Abstract
Purpose of the Review
Epigenetic modifications are heritable alterations of the genome, which can govern gene expression without altering the DNA sequence. The purpose of this review is to render an overview of the possible mechanisms of epigenetic regulation of gene expression in response to environmental pollutants leading to cardiovascular diseases (CVD).
Recent Findings
An era of cataloging epigenetic marks of the various diseased states has recently commenced, including those within the genes responsible for atherosclerosis, ischemia, hypertension and heart failure. From varied study approaches directed either towards the general understanding of the key pathway regulatory genes, or sampling population cohorts for global and gene-specific changes, it has been possible to identify several epigenetic signatures of environmental exposure relevant to CVD. Signatures of epigenetic dysregulation can be detected in peripheral blood samples, even within few hours of environmental exposure. However, the field now faces the demand for thorough, systematic, rationalized approaches to establish the relation of an exposure-driven epigenetic changes to clinical outcomes, by using sophisticated and reliable research designs and tools.
Summary
An understanding of chromatin remodeling in response to environmental stimuli conducive to CVD is emerging, with the promise of novel diagnostic and therapeutic candidates.
Keywords: Environment, Cardiovascular, Epigenetics, DNA methylation, Histone modifications, Biomarkers
Introduction
According to the 2012 update on Heart Disease and Stroke Statistics by the American Heart Association, cardiovascular diseases (CVDs) represent a substantial disease burden, and accounted for 32% of the total mortality in the United States in the most recent 2008 data (1). Although this represents a drop in the mortality rate since 1998, it nonetheless is a huge burden, both in terms of morbidity and cost. Especially, as a result of the obesity epidemics, CVD incidence remains to be on the rise, thus leading to increased prevalence of individuals in the U.S. population and elsewhere with chronic disease requiring a continuous augmentation of long term resources. Heritability estimates for most CVDs are around 30%. However, dietary and exercise habits, as well as environmental exposure to toxic agents determine the cardiovascular health of an individual to a large extent, such that CVDs are often loosely termed environmental disease.
How epigenetic regulation works
Epigenetic alterations occur via altering the chromatin packaging and the accessibility of DNA to regulatory proteins, with potential influences on gene expression. Epigenetic changes maybe rapid or slow, they are thought to be stable and once established are potentially heritable.
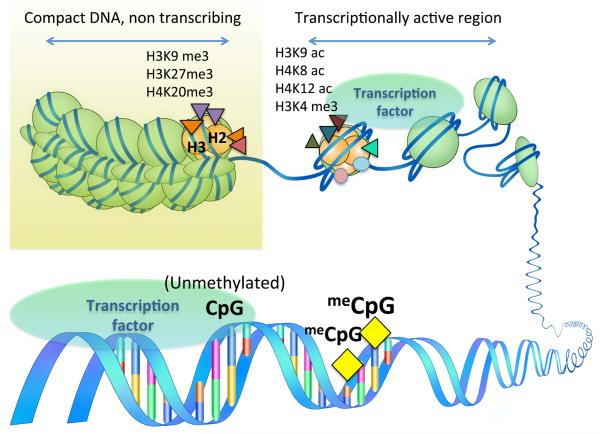
DNA methylation represents an extensively investigated well-understood epigenetic mechanism. Methylation occurs in the 5-position of the cytosine in a CpG dinucleotide. CpG dinucleotides are often clustered within the promoter regions of genes, but are also present in the gene bodies and other gene-associated regions. In general, increased methylated CpG within the promoters is associated with transcriptional repression as it interferes with the binding of transcription factors (Figure 1). Almost paradoxically, 90% of the methylated CpG lies outside the coding regions, possibly to serve as repressors of transposons or viral like transcripts. A global assessment of the changes in the DNA methylation status in the genome can be achieved by interrogating the proportion of methylated CpGs in the highly abundant and repetitive sequences including the retrotransposons such as Long Interspersed Nuclear Element-1 (LINE-1) and Alu sequences. Cancer tissues nearly universally show global hypomethylation of CpG sites, particularly in the repetitive sequences, which is associated with genomic instability. DNA methylation patterns are generated and maintained by enzymes called DNA methyl transferases (DNMTs: DNMT1, DNMT3a and 3b) that use S-adenosylmethionine (SAM), a universal methyl donor also used for various other cellular processes like RNA and protein methylation, detoxification and biochemical synthesis. SAM is converted to S-adenosylhomocysteine (SAH) after donation of the methyl group. A ratio of SAM and SAH represents the methylation capacity. A decrease in the ratio represents poor methylation capacity. In addition, SAH binds to DNA methyl transferases with higher affinity than SAM and acts as an inhibitor of methylation reactions. Therefore, hyperhomocysteinemia can disturb methylation homeostasis and potentially lead to epigenetic alterations.
Figure 1.
Epigenetic modifications of the nucleosomal histones and DNA, regulating gene expression. Histone modifications of compactly coiled heterochromatic regions, or relaxed transcriptionally active chromatin. H, histone; K, lysine; me, methylation; me3, trimethylation; ac, acetylation; DNA methylation silencing the gene; meCpG, methylated Cytosine in Cytosine-Guanine dinucleotide sequences within the DNA.
In the nucleosomes, the DNA is intimately wound around the octamer of four histone proteins, H2A, H2B, H3 and H4 with H1 as a linking unit. Histones undergo various post-translational modifications like methylation, phosphorylation, acetylation, ubiquitination, and others (2). These post-translational modifications have specific effects in gene transcription, for example, acetylation of the K9 residue of H3 in the promoter is associated with gene expression, whereas, tri-methylation leads to repression (3). Histone acetylation is mediated by histone acetyl transferases (HATs), which relaxes the nucleosome and enables transcriptional activation, and removal of acetyl groups is mediated by deacetylases (HDACs) (Figure 1). Apart from deacetylase activity, HDAC II family members contain an amino terminal region capable of protein-protein interaction. Chemical inhibitors of HDACs like valproic acid (Phase II clinical trial) and FK228 (FDA approved) are anticancer therapeutics (4).
In addition to remodeling of the chromatin, regulatory non-coding RNA such as microRNA plays a crucial role in regulation of gene expression. MicroRNAs lead to the suppression of target genes through Dicer mediated cleavage into 22 nucleotide small interfering RNA in the cytoplasm which activates the RNA-induced silencing (RISC) complex to degrade specific mRNA.
Environmental exposure and cardiovascular outcome
Epidemiological research has established the strong association of different environmental exposures to cardiovascular diseases (5). Recent studies are exploring the epigenetic changes in various exposure scenarios, and we will concentrate on the research relating the epigenetic mechanisms of the diseases. An extensive tabulation of chemical exposure related epigenetic changes and disease outcome may be found in our earlier review (6).
Global DNA methylation changes, and LINE1 and Alu methylation status in toxicoepigenetics of cardiovascular diseases
Movassagh et al. reported altered DNA methylation in heart failure patients (7). A large population of CpG islands within gene promoters had significant hypomethylation at end-stage cardiomyopathic hearts and irrespective of the etiology of the heart failure, had convergent methylation patterns in genes relevant to myocyte apoptosis, fibrosis and altered contractility. In addition, immunoprecipitation of the genome wide H3K36m3 sites and DNA sequence analysis showed distinct enrichment of the gene related regions (i.e., a region corresponding to a gene) of the genome in end-stage cardiomyopathy left ventricular samples, compared to normal controls (8). Elevated plasma homocysteine level is positively associated with atherosclerosis and CVD risk (9), although, failure to prevent CVD using homocysteine-lowering treatment overrides this long standing claim, suggesting that homocysteinemia cannot be directly associated as a cause of CVD (10). The alternative hypothesis favors that reduced methylation capacity, as determined by the SAM/SAH ratio, is a greater predictor of CVD risk than absolute homocysteine levels. One of the most extensively used surrogate markers for genome wide epigenetic changes is the interspersed repetitive elements, (LINE-1) and Alu. The LINE-1 retrotransposons constitute 17% of the human genome, are GC poor stretches of sequences about 6–8 kb long. Alu elements are shorter, about 300 bp long and make up nearly 10% of the genome. Baccarelli et al investigated the role of recent exposure to ambient particulate pollutants in relation to DNA methylation in a cohort of 718 elderly subjects from the Normative Aging Study. They showed that elevated black carbon content and PM2.5 exposure was significantly associated with decreased LINE-1 methylation (β = −0.11, 95% CI, P=0.002, and β = −0.13, 95% CI, P=0.001) in repeated temporal windows ranging between 4 hours to 7 days before blood drawing. Lower LINE-1 methylation correlated with higher systolic, diastolic and mean arterial blood pressures along with higher VCAM-1 expression in serum (11). LINE-1 methylation also inversely correlated with existing diagnosis of hypertension, and preexisting ischemic heart disease and stroke (12). Increased Alu methylation is related to increase risk for stroke (13). Differential dietary pattern had their signatures on peripheral blood leukocyte LINE 1 methylation as observed from the North Texas Healthy Heart study (14). Participants with high vegetable diets had lower prevalence of DNA hypomethylation compared to those on Western diet, composed of high meat intake and carbohydrates.
Polymorphisms in methionine cycle genes and CVD
Polymorphisms in the epigenetic modifier enzymes of the methionine cycle modified the effect of the environment on heart rate variability. The enzyme methylene tetrahydrofolate reductase (MTHFR) is required for the conversion of 5-10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is the methyl donor for the conversion of homocysteine to methionine. The CT and TT genotypes (compared to CC) of the C677T polymorphism in MTHFR are associated with reduced enzyme activity, and are associated with increased CVD. Baccarelli et al. (15) showed that CT/TT MTHFR carriers showed a statistically significant reduction in heart rate variability associated with PM2.5 exposure, thereby posing increased CVD risk. The TT genotype of the C1420T polymorphism of the cytoplasmic serine hydroxymethyl transferase (cSHMT) was associated with higher homocysteine levels. A diet rich in folate, vitamin B6, B12 or methionine abrogated the effects of PM on heart rate variability.
Epigenetic regulation of vascular endothelial genes in atherosclerosis and ischemia
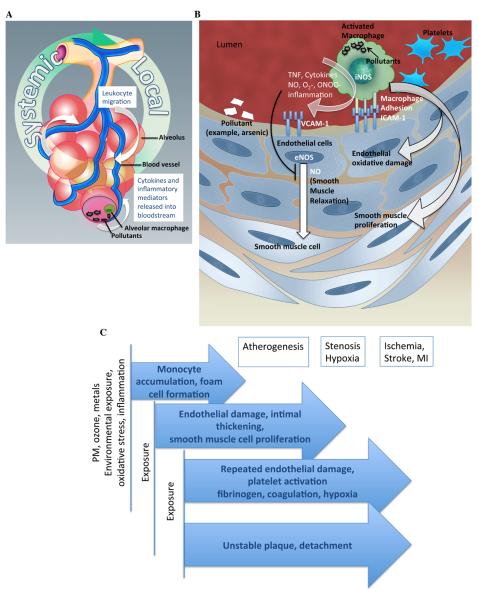
Inflammatory gene activation is the hallmark of atherogenesis and can be triggered by various environmental exposures including inhaled toxic and particulate matter (PM) (Figure 2). Pollutants primarily act as inducers of oxidative stress, and carbon and transition metals in inhaled pollutants contribute largely to this effect. Heavy metal arsenic (As) has strong atherogenic potential, which is mediated through direct deposition of on the endothelial wall. In vitro studies reported induction of NF- kappa B activation and reactive oxygen species generation and apoptosis of endothelial cells by arsenite (16). NF-kappa B mediated induction of oxidative stress response genes, cytokines such as Tumor Necrosis Factor alpha (TNF) and adhesion molecules such as ICAM-1 and VCAM-1 on the endothelial surface promote leukocyte transmigration, monocyte accumulation, smooth muscle cell proliferation (Figure 2), leading to formation of the atherosclerotic plaques. Elevation in procoagulant factors like fibrinogen promotes atherothrombosis, and is a known marker of CVD.
Figure 2.
Mechanism of environmental exposure mediated cardiovascular outcome. A. Air pollution induces release of cytokines and chemokines, causing inflammatory cellular recruitment and local inflammation and cyclical systemic impact through the vasculature. B. Inflammatory and oxidative stress induces atherosclerotic processes. Ingestion of particulate matter activates macrophages, induces reactive oxygen species, monocyte adhesion molecules and accumulation of monocytes on endothelial layer, foam cell transformation. Subsequently, endothelial cell dysfunction and smooth muscle cell proliferation take place. TNF, tumor necrosis factor; iNOS, inducible nitric oxide synthase; NO-, nitric oxide, O2-, superoxide; ONOO-, peroxynitrile; ICAM-1 Intercellular adhesion molecule 1; VCAM-1, Vascular cell adhesion molecule 1; eNOS, endothelial nitric oxide synthase. C. Exposure to environmental pollutants causes oxidative stress and inflammation, which triggers onset of, or exacerbates cardiovascular disease process at any stage of progression.
Most recent studies by Bind et al. reported that subjects with Alu hypermethylation had higher increase in CVD-related marker fibrinogen in the peripheral blood upon exposure to traffic air pollution (6.4% increase in fibrinogen per interquartile range increase in NO2), compared to those with lower methylation (2.4% increase in fibrinogen) (17). Increase in fibrinogen by air pollution was also associated with lower methylation and higher expression of tissue factor 3 (TF3).
NF-kappa B
As indicated above, NF-kappa B activation in the cells following exposure triggers the inflammatory gene expression cascade (18). It is now well known that NF-kappa B dependent transcriptional activation is regulated by histone methylation and acetylation events. The co-activator PCAF (p300/CBP associated factor) has intrinsic lysine acetyl transferase (KAT) activity, which acetylated the ε-group on lysine residues on the H3 and H4 histones at the NF-kappa B binding promoter regions, thereby allowing NF-kappa B binding and activity. A polymorphic association of PCAF promoter with coronary heart disease mortality suggests that it may play a regulatory role in CVD. The G genotype of the G/C polymorphism at the −2481 site of the promoter is believed to increase its expression and is associated with increased CHD mortality (19). Histone deacetylases HDAC2 and 3, on the other hand inhibit NF-kappa B binding. Expression of HDAC2 is reduced in cigarette smokers, thereby leading to unrestrained NF-kappa B activation and higher activation of pro-inflammatory genes (20).
Inducible nitric oxide synthase (iNOS)
iNOS is the mediator of acute inflammatory response in tissue. Methylation at the CpG sites of the iNOS promoter was found to be lower in the peripheral blood of the furnace steel plant workers following exposure to work environment (21).
Endothelial nitric oxide synthase (eNOS)
Vasomotor responses to several vasodilator agents was attenuated in normal healthy volunteers as early as 2 h after diesel exhaust exposure containing 300 μg/m3 PM. Endothelial nitric oxide synthase (eNOS) is responsible for vascular NO production and smooth muscle relaxation. Inhalation of CAP plus ozone led to inhibition of NO production (22). It is well known that eNOS is subject to strong epigenetic regulation. eNOS is readily activated by the HDAC family member, SIRTUIN 1 (SIRT1), activated by resveratrol, a component of red wines. SIRT 1 deacetylates eNOS at the calmodulin binding region of the catalytic domain and increases its catalytic activity (23). In addition, a 27-nucleotide piece of double stranded RNA produced at the intron 4 is capable of regulating eNOS expression.
Bollati et al. investigated the effects of PM exposure on miR-222, miR-21 and miR-146a in foundry workers (24). Relative expressions of miR-222 and miR-21 after 3 days of work compared to a baseline of first day of the week increased by 3.17 and 1.13 fold respectively. miR-222 increase positively correlated with lead exposure (β=0.41). miR-146a increase was not statistically significant in this study. Overexpression of miR-222 was shown to reduce the expression of eNOS (25). A list of miRNA altered by environmental stress and implicated in cardiovascular diseases is provided (Table 1).
Table 1.
MicroRNAs in response to different environmental exposures and relation to cardiovascular disease
| Exposure | miRNA/miRNA regulatory gene | Change/Effect of | Target/Function | CVD relevance | References |
|---|---|---|---|---|---|
| PM, carbon black | Dicer polymorphism rs13078 | Minor allele A | miRNA biogenesis | Correlated with higher serum sICAM-1 and sVCAM-1 levels | (26) |
| GEMIN 4 polymorphism rs1062923 | Minor allele C | miRNA biogenesis | Higher sVCAM-1 levels | ||
| Air pollution, metal pollutants | miR 222 | Increased in peripheral blood | cKit, p57 (Kip2) | Induce vascular smooth muscle cell growth, angiogenesis (27); reduction in eNOS, vasoconstriction (25) | (24) |
| miR 21 | Phosphatase PTEN, PI3 Kinase pathway | Prevents cardiomyocyte apoptosis in MI (28) | |||
| Aluminum | miR 146a | Increased, in vitro experimental model | NF-kappa B dependent, oxidoreductive pathway, ErbB pathway | Cardiomyocyte apoptosis cardiac hypertrophy (29) | (30) |
| Bisphenol A | miR 146a | Increased in placental cells | (31) | ||
| Alcohol | miR199a | Increased in liver sinusoidal endothelial cells | Hypoxia Inducible Factor HIF-1 α, Sirtuin 1. | Prevents hypoxia injury | (32) |
Hypoxia Inducible Factor (HIF1)
Atherosclerotic and stenotic conditions induce hypoxia inducible factor (HIF1), expression, which initiates a self-sustaining vicious cycle by binding to the promoters and inducing more pro-inflammatory genes, the inducible NOS (iNOS), vascular endothelial growth factor (VEGF) and others. HIF-1 recruits histone demethylases, Jumonji Domain 1 A (JMJD1A) and 2 A, which remove the H3K9 and H2K9 methyl groups respectively at the promoter regions (33).
Epigenetic regulation of dilated cardiomyopathy and heart failure
Epigenetic mechanisms underlie diet induced CVD. Individuals conceived during the period of acute food shortage at the end of World War II known as Dutch Hunger Winter had multiple complications including increased risk of coronary heart disease. In a comparison for CpG methylation in early gestational famine exposed versus non-exposed siblings, DNA methylation was significantly increased in imprinted loci of signaling G-protein alpha subunit gene GNAS-antisense 1 (GNASAS1), maternally expressed protein MEG3, and in non-imprinted IL10 promoters (34). On the other hand there was a decrease in insulin like growth factor (INSIGF). High glucose diet in is associated with decreased expression of insulin like growth factor receptor IGF-1R via p53 activation of HDAC1. HDAC1 deacetylates H4 histones in the IGF-1R promoter. IGF-1 is a promoter of cell survival, lack of which appears to be the basis of p53 mediated cardiomyocyte apoptosis in diabetic hyperglycemia in humans, as shown in the rat model of the disease (35).
Epigenetic regulation of neurohormonal axis regulating hypertension
The enzyme corticosteroid 11-β-hydrogenase isozyme 2 (HSD11B2) is an NAD+-dependent enzyme that oxidizes glucocorticoids to the inactive metabolite cortisone. Decreased HSD11B2 activity is related to hypertension. Friso et al. showed that the promoter methylation of HSD11BD2 in the peripheral blood mononuclear cells was upregulated in hypertensive patients, concurrent with the increase in their urinary THFs/THE ratio (tetrahydrocorticosol vs tetrahydocortisone ratio) (36). Function of catecholaminergic neurons in the vasomotor centers of the brainstem may be epigenetically regulated by environmental changes through histone acetylation inducer melatonin (37).
Conclusion
Epigenetics is anticipated to greatly advance our current understanding in cardiovascular diseases research. Several epigenetic biomarkers are emerging. The LINE1 methylation status or SAM/SAH ratio in peripheral blood are indicators of the methylation perturbations and may serve as predictors of exposure-related future CVD risk. Known polymorphisms such as those in the methionine-cycle genes, for example MTHFR, can also serve as predictors of disease risk. Circulating levels of miR 126, miR 17, miR 145 and miR 222 have emerged as prospective indicators of exposure related cardiovascular risk. Future directions in epigenetic research will be able to link universal and unique epigenetic markers of exposure related CVD outcomes and will bring about major advances in CVD theragnostics.
Key points
Acute and chronic environmental stresses induce epigenetic alterations driving gene expression relevant to the onset or progression of CVD.
Epigenetic changes can be monitored in peripheral blood and therefore can serve as important biomarkers.
Epigenetic targets offer therapeutic potential in post-exposure CVD.
Acknowledgements
The authors would like to acknowledge Arunabha Sengupta for the digital artwork.
Disclosure of funding received for this work:
Dr. Baccarelli receives salary support from the following grants from the National Institute of Health: P30ES000002; R21ES020010, R21ES019773; R01ES020268
Footnotes
Conflict of Interest:
The authors have no conflict of interests.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Heart Disease and Stroke Statistics--2012 Update: A Report From the American Heart Association. Circulation. 2012 Jan;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007 Feb;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- **3.Shirodkar AV, Marsden PA. Epigenetics in cardiovascular disease. Curr Opin Cardiol. 2011 May;26(3):209–15. doi: 10.1097/HCO.0b013e328345986e. [DOI] [PMC free article] [PubMed] [Google Scholar]; Very helpful review, with lysine methylations on histones and their effect at a glance, and extensive information on epigenetic alterations in endothelial cells in CVD.
- 4.Chen S, Sang N. Histone deacetylase inhibitors: the epigenetic therapeutics that repress hypoxia-inducible factors. J Biomed Biotechnol. 2011;2011:197946. doi: 10.1155/2011/197946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *5.Bhatnagar A. Environmental cardiology: studying mechanistic links between pollution and heart disease. Circulation research. 2006 Sep;99(7):692–705. doi: 10.1161/01.RES.0000243586.99701.cf. [DOI] [PubMed] [Google Scholar]; Very good overview of the topic.
- *6.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012 Feb;41(1):79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extensive catalog of environmental exposure and epigenetic changes
- 7.Movassagh M, Vujic A, Foo R. Genome-wide DNA methylation in human heart failure. Epigenomics. 2011 Feb;3(1):103–9. doi: 10.2217/epi.10.70. [DOI] [PubMed] [Google Scholar]
- **8.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011 Nov;124(22):2411–22. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors have mapped the DNA and histone methylations in failing versus normal hearts. Model discovery approach.
- 9.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, et al. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003 Aug;49(8):1292–6. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 10.Bonaa KH, Njolstad I, Ueland PM, Schirmer H, Tverdal A, Steigen T, et al. Homocysteine lowering and cardiovascular events after acute myocardial infarction. N Engl J Med. 2006 Apr;354(15):1578–88. doi: 10.1056/NEJMoa055227. [DOI] [PubMed] [Google Scholar]
- 11.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009 Apr;179(7):572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics. 2010;5(3) doi: 10.4161/epi.5.3.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5(3):e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, et al. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr. 2011 Jun;141(6):1165–71. doi: 10.3945/jn.110.134536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccarelli A, Cassano PA, Litonjua A, Park SK, Suh H, Sparrow D, et al. Cardiac autonomic dysfunction: effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation. 2008 Apr;117(14):1802–9. doi: 10.1161/CIRCULATIONAHA.107.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsou TC, Tsai FY, Wu MC, Chang LW. The protective role of NF-kappaB and AP-1 in arsenite-induced apoptosis in aortic endothelial cells. Toxicol Appl Pharmacol. 2003 Sep;191(2):177–87. doi: 10.1016/s0041-008x(03)00239-4. [DOI] [PubMed] [Google Scholar]
- 17.Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, et al. Air Pollution and Markers of Coagulation, Inflammation, and Endothelial Function: Associations and Epigene-environment Interactions in an Elderly Cohort. Epidemiology. 2012 Mar;23(2):332–40. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churg A, Xie C, Wang X, Vincent R, Wang RD. Air pollution particles activate NF-kappaB on contact with airway epithelial cell surfaces. Toxicol Appl Pharmacol. 2005 Oct;208(1):37–45. doi: 10.1016/j.taap.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Pons D, Trompet S, de Craen AJ, Thijssen PE, Quax PH, de Vries MR, et al. Genetic variation in PCAF, a key mediator in epigenetics, is associated with reduced vascular morbidity and mortality: evidence for a new concept from three independent prospective studies. Heart. 2011 Jan;97(2):143–50. doi: 10.1136/hrt.2010.199927. [DOI] [PubMed] [Google Scholar]
- 20.Yang SR, Chida AS, Bauter MR, Shafiq N, Seweryniak K, Maggirwar SB, et al. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006 Jul;291(1):L46–57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 21.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009 Feb;117(2):217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai Y, Suzuki AK, Sagai M. The cytotoxic effects of diesel exhaust particles on human pulmonary artery endothelial cells in vitro: role of active oxygen species. Free Rad Biol & Med. 2001;30(5):555–62. doi: 10.1016/s0891-5849(00)00499-8. [DOI] [PubMed] [Google Scholar]
- 23.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci USA. 2007;104(37):14855–60. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect. 2010 Jun;118(6):763–8. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007 Apr;100(8):1164–73. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 26.Wilker EH, Alexeeff SE, Suh H, Vokonas PS, Baccarelli A, Schwartz J. Ambient pollutants, polymorphisms associated with microRNA processing and adhesion molecules: the Normative Aging Study. Environ Health. 2011;10:45. doi: 10.1186/1476-069X-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009 Feb 27;104(4):476–87. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009 Oct;284(43):29514–25. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horie T, Ono K, Nishi H, Nagao K, Kinoshita M, Watanabe S, et al. Acute doxorubicin cardiotoxicity is associated with miR-146a-induced inhibition of the neuregulin-ErbB pathway. Cardiovas Res. 2010 Sep;87(4):656–64. doi: 10.1093/cvr/cvq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TP, Percy ME, et al. Characterization of an NF-kappaB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009 Nov;103(11):1591–5. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, et al. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol. 2010 Jul;29(4):401–6. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J Immunol. 2009 Oct;183(8):5232–43. doi: 10.4049/jimmunol.0901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brigati C, Banelli B, di Vinci A, Casciano I, Allemanni G, Forlani A, et al. Inflammation, HIF-1, and the Epigenetics that follows. Mediators Inflamm. 2010;2010:263914. doi: 10.1155/2010/263914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009 Nov;18(21):4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu XY, Geng YJ, Liang JL, Lin QX, Lin SG, Zhang S, et al. High levels of glucose induce apoptosis in cardiomyocyte via epigenetic regulation of the insulin-like growth factor receptor. Exp Cell Res. 2010 Oct;316(17):2903–9. doi: 10.1016/j.yexcr.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Friso S, Pizzolo F, Choi SW, Guarini P, Castagna A, Ravagnani V, et al. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008 Aug;199(2):323–7. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 37.Millis RM. Epigenetics and hypertension. Curr Hypertens Rep. 2011 Feb;13(1):21–8. doi: 10.1007/s11906-010-0173-8. [DOI] [PubMed] [Google Scholar]