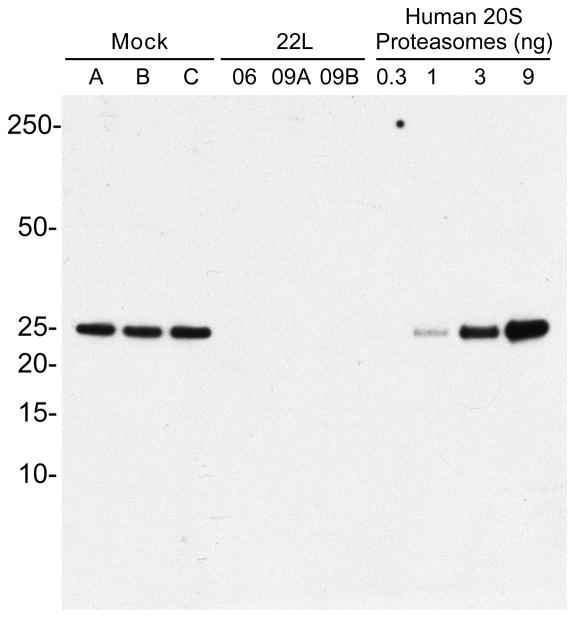
Approximately 30% of the total protein identifications were shared between the mock and PrPSc samples, as shown in the Venn diagram in Figure 1C. These shared proteins were either oligomeric (e.g, ferritin, proteasomes), polymeric (e.g, collagen, actin) or highly abundant in the brain (e.g, calcium/calmodulin-dependent protein kinase). The majority of proteins unique to the mock preparations were from various proteasomal subunits. In fact, only 2 of the 283 MS/MS spectra corresponding to proteasomal subunits were found with PrPSc (see CH-04, Table 2). An immunoblot for proteasomes confirmed the low abundance of proteasomal proteins in infected PrPSc samples compared to the mock PrPSc samples. Thus, proteasomal fragments appear to be overrepresented in uninfected versus infected samples enriched for PrPSc (Figure 2).
Table 2.
Relative apparent abundance of proteins between PrPSc samples. The sequence coverage and spectral counts were calculated based upon individual samples. The relative abundance for each protein was calculated based upon spectral count and is presented as a percentage of the total number of assigned spectra for each protein within each sample.
| Sample | Accession (Swiss-prot) | mw (Da) | Sequence Coverage | Spectral Count | Relative Abundance |
|---|---|---|---|---|---|
| 22L-06 | Prion protein (P04925) | 27,977 | 38.6 | 307 | 83.4 |
| Ferritin heavy chain (P09528) | 21,067 | 34.1 | 27 | 7.3 | |
| Ferritin light chain 1 (P29391) | 20,802 | 33.3 | 19 | 5.2 | |
| Versican core protein (Q62059) | 366,788 | 1.3 | 9 | 2.4 | |
| Apolipoprotein E (P08226) | 35,867 | 7.7 | 3 | 0.8 | |
| 60S ribosomal protein L35 (Q6ZWV7) | 14,553 | 8.1 | 2 | 0.5 | |
| Hyaluronan and proteoglycan link protein 2 (Q9ESM3) | 37,925 | 3.5 | 1 | 0.3 | |
| 22L-09A | Prion protein (P04925) | 27,977 | 53.1 | 414 | 59.8 |
| Ferritin light chain 1 (P29391) | 20,802 | 49.2 | 93 | 13.4 | |
| Ferritin heavy chain (P09528) | 21,067 | 37.4 | 78 | 11.3 | |
| Myelin-associated oligodendrocyte basic protein (Q9D2P8) | 19,197 | 18.2 | 27 | 3.9 | |
| Apolipoprotein E (P08226) | 35,867 | 22.2 | 23 | 3.3 | |
| Collagen alpha-1(I) chain (P11087) | 138,033 | 1.5 | 20 | 2.9 | |
| CAMK-II subunit alpha (P11798) | 54,115 | 13.4 | 15 | 2.2 | |
| Myelin proteolipid protein (P60202) | 30,077 | 13.4 | 10 | 1.4 | |
| Myelin-oligodendrocyte glycoprotein (Q61885) | 28,271 | 9.8 | 4 | 0.6 | |
| 60S ribosomal protein L35 (Q6ZWV7) | 14,553 | 18.7 | 4 | 0.6 | |
| 60S ribosomal protein L13a (P19253) | 23,464 | 5.4 | 2 | 0.3 | |
| Claudin-11 (Q60771) | 22,114 | 7.7 | 1 | 0.1 | |
| CAMK-II subunit beta (P28652) | 60,461 | 2.0 | 1 | 0.1 | |
| 22L-09B | Prion protein (P04925) | 27,977 | 46.9 | 525 | 70.8 |
| Ferritin heavy chain (P09528) | 21,067 | 52.2 | 60 | 8.1 | |
| Ferritin light chain 1 (P29391) | 20,802 | 49.2 | 60 | 8.1 | |
| Apolipoprotein E (P08226) | 35,867 | 19.3 | 31 | 4.2 | |
| Myelin-associated oligodendrocyte basic protein (Q9D2P8) | 19,197 | 18.2 | 16 | 2.2 | |
| Collagen alpha-1(I) chain (P11087) | 138,033 | 1.5 | 15 | 2.0 | |
| CAMK-II subunit alpha (P11798) | 54,115 | 18.6 | 12 | 1.6 | |
| Myelin proteolipid protein (P60202) | 30,077 | 13.4 | 8 | 1.1 | |
| Myelin-oligodendrocyte glycoprotein (Q61885) | 28,271 | 9.8 | 4 | 0.5 | |
| 60S ribosomal protein L35 (Q6ZWV7) | 14,553 | 18.7 | 4 | 0.5 | |
| 60S ribosomal protein L13a (P19253) | 23,464 | 11.3 | 3 | 0.4 | |
| CAMK-II subunit beta (P28652) | 60,461 | 4.4 | 3 | 0.4 | |
| Junction plakoglobin (Q02257) | 81,801 | 2.4 | 1 | 0.1 | |
| CH-04 | Prion protein (P04925) | 27,977 | 27.2 | 107 | 19.8 |
| Ferritin light chain 1 (P29391) | 20,802 | 64.8 | 106 | 19.6 | |
| Hyaluronan and proteoglycan link protein 2 (Q9ESM3) | 37,925 | 45.7 | 79 | 14.6 | |
| Versican core protein (Q62059) | 366,788 | 4.9 | 71 | 13.1 | |
| Ferritin heavy chain (P09528) | 21,067 | 41.0 | 53 | 9.8 | |
| Myelin-associated oligodendrocyte basic protein (Q9D2P8) | 19,197 | 18.2 | 46 | 8.5 | |
| Hyaluronan and proteoglycan link protein 1 (Q9QUP5) | 40,478 | 32.3 | 21 | 3.9 | |
| Collagen alpha-1(I) chain (P11087) | 138,033 | 1.5 | 13 | 2.4 | |
| CAMK-II subunit alpha (P11798) | 54,115 | 7.7 | 11 | 2.0 | |
| CAMK-II subunit beta (P28652) | 60,461 | 6.8 | 10 | 1.9 | |
| Myelin proteolipid protein (P60202) | 30,077 | 13.4 | 8 | 1.5 | |
| Ras-related protein Rab-3D (P35276) | 24,416 | 10.0 | 4 | 0.7 | |
| 60S ribosomal protein L35 (Q6ZWV7) | 14,553 | 8.1 | 3 | 0.6 | |
| Junction plakoglobin (Q02257) | 81,801 | 5.8 | 3 | 0.6 | |
| Proteasome subunit alpha type-7 (Q9Z2U0) | 27,855 | 11.7 | 2 | 0.4 | |
| Claudin-11 (Q60771) | 22,114 | 7.7 | 2 | 0.4 | |
| 60S ribosomal protein L13a (P19253) | 23,464 | 5.9 | 1 | 0.2 | |
| CH-06 | Prion protein (P04925) | 27,977 | 39.8 | 286 | 82.7 |
| Ferritin light chain 1 (P29391) | 20,802 | 55.7 | 18 | 5.2 | |
| Versican core protein (Q62059) | 366,788 | 2.9 | 14 | 4.0 | |
| Ferritin heavy chain (P09528) | 21,067 | 26.9 | 8 | 2.3 | |
| 60S ribosomal protein L35 (Q6ZWV7) | 14,553 | 11.4 | 6 | 1.7 | |
| Apolipoprotein E (P08226) | 35,867 | 9.3 | 6 | 1.7 | |
| CAMK-II subunit alpha (P11798) | 54,115 | 2.5 | 4 | 1.2 | |
| Junction plakoglobin (Q02257) | 81,801 | 4.3 | 2 | 0.6 | |
| Actin, cytoplasmic 1 (P60710) | 41,737 | 4.5 | 1 | 0.3 | |
| CAMK-II subunit beta (P28652) | 60,461 | 2.4 | 1 | 0.3 |
Figure 2.
Proteasomal subunit 5 is present at higher levels in mock versus 22L-derived PrPSc samples. Proteins were subjected to western blot with anti-proteasome antibody MCP196 as described in the experimental section. Human 20 S proteasomes were loaded as a positive control.
3.3 Sample variation between PrPSc preparations is not strain dependent
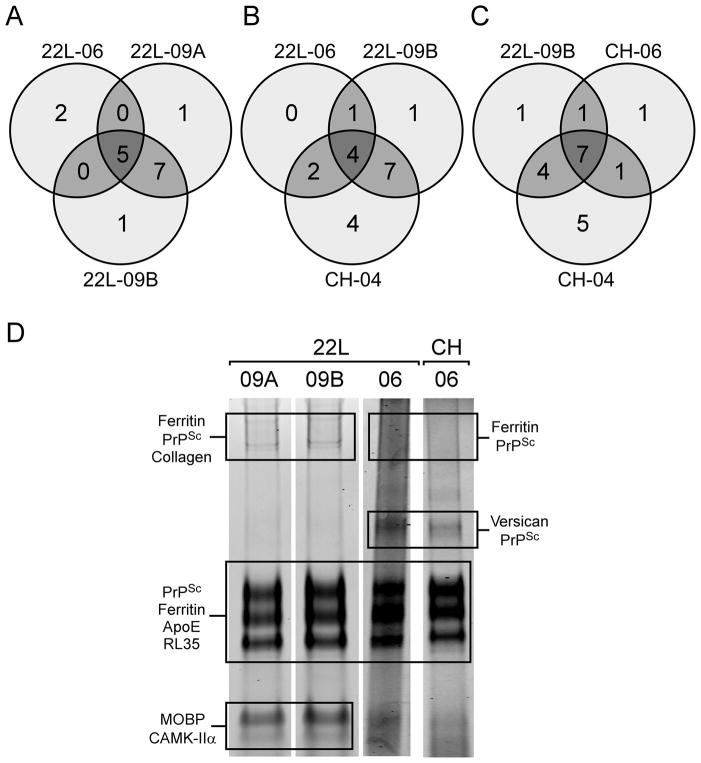
The protein identifications derived from our five PrPSc samples were compared based on whether any of the identified proteins displayed a strain preference. Ferritin and 60S ribosomal protein L35 were identified in all samples, including mock. Apolipoprotein E (ApoE), which is known to associate with amyloid plaque in vivo [38,39], was also found in all but one (CH-04) of our mouse PrPSc samples but not in the mock preparations. The remaining protein identifications were more variable and did not appear to be strain-specific. Samples that were prepared side by side (e.g., 22L-09A and B) gave markedly more similar protein profiles when compared to samples that had been prepared separately (Figure 3A). Infected samples from 2 different strains (22L-09B and CH-04) shared a higher percentage of the total number of identified proteins than did 2 samples prepared independently from the same strain (22L-09B and 22L-06, Figure 3B). Examination of selected Coomassie blue stained gels from which the in-gel digestions were done (Figure 3D) further illustrates the lack of strain specificity in the non-PrPSc protein components. The Chandler and 22L samples from 2006 displayed a similar pattern of banding and shared more common protein identifications (e.g, versican core protein at 60 kDa) than 22L samples compared between 2006 and 2009. Thus, the data suggest that the protein composition of a particular PrPSc mixture is not necessarily dictated by strain characteristics, but rather by variations in the enrichment protocol.
Figure 3.
Variations in protein identifications from PrPSc preparations do not display strain specificity. A) Samples of 22L that were prepared in 2009 share more protein identifications with each other than they do with 22L that was prepared in 2006. B) Sample 22L-09B shares more protein identifications with CH-04 than it does with 22L-06. C) CH-04 shares more protein identifications with 22L-09B than it does with CH-06 and D) The Coomassie blue-stained SDS-PAGE fingerprint of each sample appears to be more similar based upon when it was prepared rather than what strain PrPSc was derived from. Samples 22L-09A and B contain discernable lower molecular weight bands for myelin-associated oligodendrocyte basic protein (MOBP) and calcium/calmodulin-dependent protein kinase (CAMK-IIα), while 22L-06 and CH-06 display bands at ~50 kDa that are specific for versican core protein and an apparent PrPSc dimer.
3.4 Relative protein abundance in different PrPSc samples
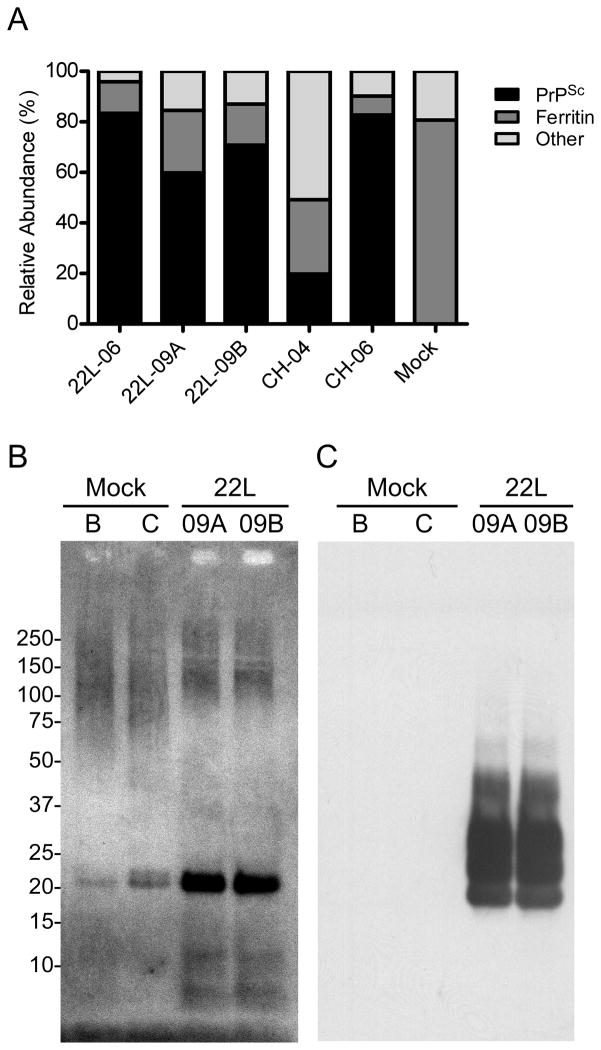
We estimated the relative apparent abundance of PrPSc in our samples by tabulating the number of spectra [40] for PrPSc and other proteins in each sample and then representing them as a percentage of the total assigned spectra (Table 2). Ferritin and PrPSc were the most abundant proteins both identified with similar sequence coverage and represented by approximately 60 unique peptides (Table 1). The data show that the amount of PrPSc found in a particular preparation was variable, ranging from approximately 20 to 83% relative apparent abundance of the proteins measured (Figure 4A). The low abundance of PrPSc relative to all of the other proteins in CH-04 was particularly revealing and is consistent with the silver-stained image in Figure 1A and the Coomassie blue stained images in Supplementary Figure 1. The proportion of ferritin was also higher in CH-04 than in the other samples as were the significantly elevated amounts of proteoglycan link and versican core protein, which were found at roughly 14% and 13%, respectively.
Figure 4.
Ferritin is the most abundant non-PrPSc protein found in all of the samples. A) The amount of PrPSc relative to ferritin and other proteins was estimated by spectral counting and was found to vary between samples. Lower relative abundances of PrPSc were associated with higher abundances of ferritin. In the case of CH-04, the amounts of ferritin, versican, and proteoglycan link proteins were much higher than in any of the other PrPSc samples. An immunoblot from representative mock and PrPSc samples were separated by SDS-PAGE on a 4–12% NuPAGE gradient gel and probed with B) anti-ferritin rabbit antibody, washed with TBST buffer and then probed with C) 6D11 anti-PrP mouse monoclonal antibody.
Ferritin has been repeatedly implicated as a component of PrPSc preparations [17,24] and a PrPSc enrichment protocol has even been used to isolate ferritin itself [41]. Like PrPSc, ferritin is oligomeric [42] and resists protease digestion [43]. A ferritin-specific western blot confirmed the presence of ferritin at approximately 20 kDa in each of the samples tested as well as apparent higher molecular weight isoforms (Figure 4B). Since PrPSc is also known to form high molecular weight aggregates similar to that shown in Figure 4B, we were prompted to confirm that all of the banding on that blot was from ferritin rather than cross-reaction of PrPSc with the ferritin antibody. When the ferritin blot was washed and subsequently probed with PrP-specific antibody 6D11, the characteristic 20 – 30 kDa isoforms of PrPSc were detected, but PrPSc was not detected at higher molecular weight (Figure 4C). It is thus likely that the high molecular weight material on the ferritin western blot is composed primarily of ferritin and is not due to significant cross-reactivity with PrPSc aggregates. Thus, the data show that ferritin is typically the most abundant contaminant in standard PrPSc preparations and that the relative amounts of other contaminants can also be variable.
4 Discussion
Our analysis of 5 PrPSc samples from two different mouse scrapie strains, as well as 3 samples prepared from uninfected mouse brain did not reveal any non-PrPSc proteins that appeared to be strain-specific. While the 22L-09 A and B samples were similar with respect to the number of shared protein identifications, only ~30% of the identified proteins were shared with a different 22L preparation, 22L-06 (Figure 3A). Furthermore, the 22L-09 samples shared more proteins with PrPSc isolated from Chandler scrapie than from 22L-06 (Figure 3B). Since these results are the opposite of what one might expect based upon assumed strain-associated similarities, the implication is that many of these proteins were present due to the variable nature of the enrichments. Thus, our data suggest that either strain-specific protein co-factors are not required to facilitate prion transmissibility or that they are present in amounts below the detectable limits of the current study.
Over 30% of the proteins that became enriched with PrPSc were also found in the mock preparations (Table 1). When these non-PrPSc-specific proteins were found in the infected samples, they were present in stoichiometrically lower concentrations than PrPSc. These proteins were likely isolated during the enrichment protocol because they share common biochemical properties with PrPSc such as detergent insolubility, aggregation, and protease resistance. For example, ferritin, proteasomes, and collagen are either oligomeric or very large molecules that are likely to sediment with aggregates. Highly abundant proteins, such as myelin associated protein, may be present due to incompletely dispersed cellular membrane components. Taken together, the data suggest that the pathological relevance of proteins that co-purify with PrPSc from enrichment protocols based primarily upon centrifugation and PK treatment should be interpreted with caution.
Using an approach similar to ours, two recent studies [27,28] have also identified co-purifying proteins present in PrPSc preparations. Two non-PrPSc proteins consistently identified in the current study as well as the previous work [27,28] were ferritin and calcium calmodulin-dependent protein kinase II alpha (CAMK-IIα). In fact, CAMK-II genes have been shown to be differentially regulated in sporadic CJD brains relative to controls [44]. It is important to note, however, that CAMK-IIα was also found in our non-infected mock preparations and that this protein has been reported to constitute up to 0.74% of total brain protein [45] with an apparent molecular weight of 650,000 [46]. Thus, its presence in PrPSc preparations may be due more to its abundance and molecular mass than to any pathologically relevant association with PrPSc. Interestingly, a comparison of two recently published mass spectrometry PrPSc identification studies [27,28] reveals that 18 out of the 19 protein identifications (other than PrPSc or ferritin) were different, again raising the possibility that the enrichment protocols that they used [20,47] might have a significant effect upon the resulting protein composition of the sample.
We estimated the relative apparent abundance of PrPSc to other proteins in our samples by spectral counting [40,48]. This technique is often used to estimate the relative protein abundances in a mixture since the number of assigned spectra has been shown to correlate well with protein concentration [49,50]. The spectral counting data was consistent with silver-stained protein patterns in Figure 1A and showed that the amount of PrPSc isolated from a particular preparation can be variable, ranging from approximately 20% in CH-04 to 83% of the total protein content in CH-06 (Figure 4a). While impurities are unlikely to have an impact upon TSE disease transmission in laboratory animals [51] or in the growing use of in vitro reactions where brain homogenate is used as the reaction medium [9], there are other types of studies (e.g., structural studies or protein-specific labeling) in which PrPSc purity is more critical. Our results suggest that before using PrPSc preparations for such applications it will be important to consider the concentration of PrPSc relative to other protein contaminants.
It is unlikely that we have identified all of the proteins in our PrPSc preparations. Our samples had been PK-treated prior to denaturation and in-gel trypsin digestion, making lower molecular weight components more difficult to identify. It is possible that many of the proteotypic peptides that are the most amenable to identification by tandem mass spectrometry [52] were replaced by harder to identify protein remnants and non-tryptic fragments. PK digestion may also explain why some proteins identified by MS/MS could not be confirmed by immunoblot. For example, versican core protein, which has a molecular weight of 367 kDa, was identified in an SDS-PAGE band at approximately 50 kDa. Only N-terminal peptide fragments were identified and none of them were reactive with a commercially available antibody that detects sequences in the C-terminus of versican (data not shown). Thus, due to the PK treatment step of the enrichment protocol, proteins in our preparations were often observed as truncated versions (e.g., Versican and PrPSc) of their original sequence.
We also cannot rule out the possibility that some of the co-purifying proteins in our preparations have significance to TSE disease. Apolipoprotein E (ApoE) in particular has been consistently implicated across interdisciplinary lines of study as being associated with various types of amyloid plaque [38,39], with PrPSc deposits [53], and in the cerebrospinal fluid of TSE-affected humans and cattle [54,55]. Gene profiling studies have shown that ApoE is upregulated in prion-infected versus non-infected mice in response to multiple TSE strains, including 22L and Chandler [56,57]. ApoE was also recently identified in hamster-adapted PrPSc and shown to localize with PrPSc deposits in vivo [28]. We also identified ApoE in four out of our five PrPSc samples, although Petrakis and co-workers did not report this protein in their single replicate study [27]. It remains plausible that important components related to TSE infectivity will be found in more concentrated PrPSc preparations, especially after the removal of various contaminants that we and others have identified. Moving forward, it should be most useful to expand the breadth of the protein profiling approach to include a wider range of PrPSc-associated molecules given that metal ions [58], nucleic acids [11], polysaccharides [25], and lipids [59] might also play important roles in PrPSc formation.
Supplementary Material
Supplementary Figure 1. Coomassie blue stained images from 10% NuPAGE gels of the PrPSc preparations used for LC-MS/MS analysis. At least 12 Coomassie blue stained bands from each of the samples shown were subjected to in-gel digestion and LC-MS/MS analysis. This process was repeated 3 or more times for each sample. These same samples are also shown in Figure 1.
Acknowledgments
The authors would like to thank Dr. Byron Caughey for providing 22L-06, CH-06, and CH-04 PrPSc and Andy Hughson for purifying those samples. The authors would also like to thank Drs. Jay Carroll, Karin Peterson, and Mikael Klingeborn for critically evaluating the manuscript and Anita Mora for technical assistance in preparation of the figures. The authors also thank Charlie Roberts from Proteome Software for helpful discussions. This research was supported by intramural research program of the National Institute of Allergy & Infectious Disease, National Institutes of Health (Project #1-Z01-AI000752-12). PAW was supported by National Institutes of Health grant EY007755.
Abbreviations
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- CH
Chandler
- TSE
Transmissible Spongiform Encephalopathy
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Chesebro B. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br Med Bull. 2003;66:1–20. doi: 10.1093/bmb/66.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Caughey B, Baron GS, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughey BW, Dong A, Bhat KS, Ernst D, et al. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 4.Pan KM, Baldwin M, Nguyen J, Gasset M, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion protein. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 6.Bruce ME. TSE strain variation. Br Med Bull. 2003;66:99–108. doi: 10.1093/bmb/66.1.99. [DOI] [PubMed] [Google Scholar]
- 7.Moore RA, Taubner LM, Priola SA. Prion protein misfolding and disease. Curr Opin Struct Biol. 2009;19:14–22. doi: 10.1016/j.sbi.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 9.Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Makarava N, Kovacs GG, Bocharova O, Savtchenko R, et al. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 2010;119:177–187. doi: 10.1007/s00401-009-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci U S A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diringer H, Hilmert H, Simon D, Werner E, et al. Towards purification of the scrapie agent. Eur J Biochem. 1983;134:555–560. doi: 10.1111/j.1432-1033.1983.tb07602.x. [DOI] [PubMed] [Google Scholar]
- 13.Prusiner SB, Hadlow WJ, Garfin DE, Cochran SP, et al. Partial purification and evidence for multiple molecular forms of the scrapie agent. Biochemistry. 1978;17:4993–4999. doi: 10.1021/bi00616a021. [DOI] [PubMed] [Google Scholar]
- 14.Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 15.Prusiner SB, Bolton DC, Groth DF, Bowman KA, et al. Further purification and characterization of scrapie prions. Biochemistry. 1982;21:6942–6950. doi: 10.1021/bi00269a050. [DOI] [PubMed] [Google Scholar]
- 16.Hilmert H, Diringer H. A Rapid and Efficient Method to Enrich Saf-Protein from Scrapie Brains of Hamsters. Biosci Rep. 1984;4:165–170. doi: 10.1007/BF01120313. [DOI] [PubMed] [Google Scholar]
- 17.Diringer H, Beekes M, Ozel M, Simon D, et al. Highly infectious purified preparations of disease-specific amyloid of transmissible spongiform encephalopathies are not devoid of nucleic acids of viral size. Intervirology. 1997;40:238–246. doi: 10.1159/000150553. [DOI] [PubMed] [Google Scholar]
- 18.Bolton DC, Bendheim PE, Marmostein AD, Potempska A. Isolation and structural studies of the intact scrapie agent protein. Arch Biochem Biophys. 1987;258:579–590. doi: 10.1016/0003-9861(87)90380-8. [DOI] [PubMed] [Google Scholar]
- 19.Raymond GJ, Chabry J. Purification of the pathological isoform of prion protein (PrPSc or PrPres) from transmissible spongiform encephalopathy-affected brain tissue. 2004:16–26. [Google Scholar]
- 20.Sklaviadis TK, Manuelidis L, Manuelidis EE. Physical properties of the Creutzfeldt-Jakob disease agent. J Virol. 1989;63:1212–1222. doi: 10.1128/jvi.63.3.1212-1222.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabizon R, McKinley MP, Groth D, Prusiner SB. Immunoaffinity purification and neutralization of scrapie prion infectivity. Proc Natl Acad Sci USA. 1988;85:6617–6621. doi: 10.1073/pnas.85.18.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Multhaup G, Diringer H, Hilmert H, Prinz H, et al. The protein component of scrapie-associated fibrils is a glycosylated low molecular weight protein. EMBO J. 1985;4:1495–1501. doi: 10.1002/j.1460-2075.1985.tb03808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akowitz A, Sklaviadis T, Manuelidis EE, Manuelidis L. Nuclease-resistant polyadenylated RNAs of significant size are detected by PCR in highly purified Creutzfeldt-Jakob disease preparations. Microbial Pathogenesis. 1990;9:33–45. doi: 10.1016/0882-4010(90)90038-r. [DOI] [PubMed] [Google Scholar]
- 24.Kascsak RJ, Rubenstein R, Merz PA, Carp RI, et al. Biochemical differences among scrapie- associated fibrils support the biological diversity of scrapie agents. J Gen Virol. 1985;66:1715–1722. doi: 10.1099/0022-1317-66-8-1715. [DOI] [PubMed] [Google Scholar]
- 25.Appel TR, Dumpitak C, Matthiesen U, Riesner D. Prion rods contain an inert polysaccharide scaffold. Biol Chem. 1999;380:1295–1306. doi: 10.1515/BC.1999.165. [DOI] [PubMed] [Google Scholar]
- 26.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 27.Petrakis S, Malinowska A, Dadlez M, Sklaviadis T. Identification of proteins co-purifying with scrapie infectivity. J Proteomics. 2009;72:690–694. doi: 10.1016/j.jprot.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Giorgi A, Di Francesco L, Principe S, Mignogna G, et al. Proteomic profiling of PrP27–30-enriched preparations extracted from the brain of hamsters with experimental scrapie. Proteomics. 2009;9:3802–3814. doi: 10.1002/pmic.200900085. [DOI] [PubMed] [Google Scholar]
- 29.Silveira JR, Raymond GJ, Hughson AG, Race RE, et al. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamguney G, Giles K, Bouzamondo-Bernstein E, Bosque PJ, et al. Transmission of elk and deer prions to transgenic mice. J Virol. 2006;80:9104–9114. doi: 10.1128/JVI.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Wilmarth PA, Riviere MA, David L. L, Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. Journal of Ocular Biology, Diseases, and Informatics. 2009;2:223–234. doi: 10.1007/s12177-009-9042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Bruce ME, Fraser H. Scrapie strain variation and its implications. Curr Top Microbiol Immunol. 1991;172:125–138. doi: 10.1007/978-3-642-76540-7_8. [DOI] [PubMed] [Google Scholar]
- 36.Kascsak RJ, Rubenstein R, Carp RI. Evidence for Biological and Structural Diversity Among Scrapie Strains. 1991;1:139–152. doi: 10.1007/978-3-642-76540-7_9. [DOI] [PubMed] [Google Scholar]
- 37.Diringer H, Gelderblom H, Hilmert H, Ozel M, et al. Scrapie infectivity, fibrils and low molecular weight protein. Nature. 1983;306:476–478. doi: 10.1038/306476a0. [DOI] [PubMed] [Google Scholar]
- 38.Lashley T, Holton JL, Verbeek MM, Rostagno A, et al. Molecular chaperons, amyloid and preamyloid lesions in the BRI2 gene-related dementias: a morphological study. Neuropathol Appl Neurobiol. 2006;32:492–504. doi: 10.1111/j.1365-2990.2006.00747.x. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi H, Ishiguro K, Sugihara S, Nakazato Y, et al. Presence of apolipoprotein E on extracellular neurofibrillary tangles and on meningeal blood vessels precedes the Alzheimer beta-amyloid deposition. Acta Neuropathol. 1994;88:413–419. doi: 10.1007/BF00389492. [DOI] [PubMed] [Google Scholar]
- 40.Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev Proteomics. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 41.Kaneko Y, Kitamoto T, Tateishi J, Yamaguchi K. Ferritin immunohistochemistry as a marker for microglia. Acta Neuropathol. 1989;79:129–136. doi: 10.1007/BF00294369. [DOI] [PubMed] [Google Scholar]
- 42.Bjork I, Fish WW. Native and subunit molecular weights of apoferritin. Biochemistry. 1971;10:2844–2848. doi: 10.1021/bi00791a007. [DOI] [PubMed] [Google Scholar]
- 43.Coux O, Camoin L, Nothwang HG, Bey F, et al. The protein of M(r) 21,000 constituting the prosome-like particle of duck erythroblasts is homologous to apoferritin. Eur J Biochem. 1992;207:823–832. doi: 10.1111/j.1432-1033.1992.tb17113.x. [DOI] [PubMed] [Google Scholar]
- 44.Xiang W, Windl O, Westner IM, Neumann M, et al. Cerebral gene expression profiles in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 2005;58:242–257. doi: 10.1002/ana.20551. [DOI] [PubMed] [Google Scholar]
- 45.Hanson PI, Schulman H. Neuronal Ca2+/calmodulin-dependent protein kinases. Annu Rev Biochem. 1992;61:559–601. doi: 10.1146/annurev.bi.61.070192.003015. [DOI] [PubMed] [Google Scholar]
- 46.Bennett MK, Erondu NE, Kennedy MB. Purification and characterization of a calmodulin- dependent protein kinase that is highly concentrated in brain. J Biol Chem. 1983;258:12735–12744. [PubMed] [Google Scholar]
- 47.Silvestrini MC, Cardone F, Maras B, Pucci P, et al. Identification of the prion protein allotypes which accumulate in the brain of sporadic and familial Creutzfeldt-Jakob disease patients. Nature/Medicine. 1997;1:1. doi: 10.1038/nm0597-521. [DOI] [PubMed] [Google Scholar]
- 48.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. Journal of Proteome Research. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, Sadygov RG, Yates JR. III, A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 50.Mosley AL, Florens L, Wen ZH, Washburn MP. A label free quantitative proteomic analysis of the Saccharomyces cerevisiae nucleus. Journal of Proteomics. 2009;72:110–120. doi: 10.1016/j.jprot.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKinley MP, Bolton DC, Prusiner SB. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 52.Mallick P, Schirle M, Chen SS, Flory MR, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 53.Rangon CM, Haik S, Faucheux BA, Metz-Boutigue MH, et al. Different chromogranin immunoreactivity between prion and a-beta amyloid plaque. Neuroreport. 2003;14:755–758. doi: 10.1097/00001756-200304150-00019. [DOI] [PubMed] [Google Scholar]
- 54.Choe LH, Green A, Knight RS, Thompson EJ, et al. Apolipoprotein E and other cerebrospinal fluid proteins differentiate ante mortem variant Creutzfeldt-Jakob disease from ante mortem sporadic Creutzfeldt-Jakob disease. Electrophoresis. 2002;23:2242–2246. doi: 10.1002/1522-2683(200207)23:14<2242::AID-ELPS2242>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 55.Hochstrasser DF, Frutiger S, Wilkins MR, Hughes G, et al. Elevation of apolipoprotein E in the CSF of cattle affected by BSE. FEBS Lett. 1997;416:161–163. doi: 10.1016/s0014-5793(97)01191-5. [DOI] [PubMed] [Google Scholar]
- 56.Skinner PJ, Abbassi H, Chesebro B, Race RE, et al. Gene expression alterations in brains of mice infected with three strains of scrapie. BMC Genomics. 2006;7:114. doi: 10.1186/1471-2164-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorensen G, Medina S, Parchaliuk D, Phillipson C, et al. Comprehensive transcriptional profiling of prion infection in mouse models reveals networks of responsive genes. BMC Genomics. 2008;9:114. doi: 10.1186/1471-2164-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wadsworth JD, Hill AF, Joiner S, Jackson GS, et al. Strain-specific prion-protein conformation determined by metal ions. Nat Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 59.Sanghera N, Swann MJ, Ronan G, Pinheiro TJ. Insight into early events in the aggregation of the prion protein on lipid membranes. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamem.2009.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Coomassie blue stained images from 10% NuPAGE gels of the PrPSc preparations used for LC-MS/MS analysis. At least 12 Coomassie blue stained bands from each of the samples shown were subjected to in-gel digestion and LC-MS/MS analysis. This process was repeated 3 or more times for each sample. These same samples are also shown in Figure 1.