Abstract
The ubiquitin proteasome system (UPS) regulates many biological pathways by post-translationally ubiquitylating proteins for degradation. Although maintaining a dynamic balance between free ubiquitin and ubiquitylated proteins is key to UPS function, the mechanisms that regulate ubiquitin homeostasis in different tissues through development are not clear. Here we show, via analysis of the magellan (magn) complementation group, that loss of function of the Drosophila polyubiquitin Ubi-p63E results specifically in meiotic arrest sterility in males. Ubi-p63E contributes predominantly to maintaining the free ubiquitin pool in testes. The function of Ubi-p63E is required cell-autonomously for proper meiotic chromatin condensation, cell cycle progression and spermatid differentiation. magn mutant germ cells develop normally to the spermatocyte stage but arrest at the G2/M transition of meiosis I, with lack of protein expression of the key meiotic cell cycle regulators Boule and Cyclin B. Loss of Ubi-p63E function did not strongly affect the spermatocyte transcription program regulated by the testis TBP-associated factor (tTAF) or meiosis arrest complex (tMAC) genes. Knocking down proteasome function specifically in spermatocytes caused a different meiotic arrest phenotype, suggesting that the magn phenotype might not result from general defects in protein degradation. Our results suggest a conserved role of polyubiquitin genes in male meiosis and a potential mechanism leading to meiosis I maturation arrest.
Keywords: Ubiquitin, Drosophila, Spermatogenesis, Meiosis, Spermatocyte
INTRODUCTION
In eukaryotic cells, proteins destined for degradation by the 26S proteasome are marked with covalently linked chains of ubiquitin (Ub) (Hochstrasser, 1996). The crucial role of Ub in protein degradation necessitates that cellular Ub is maintained at a homeostatic balance between free Ub monomer and Ub-conjugated proteins (Kimura and Tanaka, 2010). Although Ub can be recycled from Ub chains attached to protein substrates by deubiquitylating enzymes (DUBs) before tagged proteins enter the 26S proteasome for degradation (Reyes-Turcu et al., 2009), levels of free Ub are maintained primarily by de novo synthesis from ubiquitin genes (Ryu et al., 2008).
Conserved from yeast to mammals, ubiquitin genes are of two structural types: polyubiquitin genes and monomeric ubiquitin fusion genes. Polyubiquitin genes encode a precursor protein with many head-to-tail tandem Ub repeats, which is cleaved by the ubiquitin C-terminal isopeptidase activities of DUBs to produce monomeric Ub molecules (D’Andrea and Pellman, 1998; Lee et al., 1988; Özkaynak et al., 1984). Mono-ubiquitin genes encode a single Ub moiety fused directly at its C-terminus to ribosomal protein subunits, either RpS27 or RpL40 (Cabrera y Poch et al., 1990; Finley et al., 1989; Lee et al., 1988; Redman and Rechsteiner, 1989), with the single Ub moiety released post-translationally by ubiquitin C-terminal isopeptidase activities. Studies from S. cerevisiae suggest that the mono-ubiquitin genes are the primary contributors to cellular Ub synthesis under normal conditions, whereas expression of the polyubiquitin gene is highly stress inducible (Finley et al., 1987). In mouse, however, the polyubiquitin gene Ubc is required for normal embryonic development (Ryu et al., 2007).
Here we show that one of the three polyubiquitin genes of Drosophila, Ubi-p63E, is required specifically for meiotic cell cycle progression and spermatid differentiation in the male germ line. Males null for Ubi-p63E are viable, but show striking phenotypic similarities to the clinical pathology of meiosis I maturation arrest azoospermia, a common form of idiopathic male infertility in humans (Meyer et al., 1992). In both mammals and Drosophila, male germ cells descended from germline stem cells (GSCs) go through spermatogonial mitotic divisions and then switch to differentiation as spermatocytes. Primary spermatocytes grow in volume, turn on cell type-specific transcription and then undergo meiotic divisions before initiating spermatid differentiation (Fuller, 1993; Kierszenbaum and Tres, 1978).
Much of our knowledge of the molecular mechanisms coordinating the gene expression program for spermatid differentiation and meiosis in Drosophila stems from analysis of meiotic arrest mutants. Most of the previously identified meiotic arrest genes fall into two functional classes: genes that encode testis-specific paralogs of TBP-associated factors (tTAFs) or genes that encode protein components of the testis meiosis arrest complex (tMAC) (Beall et al., 2007; Hiller et al., 2004). The functions of both classes of genes are required to establish the spermatocyte transcription program that drives the expression of hundreds of spermatid differentiation genes. Meiotic arrest genes involved in other cellular processes, such as nucleolar integrity (Moon et al., 2011), are also beginning to emerge.
We found that a complementation group of Drosophila meiotic arrest mutants, magellan (magn), required for meiotic cell cycle progression and spermatid differentiation in vivo, map to the polyubiquitin gene Ubi-p63E, which is a major source of ubiquitin expression in spermatocytes. Consistent with the viable, female fertile but male sterile magn phenotype, Ubi-p63E appeared necessary for Ub homeostasis in testes but not in the adult body or ovary. The function of Ubi-p63E is required cell-autonomously in germ cells for normal chromatin condensation during meiotic prophase and for progression of the meiotic cell cycle through the G2/M transition of the first meiotic division. However, unlike the previously studied tTAF and tMAC meiotic arrest mutants, the spermatocyte transcription program was largely unaffected in magn mutants. The defects observed in magn mutants were unlikely to be due to general defects in protein degradation by the proteasome, as knockdown of proteasome function in male germ cells by RNAi had much more severe effects on the spermatocyte transcription program.
MATERIALS AND METHODS
Fly husbandry
Drosophila stocks were raised on cornmeal/dextrose or cornmeal/molasses media at 25°C. Fly strains were obtained from the Bloomington Stock Center and the Vienna Drosophila RNAi Center. Wild-type control flies were y,w unless otherwise stated.
RNA interference (RNAi)
Virgin UAS-Dicer2;;BAM-GAL4 females were crossed to y,w males (control) or males carrying RNAi hairpin against each of the proteasome subunits: Rpt2, Prosα3, Prosα7 or Prosβ7. Crosses were grown at 29°C. Effectiveness of the RNAi knockdown of individual proteasome subunits was assessed by directly comparing the mRNA level with that of control testes samples by in situ hybridization (supplementary material Fig. S4).
Deficiency mapping, cloning of magn and generation of magn null alleles
The original magn alleles, magnZ3-4216, magnZ3-3969, magnZ3-2534 and magnZ3-5802, were identified in the M.T.F. laboratory from the Zuker collection of viable but male sterile lines (Wakimoto et al., 2004). magnZ3-5802 was mapped by deficiency complementation to the 11.94 kb gap region (3L: 3905091-3893148) between two adjacent non-overlapping end point-defined deficiencies Df(3L)ED208 and Df(3L)ED4341. Df(3L)Exel6098, which uncovered this gap region, failed to complement all Zuker alleles. The Zuker alleles were sequenced from 1 kb upstream of the start codon to the end of the first 5′ ubiquitin unit and from the beginning of the last 3′ ubiquitin to 580 bp after the stop codon. No mutations were identified in these regions in any of the magn Zuker alleles.
Excision of the p[EPGY2]Ubi63EEY07341 insertion was carried out by crossing p[EPGY2]Ubi63EEY07341/TM3Δ2-3 males to w;;TM3/TM6B females, collecting TM6B balanced white-eyed male progenies and individually crossing to w;;TM3/TM6B females to establish stock before backcrossing to p[EPGY2]Ubi63EEY07341/TM6B females to examine the meiotic arrest phenotype in progeny. Two imprecise excision null mutations, magn12c and magn23b, genetically behaved the same as Df(3L)Exel6098 when placed in trans to p[EPGY2]Ubi63EEY07341. The lesions in the Ubi-p63E region were sequence verified for both alleles (Fig. 1C). magn12c and magn23b were both homozygous lethal due to additional lesions other than loss of function of Ubi-p63E. The exact cause of magn23b lethality was not identified. The lethality of magn12c was fully rescued by an Sc2 genomic rescue construct.
Fig. 1.
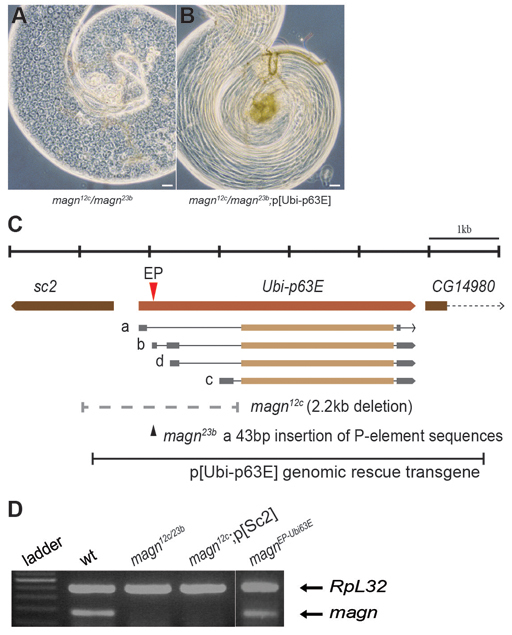
The magn meiotic arrest locus encodes the Drosophila polyubiquitin Ubi-p63E. (A,B) Phase contrast images of testis from (A) magn12c/23b and (B) magn12c/23b;p[Ubi-p63E] flies. Scale bars: 25 μm. (C) Genomic region showing the four magn transcripts (a, b, c and d). Red triangle, P-element insertion site in magnEP; black triangle and dotted line, nature of the magn null mutations. The Ubi-p63E genomic rescue construct is illustrated beneath. The Sc2 gene is upstream of Ubi-p63E and is transcribed in the opposite orientation. CG14890 starts ∼200 bp downstream of Ubi-p63E. (D) RT-PCR to detect magn mRNA from wild-type and magn null mutant testes. RpL32 RT-PCR provided a loading control.
To construct the Ubi-p63E rescuing plasmid, a PstI-BamHI fragment containing the Ubi-p63E coding region including 1.3 kb of promoter and 1 kb of downstream sequences, was released from pUB#3 (Lee et al., 1988) and inserted into pCaSpeR4 (Thummel and Pirrotta, 1992). The Sc2 rescuing plasmid was constructed by inserting PCR-amplified Sc2 genomic sequences into the XbaI-XhoI sites of pCaSpeR4. Primers used were: forward, 5′-CAAGGCTTGTTAGGATTGGTT; reverse, 5′-TGGCGCGTTGTTTCAAAATG.
RT-PCR
Total RNA was extracted from dissected testes with Trizol reagent (Invitrogen) and DNase I treated. First-strand cDNA was generated with Ready-To-Go You-Prime First-Strand Beads (GE Healthcare) using oligo(dT) primers. cDNA derived from 30 pairs of testes was used for each PCR reaction, with 25 amplification cycles. A magn-specific cDNA fragment was amplified with the following primers: 5′-ATCAGCAGCGTCTGATCTTC and 5′-ATTTAGAGTGAAGTGCGTGC. boule cDNA was amplified with 5′-CAAAAAGCAGCCCAATCCTC and 5′-TGCGAACTGATCCATGGG. RT-PCR of RpL32 provided a loading control across all samples. magn RT-PCR and RpL32 RT-PCR were performed in the same tube.
In situ hybridization of adult testis
RNA in situ hybridization was performed following a standard protocol (Morris et al., 2009). Primer pairs used to generate gene and isoform-specific probes are listed in supplementary material Table S1. Primer pairs amplifying full-length cDNA were used for gdl and CG9975. PCR-amplified fragments were subcloned into TOPO-pCR4 (Invitrogen) or pBluescript II SK+ for in vitro transcription and DIG labeling (Roche). Antisense probes for Cyclin A, Cyclin B, twine, fzo and aly were generated as previously described (White-Cooper et al., 1998).
Western blotting
Testis or ovaries were dissected in cold PBS containing Protease Inhibitor Cocktail (Roche). Gonadoectomized male flies were generated by surgical removal of the entire male reproductive tissue. Samples were homogenized in 50 μl cold PBS containing 1×SDS sample buffer, boiled and loaded onto a 10% SDS-PAGE gel (Bio-Rad). Primary antibodies anti-Ub (clone 6C1, Sigma), anti-uH2A (clone D27C4, Cell Signaling), anti-uH2B (clone D11, Cell Signaling), anti-actin (clone C4, Millipore) and anti-Boule (Cheng et al., 1998) were used at 1:2000 dilution.
Microscopy and immunofluorescence
For live squash, dissected testes were cut open in PBS containing 2 μg/ml Hoechst 33342, squashed with a coverslip and examined on a Zeiss Axioplan microscope. Immunofluorescence was performed as previously described (Chen et al., 2005), using the following antibodies: mouse anti-GFP (1:200, Roche), rabbit anti-HA (1:200, Bethyl Laboratories), rabbit anti-Myc (1:1000, Santa Cruz Biotechnology), anti-Topi (1:1000) (Perezgasga et al., 2004), anti-Cyclin B (1:50, Developmental Studies Hybridoma bank) and anti-Fibrillarin (1:200, Cytoskeleton). Anti-uH2A (Millipore) staining was performed as described (Rathke et al., 2007). Alexa Fluor-conjugated goat or donkey secondary antibodies were used (Molecular Probes). Images were captured with a Leica SP2 confocal microscope and processed with Adobe Photoshop software. Stem cell numbers were scored as described (Srinivasan et al., 2012).
Clonal analysis
Homozygous mutant clones in a heterozygous background were induced using Flippase (FLP)-mediated mitotic recombination. hs-flp122;;FRT2A, Ubi-GFP virgin females were crossed to p[sc-2];FRT2A or p[Sc-2];FRT2A, magn12c males. The progeny were grown at 25°C and heat shocked at 37°C for 2 hours each on day 8 and 9. Testes were dissected on the indicated days after the second day of heat shock and examined for the presence of GFP-negative clones in germ cells. Clonal analyses were repeated independently three times and the data are shown as the average of each of the experiments.
Microarray
magn, red,e, aly and sa testes without seminal vesicle and accessory glands were dissected in ice-cold PBS containing 0.1% DEPC. Total RNA was extracted using TRIzol (Invitrogen). Affymetrix 2.0 Drosophila chips were processed by the Stanford Protein and Nucleic Acid (PAN) Facility. Data on three replicates of red,e, aly2/5p and sa1/2 samples were from previous studies (X. Chen and M.T.F., unpublished). Two replicates of magn12c/23b and two replicates of magn12c;[pSC2] were prepared for this study. Data were normalized using the R program (v2.13.2) with the BioConductor package GCRMA (Bolstad et al., 2003). 4306 probe sets without gene annotation were discarded. To obtain differentially expressed genes, two-sample t-tests were performed and false discovery rate (FDR)-adjusted P-values were calculated with the Benjamini-Hochberg (BH) procedure. Genes showing more than 23-fold (i.e. 8-fold) differences and BH FDR-adjusted P<0.05 were chosen as differentially expressed genes in each comparison. Statistical tests and plots were performed with MATLAB software (MathWorks, R2011b). Microarray data are available at GEO with accession number GSE48837.
RESULTS
The magn meiotic arrest locus encodes the Drosophila polyubiquitin Ubi-p63E
Four mutants from the C. Zuker collection of EMS-induced viable but male sterile Drosophila strains (Wakimoto et al., 2004) formed a single complementation group, in which males transheterozygous for any pair of the Zuker alleles displayed a meiotic arrest phenotype. In testis of mutant males, germ cells developed into mature spermatocytes but cell cycle progression was blocked prior to entering the first meiotic division (Fig. 1A). The locus identified by the complementation group was termed magellan (magn), after the effect of loss of function on meiotic chromosome condensation in spermatocytes (as described below).
Genetic mapping and complementation tests identified magn as the Drosophila polyubiquitin gene Ubi-p63E. The mutant allele magnz3-5802 was mapped by deficiency complementation to an 11.94 kb genomic region (see Materials and methods). An EP transgene insertion in this genomic region (p[EPGY2]Ubi63E), located in the first intron of the longest transcript of Ubi-p63E (Fig. 1C), showed the male meiotic arrest phenotype when in trans to any of the Zuker magn alleles or Df(3L)Exel6098, which removes the Ubi-p63E locus. Although flies homozygous for the EP insertion were male sterile, they did not show meiotic arrest, suggesting that the EP line was a weak allele. Consistent with this, RT-PCR showed that mRNA was still produced from the Ubi-p63E locus in testes from EP/EP homozygotes (Fig. 1D). Mobilization of the P-element in the EP line (see Materials and methods) produced two null magn alleles, magn12c and magn23b. PCR from genomic DNA revealed that the magn12c allele had a 2.2 kb deletion that removed the transcription start site of all four Ubi-p63E mRNAs plus most of the 5′UTR, stopping at 34 bp before the ATG. The 2.2 kb deletion also removed the promoter and first 583 bp of protein coding sequence of a neighboring essential gene, Sc2 (Fig. 1C) (Wohlwill and Bonner, 1991). However, when a genomic fragment containing the entire Sc2 gene was introduced, magn12c/magn12c;[pSc2] flies were completely viable but male sterile and displayed the magn meiotic arrest phenotype, indicating that the lethality of magn12c homozygotes was due to loss of function of Sc2 rather than Ubi-p63E. The magn23b allele retained a 43 bp insertion in the Ubi-p63E 5′UTR consisting of a fragment of the starting P-element remaining after incomplete excision (Fig. 1C). No Ubi-p63E mRNA was detected by RT-PCR from magn12c;[pSc2] or magn12c/magn23b testes (Fig. 1D) or whole mutant flies (data not shown), indicating that both of the new alleles were null. The highly repetitive nature of the polyubiquitin gene, which encodes ten tandem copies of the Ub monomer, prevented PCR amplification and sequencing of the polyubiquitin coding region of Ubi-p63E from the Zuker alleles. All four Zuker alleles behaved genetically as nulls in terms of their meiotic arrest phenotype, both in combination with each other and with the new null mutations induced by imprecise excision, although low levels of magn transcript were detected in testis of the Zuker alleles (supplementary material Fig. S1). Finally, confirming the identity of magn as Ubi-p63E, a transgene containing the Ubi-p63E genomic region (Fig. 1C) completely rescued the meiotic arrest and the male sterility when introduced into any magn allele (Fig. 1B).
Ubi-p63E is the main ubiquitin gene expressed in spermatocytes
Surprisingly, magn12c/magn23b and magn12c;p[Sc2] null flies were fully viable, developed at a normal pace into adulthood, and females were robustly fertile (data not shown), indicating that under normal non-stress growth conditions Ubi-p63E gene function is predominantly required for male fertility. Consistent with this requirement, analysis of the expression patterns of the genes encoding mono-ubiquitin and polyubiquitin in Drosophila suggested that Ubi-p63E is the major ubiquitin gene expressed in spermatocytes. In situ hybridization using isoform-specific RNA probes revealed that the four annotated transcripts of Ubi-p63E were all expressed in adult testes (Fig. 2A,C,E), with highest expression in primary spermatocytes. Of the four predicted mRNA isoforms, Ubi-p63E-a (Fig. 2A) and Ubi-p63E-b/Ubi-p63E-d (Fig. 2C), as detected by a common probe, were expressed at high levels in spermatocytes, whereas the level of the Ubi-p63E-c isoform was much lower (Fig. 2E).
Fig. 2.
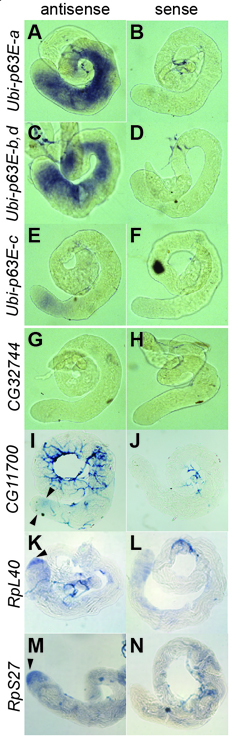
Ubi-p63E is the major ubiquitin gene expressed in spermatocytes. In situ hybridization on wild-type testes using transcript-specific antisense and sense RNA probes for (A-F) the four isoforms of Ubi-p63E, (G,H) CG32744, (I,J) CG11700, (K,L) RpL40 and (M,N) RpS27 mRNA. Arrowheads indicate early spermatocytes (I) or the tip of the testis where GSCs and spermatogonia reside (K,M). Tip of the testis is to the left in all panels.
Three loci in D. melanogaster encode polyubiquitin: Ubi-p63E, with ten consecutive Ub repeats; and two closely linked polyubiquitin genes on the X chromosome, namely CG32744 (Ubi-p5E, previously named Ubi-p5E4) and CG11700 (previously named Ubi-p5E5), with seven and four consecutive Ub repeats, respectively. In situ hybridization with probes designed to best distinguish the two X-linked transcripts (see Materials and methods) detected no transcripts from CG32744 (Fig. 2G,H) and possibly only very low levels of transcripts from CG11700 in early spermatocytes (Fig. 2I,J, arrowheads). Consistently, although RNA-Seq data from ModENCODE (source: FlyBase) showed that CG11700 was expressed primarily in testis, imaginal discs and larval gut, expression of Ubi-p63E was 32-fold higher than CG11700. D. melanogaster, similar to other higher eukaryotes, also has two mono-ubiquitin genes, RpL40 and RpS27, which encode fusion proteins containing a single Ub moiety fused directly upstream of the ribosomal proteins L40 and S27, respectively. Interestingly, both the mono-ubiquitin RpL40 and the mono-ubiquitin RpS27 transcripts were detected by in situ hybridization in the tip spermatogonial region of adult testes, but were at much lower levels or not detected in the spermatocyte region of wild-type testes (Fig. 2K-N). Thus, Ubi-p63E might serve a crucial role in spermatocytes as the main source of newly synthesized Ub.
Ubi-p63E is required for ubiquitin homeostasis in the testis
Consistent with the expression patterns of the D. melanogaster ubiquitin genes and the specificity of the magn null mutant phenotype, loss of function of Ubi-p63E drastically reduced the pool of free Ub protein in testes, but not in ovaries or the bodies of gonadoectomized adult males (Fig. 3). In western blots of protein extracts from testes dissected from wild-type versus magn mutant flies probed with an antibody against Ub, the amount of free Ub (running as an ∼8 kDa band) dropped to almost undetectable levels in the magn null mutant samples (Fig. 3A, lanes 2 and 3) under conditions in which free Ub was clearly seen in parallel samples of wild-type testes (Fig. 3A, lane 1). High molecular weight ubiquitylated proteins were still abundant in the extracts from magn null mutant adult testes, suggesting that lack of Ubi-p63E function had its greatest effect on the levels of the free Ub pool. This effect was not due to the lack of post-meiotic stages in magn mutant testes, as free Ub levels were not reduced in sa mutant testis, which also exhibit meiotic arrest and lack of spermatid stages (Fig. 3, lane 6). Likewise, lack of spermatids caused by RNAi against the proteasome subunit Rpt2 did not result in dramatic reduction of free Ub levels (Fig. 3, lane 5). Testes from flies homozygous for the magnEP allele had moderately decreased levels of free Ub (Fig. 3A, lane 4), consistent with the behavior of magnEP as a weak allele. Male germ cells in magnEP homozygotes progressed through meiosis and spermatid elongation but did not produce functional sperm, suggesting that lowered free Ub levels either directly affected a late differentiation event or affected the accumulation, in spermatocytes, of particular transcripts needed for spermatid differentiation. By contrast, the levels of free Ub and ubiquitylated proteins in gonadoectomized males or ovaries were similar in extracts from magn null animals and wild type (Fig. 3B,C), suggesting that under non-stress conditions Ubi-p63E function is not required to maintain Ub homeostasis in tissues other than the testis, consistent with the viable, but male sterile, phenotype of magn null mutant flies.
Fig. 3.
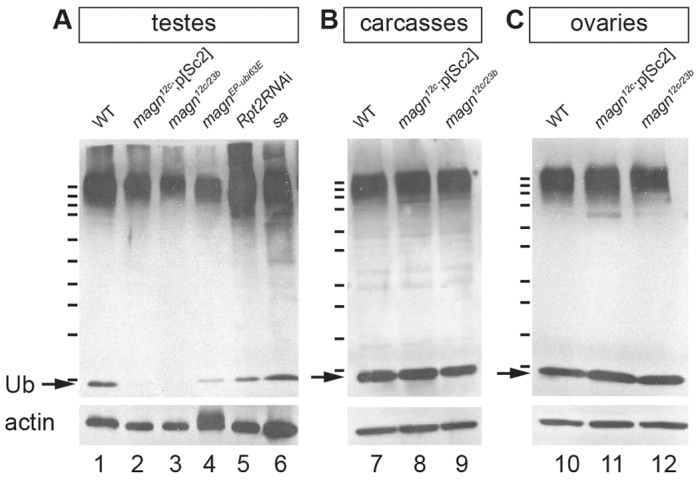
Ubiquitin homeostasis in the testis is affected in magn mutants. Anti-ubiquitin western blot showing levels of free ubiquitin (arrows) and ubiquitylated proteins in (A) testes, (B) gonadoectomized males (carcasses) and (C) ovaries. Anti-actin provided a loading control. Thirty pairs of testes (A), five male carcasses (B) and ten pairs of ovaries (C) were used as starting material across the different genotypes. Molecular weight markers, from top to bottom: 250, 150, 100, 75, 50, 37, 25, 20, 15 and 10 kDa. Ub, ubiquitin.
Ubi-p63E is required in male germ cells for meiotic chromatin condensation and cell cycle progression
The early stages of male germ cell differentiation appeared to proceed normally in magn null mutant testes, consistent with the expression of mono-ubiquitin-RpL40 and mono-ubiquitin-RpS27 during the spermatogonial stages. Testes from magn null mutant males had similar numbers of GSCs to wild-type controls (supplementary material Fig. S2). Likewise, mature spermatocyte cysts had the normal number of 16 germ cells per cyst in magn null mutant testes, indicating that spermatogonia proceeded through the correct number of transit-amplifying divisions prior to entering the spermatocyte differentiation program. However, mature spermatocytes from magn mutant males were all arrested at the G2/M transition of meiosis I and failed to initiate spermatid differentiation (Fig. 4A,A′).
Fig. 4.
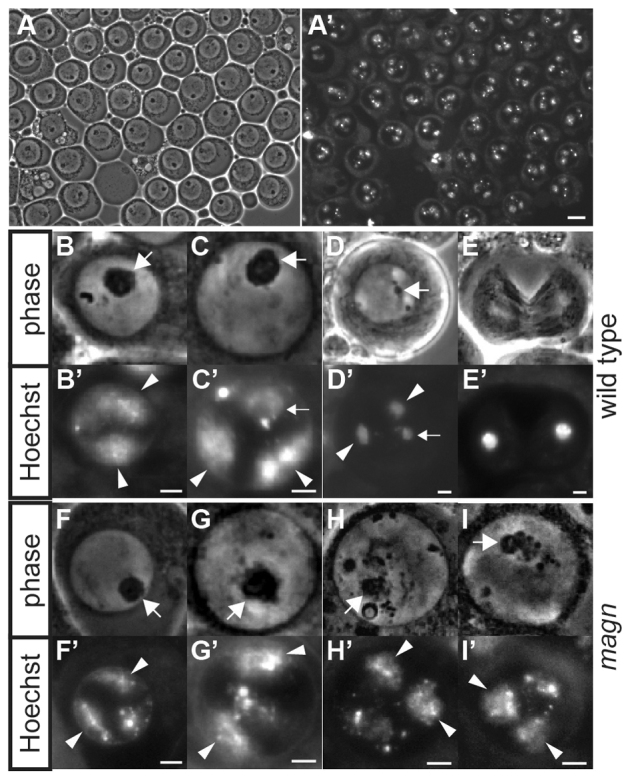
magn mutant spermatocytes arrest at the G2/M transition with chromatin condensation defects. (A,A′) Arrested magn mature spermatocytes: (A) phase contrast, (A′) Hoechst. (B-I′) Stage-matched spermatocytes from (B-E′) wild-type and (F-I′) magn mutant testes squashes. (B-I) Phase contrast, (B′-I′) Hoechst. Large arrows, nucleolus; small arrows, sex bivalents; arrowheads, condensing autosomal bivalents. (B,B′,F,F′) Wild-type and magn young spermatocytes with autosomal bivalents starting to condense. (C,C′,G,G′) Wild-type and magn mature spermatocytes with autosomal bivalents forming crescents at the nuclear periphery. (D,D′) Wild-type spermatocytes entering meiosis, with condensed bivalents moving towards the middle of the nucleus and nucleolus breaking down. (H-I′) Arrested magn spermatocytes with partially fragmented nucleolus and defectively condensed sphere-shaped chromatin, resembling Magellanic clouds. (E,E′) Wild-type spermatocyte undergoing meiosis I; the corresponding stage was never observed in magn mutant testes. Scale bars: 10 μm in A,A′; 2 μm in B-I′.
The first defects observed in magn mutant male germ cells were abnormalities in chromatin condensation during the spermatocyte growth stages. In wild-type growing spermatocytes, the major autosomal bivalents begin to condense, forming distinct domains near the nuclear periphery as nuclear volume increases (Fig. 4B,B′). By contrast, in magn mutant spermatocytes, the autosomal bivalents had subtle defects in organization even at such immature stages (Fig. 4F,F′). These defects became more apparent as spermatocyte growth and maturation progressed. In wild-type mature spermatocytes, the autosomal bivalents formed a pair of crescent-shaped structures near the nuclear periphery, and the sex bivalent associated with the nucleolus (Fig. 4C,C′). In magn mature spermatocytes, however, although the major autosomal and sex bivalents occupied separate domains of the nucleus, overall the chromatin appeared less uniformly condensed, forming variable structures lacking clear boundaries (Fig. 4G,G′).
In wild-type primary spermatocytes entering the first meiotic division, the spermatocyte nucleolus breaks down as the compacted bivalents move towards the middle of the nucleus (Fig. 4D,D′), prior to clustering tightly at the metaphase plate. magn null mutant spermatocytes never reached this state, but arrested with partially fragmented nucleoli and incompletely condensed chromatin, forming abnormal structures resembling the Magellanic clouds (Fig. 4G-H′) - hence the mutant name magellan. Whereas spermatocytes undergoing meiotic division were observed in wild-type testis squashes (Fig. 4E,E′), spermatocytes undergoing meiotic division were never observed in magn mutant testes. Likewise, magn mutant testes never displayed any signs of spermatid differentiation visible by phase contrast imaging of live squashed preparations. Instead, arrested spermatocytes accumulated throughout the basal end of magn null testes and eventually began to degenerate (Fig. 1A).
Clonal analysis revealed that Ubi-p63E function is required cell-autonomously in germ cells for spermatocytes to progress to spermatid differentiation. Negatively marked clones of germ cells homozygous for the null allele magn12c were generated in heterozygous males using the FLP-FRT-based mitotic recombination system, and the percentage of testes showing GFP-negative spermatocyte or post-meiotic spermatid clones was scored at various time points post-clone induction (PCI) (Fig. 5A,B). By day 4 PCI, 100% of both FRT control and magn testes had GFP-negative homozygous spermatocyte cysts. However, whereas wild-type control clonally marked germ cysts progressed to spermatid stages, such that 56.5% of control testes at day 8 PCI and 43.9% at day 12 PCI had at least one spermatid clone (Fig. 5A), absolutely no magn null mutant germ cell clones were observed at the spermatid stage, even at 12 days PCI (Fig. 5B). Instead, magn null mutant spermatocyte clonal cysts accumulated, with the characteristic chromatin condensation defects, failure to reach metaphase and failure of complete nucleolar breakdown, and eventually degenerated (data not shown).
Fig. 5.
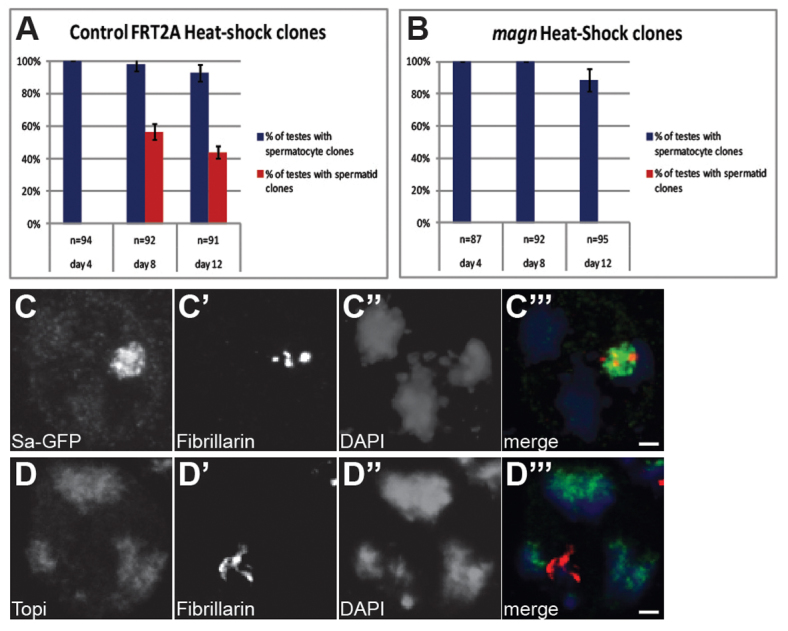
Ubi-p63E is required cell-autonomously for spermatocyte cell cycle progression and spermatid differentiation without affecting tTAF or tMAC localization. (A,B) Ubi-p63E function is required cell-autonomously in germ cells for cell cycle progression. Germline clonal analysis of (A) FRT2A controls compared with (B) magn12c. Blue columns indicate the percentage of testis with GFP-negative germ cells reaching the spermatocyte stage; red columns indicate the percentage of testis with GFP-negative germ cells reaching post-meiotic spermatid stages. Error bars indicate s.d. from three independent clone inductions. (C-D′′) Indirect immunofluorescence of arrested magn12c/23b spermatocytes stained for (C) the testis TAF fusion protein Sa-GFP, (D) the tMAC protein Topi or (C′,D′) Fibrillarin and stained with (C′,D′) DAPI. (C′′,D′′) Merge, showing Sa-GFP (green in C′′) or Topi (green in D′′), Fibrillarin (red) and DAPI (blue). Scale bars: 2 μm.
The spermatocyte transcription program is largely unaffected in magn mutant testis
Although the meiotic arrest observed in magn mutant testes resembled the G2/M meiotic arrest previously described for loss of function of either the tTAF or tMAC genes, the arrest in magn mutants was unlikely to be due to loss of function of tTAF or testis-specific tMAC components. The tTAF proteins Sa and Can, as marked by Sa-GFP or Myc-Can (Chen et al., 2005), respectively, were still expressed and localized normally in magn mutant spermatocytes, appearing weakly on the condensing chromatin and strongly within a subcompartment of the nucleolus interdigitating with the nucleolar marker Fibrillarin, as in wild type (Fig. 5C-C′′, Fig. 8E,F). Likewise, staining of magn testis with antibodies against Topi or Aly, the protein products of two tMAC genes (Perezgasga et al., 2004; White-Cooper et al., 2000), showed that they were expressed in spermatocytes and localized to the condensing chromatin, as in wild type (Fig. 5D-D′′; data not shown). In addition, mono-ubiquitylated histone H2A (uH2A) was present and localized to a subnuclear structure in magn mutant testis, as in wild type (supplementary material Fig. S3A-C). Likewise, mono-ubiquitylated histone H2B (uH2B) was also present at similar levels in magn mutant testis and wild type (supplementary material Fig. S3D).
Fig. 8.
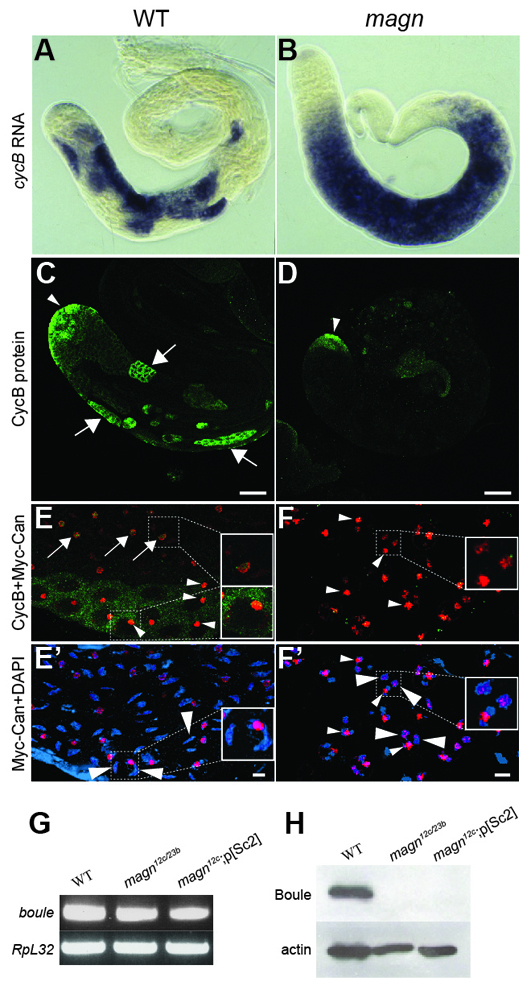
Protein, but not transcripts, of key cell cycle regulators fails to accumulate in magn spermatocytes. (A,B) In situ hybridization to (A) magn mutant and (B) wild-type testes with antisense RNA probes for CycB transcripts. (C,D) CycB protein stain in wild-type (C) and magn (D) testis. Arrowheads, CycB protein expression in spermatogonia; arrows, CycB protein expression in wild-type mature spermatocytes. (E,E′) Timing of CycB protein accumulation in wild-type mature spermatocytes. (F,F′) magn spermatocytes arrest at a stage past that of normal spermatocyte CycB accumulation. (E,F) Merge of the Myc-Can and CycB channels. Green, CycB; red, Myc-Can. (E′,F′) Merge of Myc-Can and DAPI channels. Red, Myc-Can; blue, DAPI. Large arrowheads indicate crescent-shaped autosomal bivalents in wild-type mature spermatocytes (E′) or globular arrested magn chromatin (F′). Arrows (E) indicate the less compacted circular Myc-Can nucleolar stain associated with low levels of CycB. Small arrowheads indicate the solid nucleolar Myc-Can stain associated with high levels of CycB (E) or compacted nucleolar Myc-Can stain as solid dots (F,F′). (G) RT-PCR showing mRNA expression of boule in wild-type and magn null mutant testes. RpL32 RT-PCR provided a loading control. (H) Western blot of Boule protein in wild-type and magn testes. Anti-actin provided a loading control. Thirty pairs of testes were used as starting material across the different genotypes in G and H. Scale bars: 10 μm in C,D; 4 μm in E′,F′.
Consistent with the normal expression and localization of tTAFs, Aly and Topi in the mutant spermatocytes, magn mutant testes did not display the widespread defects in the transcription of spermatid differentiation genes characteristic of tTAF or tMAC mutants. In situ hybridization analysis showed that the representative differentiation genes fzo and gdl, which are expressed robustly in wild-type spermatocytes but only at low levels in tTAF or tMAC mutant spermatocytes (Hiller et al., 2004; White-Cooper et al., 1998), were expressed normally in magn mutant spermatocytes (Fig. 6A-D). Transcripts of twine and Cyclin B (CycB), which are expressed in wild-type spermatocytes dependent on the function of tMAC genes, were also expressed at levels comparable to wild type in magn mutant spermatocytes (Fig. 6E,F, Fig. 8A,B). In addition, genes such as aly, Cyclin A (CycA), Rbp4 and CG9975, which are upregulated in wild-type spermatocytes and independent of either the tMAC or the tTAFs, were expressed normally in magn spermatocytes (Fig. 6G-N).
Fig. 6.
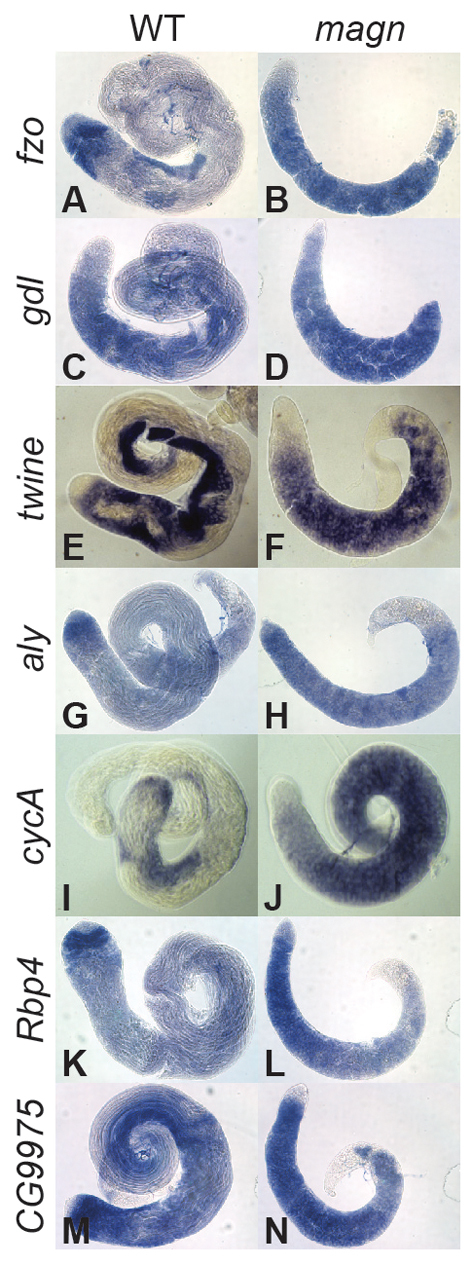
Ubi-p63E is not required for the expression of tTAF- and tMAC-dependent transcripts. In situ hybridization on wild-type and magn null mutant testes processed in the same tube using antisense RNA probes for (A,B) fzo, (C,D) gdl, (E,F) twine, (G,H) aly, (I,J) CycA, (K,L) Rbp4 and (M,N) CG9975 transcripts. The testis apical tip is to the left in all panels.
Global gene expression analysis also indicated that the majority of transcripts that are dependent on either aly or sa for normal expression in testis did not require Ubi-p63E function. Using a 23-fold cut-off, microarray data showed 1476 genes downregulated in aly compared with wild type (Fig. 7A, red). Of those, 82.5% (1217/1476) were not dependent on Ubi-p63E (Fig. 7B). Similarly, of the 766 genes downregulated in sa compared with wild type (Fig. 7C, magenta), 67.5% (517/766) were not dependent on Ubi-p63E (Fig. 7D). These numbers suggest that Ubi-p63E function is, for the most part, not required for regulation of transcription by tMAC or the tTAFs.
Fig. 7.
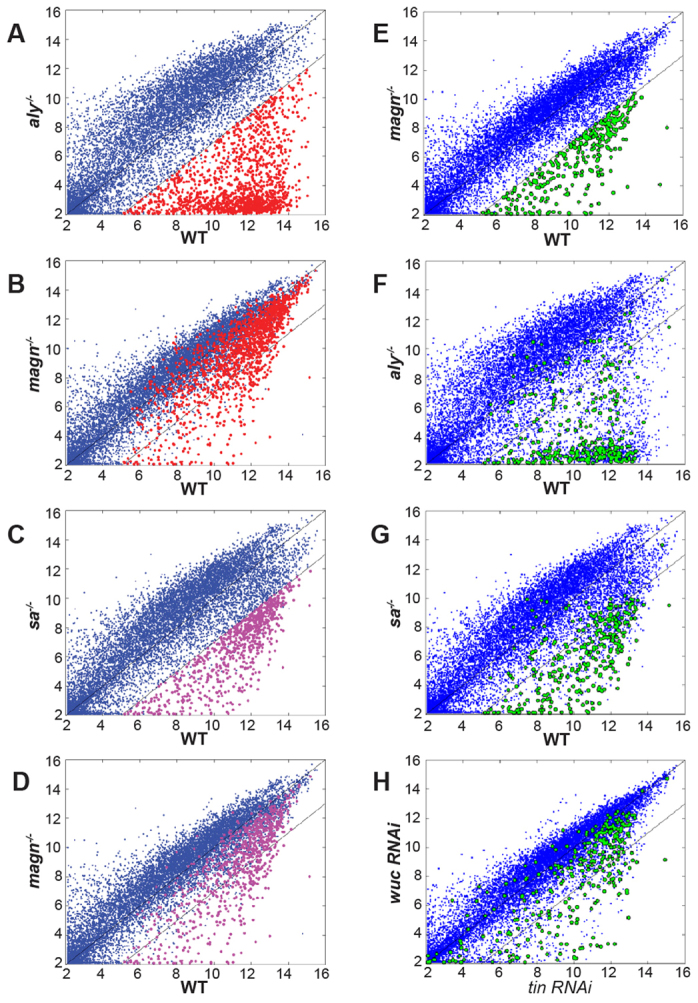
Ubi-p63E is dispensable for the tMAC- and tTAF-dependent spermatocyte transcription program. (A-G) Scatter plots comparing transcript levels in wild-type testes for (A,F) aly, (B,D,E) magn and (C,G) sa. (H) Scatter plot comparing transcript levels between wuc RNAi and tin RNAi [data from Doggett et al. (Doggett et al., 2011)]. x and y axes show log2 transformed gene expression values. Red, 1476 aly-dependent transcripts (≥23-fold higher in wild type than in aly-/-); magenta, 766 sa-dependent transcripts (≥23-fold higher in wild type than in sa-/-); green, 309 magn-dependent transcripts (≥23-fold higher in wild type than in magn-/-). FDR adjusted to P<0.05 for all samples.
The majority of the genes downregulated in magn mutant testes were also downregulated in aly or sa. Only 309 genes were more than 23-fold lower in magn testis than in wild type (Fig. 7E, green). Of those, 84% (260/309) and 79% (244/309) were also aly and sa dependent, respectively (Fig. 7F,G). No significant gene ontology (GO) terms were associated with Ubi-p63E-dependent genes that did, versus those that did not, depend on the function of aly and sa. The 225 transcripts that were downregulated in all three genotypes (magn, aly and sa) are likely to include mRNAs that are either expressed or stabilized at later stages of spermatogenesis that are not present in magn or the other meiotic arrest mutant testes.
Loss of function of a testis-enriched lin-52 homolog, wake-up-call (wuc), by RNAi also showed meiotic arrest but had very mild effects on the spermatocyte transcription program when knocked down specifically in spermatocytes (Doggett et al., 2011). Only 28% (88/309) of the transcripts that were ≥23-fold downregulated in magn compared with wild type were also ≥23-fold downregulated in wuc (Fig. 7H), suggesting that Ubi-p63E and wuc are unlikely to act together, or in the same pathway, to control gene expression in spermatocytes.
Ubi-p63E is required for expression of key G2/M cell cycle regulatory proteins in spermatocytes
Key cell cycle regulatory proteins that are normally expressed in advance of the G2/M transition in mature spermatocytes were not expressed in magn null mutant spermatocytes, suggesting a mechanism underlying the arrest in meiotic cell cycle progression in the mutant. Although transcripts of the cell cycle regulators CycB and boule were expressed in magn mutant spermatocytes at levels comparable to wild type (Fig. 8A,B,G), Cyclin B and Boule proteins did not accumulate in magn spermatocytes. Immunofluorescence staining of wild-type testes revealed that Cyclin B protein was expressed at the tip (Fig. 8C, arrowhead), where the stem cells and spermatogonia undergo mitotic divisions, declined to very low levels in newly formed and growing spermatocytes, and accumulated again in mature spermatocytes preceding the onset of the G2/M transition (Fig. 8C, arrows), as described previously (White-Cooper et al., 1998). Although expression of Cyclin B protein at the testis tip was not affected in magn null testes (Fig. 8D, arrowhead), Cyclin B protein was not detected by immunofluorescence in the mature spermatocytes that accumulate to fill magn mutant testes (Fig. 8D). Two lines of evidence indicate that the lack of expression of Cyclin B protein in magn mutant spermatocytes was not simply a consequence of the magn cell cycle arrest but that the magn mutant spermatocytes arrest at a stage after the point when Cyclin B would have accumulated in wild type. First, when Cyclin B levels became high in wild-type spermatocytes, the condensing autosomal bivalents were still crescent shaped and localized to the nuclear periphery (Fig. 8E′, large arrowheads), a stage much earlier than the magn arrest point, when autosomal bivalents were more condensed, adopting smaller spherical shapes inside the nucleus (Fig. 4H′,I′; Fig. 8F′, large arrowheads). Second, wild-type testes co-stained for Cyclin B and Myc-Can (Chen et al., 2005) showed the accumulation of Cyclin B protein in mature spermatocytes coinciding with compaction of the Myc-Can nucleolar stain from small circles into solid dots (Fig. 8E, compare arrows and small arrowheads). By contrast, Cyclin B expression was still not observed in the arrested magn mutant spermatocytes even though the Myc-Can nucleolar stain had already compacted (Fig. 8F,F′, small arrowheads).
Likewise, magn loss of function blocked the accumulation of Boule, an RNA-binding protein required for translation of the G2/M cell cycle regulator twine in spermatocytes. RT-PCR showed the presence of boule transcript in testes from flies null mutant for magn (Fig. 8G). However, no Boule protein was detected on western blots of magn mutant testes (Fig. 8H). In wild type, Boule protein expression normally begins in early spermatocytes, well before the arrest point of magn spermatocytes. Unlike boule or twine mutant germ cells, which eventually skip meiosis and initiate spermatid differentiation (Maines and Wasserman, 1999), magn mutant spermatocytes never displayed any sign of spermatid differentiation, suggesting that Ubi-p63E function is required for more than just meiotic cell cycle progression.
The meiotic arrest in magn mutants does not resemble defects caused by knocking down proteasome subunits
Interfering with the function of the proteasome in spermatocytes did not cause the same phenotype as magn loss of function, suggesting that the effects observed in magn mutants might not be due to general defects in protein degradation of unwanted proteins. Proteasome function was impaired by expressing UAS-RNAi hairpins against each of several key proteasome subunits, Rpt2, Prosα3, Prosα7 or Prosβ7, specifically in late spermatogonia and spermatocytes under control of the Bam-GAL4 expression driver. In situ hybridization with gene-specific probes showed that this RNAi method effectively abolished messenger expression of the targeted proteasome subunits in spermatocytes (supplementary material Fig. S4G-L). Although germ cell-specific knockdown of each generally expressed proteasome subunit tested resulted in a meiotic arrest phenotype with lack of spermatid stages (supplementary material Fig. S4A-D), the arrested spermatocytes displayed phenotypes that were strikingly different from the magn null phenotype. For example, knocking down proteasome function resulted in spermatocytes with a largely intact nucleolus (supplementary material Fig. S4E,F, arrows) and poorly condensed chromatin adjacent to the nuclear periphery (supplementary material Fig. S4E′,F′, arrowheads), suggesting an earlier arrest than in magn mutants, in which the spermatocyte nucleolus had begun to break down and chromatin condensation had proceeded to a spherical shape (Fig. 4H-I′).
Consistent with this difference, analysis of transcript expression by in situ hybridization revealed that knockdown of the key proteasome component Rpt2 had very different effects on the spermatocyte transcription program than magn loss of function. For example, transcripts for the differentiation genes fzo and gdl were not detected and transcripts of the cell cycle genes CycB and twine remained at background levels in Rpt2 knockdown spermatocytes (Fig. 9A-H), whereas they were expressed in magn null spermatocytes (Fig. 6A-F). The tTAF- and tMAC-independent transcripts aly, CycA, Rbp4 and CG9975 were expressed at either lower (CycA) (Fig. 9I,J) or at wild type-comparable levels (aly, Rbp4 and CG9975) in Rpt2 RNAi knockdown spermatocytes (Fig. 9K-P), indicating that only the later events of the spermatocyte transcription program were affected in Rpt2-depleted spermatocytes. Notably, knockdown of Rpt2 in spermatocytes did not deplete the pool of free Ub (Fig. 3A, lane 5). Together, these findings suggest that magn might affect germ cell differentiation due to defects in Ub homeostasis rather than through disrupted protein degradation.
Fig. 9.
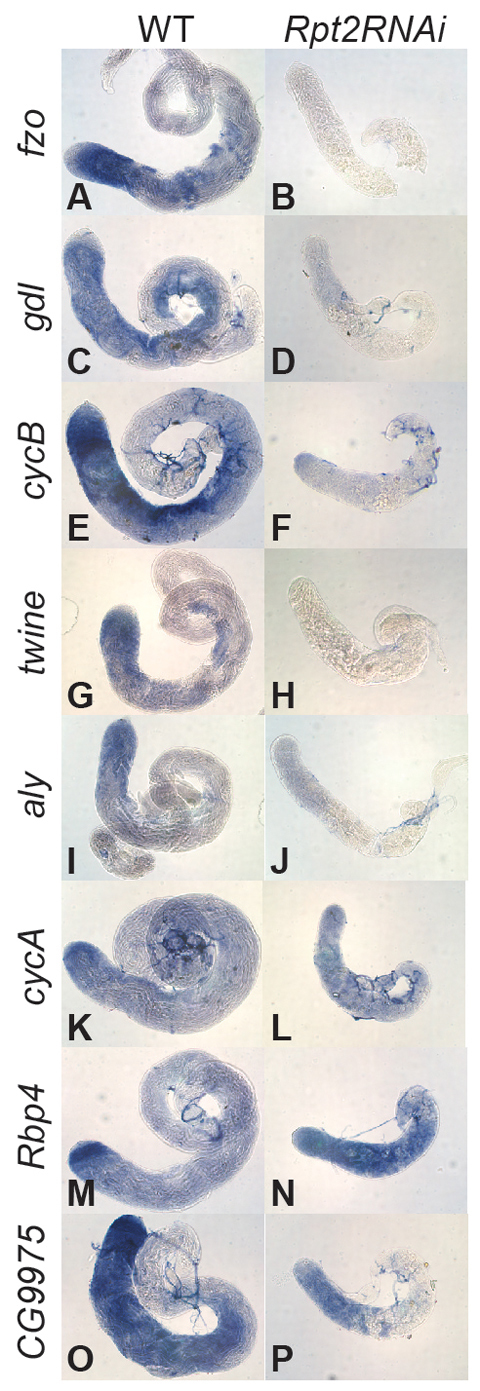
Function of the 26S proteasome is required for the spermatocyte transcription program. In situ hybridization on wild-type and Rpt2 RNAi knockdown testes processed in the same tube with antisense RNA probes for (A,B) fzo, (C,D) gdl, (E,F) CycB, (G,H) twine, (I,J) CycA, (K,L) aly, (M,N) Rbp4 and (O,P) CG9975. The tip of the testis is to the left in all panels.
DISCUSSION
The Drosophila polyubiquitin gene Ubi-p63E, identified here as the magellan (magn) locus needed specifically for male fertility, is required in spermatocytes for normal chromatin condensation, cell cycle progression through the G2/M transition of meiosis I and for the onset of spermatid differentiation. magn loss of function causes a male meiotic arrest phenotype similar to meiosis I maturation arrest, the most common form of idiopathic male infertility in humans (Meyer et al., 1992): testes fill with mature spermatocytes that do not progress into meiotic division and lack all spermatid stages.
The cell type-specific expression patterns of Drosophila ubiquitin genes may underlie the male germ cell-specific phenotype of magn loss of function, as Ubi-p63E was by far the most abundantly expressed ubiquitin gene in spermatocytes. Although Ubi-p63E is generally expressed in all fly tissues (Lee et al., 1988), its function is not required for viability or female fertility. It is likely that the two mono-ubiquitin genes and the other polyubiquitin genes provide a sufficient cellular Ub supply throughout development and in other adult tissues, including the female germ line. However, we expect that because Ubi-p63E is highly stress inducible (Lee et al., 1988), it might be required for the stress response in most fly tissues.
The requirement for polyubiquitin gene function for meiosis is evolutionarily conserved. The fission yeast polyubiquitin ubi4+ and budding yeast polyubiquitin Ubi4 are both highly expressed in meiotic cells (Okazaki et al., 2000; Treger et al., 1988), although the level of free Ub was not raised in meiotic cells. Loss of function of polyubiquitin caused defects specifically in meiosis (fission yeast) or sporulation (budding yeast) (Okazaki et al., 2000; Treger et al., 1988). As with magn mutant testes in Drosophila, the level of free Ub was greatly reduced in cells entering meiosis in the yeast ubi4 mutant. The mouse genome has two polyubiquitin genes: Ubb and Ubc (Wiborg et al., 1985). Whereas Ubc is needed for embryonic liver development, Ubb null mice grow normally into adulthood but are male and female infertile (Ryu et al., 2007; Ryu et al., 2008). Ubb mutant male germ cells arrest in meiotic prophase, resembling the magn arrest point. The level of free Ub was also significantly lower in testes from Ubb null mice. The similarity of the phenotypes in yeast, flies and mammals suggests that a conserved, but unknown, mechanism consumes free Ub during meiosis, generating a requirement for compensation from induced polyubiquitin gene expression to maintain Ub homeostasis in meiotic prophase. The specific events that demand high levels of free ubiquitin expression remain to be determined. However, our results indicate that the magn phenotype does not simply reflect defects in proteasome-mediated protein degradation, but a depleted reservoir of free Ub.
It is possible that the conserved requirement for polyubiquitin might arise from a stress response mechanism that is unique to male meiosis. A large number of genes required for spermatid differentiation are transcribed in spermatocytes but translation of their mRNAs is delayed until after meiosis. The delayed translation of so many terminal differentiation genes in spermatocytes might trigger stress conditions demanding high levels of free ubiquitin. In yeast, it was shown that ubiquitin overexpression confers resistance to overall inhibition on translation (Hanna et al., 2003).
It has been suggested that high activity of the ubiquitin proteasome system (UPS) during the dramatic events of cellular differentiation, such as the replacement of histones by protamines, towards the end of spermatogenesis might stimulate the expression of polyubiquitin genes in the testis (Baarends et al., 2000). However, magn mutant spermatocytes arrested well before spermatid differentiation or translation of protamines, suggesting that the meiotic arrest phenotype is not due to defects in these later events. The meiotic chromatin condensation defects observed in growing spermatocytes raised the possibility that some features of chromatin compaction and rearrangement during meiosis are sensitive to free Ub levels.
Most of the previously identified meiotic arrest mutants characterized in Drosophila affected the spermatocyte transcription program that controls the expression of a large number of spermatid differentiation gene transcripts. Most of the meiotic arrest genes characterized to date encode either tTAFs or components of the tMAC complex. Ubi-p63E does not seem to belong to either of these classes of transcriptional regulators, as many tTAF-dependent and tMAC-dependent genes were expressed at relatively normal levels in magn mutant testes (Figs 6, 7).
The arrest in meiotic cell cycle progression in magn mutant spermatocytes might be due to failure of the key G2/M regulators Boule and Cyclin B to accumulate. One possibility is that defects in chromatin condensation or other events in magn mutant spermatocytes trigger a checkpoint mechanism that blocks the translation of Boule and Cyclin B. In Drosophila, spermatid differentiation does not require prior completion of the meiotic divisions, as germ cells mutant for the cell cycle-activating phosphatase cdc25/twine or for the CDK-1 kinase fail to undergo the meiotic divisions but proceed with extensive spermatid differentiation with 4N nuclei (Alphey et al., 1992; Sigrist et al., 1995). It is possible that high levels of free Ub are required to pass through a checkpoint that blocks both meiotic cell cycle progression and the onset of spermatid differentiation. In mammals, a checkpoint elicited by transcriptionally active unpaired chromatin in mature spermatocytes blocks both cell cycle progression at the onset of the first meiotic division and the initiation of spermatid differentiation (Burgoyne et al., 2009).
Strikingly, analysis of testis biopsies of 32/32 patients with meiosis I maturation arrest male infertility showed lack of Boule protein expression in testes, although no defects in the boule gene itself were identified (Luetjens et al., 2004), suggesting a shared arrest mechanism involving regulation of Boule protein translation or stability. The similarity in the meiosis I arrest phenotype of magn mutant Drosophila males, Ubb mutant mice and human meiosis I maturation arrest patients (Luetjens et al., 2004; Ryu et al., 2008) raises the possibility that Ub homeostasis and the regulatory pathways that depend on a robust pool of free Ub might be compromised in some patients with meiosis I maturation arrest idiopathic human azoospermia.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center and the Vienna Drosophila RNAi Center for fly stocks; the Developmental Studies Hybridoma Bank for antibodies; John Lis for the plasmid containing Ubi-p63E genomic sequences; Helen White-Cooper for wuc microarray data and Topi and Aly antibodies; Steven Wasserman for the Boule antibody; members of the M.T.F. laboratory for helpful discussions and advice; and Xin Chen for sharing unpublished microarray data.
Footnotes
Funding
C.L. was partly supported by a Stanford Dean’s Fellowship. J.K. was supported by a Bio-X Stanford Interdisciplinary Graduate Fellowship. This research was funded by a National Institutes of Health grant [NIH 3 R01 GM061986] to M.T.F. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
C.L. performed the experiments. J.K. analyzed and discussed the microarray data. C.L. and M.T.F. designed the study, interpreted data and wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.098947/-/DC1
References
- Alphey L., Jimenez J., White-Cooper H., Dawson I., Nurse P., Glover D. M. (1992). twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69, 977–988 [DOI] [PubMed] [Google Scholar]
- Baarends W. M., van der Laan R., Grootegoed J. A. (2000). Specific aspects of the ubiquitin system in spermatogenesis. J. Endocrinol. Invest. 23, 597–604 [DOI] [PubMed] [Google Scholar]
- Beall E. L., Lewis P. W., Bell M., Rocha M., Jones D. L., Botchan M. R. (2007). Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 21, 904–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- Burgoyne P. S., Mahadevaiah S. K., Turner J. M. (2009). The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10, 207–216 [DOI] [PubMed] [Google Scholar]
- Cabrera y Poch H. L., Arribas C., Izquierdo M. (1990). Sequence of a Drosophila cDNA encoding a ubiquitin gene fusion to a 52-aa ribosomal protein tail. Nucleic Acids Res. 18, 3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hiller M., Sancak Y., Fuller M. T. (2005). Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310, 869–872 [DOI] [PubMed] [Google Scholar]
- Cheng M. H., Maines J. Z., Wasserman S. A. (1998). Biphasic subcellular localization of the DAZL-related protein boule in Drosophila spermatogenesis. Dev. Biol. 204, 567–576 [DOI] [PubMed] [Google Scholar]
- D’Andrea A., Pellman D. (1998). Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33, 337–352 [DOI] [PubMed] [Google Scholar]
- Doggett K., Jiang J., Aleti G., White-Cooper H. (2011). Wake-up-call, a lin-52 paralogue, and Always early, a lin-9 homologue physically interact, but have opposing functions in regulating testis-specific gene expression. Dev. Biol. 355, 381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Özkaynak E., Varshavsky A. (1987). The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48, 1035–1046 [DOI] [PubMed] [Google Scholar]
- Finley D., Bartel B., Varshavsky A. (1989). The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401 [DOI] [PubMed] [Google Scholar]
- Fuller M. T. (1993). Spermatogenesis. In The Development of Drosophila Melanogaster, Vol. I (ed. Bate M., Arias A. M.), pp. 71–147 Plainview, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Hanna J., Leggett D. S., Finley D. (2003). Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M., Chen X., Pringle M. J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T. Y., Marino S., Fuller M. T. (2004). Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131, 5297–5308 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996). Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 [DOI] [PubMed] [Google Scholar]
- Kierszenbaum A. L., Tres L. L. (1978). RNA transcription and chromatin structure during meiotic and postmeiotic stages of spermatogenesis. Fed. Proc. 37, 2512–2516 [PubMed] [Google Scholar]
- Kimura Y., Tanaka K. (2010). Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147, 793–798 [DOI] [PubMed] [Google Scholar]
- Lee H. S., Simon J. A., Lis J. T. (1988). Structure and expression of ubiquitin genes of Drosophila melanogaster. Mol. Cell. Biol. 8, 4727–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetjens C. M., Xu E. Y., Rejo Pera R. A., Kamischke A., Nieschlag E., Gromoll J. (2004). Association of meiotic arrest with lack of BOULE protein expression in infertile men. J. Clin. Endocrinol. Metab. 89, 1926–1933 [DOI] [PubMed] [Google Scholar]
- Maines J. Z., Wasserman S. A. (1999). Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell Biol. 1, 171–174 [DOI] [PubMed] [Google Scholar]
- Meyer J. M., Maetz J. L., Rumpler Y. (1992). Cellular relationship impairment in maturation arrest of human spermatogenesis: an ultrastructural study. Histopathology 21, 25–33 [DOI] [PubMed] [Google Scholar]
- Moon S., Cho B., Min S. H., Lee D., Chung Y. D. (2011). The THO complex is required for nucleolar integrity in Drosophila spermatocytes. Development 138, 3835–3845 [DOI] [PubMed] [Google Scholar]
- Morris C. A., Benson E., White-Cooper H. (2009). Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat. Protoc. 4, 1807–1819 [DOI] [PubMed] [Google Scholar]
- Okazaki K., Okayama H., Niwa O. (2000). The polyubiquitin gene is essential for meiosis in fission yeast. Exp. Cell Res. 254, 143–152 [DOI] [PubMed] [Google Scholar]
- Özkaynak E., Finley D., Varshavsky A. (1984). The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature 312, 663–666 [DOI] [PubMed] [Google Scholar]
- Perezgasga L., Jiang J., Bolival B., Jr, Hiller M., Benson E., Fuller M. T., White-Cooper H. (2004). Regulation of transcription of meiotic cell cycle and terminal differentiation genes by the testis-specific Zn-finger protein matotopetli. Development 131, 1691–1702 [DOI] [PubMed] [Google Scholar]
- Rathke C., Baarends W. M., Jayaramaiah-Raja S., Bartkuhn M., Renkawitz R., Renkawitz-Pohl R. (2007). Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J. Cell Sci. 120, 1689–1700 [DOI] [PubMed] [Google Scholar]
- Redman K. L., Rechsteiner M. (1989). Identification of the long ubiquitin extension as ribosomal protein S27a. Nature 338, 438–440 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K. Y., Maehr R., Gilchrist C. A., Long M. A., Bouley D. M., Mueller B., Ploegh H. L., Kopito R. R. (2007). The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 26, 2693–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K. Y., Sinnar S. A., Reinholdt L. G., Vaccari S., Hall S., Garcia M. A., Zaitseva T. S., Bouley D. M., Boekelheide K., Handel M. A., et al. (2008). The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol. Cell. Biol. 28, 1136–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S., Ried G., Lehner C. F. (1995). Dmcdc2 kinase is required for both meiotic divisions during Drosophila spermatogenesis and is activated by the Twine/cdc25 phosphatase. Mech. Dev. 53, 247–260 [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Mahowald A. P., Fuller M. T. (2012). The receptor tyrosine phosphatase Lar regulates adhesion between Drosophila male germline stem cells and the niche. Development 139, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. S., Pirrotta V. (1992). New pCaSpeR P element vectors. Drosophila Inf. Service 71, 150 [Google Scholar]
- Treger J. M., Heichman K. A., McEntee K. (1988). Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol. Cell. Biol. 8, 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B. T., Lindsley D. L., Herrera C. (2004). Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H., Schäfer M. A., Alphey L. S., Fuller M. T. (1998). Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 125, 125–134 [DOI] [PubMed] [Google Scholar]
- White-Cooper H., Leroy D., MacQueen A., Fuller M. T. (2000). Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development 127, 5463–5473 [DOI] [PubMed] [Google Scholar]
- Wiborg O., Pedersen M. S., Wind A., Berglund L. E., Marcker K. A., Vuust J. (1985). The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 4, 755–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlwill A. D., Bonner J. J. (1991). Genetic analysis of chromosome region 63 of Drosophila melanogaster. Genetics 128, 763–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


