Abstract
Alternative reproductive cycles make use of different strategies to generate different reproductive products. In Escherichia coli, recA and several other rec genes are required for the generation of recombinant genomes during Hfr conjugation. During normal asexual reproduction, many of these same genes are needed to generate clonal products from UV-irradiated cells. However, unlike conjugation, this latter process also requires the function of the nucleotide excision repair genes. Following UV irradiation, the recovery of DNA replication requires uvrA and uvrC, as well as recA, recF, and recR. The rec genes appear to be required to protect and maintain replication forks that are arrested at DNA lesions, based on the extensive degradation of the nascent DNA that occurs in their absence. The products of the recJ and recQ genes process the blocked replication forks before the resumption of replication and may affect the fidelity of the recovery process. We discuss a model in which several rec gene products process replication forks arrested by DNA damage to facilitate the repair of the blocking DNA lesions by nucleotide excision repair, thereby allowing processive replication to resume with no need for strand exchanges or recombination. The poor survival of cellular populations that depend on recombinational pathways (compared with that in their excision repair proficient counterparts) suggests that at least some of the rec genes may be designed to function together with nucleotide excision repair in a common and predominant pathway by which cells faithfully recover replication and survive following UV-induced DNA damage.
Genetic recombination is universal to all organisms and it is clearly an important, sometimes essential, component of many reproductive cycles. However, not all recombination is beneficial for the cell or organism in which it occurs. When it occurs at the wrong time in the cell cycle or the wrong place in the genome, it can create the rearrangements, duplications, and deletions found in cancer cells, or it can create the deregulated overreplication of genomic material that occurs during lytic viral replication. Thus, in trying to understand the biological role of recombination in a given cellular context, it is important to consider the strategy and products of the reproductive cycle being studied.
Different reproductive cycles use different strategies to produce their own distinct products, and in some reproductive cycles recombination plays a very clear role. Of the reproductive cycles that do rely prominently on recombination, many appear to tolerate or even promote variation among their progeny. Among eukaryotic organisms, sexual reproductive cycles produce variation and create progeny that are genetically distinct from their parents. That variation is achieved in part because genetic material from more than one cell is used, and in part, because homologous strand exchanges occur during the reproductive cycle. Following chromosomal replication in meiotic cells, the homologous chromosomes are aligned and paired, and numerous strand exchanges occur.
Recombinational strategies are also prominent in many viral reproductive cycles. In Herpes simplex virus, the progeny are largely comprised of recombinant genomes (1, 2). Here, the recombinational exchanges are intimately linked with the onset of lytic viral replication and have been proposed to be used as substrates to initiate replication, thereby allowing the virus to rapidly produce more of its genetic material than it might if it relied on a unique origin of replication (1, 2). However, the recombinational replication produces a large, intricate meshwork of branched, genomic concatamers rather than discrete viral chromosomes. Although much of the genetic material produced will neither form complete genomes nor be packaged into particles, this strategy is effective because the virus relies more on mass production than precision in reproducing its genetic message. Thus, viruses like Herpes use recombination as a mechanism to escape from the limited genomic doubling that occurs during processive replication in the host cell. In fact, many viruses inactivate cellular proteins, such as p53 and Rb, which otherwise suppress recombinational replication and prevent genomic rearrangements from occurring (3–6).
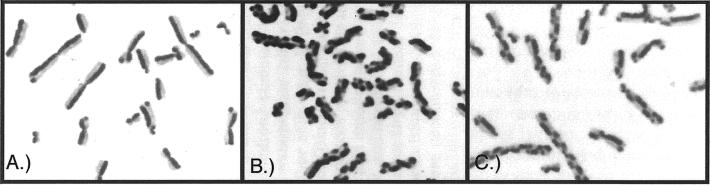
Unlike the sexual and viral reproductive cycles noted above, recombination is not clearly advantageous for all reproductive strategies. The mitotic (asexual) reproductive cycles of eukaryotes produce precisely two genetically identical clones through processive replication of the parental genome. In general, recombination is not detected. On the large chromosomes of mammals and insects, the number of exchange events between sister chromatids can be observed directly by labeling them differentially (reviewed in ref. 7). What is perhaps most striking about this type of analysis is precisely how few exchanges are observed in mitotic cells (Fig. 1A). In fact, the few observed exchanges are primarily caused by the nucleoside analogs or isotopes (e.g., BrdUrd or [3H]thymidine) used to label the chromatids for analysis (8, 9). In one study using reduced concentrations of BrdUrd, no strand exchanges were detected in a sample of 361 Drosophila cells (8). By analogy, this strand exchange frequency (i.e., zero) would amount to less than 1 exchange for every 20 cell divisions in human cells and less than 1 for over 10,000 cell divisions in Escherichia coli (9). Although such a direct comparison is clearly flawed, the point should not be lost that recombinational exchanges during asexual cell cycles are extremely infrequent. When higher frequencies of strand exchange occur, they appear to be detrimental to the reproductive success of the cell (Fig. 1 B and C). In mammalian cells, the frequency of strand exchange correlates quite directly with genomic instability, cell death, and carcinogenic transformation and, in fact, is often used as a diagnostic and prognostic marker in cancer patients (10–13). Thus, whereas the role of recombination is quite clear in reproductive cycles that tolerate variations among the quantity or genetic makeup of the progeny, when recombination occurs during asexual reproductive cycles the effects often appear to be detrimental.
Figure 1.
Visualization of sister chromatid exchanges during genomic replication in human cells. By growing cells in the presence of the thymine analog, 5-BrUra, for two generations, sister chromatids can be differentiated by using Giemsa stain. Exchanges between sister chromatids can then be observed directly by the staining pattern. (A) In normal human cells, semiconservative replication is maintained and few, if any, exchanges are observed. (B) However, cells from patients with the cancer-prone disorder of Bloom's syndrome lack a RecQ homolog that allows recombination to occur more frequently between sister chromatids. (C) Sister chromatid exchanges can also be induced in normal cells by treatment with carcinogenic agents such as MMS. [Photos taken from Sister Chromatid Exchange, S. Wolff, ed. (Copyright 1982; this material is used by permission of John Wiley & Sons, Inc.; ref. 7) and from The Chromosomes in Human Cancer and Leukemia, A. A. Sandberg (Copyright 1990; McGraw–Hill Companies, Inc.; ref. 62).]
Considering these examples, can analogies be drawn between the different reproductive cycles of eukaryotes and prokaryotic organisms such as E. coli? In our opinion, the different prokaryotic cycles appear quite analogous to those of eukaryotes. Several aspects of bacterial conjugation are similar to the sexual cycles of eukaryotes (reviewed in refs. 14 and 15). During conjugational reproduction, new genetic material is transferred from one parent cell to another and recombinant products can be isolated, thereby producing progeny that are genetically distinct from the original parent cells. Furthermore, many bacteriophage, including T4 and λ, use recombination as a mechanism to amplify their own genetic material during lytic replication, similar to the amplification strategies of mammalian viruses (reviewed in refs. 16 and 17). However during asexual reproduction, as with eukaryotic cells, the frequency of recombination is much less. Using an analysis based on chromosome linkage during replication, Peter Kuempel (18) estimated that an exchange event occurs in only 15% of E. coli replication cycles. The actual value could be significantly lower considering that the experimental analysis required labeling cells with both BrdUrd and [3H]thymidine, the same agents known to stimulate sister chromatid exchanges in mammalian cells. In E. coli, 5-BrUra (5-BU) is also associated with toxicity and increased levels of recombination. The bromine group of 5-BU is labile and leads to the initiation of uracil glycolyase-induced nicks in the DNA (19). Furthermore, strains that incorporate 5-BU fail to replicate beyond roughly two generations. Extended growth causes a lethality that resembles a “thymineless death-like” phenotype that is also associated with an aberrant hyperrecombinational form of replication (19, 20). The introduction of the thy-mutation, which is required so that E. coli will use 5-BU, has also been demonstrated to increase the recombinational frequencies and cause abnormal replication patterns in E. coli (20, 21). Clearly, chromosomal replication is not completely normal in 5-BrUra containing media. Additionally, several observations (discussed below) suggest that when recombination does occur in the asexual cycle of prokaryotes, it is often detrimental to the genomic stability and reproductive success of the cell.
The high frequencies of recombination that occur during conjugation and phage replication have made these systems extremely useful for dissecting the mechanism by which recombination occurs. However, these recombinational reproductive cycles have very different strategies and products from those of the asexual reproductive cycle, and thus, in trying to understand the role of recombination in asexual reproduction, it is useful to consider the differences between the strategies and products of these processes.
The fundamental mechanism by which cellular replication occurs (i.e., processive duplication of the genomic template) provides several intuitive reasons to suspect that strand exchanges may confound rather than promote the generation of the desired clonal products during an asexual reproductive cycle. Although there are clearly special situations, such as double-strand breaks or interstrand crosslinks, for which recombination may be essential for the repair process, the basic observations stated above suggest that strand exchange may be neither a frequent nor productive DNA transaction during the asexual process of genomic replication. In marked contrast to this view, however, is the prevailing hypothesis that recombination serves as an efficient and nearly essential “repair” mechanism, required to maintain genomic stability during cellular reproduction (22–26).
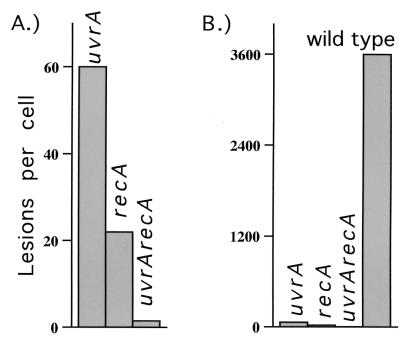
The concept of recombination as a mechanism of DNA repair developed from the founding study by Clark and Margulies (27) in which they identified recA of E. coli as a gene that was required for genetic recombination. In recognizing the sexual nature of recombination, the authors defined the process as “the inheritance by recombinant progeny of double-stranded elements of DNA derived from two parents” and they used an Hfr conjugation assay in which they could select for recombinant genomes based on the inheritance of growth properties from both parents. Using this technique, they identified recA and, subsequently, several additional genes involved in the formation of recombinant molecules during bacterial sex. Clark and Margulies also made the important observation that many of their recombination-deficient mutants were hypersensitive to UV light and ionizing radiation during the normal asexual reproductive cycle. Based on these observations, Howard-Flanders and Theriot (28) speculated that recombination may additionally operate during asexual reproduction as a mechanism for repairing DNA lesions such as those generated by UV light. To test this possibility, Howard-Flanders and his colleagues (29) reasoned that if recA promotes recombinational DNA repair, then that should be most easily revealed in a uvrA mutant that cannot remove UV-induced lesions from its genome because of a defect in nucleotide excision repair. Indeed, they found that a recA mutation further sensitized uvrA mutants to UV irradiation and thus concluded that the survival promoted by recA could represent a recombinational mechanism of repair, independent of excision repair (Fig. 2A). Following this initial interpretation, it was assumed in many subsequent studies that recA promotes recombinational repair. Investigators then went on to characterize the molecular events occurring in uvr mutants with the belief that the observed phenotypes must have resulted from this new repair pathway. These subsequent studies in uvr mutants revealed that the very limited replication occurring after UV irradiation was fragmentary and accompanied by high frequencies of strand exchanges (30–33). Because these events were assumed to represent repair, a model was proposed in which RecA promoted recombination as a mechanism to reconstruct genomes from the partially replicated sequences of undamaged regions. Because of the extreme hypersensitivity of recA mutants to UV, the general view evolved that the proposed recombinational function must represent a major repair pathway, required for cellular survival and genomic stability (30–33).
Figure 2.
The average number of UV-induced lesions that results in lethality is plotted for each E. coli strain. (A) Focusing on the question, Can recombination promote recovery following UV irradiation?, it has been suggested that because a uvrA mutant is more resistant to UV irradiation than a uvrArecA mutant, recA may promote survival in the uvrA mutant by recombining around DNA lesions. (B) However, focusing on the question, How do cells recover following UV irradiation?, one might conclude that the recombination does not significantly contribute to the recovery seen in wild-type cells (lesion numbers plotted: uvrA, 60; recA, 22; uvrArecA, 1.5; wild type, 3,600; as reported in ref. 28).
Considering that recA was discovered through a recombination assay and that recombination was the only known phenotype associated with recA, the proposal was very reasonable, and it led to a line of investigation that has unquestionably provided an immense amount of information about the genetic elements involved in recombination. However, today we know much more about the different strategies of reproductive cycles and more specifically about RecA function itself, which allows for additional and alternative possibilities. For example, we now know that RecA itself is central to the induction and regulation of over three dozen genes (termed the SOS response) in response to the arrest of replication (reviewed in ref. 34; see also ref. 35). When one examines the functions of the genes that are induced, one is struck by the observation that most of them have nothing to do with recombination, but instead center on the task of restoring processive replication. Among the genes up-regulated by RecA are the uvrA and uvrB genes, which are required for removing UV-induced lesions from the DNA template. Recently, this induction was shown to be critical for efficient DNA repair and survival of E. coli following moderate UV doses (36). Indeed, in the absence of the SOS response, there is very poor repair of the predominant UV-photoproduct, the cyclobutane pyrimidine dimer. Although the SOS-controlled excision repair is recA-dependent, it would be expected to reduce rather than increase the need for recombination to deal with the problems of UV-induced genomic stress. Other genes up-regulated by RecA include at least three DNA polymerases, polB, dinB, and umuCD (Pol II, Pol IV, and Pol V), thought to operate in translesion DNA synthesis, a process that by its nature does not require recombination. It helps the replication machinery processively overcome DNA lesions that otherwise block replication (37–39).
Based on these recent advances, we have reexamined the original question, Is recombination an efficient mechanism for repairing DNA damage? (40). A simple alternative that the initial speculation and experiments could not address, and that may be worth reconsidering now, is that recA function may not be specific to recombinational processes. Perhaps recA is required for a fundamental step common to both types of reproductive cycles; one that is necessary to achieve the recombinant products of Hfr conjugation and the clonal products of asexual replication. Even though the end products of each reproductive cycle are unique, the same recA function may be needed to successfully complete both processes, a perspective that we have recently considered in greater depth (40).
If the authors of these original studies had the benefit of what is now known about the different reproductive cycles, recA function, and genomic replication, they might have alternatively suggested that recombination is not a predominant repair pathway—a reasonable conclusion considering the dramatically higher UV resistance of wild-type cells compared with that of either uvrA or recA mutants (Fig. 2B). Clearly, the survival promoted by recA is synergistically increased in the presence of excision repair, a phenomenon that was not addressed in the original study, nor in most subsequent studies that have focused on recombinational pathways of recovery. Fundamental genetics would argue that the contributions of truly independent pathways should be additive. Yet, the UV survival of wild-type cells reduces to almost zero in the absence of either gene, suggesting that the recovery in wild-type cells requires that both genes are functional. From a practical point of view, the survival curves suggest that the ability of recA to promote recombination is virtually useless for cellular survival. Considering the survival of uvr mutants, the potentially minor contribution from recombination is further diminished if one considers that in the presence of recA, the polmerases Pol II, Pol IV, and Pol V are promoting survival by bypassing some lesions. Furthermore, it should be noted that RecA is a direct participant in the translesion synthesis carried out by Pol V, a process that clearly does not depend on recombination. One cannot avoid the conclusion that the vast majority of RecA-catalyzed events are, in fact, epistatic with excision repair. Although strand exchanges and some limited fragmentary replication are observed when uvr mutants are UV-irradiated, these populations also contain high levels of lethality, mutagenesis, and genomic rearrangements. This leads to the possibility that the few surviving cells could represent “lucky individuals” that either survived the recombinational exchanges or did not undergo extensive strand exchanges at all. The fact that significant levels of strand exchanges are observed in the uvr− populations in which replication does not recover normally could suggest that these strand exchanges represent a potentially lethal consequence of DNA lesions that cannot be repaired normally.
It seems reasonable that the phenotypes classically associated with recombinational repair could also be generated when blocked replication forks are prevented from recovering normally. Again, looking to analogies in eukaryotes, it is known that defects in human cells that prevent the recovery of replication produce similar phenotypes. Cells from the xeroderma pigmentosum variant (XPV) complementation group exhibit the same clinical features as classical, excision-repair-deficient, XP patients (sunlight sensitivity and cancer predisposition). However, following UV irradiation the XPV cells are proficient in repairing UV lesions. Yet, despite their ability to repair DNA lesions, replication does not recover and limited fragments of nascent DNA are generated. Additionally, the abnormally high frequencies of strand exchanges that are associated with repair-deficient XP complementation groups are not observed in XPV cells (41–43). Although such observations initially led to the interpretation that XPV cells were defective in recombinational repair, the results of more recent studies and the cloning of the XPV gene have shown that these phenotypes are produced by a homolog of the E. coli dinB gene that allows replication to continue processively through the otherwise blocking DNA lesions (41, 44). Thus, recombination is neither necessary nor desirable in that case.
A Common Pathway Involving Both Nucleotide Excision Repair and rec Genes
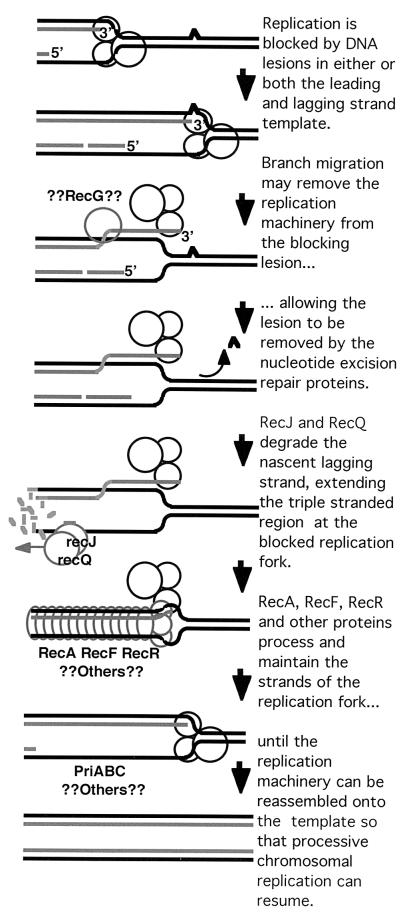
Several studies, including our own, have linked the synergism between recA function and excision repair, to an effect on the recovery of replication following UV irradiation (21, 45–47). Furthermore, other studies have observed a strong correlation between the time at which replication recovers and the time at which DNA repair is almost complete (45, 48, 49). To explain these observations, we have considered the possibility that during the asexual reproductive cycle, RecA function does not normally promote recombination to circumvent DNA lesions, but rather that it is needed to maintain and process replication forks that are blocked by DNA lesions so that those lesions can be removed as shown in Fig. 3 (21, 40, 45). Of course, if the offending lesion can be repaired, then there is no need for recombination to occur.
Figure 3.
Working model for the recovery of replication at a blocking DNA lesion.
A connection between the rec genes, nucleotide excision repair, and the recovery of replication was also made by P. K. Cooper and P.C.H. (48, 50) while characterizing a phenomenon they termed long-patch excision repair. The authors originally reported that the size distribution of the nucleotide excision repair patches in UV-irradiated E. coli was bimodal. Short patches appeared at early times and were shown to be due to normal nucleotide excision repair. At later times, correlating with the recovery of DNA replication, long patches were observed that were dependent on recA, recF, and the nucleotide excision repair genes. Furthermore, the long patches were shown to localize primarily at DNA replication forks and were found to be either 1,500 bp or greater than 9,000 bp in size (48, 51), corresponding to the DNA sizes expected for Okazaki fragments on the lagging-strand and leading-strand DNA synthesis, respectively. At the time, the authors simply noted that the long patches were important for the recovery of replication. In light of present knowledge however, several of these features are remarkably consistent with a mechanism for the processive recovery of replication as presented in Fig. 3.
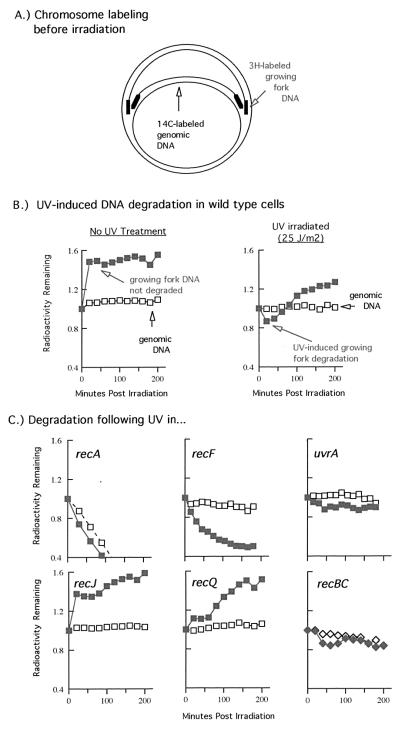
Using different techniques to identify genes that are required for the recovery of chromosomal replication when it is blocked by UV-induced DNA damage, we observe that replication does not recover in the absence of the rec genes, recA, recF, and recR, or the nucleotide excision repair genes, uvrA or uvrC (Fig. 4 and ref. 21 and 45). It remains likely that additional genes will be required in this process, but many have not yet been examined directly. One of the earlier suggestions for an alternative role of the rec genes during asexual replication came from the studies by Hori and Suzuki (52). By examining the fate of the genomic DNA after UV irradiation, they found that a rapid and complete degradation of the entire genome occurred in the recA mutants, something that did not occur in either wild-type or uvrA mutants. Furthermore, the genomic degradation only occurred if the recA mutants were actively replicating DNA at the time of irradiation. Through pulse–chase labeling techniques, they also found that the degradation initiated from the replication forks and processively degraded back from these points, leading them to initially propose that RecA plays a role in protecting the DNA at replication forks when they are blocked by DNA damage.
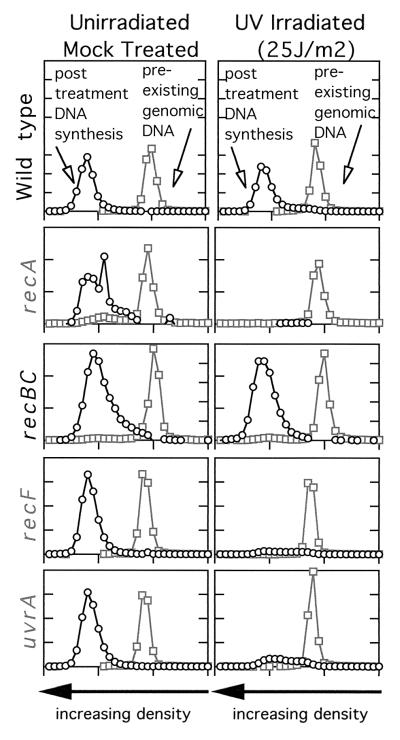
Figure 4.
The recovery of replication following UV irradiation requires both the nucleotide excision repair genes and the recA and recF genes. The amount of replication occurring within 1 h postirradiation was analyzed by alkaline CsCl density gradients. Cells prelabeled with [14C]thymine were either UV-irradiated or mock-treated. Cells were then filtered and grown in media containing [3H]BrdUrd for 1 h to density label the replication occurring during this period. [Adapted with permission from ref. 21 (Copyright 1997, PNAS) and ref. 45 (Copyright 1999, Am Soc. Microbiol.).] Whereas little replication occurs in irradiated recA, recF, or uvrA mutants, wild-type cells and recBC mutants recover replication within this time period.
Using a similar technique, we examined the degradation of both the genomic DNA and the nascent DNA at the blocked replication forks and observed that limited degradation of the nascent DNA at blocked replication forks occurs in wild-type cells at times before the recovery of replication. In the absence of either recF or recR (when replication does not recover), the nascent DNA degradation is much more extensive (Fig. 5 and ref. 21). We interpret this to mean that recF and recR are needed to protect and maintain the DNA strands of the replication fork when it is blocked by DNA damage. Interestingly, although uvrA and uvrC mutants also fail to recover replication, the nascent DNA of the blocked replication forks remains protected (45). This observation we interpret to suggest that although the rec machinery can function to protect and maintain the replication fork, replication cannot efficiently resume because the blocking lesion has not been removed.
Figure 5.
Degradation or processing of the nascent DNA at blocked replication forks following UV irradiation in various mutants. (A) A 10 s pulse of [3H]thymidine is added to [14C]thymine-prelabeled cells immediately before the cells are filtered and irradiated with 25 J/m2 in nonlabeled medium. To assay for degradation, the fraction of radioactivity remaining in the DNA, as measured by TCA precipitation, is plotted against time. (B) The loss (or degradation) of 14C genomic DNA (open symbols) can be compared with the loss of the 3H DNA synthesized at the growing fork just before irradiation (closed symbols). In wild-type cells, no DNA degradation is detected in the absence of UV irradiation. Following UV irradiation, limited degradation at the growing fork is detected at times before the recovery of replication. (C) In recA mutants, both the nascent DNA and the entire genomic DNA is rapidly degraded. In recF mutants, approximately half of the pulse-labeled nascent DNA is degraded. In uvrA mutants, the nascent DNA degradation is similar in extent to that in wild-type cells. In recJ or recQ mutants, no detectable degradation of nascent DNA occurs. In recBC mutants, the nascent DNA degradation is similar in extent to that in wild-type cells. [Adapted with permission from ref. 45 (Copyright 1999, Am. Soc. Microbiol.) and from Mol. Gen. Genet., “RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli,” J. Courcelle & P. C. Hanawalt, 262, pp. 543–551, figures 2 and 3 (Copyright 1999, Springer-Verlag; ref. 53).]
In characterizing the nascent DNA degradation, we demonstrated that other recF pathway genes, recJ and recQ, are responsible for the degradation and appear to preferentially degrade the nascent lagging strand of the blocked replication forks before their resumption (Fig. 4 and ref. 53). Interestingly, RecQ homologs in yeast, Drosophila, and humans have been found to play critical roles in maintaining processive replication and suppressing strand exchanges from occurring (Fig. 1B; reviewed in ref. 54). In E. coli, RecQ acts by increasing the single-strand region at blocked replication forks, creating a much larger substrate on which the RecA protein may bind and stabilize. Perhaps as is seen in other organisms, this action may help prevent recombination from occurring during replication. Potentially reflective of this role, RecQ has recently been shown to reduce the frequency of illegitimate recombination in E. coli as well (55).
Several aspects of the mechanism by which excision repair synergistically enhances the recovery promoted by these rec genes remain to be determined. One question that arises from transcriptional studies is, How does the excision repair machinery gain access to a lesion that may be hidden or obstructed by a blocked polymerase? In the case of transcription, an RNA polymerase blocked at a DNA lesion is known to prevent access and lesion recognition by the excision repair machinery (56, 57). Before repair can occur in E. coli, a special helicase-like protein, encoded by mfd, is needed to displace the RNA polymerase and transcript. In mfd mutants, transcription-blocking lesions are not rapidly repaired and the cells are moderately UV sensitive. One might expect that for excision repair machinery to access and repair the DNA lesion, the replication machinery and nascent DNA will also have to be transiently displaced. By analogy, perhaps a helicase, such as RecG, is required to displace the replication machinery and nascent DNA to effect repair. It is worth noting that RecG catalyzes branch migration on three-stranded branched substrates with a polarity that would be required for this function (58). Also of interest is the reported observation that recG shares a significant degree of homology with mfd and confers a moderate degree of hypersensitivity to UV irradiation when mutated (59). At present however, no direct experimental test of RecG action in replicational recovery has been reported.
Additional genes, of which many display poor viability or confer a UV-sensitive phenotype when mutated, have been speculated to be involved in the recovery of blocked replication forks, but have yet to be examined experimentally. Because any number of critical biological steps might be responsible for these viability problems, it will be important to ascertain where and when the defects in each of these mutants are manifested in the reproductive cycle. One surprising example of the need to carry out this rigorous scrutiny comes from the study of recBC mutants, which are deficient in double-strand-break repair. Based on their UV-sensitive phenotype and poor viability during growth, it was initially speculated that replication forks may frequently collapse to form double-strand breaks at DNA lesions and that RecBC was then required for replication recovery (60). However, multiple studies, including our own, have reported that the recovery of replication occurs quite normally in recBC mutants following UV irradiation, despite their hypersensitivity (21, 53, 61). Thus, in the case of recBC, and possibly other genes as well, survival analysis in itself does not provide adequate information to conclude that a gene is involved in the recovery of replication forks that are blocked by UV-induced lesions.
In our opinion, the poor survival and recovery of populations dependant on recombinational repair pathways (compared with that in their uvr+ counterparts) suggests that the rec and uvr genes may be designed to cooperate in a common and predominant pathway by which cells faithfully recover replication following DNA damage. Although recombination is vital to many cellular processes and is certainly essential to the generation of diversity, adaptation, and evolution, perhaps during asexual replication it does not efficiently promote cellular survival. Genomic replication appears to accomplish its monumental task of duplicating multimegabase chromosomes at high fidelity by maintaining a symmetry throughout this process. Although the recombination proteins are indeed central to this process, it is worth reflecting more broadly on their possible roles to accurately describe how they function; taking a step back from the molecular level to the cellular level, to appreciate the symmetry and stability of the overall replication process.
Acknowledgments
We thank Gerald Smith, Allan Campbell, and our long-standing colleague Ann Ganesan for helpful discussions throughout the course of our study. This work was supported in part by Outstanding Investigator Grant CA44349 from the National Institutes of Health.
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Bataille D, Epstein A L. Biochimie. 1995;77:787–795. doi: 10.1016/0300-9084(96)88197-1. [DOI] [PubMed] [Google Scholar]
- 2.Severini A, Scraba D G, Tyrrell D L. J Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lepik D, Ilves I, Kristjuhan A, Maimets T, Ustav M. J Virol. 1998;72:6822–6831. doi: 10.1128/jvi.72.8.6822-6831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staib C, Pesch J, Gerwig R, Gerber J K, Brehm U, Stangl A, Grummt F. Virology. 1996;219:237–246. doi: 10.1006/viro.1996.0241. [DOI] [PubMed] [Google Scholar]
- 5.Ridgway P J, Hall A R, Myers C J, Braithwaite A W. Virology. 1997;237:404–413. doi: 10.1006/viro.1997.8782. [DOI] [PubMed] [Google Scholar]
- 6.Zacny V L, Wilson J, Pagano J S. J Virol. 1998;72:8043–8051. doi: 10.1128/jvi.72.10.8043-8051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolff S, editor. Sister Chromatid Exchange. New York: Wiley; 1982. pp. 41–57. [Google Scholar]
- 8.Gatti M, Santini G, Pimpinelli S, Olivieri G. Genetics. 1979;91:255–274. doi: 10.1093/genetics/91.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Latt S A. In: Sister Chromatid Exchange. Wolff S, editor. New York: Wiley; 1982. pp. 121–128. [Google Scholar]
- 10.Dhar P K, Devi S, Rao T R, Kumari U, Joseph A, Kumar M R, Nayak S, Shreemati Y, Bhat S M, Bhat K R. Cancer Genet Cytogenet. 1996;89:105–108. doi: 10.1016/0165-4608(95)00071-2. [DOI] [PubMed] [Google Scholar]
- 11.Donmez H, Ozkul Y, Ucak R. Mutat Res. 1996;361:129–132. doi: 10.1016/s0165-1161(96)90247-2. [DOI] [PubMed] [Google Scholar]
- 12.Husain S A, Balasubramanian S, Bamezai R. Cancer Genet Cytogenet. 1992;61:142–146. doi: 10.1016/0165-4608(92)90077-l. [DOI] [PubMed] [Google Scholar]
- 13.Murthy M K, Bhargava M K, Augustus M. Indian J Cancer. 1997;34:49–58. [PubMed] [Google Scholar]
- 14.Jacob F, Wollman E L. Sexuality and the Genetics of Bacteria. New York: Academic; 1961. [Google Scholar]
- 15.Lederberg J, Tatum E L. Science. 1953;118:169–175. doi: 10.1126/science.118.3059.169. [DOI] [PubMed] [Google Scholar]
- 16.Mosig G. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 17.Stahl F W. Gene. 1998;223:95–102. doi: 10.1016/s0378-1119(98)00246-7. [DOI] [PubMed] [Google Scholar]
- 18.Steiner W W, Kuempel P L. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanawalt P, Grivell A, Nakayama H. Basic Life Sci. 1975;5A:47–50. doi: 10.1007/978-1-4684-2895-7_7. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama K, Kusano K, Irino N, Nakayama H. J Mol Biol. 1994;243:611–620. doi: 10.1016/0022-2836(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 21.Courcelle J, Carswell-Crumpton C, Hanawalt P C. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox M M. Prog Nucleic Acid Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 23.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 24.Kogoma T. Proc Natl Acad Sci USA. 1997;94:3483–3484. doi: 10.1073/pnas.94.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzminov A. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marians K J. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 27.Clark A J, Margulies A D. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard-Flanders P, Theriot L. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard-Flanders P, Theriot L, Stedeford J B. J Bacteriol. 1969;97:1134–1141. doi: 10.1128/jb.97.3.1134-1141.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesan A K, Smith K C. Mol Gen Genet. 1971;113:285–296. doi: 10.1007/BF00272328. [DOI] [PubMed] [Google Scholar]
- 31.Ganesan A K. J Mol Biol. 1974;87:103–119. doi: 10.1016/0022-2836(74)90563-4. [DOI] [PubMed] [Google Scholar]
- 32.Rupp W D, Howard-Flanders P. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 33.Rupp W D, Wilde C E I, Reno D L, Howard-Flanders P. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 34.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 35.Courcelle J, Khodursky A, Peter B, Brown P O, Hanawalt P C. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crowley D J, Hanawalt P C. J Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rangarajan S, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner J, Gruz P, Kim S R, Yamada M, Matsui K, Fuchs R P, Nohmi T. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 40.Courcelle J, Ganesan A K, Hanawalt P C. BioEssays. 2001;23:463–470. doi: 10.1002/bies.1065. [DOI] [PubMed] [Google Scholar]
- 41.Cordeiro-Stone M, Zaritskaya L S, Price L K, Kaufmann W K. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 42.De Weerd-Kastelein E A, Keijzer W, Rainaldi G, Bootsma D. Mutat Res. 1977;45:253–261. doi: 10.1016/0027-5107(77)90025-2. [DOI] [PubMed] [Google Scholar]
- 43.Mamada A, Kondo S, Satoh Y. Photodermatology. 1989;6:124–130. [PubMed] [Google Scholar]
- 44.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 45.Courcelle J, Crowley D J, Hanawalt P C. J Bacteriol. 1999;181:916–922. doi: 10.1128/jb.181.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganesan A K, Seawell P C. Mol Gen Genet. 1975;141:189–205. doi: 10.1007/BF00341799. [DOI] [PubMed] [Google Scholar]
- 47.Rothman R H, Clark A J. Mol Gen Genet. 1977;155:279–286. doi: 10.1007/BF00272806. [DOI] [PubMed] [Google Scholar]
- 48.Cooper P K. Mol Gen Genet. 1982;185:189–197. doi: 10.1007/BF00330785. [DOI] [PubMed] [Google Scholar]
- 49.Setlow R B, Swenson P A, Carrier W L. Science. 1963;142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- 50.Cooper P K, Hanawalt P C. J Mol Biol. 1972;67:1–10. doi: 10.1016/0022-2836(72)90381-6. [DOI] [PubMed] [Google Scholar]
- 51.Cooper P K. Annual Progress Report to Department of Energy. Lawrence Berkeley Laboratory, Berkeley, CA: University of California; 1990. [Google Scholar]
- 52.Horii Z, Suzuki K. Photochem Photobiol. 1968;8:93–105. doi: 10.1111/j.1751-1097.1970.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 53.Courcelle J, Hanawalt P C. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 54.Karow J K, Wu L, Hickson I D. Curr Opin Genet Dev. 2000;10:32–38. doi: 10.1016/s0959-437x(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 55.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selby C P, Sancar A. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 57.Tornaletti S, Reines D, Hanawalt P C. J Biol Chem. 1999;274:24124–24130. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitby M C, Lloyd R G. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lloyd R G, Sharples G J. J Bacteriol. 1991;173:6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuzminov A. BioEssays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 61.Khidhir M A, Casaregola S, Holland I B. Mol Gen Genet. 1985;199:133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- 62.Sandberg A A. The Chromosomes in Human Cancer and Leukemia. New York: Elsevier; 1990. [Google Scholar]