Abstract
Suppressor of Hairy-wing [Su(Hw)] is a DNA-binding factor required for gypsy insulator function and female germline development in Drosophila. The insulator function of the gypsy retrotransposon depends on Su(Hw) binding to clustered Su(Hw) binding sites (SBSs) and recruitment of the insulator proteins Centrosomal Protein 190 kD (CP190) and Modifier of mdg4 67.2 kD (Mod67.2). By contrast, the Su(Hw) germline function involves binding to non-clustered SBSs and does not require CP190 or Mod67.2. Here, we identify Su(Hw) target genes, using genome-wide analyses in the ovary to uncover genes with an ovary-bound SBS that are misregulated upon Su(Hw) loss. Most Su(Hw) target genes demonstrate enriched expression in the wild-type CNS. Loss of Su(Hw) leads to increased expression of these CNS-enriched target genes in the ovary and other tissues, suggesting that Su(Hw) is a repressor of neural genes in non-neural tissues. Among the Su(Hw) target genes is RNA-binding protein 9 (Rbp9), a member of the ELAV/Hu gene family. Su(Hw) regulation of Rbp9 appears to be insulator independent, as Rbp9 expression is unchanged in a genetic background that compromises the functions of the CP190 and Mod67.2 insulator proteins, even though both localize to Rbp9 SBSs. Rbp9 misregulation is central to su(Hw)-/- sterility, as Rbp9+/-, su(Hw)-/- females are fertile. Eggs produced by Rbp9+/-, su(Hw)-/- females show patterning defects, revealing a somatic requirement for Su(Hw) in the ovary. Our studies demonstrate that Su(Hw) is a versatile transcriptional regulatory protein with an essential developmental function involving transcriptional repression.
Keywords: Chromatin insulator, Drosophila oogenesis, Neural gene expression, Rbp9, Su(Hw), Transcriptional regulation
INTRODUCTION
Eukaryotic gene expression depends upon the integration of regulatory inputs from multiple classes of elements. Enhancers and silencers represent two classes of regulatory elements, both having the capacity to modulate the activity of target promoters separated by large linear distances (Blackwood and Kadonaga, 1998; Bulger and Groudine, 2010). Enhancers and silencers display limited promoter specificity (Kermekchiev et al., 1991; Schoenherr et al., 1996; Dellino et al., 2004), requiring the presence of a third class of regulatory elements to achieve transcriptional fidelity. This class, called insulators, limits enhancer and silencer action to prevent inappropriate interactions with non-target promoters (Kuhn and Geyer, 2003; Raab and Kamakaka, 2010; Yang and Corces, 2011; Ghirlando et al., 2012). Insulators block enhancer and promoter interactions when positioned between these elements (enhancer-blocking activity) and define boundaries of repressive and active chromatin (barrier activity). Insulator-binding proteins have been identified in most eukaryotes, emphasizing the importance of this regulatory class in establishing transcriptional integrity.
One of the best-characterized insulator-binding proteins is the Drosophila Suppressor of Hairy wing [Su(Hw)] zinc-finger (ZF) protein. This DNA-binding protein establishes the chromatin insulator of the gypsy retrotransposon (Geyer et al., 1986; Parkhurst et al., 1988; Spana et al., 1988; Geyer and Corces, 1992; Dorsett, 1993). The function of the gypsy insulator requires Su(Hw) recruitment of Centrosomal Protein 190 kD (CP190) (Pai et al., 2004), Modifier of mdg4 67.2 kD isoform (Mod67.2) (Georgiev and Kozycina, 1996) and Enhancer of yellow 2 [E(y)2; also known as ENY2] (Kurshakova et al., 2007a). Mod67.2 and CP190 are required for enhancer blocking by the gypsy insulator (Georgiev and Gerasimova, 1989; Pai et al., 2004). Both proteins carry a Broad-complex, Tramtrack and Bric-a-brac/Poxvirus and Zinc Finger (BTB/POZ) domain (Stogios et al., 2005), which may promote homologous and heterologous associations with other distantly separated BTB/POZ domain proteins to establish chromatin domains that limit enhancer action. E(y)2 is a subunit of the SAGA histone acetylation complex and the TREX RNA export complex (Kurshakova et al., 2007b). E(y)2 is required for barrier function of the gypsy insulator (Kurshakova et al., 2007a), possibly through its chromatin-opening activity (Kurshakova et al., 2007b). The gypsy insulator displays a versatile capacity to define transcriptional domains, protecting transgenes from enhancer and silencer action when inserted randomly throughout the genome (Roseman et al., 1993; Roseman et al., 1995a; Roseman et al., 1995b; Markstein et al., 2008).
Su(Hw) is globally expressed throughout Drosophila development. Even so, loss of Su(Hw) causes a tissue-restricted phenotype, wherein oogenesis is defective owing to oocyte apoptosis during mid-oogenesis (Klug et al., 1968; Parkhurst et al., 1988; Harrison et al., 1993; Baxley et al., 2011). Two observations suggest that the fertility and insulator functions of Su(Hw) are independent. First, a loss of Su(Hw) occupancy at ∼60% of genomic SBSs has no effect on fertility (Soshnev et al., 2012), an unexpected finding for an insulator-dependent function that has been linked to the formation of physical chromatin domains important for transcriptional regulation (Gurudatta and Corces, 2009; Yang and Corces, 2011; Hou et al., 2012). Second, fertility is unaffected by loss of CP190 and Mod67.2 (Chodagam et al., 2005; Baxley et al., 2011), two partners that are required for Su(Hw) insulator function. Together, these findings suggest that Su(Hw) functions in oogenesis extend beyond the formation of chromatin insulators.
Here, we define transcriptional changes caused by Su(Hw) loss in the ovary and identify direct targets of Su(Hw) regulation during oogenesis. Studies of these target genes show that most have increased expression upon Su(Hw) loss, revealing a major role for Su(Hw) in transcriptional repression. Su(Hw) regulation of target genes is not restricted to the ovary, as loss of Su(Hw) causes gene expression changes in other tissues. The majority of repressed target genes display enriched expression in the CNS and depleted expression in the ovary. These observations suggest that Su(Hw) represses neural genes in non-neural tissues, indicating that Su(Hw) might be a functional homolog of the mammalian repressor for element-1 silencing factor (REST; also known as neuron-restrictive silencing factor, NRSF), a transcription factor that contributes to neural phenotypes via repression of neural genes in non-neural tissues (Lakowski et al., 2006; Ooi and Wood, 2007). Finally, we demonstrate that sterility of su(Hw)-/- females depends largely on the repression of one gene within developing germ cells. This gene is RNA-binding protein 9 (Rbp9), which encodes an ELAV/Hu family RNA-binding protein. Strikingly, decreasing the Rbp9 gene dosage restores oocyte production in su(Hw) null females. Eggs produced by the rescued Rbp9+/-, su(Hw)-/- females show morphological defects, revealing a previously unknown requirement for Su(Hw) in somatic cells of the ovary. Taken together, our studies demonstrate that Su(Hw) is a versatile transcriptional regulatory protein, with an essential developmental function as a transcriptional repressor.
MATERIALS AND METHODS
Fly stocks and culture conditions
Flies were raised at 25°C, 70% humidity on standard cornmeal/agar medium supplemented with yeast. A detailed description of strain genotypes and alleles used in these studies is provided in supplementary material Table S1.
Microarray analyses of gene expression in Drosophila ovary
Ovaries were dissected from 4- to 6-hour-old virgin females and stored at -80°C until needed. The su(Hw)+/+ strains include Canton S and Bloomington Strain 15598. The su(Hw)-/- strains include su(Hw)2/v, su(Hw)A2663/v and su(Hw)Pb/2. Total RNA was isolated from ∼150 pairs of ovaries per biological replicate using TRIzol (Invitrogen), DNaseI treatment (Qiagen DNaseI) and purification on RNeasy columns (Qiagen). Microarray hybridization was performed by the University of Iowa DNA facility using the Affymetrix Drosophila 2.0 arrays (Cat. #900532). Data were processed using Partek Genomics Suite 6.5 Gene Expression pipeline, using GeneChip Robust Multiarray Average (GCRMA) normalization (Irizarry et al., 2003), twofold change and 1% false discovery rate (FDR) cutoffs. Three to six independent biological replicates of each sample were studied. Microarray data were deposited to Gene Expression Omnibus (accession numbers GSE36528 and GSE45286).
Quantitative PCR (qPCR) analyses of gene expression
Quantitative (q)PCR analyses used RNA isolated from ∼50 ovary pairs, 50 third instar larval wing discs or 25 third instar larval brains per biological sample using TRIzol (Invitrogen). All steps were performed as described previously (Soshnev et al., 2008). Expression levels were determined using housekeeping gene RpL32 as an internal control. Primers amplified fragments between 100 and 200 bp. Primer sequences are listed in supplementary material Table S2.
Chromatin immunoprecipitation (ChIP)
Chromosome association of Su(Hw), CP190 and Mod67.2 was tested using ChIP from 50-200 ovary pairs per biological sample, as described previously (Baxley et al., 2011). The antibodies used were guinea pig polyclonal anti-Su(Hw) (Baxley et al., 2011; Soshnev et al., 2012), sheep polyclonal anti-CP190 (Baxley et al., 2011) and rabbit polyclonal anti-Mod67.2 (modEncode D1; kindly provided by K. White, University of Chicago). At least two independent biological samples were analyzed. Primer-amplified fragments between 100 and 200 bp were centered on the Su(Hw) binding consensus when possible. Primer sequences are listed in supplementary material Table S2.
Immunohistochemical analyses
Samples were fixed in 3% electron microscopy grade paraformaldehyde in PBT (PBS with 0.3% v/v Triton X-100) for 30 minutes, washed in PBT and blocked overnight in 5% w/v BSA in PBT at 4°C. Samples were then incubated with primary antibody for at least 8 hours at 4°C, washed in PBT, incubated with the corresponding Alexa Fluor-conjugated secondary antibodies (Molecular Probes), washed in PBT, DAPI stained (0.1 μg/ml, 10 minutes) and mounted in Vectashield (Vector Laboratories). Slides were imaged using a Zeiss 710 confocal microscope and processed using ImageJ. The antibodies used were polyclonal guinea pig anti-Su(Hw) (1:500) (Baxley et al., 2011), polyclonal rabbit anti-Su(Hw) (1:1000) (Parnell et al., 2003), rat anti-ELAV [1:10; 7E8A10; Developmental Studies Hybridoma Bank (DSHB), University of Iowa], mouse anti-Broad Z1 (1:200; Z1.3C11.OA1; DSHB), mouse anti-Repo (1:5; 8D12; DSHB), mouse anti-Gurken (1:200; 1D12; DSHB), polyclonal rabbit anti-Vasa (1:500; Santa Cruz), polyclonal guinea pig anti-Dpn (1:500; a kind gift from J. Skeath, Washington University, St Louis) and polyclonal rabbit anti-Rbp9 (1:3000; a kind gift from M. Buszczak, UT Southwestern) (Kim and Baker, 1993).
Scanning electron microscopy (SEM)
Embryos were collected on orange juice-agar plates, washed with distilled water and fixed overnight in 2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4. Following rinsing, embryos were stained with 1% OsO4 in 0.1 M cacodylate for 2 hours. Next, embryos were rinsed and dehydrated in a series of ethanol washes (20%, 50%, 75%, 94% and 100%) followed by critical point drying. Embryos were mounted on stubs and coated with gold-palladium using Polaron E5100 sputter coater and imaged using Hitachi S-4800 scanning electron microscope.
Analyses of fertility phenotypes
The fertility of 2- to 4-day-old mated females was determined by counting the number of eggs laid on orange juice plates carrying wet yeast paste at 25°C. Egg viability was determined by counting the number of eggs that hatched following egg laying on orange juice plates carrying wet yeast paste.
RESULTS
Identification of Su(Hw)-regulated genes
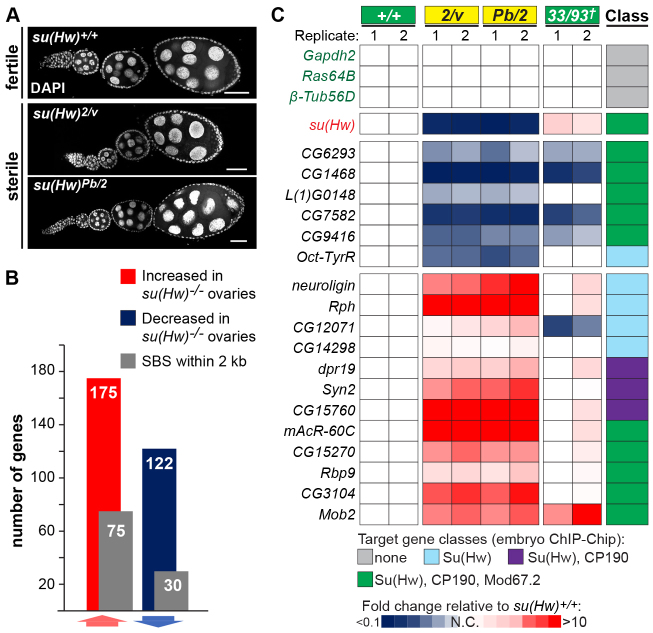
We investigated the regulatory role of Su(Hw) in the ovary using whole-genome gene expression analyses. We measured gene expression changes in ovaries obtained from females of three null and two su(Hw) wild-type genotypes (supplementary material Table S1), to minimize the identification of bystander genes for which expression changes were unrelated to Su(Hw) loss. RNAs were obtained from newly eclosed ovaries, to ensure that our gene expression analyses included equivalent stages of oogenesis. Newly eclosed ovaries lack late-stage egg chambers, establishing a natural method to avoid the stages of oogenesis that are absent in su(Hw)-/- ovaries (Fig. 1A) (Klug et al., 1968; Baxley et al., 2011).
Fig. 1.
Identification of Su(Hw) target genes. (A) Shown are 4- to 6-hour-old DAPI-stained ovarioles isolated from fertile [su(Hw)+/+] and sterile [su(Hw)2/v and su(Hw)Pb/2] females. Scale bars: 25 μm. (B) Microarray analyses identified 175 increased (red bar) and 122 decreased (blue bar) genes, of which 75 and 30 correspond to Su(Hw) target genes, respectively (gray bars). (C) Quantitative PCR validation of target genes. Expression is normalized to the housekeeping gene RpL32 and shown as heat map of fold change values relative to su(Hw)+/+, with blue and red indicating lower and higher expression, respectively. 33/93†, heteroallelic combination of two independently generated recombinant chromosomes containing double Cp190H4-1 and mod(mdg4)u1 mutations. The gypsy insulator proteins associated with the SBSs are shown. Three non-target housekeeping genes and su(Hw) were included as controls. Two independent biological samples (1, 2) were analyzed.
Affymetrix Drosophila 2.0 microarrays were used to measure transcriptional changes resulting from Su(Hw) loss. These arrays contain over 18,500 probe sets corresponding to ∼13,000 genes. We identified 297 genes that changed expression at least twofold in su(Hw)-/- relative to su(Hw)+/+ genetic backgrounds (1% FDR; P<0.01; Fig. 1B). We reasoned that if a gene were a direct target of Su(Hw) regulation, then the gene would carry at least one SBS. To this end, we used an ovary-bound SBS dataset to identify mis-regulated genes that carried an SBS within the transcribed region or within 2 kilobases (kb) upstream or downstream (Soshnev et al., 2012). This regulatory distance was chosen for two reasons. First, 2 kb matches the distance used in other genome-wide studies in Drosophila (Schwartz et al., 2012; Sexton et al., 2012). Second, most target genes identified using the 2-kb window of flanking regulatory DNA are retained at a shorter regulatory distance of 0.5 kb (87% for repressed and 76% of activated target genes; supplementary material Fig. S1A). Using the 2-kb distance, we find that 105 (35%) of the mis-regulated genes carry SBSs compared with 1709 (13.5%) of total genes in the Drosophila genome that carry an SBS, representing a highly significant enrichment (P=6.96E-27). Of the 105 target genes, 75 display increased expression in su(Hw) mutants and correspond to Su(Hw)-repressed genes, whereas 30 display decreased expression and correspond to Su(Hw)-activated genes. Using qPCR, we compared expression of randomly selected target genes in su(Hw)+/+ and su(Hw)-/- RNAs. All but one of the tested genes had altered expression in su(Hw) mutants (Fig. 1C), validating our microarray findings. Most target genes increase expression upon Su(Hw) loss (71%, 75/105; Fig. 1B,C), indicating that Su(Hw) has a major role as a transcriptional repressor.
SBSs in repressed and activated target genes show distinct properties
Su(Hw) target genes are dispersed throughout euchromatic regions of the genome. In total, target genes contain 195 SBSs, with an average of 1.9 SBSs per repressed and 1.0 SBS per activated gene (supplementary material Fig. S1B). Previous studies showed that SBSs are distributed in the genome with no apparent enrichment for gene features (Bushey et al., 2009; Nègre et al., 2010; Soshnev et al., 2012). We used the ovary SBS dataset (Soshnev et al., 2012) to determine whether regulatory SBSs in Su(Hw) target genes display different properties from bulk SBSs and whether SBSs associated with activated or repressed target genes had distinct features. First, we assessed Su(Hw) occupancy. We found that ChIP enrichment at target gene SBSs did not statistically differ from non-target SBSs (Fig. 2A), indicating that Su(Hw) occupancy does not correlate with its effect on transcriptional regulation. Second, we defined the Su(Hw) consensus sequence of SBSs, using the MEME program (Bailey and Elkan, 1994). We found that regulatory and total SBSs possess a similar DNA-binding motif (Adryan et al., 2007; Soshnev et al., 2012) (Fig. 2B), implying that the sequence of an SBS does not predict transcriptional output. Interestingly, these studies revealed that the Su(Hw) binding motif in the gypsy insulator differs from total SBSs, as the insulator motif lacks the GC-rich 3′ extension, but possesses an AT-rich 5′ extension. These observations suggest that Su(Hw) binding at the gypsy insulator might be distinct from that at endogenous SBSs. Third, we examined the distribution of SBSs relative to the gene features. As expected, the fraction of intergenic SBSs decreased (Fig. 2C), because target genes were identified based on SBS proximity. Even so, we found that target gene SBSs show enriched localization to exons (P=3.5E-8) and transcription start sites (TSSs) (P=1.5E-11; Fig. 2C). This latter observation is particularly striking, considering that these regions included only 200 bp flanking each side of the annotated TSS. Localization of many SBSs to target gene promoters is consistent with Su(Hw) having a direct role in target gene transcription.
Fig. 2.
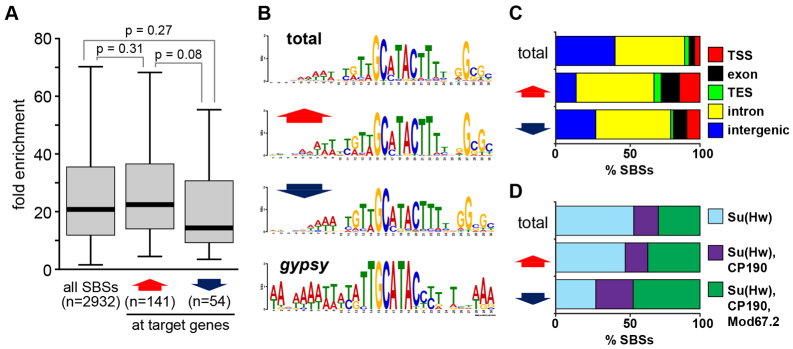
Characterization of Su(Hw) binding sites (SBSs) at target genes. (A) Shown are box plots of fold enrichment of all SBSs (total) and SBSs in activated (red arrow) and repressed (blue arrow) target genes. Within each box, the black line indicates median enrichment, boxes and whiskers represent 25-75 percentile interval and non-outlier range, respectively. P-values of Student’s t-test are indicated. (B) Weblogo of MEME-derived consensus motifs of all SBSs, SBSs at target genes, and the gypsy insulator SBSs. (C) Shown are distributions of SBSs relative to gene features. (D) Shown are enrichments of gypsy insulator proteins at SBSs.
We investigated whether regulatory SBSs were associated with the gypsy insulator proteins. To date, genome-wide mapping of CP190 and Mod67.2 has only been completed in non-ovary tissues, including a dataset obtained from embryos (Nègre et al., 2010). To assess whether the embryonic dataset predicted CP190 and Mod67.2 occupancy in the ovary, we used ChIP-qPCR to analyze the presence of CP190 and Mod67.2 at SBSs within ovary chromatin. These experiments demonstrated that 80% (28/35) and 89% (31/35) of the embryonic SBSs tested showed the expected occupancy of CP190 and Mod67.2, respectively (supplementary material Fig. S2). As these data endorse the embryonic dataset for predicting ovary binding of the gypsy insulator proteins, we examined recruitment of these proteins at target genes. We found that target genes had increased association of CP190 and Mod67.2 relative to total SBSs (Fig. 2D). To determine whether the gypsy insulator proteins were required for Su(Hw) regulation of target genes, we tested target gene expression in females carrying one of two independently generated, double mutant recombinant chromosomes (R33 and R93). Each chromosome carried the Cp190H4-1, mod(mdg4)u1 (modu1) alleles (supplementary material Table S1). Homozygous Cp190H4-1/H4-1, modu1/u1 females are viable, which allowed measurements of RNA levels in newly eclosed ovaries. As predicted, effects of Mod67.2 and CP190 loss occurred mostly at target genes that carry gypsy-like SBSs (5/6; Fig. 1C). We found that activated target genes had a stronger dependence on these gypsy insulator proteins than did repressed target genes, as expression of 80% (4/5) of activated target genes decreased in Cp190H4-1/H4-1, modu1/u1 ovaries (Fig. 1C). One target gene, CG12071, showed altered expression in Cp190H4-1/H4-1, modu1/u1 mutants, even though this gene lacks a gypsy-like SBS. However, loss of CP190 and Mod67.2 had an opposite effect to that resulting from loss of Su(Hw), implying that altered expression might reflect an indirect effect in the double mutant background. Our data demonstrate that a subset of Su(Hw) target genes require CP190 and Mod67.2 for expression.
Su(Hw) is a repressor of CNS-enriched genes in the ovary
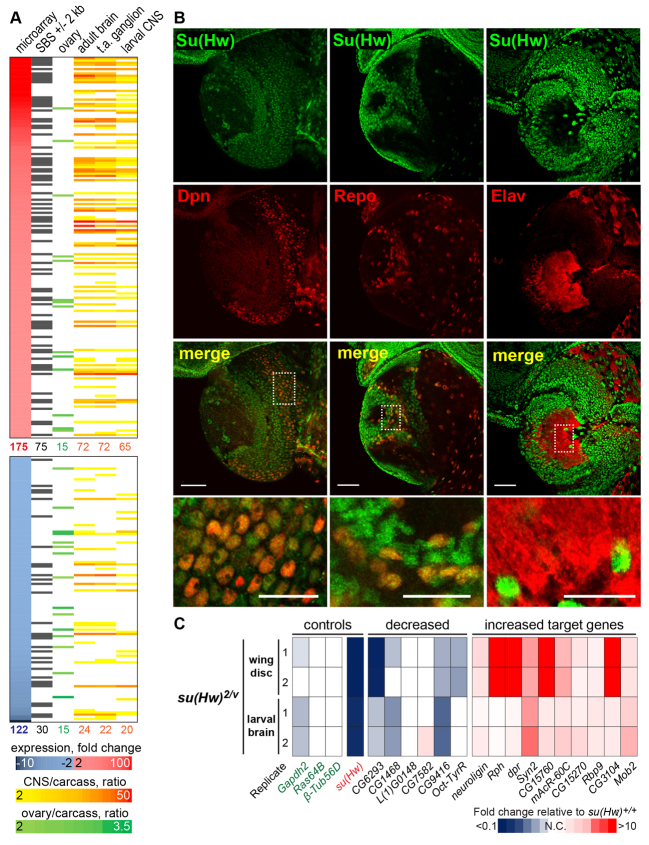
We examined the collection of target genes to identify common features of Su(Hw)-regulated genes. First, we performed Gene Ontology analysis and found no significantly over-represented developmental or signaling pathway (data not shown). Second, we determined whether target genes displayed common tissue expression patterns, using the FlyAtlas anatomical expression dataset that includes larval and adult tissues (Chintapalli et al., 2007). In our analyses, we considered that a gene showed tissue-enriched expression if RNA expression levels were twofold or higher in that tissue relative to the level in the whole fly carcass. These analyses revealed that Su(Hw) target genes showed significantly enriched and depleted gene expression in several tissues (Fig. 3A; supplementary material Tables S3, S4). We found that Su(Hw)-repressed target genes are significantly enriched for CNS expression [75% (56/75) relative to 28% of total Drosophila genes (3654/12856), P=5.3E-19], but depleted in the ovary [5% (4/75) relative to 16% (2070/12856) of total Drosophila genes, P=0.011] and in the testes [8% (6/75) relative to 22% (2778/12856) of total Drosophila genes, P=0.004]. For Su(Hw)-activated target genes, we found that expression was significantly enriched in the hindgut [40% (12/30) relative to 15% (1890/12856), P=4.80E-05]. Based on these findings, we conclude that the major role of Su(Hw) in the ovary is to repress neural genes, as more than half of all target genes show CNS-enriched, but ovary-depleted expression.
Fig. 3.
Su(Hw) is a repressor of CNS-enriched genes. (A) Left to right: target genes ranked by fold changes obtained in microarray analyses, with red corresponding to activated and blue corresponding to repressed genes; genes with SBSs; genes with ovary-enriched expression indicated by green color scale; genes with CNS-enriched expression in three CNS structures [adult brain, thoracoabdominal (t.a.) ganglion and larval CNS] indicated by the orange color scale (Chintapalli et al., 2007). (B) Confocal images of su(Hw)+/+ third instar larval brains stained for Su(Hw) (green, top) and neural markers (Dpn, Repo, ELAV; red, middle), and merged image (bottom). Scale bars: 50 μm. Magnified areas (shown below) are indicated by dotted rectangles. Scale bars: 25 μm. (C) qPCR analyses of activated and repressed target genes in RNA isolated from su(Hw)2/v larval brain and wing disc. Expression was normalized to the housekeeping gene RpL32 and is shown as heat map of fold change values relative to su(Hw)+/+, with blue and red indicating low and high expression, respectively. Three non-target housekeeping genes and su(Hw) were included as controls. Two biological samples (1, 2) were analyzed.
One implication of a role for Su(Hw) in the regulation of neural genes in the ovary is that Su(Hw) may not be globally expressed in the CNS. Such tissue-restricted expression was unexpected, as previous studies indicated that Su(Hw) accumulation was ubiquitous throughout development (Harrison et al., 1993). To investigate this, we examined Su(Hw) accumulation in third instar larval CNS and uncovered cell-type specific Su(Hw) expression (Fig. 3B). Immunohistochemical analyses showed that Su(Hw) is present in neuroblasts [Deadpan (Dpn)-positive cells (Doe and Skeath, 1996)] and glia [Reverse Polarity (Repo)-positive cells (Xiong et al., 1994)], but is absent in terminally differentiated post-mitotic neurons [Embryonic Lethal Abnormal Vision (ELAV) (Robinow and White, 1991)]. This cell type-specific expression pattern is established early in development, as Su(Hw) is absent in ELAV-positive cells in embryos (data not shown). These observations indicate that Su(Hw) accumulation is dynamic in neural lineages and is consistent with Su(Hw) acting as a repressor of neural genes in non-neural tissues.
Su(Hw) occupancy at SBSs shows little tissue specificity (Adryan et al., 2007; Soshnev et al., 2012). Based on these observations, we predicted that Su(Hw) target genes might be mis-regulated in non-ovary tissues. Previous studies using whole larvae demonstrated that loss of Su(Hw) altered expression of some of the target genes that we identified in the ovary (Adryan et al., 2007). To gain greater insight into the tissue-specificity of Su(Hw) regulation, we examined gene expression in individual tissues, performing qPCR analyses of RNAs isolated from third instar larval brains and wing discs (Fig. 3C). These studies revealed that the majority of tested target genes had altered expression in wing or brain tissue (14/16, 88%; Fig. 4B). The largest transcriptional changes were seen in the wing disc, which might reflect the more uniform expression of Su(Hw) in wing discs, but not brain tissue (Fig. 3B; data not shown). These findings demonstrate that Su(Hw) is required for the regulation of target gene expression in multiple tissues.
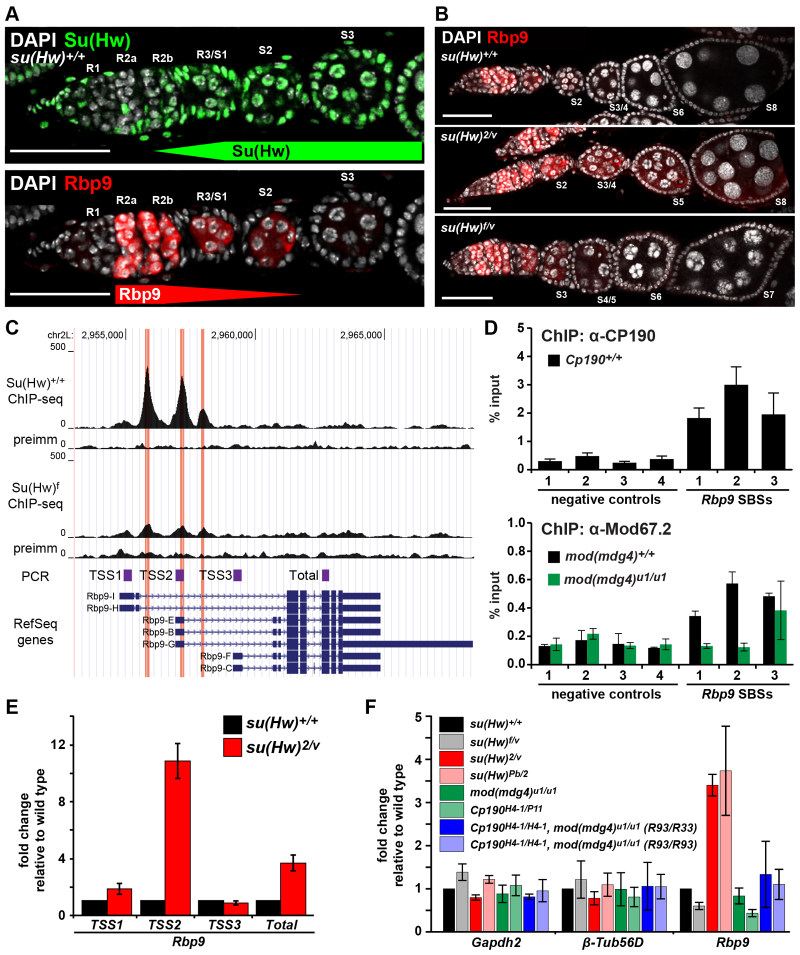
Fig. 4.
Rbp9 is repressed by Su(Hw). (A) Images of su(Hw)+/+ germarium stained for Su(Hw) (top, green), Rbp9 (bottom, red) and DAPI (white). Developmental regions (R1 to R3) of the germarium and egg chamber stages (S1 to S3) are indicated. Scale bars: 25 μm. (B) su(Hw)+/+ (top), sterile su(Hw)2/v (middle) and fertile su(Hw)f/v (bottom) ovarioles stained for Rbp9 (red) and DAPI (white). Scale bars: 25 μm. (C) UCSC genome browser view of the Rbp9 gene locus, including tracks. Top to bottom: chromosome coordinates, su(Hw)wt ChIP-Seq reads, preimmune serum IP control reads, su(Hw)f ChIP-Seq reads, preimmune serum IP control reads, fragments amplified in qPCR analyses (E), RefSeq gene annotation. (D) ChIP-qPCR analyses of ovary-bound CP190 (top) and Mod67.2 (bottom) at Rbp9 SBSs. Negative controls (1-4) were genomic regions with no SBS (Soshnev et al., 2012). ChIP from a mod(mdg4)u1 mutant background was a negative control. (E) qPCR analyses of promoter-specific Rbp9 transcripts in su(Hw)+/+ (black bars) and su(Hw)2/v (red bars) mutant background. Expression is normalized to housekeeping gene RpL32 and is shown as fold change relative to su(Hw)+/+. Error bars indicate s.d. of three biological samples. (F) qPCR analyses of gene expression changes in su(Hw), Cp190 and mod(mdg4) mutant ovaries. Expression is normalized to the housekeeping gene RpL32 and shown as a fold change relative to su(Hw)+/+. Gapdh2 and β-tubulin are negative controls. R33 and R93 indicate two independently generated recombinant chromosomes containing Cp190H4-1 and mod(mdg4)u1 mutations. Error bars indicate s.d. of two independent biological samples.
Su(Hw) represses Rbp9 expression
One Su(Hw)-repressed target gene was RNA-binding protein 9 (Rbp9), a gene first identified owing to its CNS-enriched expression (Kim and Baker, 1993). Rbp9 belongs to the ELAV/Hu gene family that encodes RNA-binding proteins (Kim and Baker, 1993; Pascale et al., 2008). Although loss of Rbp9 causes female sterility owing to an early arrest in germline development (Kim-Ha et al., 1999), ectopic Rbp9 expression causes oocyte apoptosis in mid-to-late oogenesis (Jeong and Kim-Ha, 2003). The overlap of this latter phenotype with the su(Hw) mutant phenotype prompted us to investigate Rbp9 regulation.
We studied Su(Hw) and Rbp9 protein localization in the ovary. Oogenesis begins in the germarium by asymmetric division of a germline stem cell (GSC). The resulting daughter cell, termed cystoblast, undergoes four incomplete mitotic divisions to generate a 16-cell cyst, which becomes enveloped by somatic follicle cells to form an egg chamber. Co-staining ovaries with Su(Hw) and Rbp9 antibodies revealed differences in protein accumulation (Fig. 4A). Su(Hw) is present in somatic and germ cells, whereas Rbp9 is present only in germ cells. In germ cells, Su(Hw) is found at low levels in the GSCs and daughter cystoblasts and is absent in regions of the germarium where the 16-cell cyst is formed and meiosis is initiated (regions 1 and 2a). Su(Hw) reappears in region 2b and increases during egg chamber formation (Baxley et al., 2011). By contrast, Rbp9 is found in region 2a of the germarium, remains high in region 2b, diminishes in region 3 as egg chambers form, and becomes undetectable beyond stage 3 egg chambers (Tastan et al., 2010). We reasoned that if Su(Hw) were required for transcriptional repression of Rbp9, then loss of Su(Hw) would prolong Rbp9 protein accumulation. This prediction was met. Loss of Su(Hw) is accompanied by extended presence of Rbp9 protein into late-stage egg chambers (Fig. 4B), supporting a role for Su(Hw) in Rbp9 regulation.
The Rbp9 gene contains three alternative TSSs, of which TSS3 is the most active in the ovary (Graveley et al., 2011). Interestingly, Su(Hw) binds near these TSSs, with SBS2 located 357 bp downstream of TSS2 (Fig. 4C). All Rbp9 SBSs are associated with the gypsy insulator proteins CP190 and Mod67.2 (Fig. 4D). We used transcript-specific qPCR primers to investigate how Su(Hw) loss affects Rbp9 transcription in su(Hw)2/v newly eclosed ovaries. These studies showed that Rbp9 transcription from TSS1 and TSS3 was largely unaffected, whereas transcription from TSS2 increased ∼11-fold (Fig. 4E). These data indicate that Su(Hw) repression is specific to TSS2. Such a promoter-specific de-repression was unexpected. Although TSS1 might lack binding sites responsive to ovary transcription factors, this limitation does not apply to TSS3. Based on these data, we suggest that Su(Hw) repression of Rbp9 depends on a localized action targeted to TSS2. To determine whether Rbp9 repression involves insulator formation, we determined the effects of loss of CP190 or Mod67.2 on Rbp9 transcription. Gene expression was measured using qPCR of RNAs isolated from ovaries from (1) Cp190H4-1/P11 females, (2) mod(mdg4)u1/u1 (modu1/u1) females and (3) Cp190H4-1/H4-1, modu1/u1 double mutant females. These studies showed that Rbp9 transcription is maintained in all Cp190 and modu1/u1 mutant backgrounds (Fig. 4F), implying that Su(Hw) regulation does not involve insulator formation. Taken together, these observations suggest that Su(Hw) is a direct repressor of Rbp9 transcription, through local effects on TSS2.
Suppression of sterility in su(Hw) null females
The shared mutant phenotypes between ectopic Rbp9 expression and Su(Hw) loss suggested that Rbp9 de-repression might cause sterility in su(Hw)-/- females. We reasoned that if increased transcription of Rbp9 were responsible for su(Hw)-/- sterility, then su(Hw) mutants that retain fertility should demonstrate wild-type Rbp9 regulation. To this end, we studied Rbp9 transcription and protein accumulation in ovaries obtained from su(Hw)f/v females. Importantly, su(Hw)f encodes a full-length Su(Hw) protein with a defective ZF10. Previous studies have shown that su(Hw)f/v females display wild-type fertility, even though Su(Hw)f binds only ∼40% of genomic SBSs (Baxley et al., 2011; Soshnev et al., 2012). Strikingly, in su(Hw)f/v ovaries, Rbp9 shows a near-normal accumulation during egg chamber development (Fig. 4B), corresponding to Su(Hw)f retention at Rbp9 SBSs in vivo (Fig. 4C) (Soshnev et al., 2012) and transcriptional repression of Rbp9 (Fig. 1C; Fig. 4D). Taken together, these data imply that fertility and Rbp9 regulation are linked.
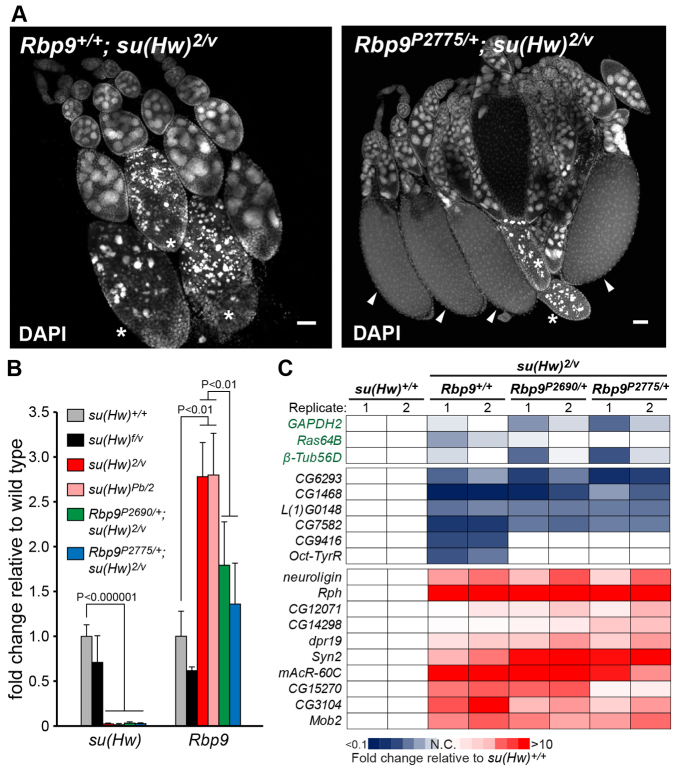
Loss of Su(Hw) increases levels of Rbp9 RNA threefold in the ovaries of newly eclosed females. We postulated that if Rbp9 de-repression caused su(Hw)-/- sterility, then loss of one gene copy of Rbp9 might reduce Rbp9 RNA to a level compatible with female fertility. To this end, we generated Rbp9+/-, su(Hw)2/v double mutants, wherein mutants carried one of four independently generated Rbp9 null alleles. These Rbp9 alleles included two P-element insertions in the Rbp9 gene (Kim-Ha, 2000) and two genomic deficiencies that removed Rbp9 (supplementary material Table S1), as well as other genes. Strikingly, Rbp9+/-, su(Hw)2/v females derived from any of the four Rbp9 null alleles produced eggs at ∼8-20% of the wild-type level (Table 1). Females carrying alleles of the large genomic deletions produced fewer eggs, a difference that might result from the larger number of genes deleted. Ovaries obtained from Rbp9+/-, su(Hw)2/v females contained late-stage egg chambers, although evidence of egg chamber apoptosis remained (Fig. 5A). RNA analyses demonstrated that Rbp9 RNA was not increased above twofold in Rbp9+/-, su(Hw)2/v ovaries relative to wild type, whereas su(Hw) RNA was undetectable (Fig. 5B). We tested whether reduced Rbp9 expression restored expression of other mis-regulated Su(Hw) target genes, as the Rbp9 protein belongs to the ELAV/Hu gene family of RNA-binding proteins and might alter mRNA stability (Pascale et al., 2008). These analyses showed that Su(Hw) target genes remained mis-regulated in the Rbp9+/-, su(Hw)2/v rescued ovaries (Fig. 5C). Strikingly, these data demonstrate that reducing the dosage of a single Su(Hw) target gene restores fertility to su(Hw) null females.
Table 1.
Egg lay analyses of su(Hw)2/v mutants carrying hemizygous mutations in target and non-target genes
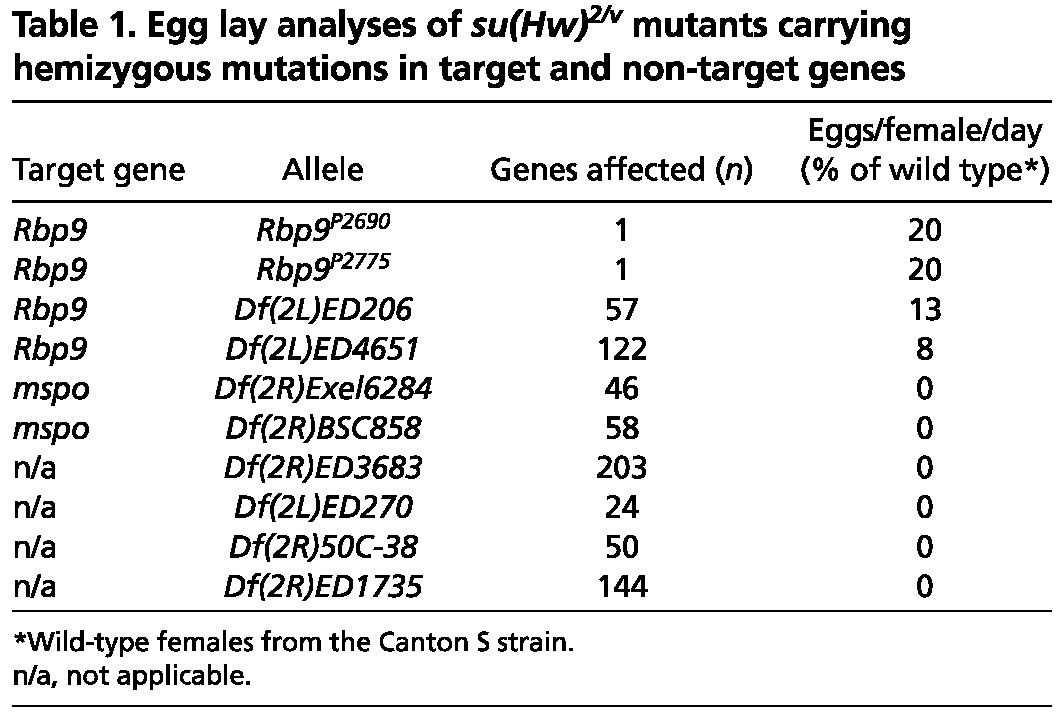
Fig. 5.
Decreased Rbp9 expression rescues female sterility of su(Hw) null mutants. (A) DAPI-stained ovarioles isolated from su(Hw)2/v and Rbp9P2775/+; su(Hw)2/v females. Asterisks indicate egg chamber apoptosis. Arrowheads indicate late-stage egg chambers. Scale bars: 50 μm. (B) qPCR analyses of su(Hw) and Rbp9 RNA levels in the fertile su(Hw)+/+ (gray), su(Hw)f/v (black), sterile su(Hw)2/v (red) and su(Hw)Pb/2 (pink) and fertile Rbp9P2690/+; su(Hw)2/v (green) and Rbp9P2775/+; su(Hw)2/v (blue) backgrounds. Expression is normalized to housekeeping gene RpL32 and is shown as fold change relative to su(Hw)+/+. Error bars indicate s.d. of three biological samples. (C) qPCR analyses of activated and repressed target genes in ovaries dissected from su(Hw)+/+, su(Hw)2/v, Rbp9P2690/+; su(Hw)2/v and Rbp9P2775/+; su(Hw)2/v females. Expression was normalized to the housekeeping gene RpL32 and is shown as heat map of fold change values relative to su(Hw)+/+, blue and red indicating low and high expression, respectively. Three non-target housekeeping genes were included as controls. Two biological samples (1, 2) were analyzed.
As a measure of the specificity of the Rbp9 rescue, we tested whether hemizygous loss of other genes restored oogenesis in su(Hw) null females. To this end, we tested a deficiency that removed mspo. This gene was chosen for two reasons. First, Su(Hw)f retains binding to mspo, potentially linking mspo de-repression to su(Hw)-/- sterility. Second, loss of Su(Hw) increases mspo transcription approximately threefold, implying that loss of one gene copy might lower their RNA level to that found in su(Hw)+/+ ovaries and might lead to rescued oogenesis. As an additional control, we tested four chromosome deficiencies that collectively remove >400 non-Su(Hw) target genes, to determine whether general decreases in gene dosage reverse the su(Hw)-/- phenotype. We found that mspo+/-, su(Hw)2/v and the Df(2)/+, su(Hw)2/v females remained sterile (Table 1). Based on these data, we conclude that de-repression of Rbp9 is the central cause of female sterility in su(Hw) null mutants.
Rescue of sterility phenotype reveals somatic function of Su(Hw) in oogenesis
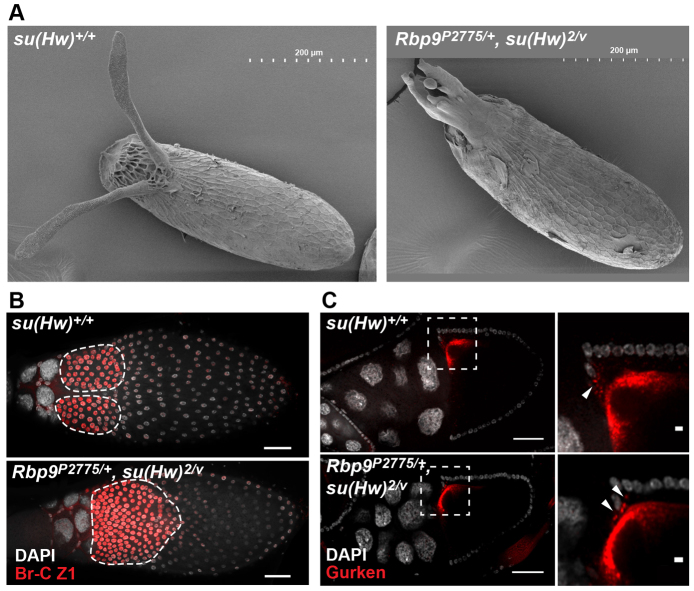
We noted that most eggs produced by Rbp9+/-, su(Hw)2/v females had fused and deformed dorsal appendages (Fig. 6A). These observations imply a second, previously unrecognized requirement for Su(Hw) during late oogenesis. Dorsal appendage formation is linked to specification of the dorsal-ventral axis of the oocyte through three classes of genes that regulate signaling pathways (Berg, 2005). The presence of a single, broad dorsal appendage in Rbp9+/-, su(Hw)2/v eggs suggested that loss of Su(Hw) might compromise the function of the midline-minus class of genes. A hallmark of disruptions in this gene class is the aberrant expression of broad, which encodes four BTB/POZ domain ZF transcription factors. Among these, the Z1 isoform is required for dorsal appendage formation, being expressed in two lateral-dorsal fields of dorsal appendage primordia (Deng and Bownes, 1997; Tzolovsky et al., 1999). A failure to define two fields is associated with expansion and fusion of the dorsal appendages (Ward and Berg, 2005). To investigate whether broad expression was altered in Rbp9+/-, su(Hw)2/v egg chambers, ovaries were stained with antibodies against Z1. Whereas wild-type stage 12 egg chambers displayed two fields of Z1 expression, Rbp9+/-, su(Hw)2/v egg chambers showed a single field (Fig. 6B). The data suggest that loss of Su(Hw) alters the regulation of broad expression in late oogenesis.
Fig. 6.
Rescue of female sterility reveals a somatic function for Su(Hw). (A) SEM image of a su(Hw)+/+ egg and a Rbp9P2775/+, su(Hw)2/v egg. (B) Stage 12 egg chambers from su(Hw)+/+ and Rbp9P2775/+, su(Hw)2/v mutant females, stained with Broad-Z1 (red) and DAPI (white). Scale bars: 50 μm. (C) Left: whole-mount egg chambers stained for Gurken (red) and DAPI (white) dissected from wild-type (top) and Rbp9P2775/+, su(Hw)-/- (bottom) females. Dashed rectangles indicate magnified areas (shown to the right). Arrowheads indicate Gurken-positive vesicles. Scale bars: 50 μm (left) and 5 μm (right).
The dorsal-ventral signaling cascade contributes to the repression of broad expression in midline cells (Berg, 2005). Activation of this cascade depends upon Gurken, a TGFα ligand that signals from the oocyte to the overlying follicle cells using the homolog of the epidermal growth factor receptor (Deng and Bownes, 1997). To test the involvement of Gurken, Rbp9+/-, su(Hw)-/- ovaries were stained with Gurken antibodies, revealing that Gurken production and localization were unaffected in the rescued ovaries (Fig. 6C). These data imply that Su(Hw) loss alters signaling events downstream of Gurken. Based on these findings, we predict that loss of Su(Hw) alters expression of one or more genes within the somatic follicle cells, leading to altered regulation of broad. We conclude that Su(Hw) has regulatory functions in somatic and germ cells during oogenesis.
DISCUSSION
Su(Hw) constitutively binds ∼3000 SBSs distributed throughout the euchromatic regions of the genome (Soshnev et al., 2012). Endogenous SBSs largely contain a single Su(Hw) binding motif and show context-specific recruitment of CP190 and Mod67.2. Here, we address how Su(Hw) contributes to gene expression in the ovary, in which Su(Hw) function is essential. These studies advance our understanding of the transcriptional role of Su(Hw), revealing an essential function as a transcriptional repressor.
Su(Hw) regulation extends beyond insulator formation
Transcriptional requirements for Su(Hw) were defined using gene expression microarrays, controlling for genetic background and the developmental differences between su(Hw)+/+ and su(Hw)-/- ovaries (Fig. 1; supplementary material Table S3). These analyses identified 297 mis-regulated genes, with over a third (105, 35%) corresponding to SBS-containing genes. Most Su(Hw)-target genes are de-repressed upon Su(Hw) loss (71%, 75/105; Fig. 1B,C), suggesting that most Su(Hw) regulation involves transcriptional repression. These data are consistent with previous findings that Su(Hw) localizes to repressive chromatin (Filion et al., 2010) and binds near genes that display low levels of transcription (Bushey et al., 2009; Roy et al., 2010).
Su(Hw) establishes an insulator when bound to the gypsy retrotransposon. This function depends upon recruitment of two non-DNA-binding proteins, CP190 and Mod67.2 (Raab and Kamakaka, 2010). Building from this well-established requirement, we investigated whether Su(Hw) regulation of target genes involved insulator formation. We find that 20% of Su(Hw) target genes lack association of the gypsy insulator proteins (supplementary material Table S3), indicating that regulation of these genes might not involve canonical insulator formation. Additionally, even though 80% of target genes bind CP190 or Mod67.2, we found that protein localization does not always predict a regulatory involvement. Of the ten target genes tested that carry SBSs associated with CP190 and Mod67.2, five displayed altered gene expression in Cp190, mod67.2 mutants (Fig. 1C). Interestingly, most of the affected target genes require the gypsy insulator proteins for transcriptional activation (Fig. 1C), suggesting that Su(Hw) might establish an insulator at some genes to prevent the spread of repressive chromatin. Alternatively, Su(Hw) might have a direct role in activation of gene expression, as CP190 shows strong association with active promoters (Bartkuhn et al., 2009). Taken together, our findings suggest that Su(Hw) regulation of transcription of most target genes is independent of insulator formation.
SBSs present in Su(Hw) target genes show a distinct distribution relative to gene features than do total SBSs. Interestingly, we observe a bias for location of regulatory SBSs with TSSs (Fig. 2C), indicating that Su(Hw) might have a direct transcriptional repression role. This prediction is supported by studies of Rbp9 regulation. Rbp9 expression involves transcription from three TSSs, with loss of Su(Hw) causing de-repression of only TSS2 (Fig. 4E). Such a limited transcriptional response is unexpected if Rbp9 mis-regulation resulted from loss of a Su(Hw)-dependent insulator, because TSS3 is an active promoter in the ovary (Graveley et al., 2011) and should also respond to an unblocked enhancer. Consistent with an insulator-independent mechanism, transcription from TSS2 is unchanged in Cp190, mod67.2 females, a genetic background that compromises gypsy insulator function (Gerasimova et al., 1995; Pai et al., 2004; Baxley et al., 2011). Notably, one Rbp9 SBS is located ∼400 bp downstream of TSS2, suggesting that Su(Hw)-dependent repression may result from a block of RNAP II recruitment or elongation from this promoter. Alternatively, Rbp9 repression might depend upon interactions between SBSs to form a chromatin loop that constrains TSS2 activity. This latter mechanism shares features of the insulator function, as insulators display an ability for long-range interactions (Yang and Corces, 2012), unifying the mechanism of gene repression and insulator function.
Su(Hw) functions as a global repressor of neural genes in non-neuronal tissues
At the beginning of our studies, we predicted that the tissue-restricted defects caused by loss of the globally expressed and constitutively bound Su(Hw) protein were due to mis-regulation of genes that are expressed primarily in the ovary. However, our data do not support this prediction. Whereas 7% (7/105) of Su(Hw) target genes display ovary-enriched expression (Fig. 3A), 65% show CNS-enriched expression (68/105). Comparisons with global gene expression in these tissues indicate that Su(Hw) target genes show ovary-depleted, CNS-enriched expression (supplementary material Table S3). Interestingly, loss of Su(Hw) causes de-repression of most target genes in multiple tissues (Fig. 1B; Fig. 3C). Based on these data, we conclude that Su(Hw) is a transcriptional repressor of neural genes in non-neural tissues. A recent publication studying Drosophila chromatin proteins using Bayesian network analysis support this conclusion, showing that Su(Hw) is associated with gene repression and neurological system processes (van Bemmel et al., 2013).
Properties of Su(Hw) are reminiscent of the REST, a mammalian transcription factor that establishes neural phenotypes owing to repression of neural genes in non-neural tissues (Chong et al., 1995). REST is an eight ZF protein that interacts with two separate co-repressor complexes, the transcriptional co-repressor CoREST and a Sin3-histone deacetylase complex (Lakowski et al., 2006; Ooi and Wood, 2007). Although REST is not conserved in drosophilids, a homolog of CoREST has been identified (Dallman et al., 2004; Yamasaki et al., 2011), implying that non-REST transcription factors direct dCoREST to chromosomes. Drosophila CoREST is a component of a newly identified transcriptional repressor complex LINT, which contains three subunits, CoREST, Drosophila lethal (3) malignant brain tumor [L(3)mbt] and Drosophila L(3)mbt interacting protein 1 (dLint-1) (Meier et al., 2012). Interestingly, L(3)mbt is a transcription factor associated with insulator elements (Richter et al., 2011). Based on this connection, we examined whether dLint-1 and L(3)mbt colocalized with Su(Hw) at SBSs in target genes. Strikingly, over a third (18/56) of CNS-enriched repressed target genes contain SBSs that colocalize with L(3)mbt (Richter et al., 2011) and >60% (35/56) contain SBSs that colocalize with dLint-1 (Meier et al., 2012). These data indicate that Su(Hw)-dependent repression might depend upon CoREST recruitment within the LINT complex. Taken together, our observations suggest that Su(Hw) might represent a third functional homolog of REST in Drosophila, with Charlatan and Tramtrack representing the other identified homologs (Dallman et al., 2004; Tsuda et al., 2006; Yamasaki et al., 2011).
Mis-regulation of a single Su(Hw) target gene is largely responsible for su(Hw)-/- sterility
Repression of one target gene, Rbp9, is central to sterility in su(Hw)-/- females. This conclusion stems from our striking observation that oogenesis is rescued in Rbp9+/-, su(Hw)2/v females, with these female producing ∼20% of the wild-type number of eggs (Fig. 6; Table 1). Rbp9 encodes a protein that belongs to the ELAV/Hu gene family of RNA-binding proteins (Kim and Baker, 1993). The ELAV family regulates multiple post-transcriptional steps in gene expression, ranging from alternative splicing to translation (Soller et al., 2010; Hilgers et al., 2012). Rbp9 is transiently expressed in germ cells of developing cysts, wherein the encoded RNA-binding protein has an essential function to repress translation of the germ cell differentiation factor, Bag of marbles (Kim-Ha et al., 1999). Su(Hw) directs repression of Rbp9 transcription after the formation of developing cysts to permit egg chamber development (Fig. 4A,B). Rescued Rbp9+/-, su(Hw)2/v females produced fewer eggs than do su(Hw)+/+ females (Table 1). We consider two possible explanations to account for this partial suppression. First, Su(Hw) target genes besides Rbp9 might contribute to su(Hw) sterility, because these genes remained mis-regulated in Rbp9+/-, su(Hw)2/v ovaries (Fig. 5). However, not all Su(Hw) target genes might contribute to defects in sterility, as oogenesis was not restored in su(Hw)-/- females carrying deletions encompassing another upregulated target gene (Table 1). Second, low levels of Rbp9 protein aberrantly accumulate in Rbp9+/-, su(Hw)2/v late-stage egg chambers. We note that Rbp9 binds U-rich RNAs and regulates translation and stability of its target RNAs (Park et al., 1998; Kim-Ha et al., 1999). As such, even low levels of ectopically produced Rbp9 might affect the function of RNAs crucial for late oogenesis, thereby triggering apoptosis. Identification of Rbp9 target RNAs may provide insight into processes involved in programmed cell death that occurs in mid-oogenesis.
In summary, we demonstrate that Su(Hw) is required for activation and repression of individual target genes, extending the known regulatory properties of this classic insulator protein. A growing body of data suggests that many insulator proteins have transcriptional functions that extend beyond insulator formation. For example, early studies documented roles for CTCF as a direct transcriptional activator and repressor (Lobanenkov et al., 1990; Burcin et al., 1997), findings supported by recent genetic analyses in transgenic mice (Heath et al., 2008; Wan et al., 2008; Ribeiro de Almeida et al., 2009; Soshnikova et al., 2010). Further studies are needed to establish the general principles that govern the interplay between genomic context and transcriptional functions of insulator proteins.
Supplementary Material
Acknowledgments
We thank Chantal Allamargot and Jean Ross at the University of Iowa Central Microscopy Research Facility for assistance with confocal and scanning electron microscopy; Mary Boes, Garry Hauser and Kevin Knudtson at the University of Iowa DNA Facility for assistance with the microarray experiments; Bing He for advice on bioinformatic analyses; and members of the Geyer Lab for critical reading of the manuscript.
Footnotes
Funding
This work was supported by a National Institutes of Health grant [GM42539 to P.K.G.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
A.A.S. and R.M.B. performed the experiments. J.R.M. and K.T. assisted with the analyses of microarray data. A.A.S. and P.K.G. designed the experiments and wrote the paper.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.094953/-/DC1
References
- Adryan B., Woerfel G., Birch-Machin I., Gao S., Quick M., Meadows L., Russell S., White R. (2007). Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 8, R167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- Bartkuhn M., Straub T., Herold M., Herrmann M., Rathke C., Saumweber H., Gilfillan G. D., Becker P. B., Renkawitz R. (2009). Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 28, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxley R. M., Soshnev A. A., Koryakov D. E., Zhimulev I. F., Geyer P. K. (2011). The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev. Biol. 356, 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg C. A. (2005). The Drosophila shell game: patterning genes and morphological change. Trends Genet. 21, 346–355 [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Kadonaga J. T. (1998). Going the distance: a current view of enhancer action. Science 281, 60–63 [DOI] [PubMed] [Google Scholar]
- Bulger M., Groudine M. (2010). Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 339, 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcin M., Arnold R., Lutz M., Kaiser B., Runge D., Lottspeich F., Filippova G. N., Lobanenkov V. V., Renkawitz R. (1997). Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol. 17, 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey A. M., Ramos E., Corces V. G. (2009). Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23, 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 [DOI] [PubMed] [Google Scholar]
- Chodagam S., Royou A., Whitfield W., Karess R., Raff J. W. (2005). The centrosomal protein CP190 regulates myosin function during early Drosophila development. Curr. Biol. 15, 1308–1313 [DOI] [PubMed] [Google Scholar]
- Chong J. A., Tapia-Ramírez J., Kim S., Toledo-Aral J. J., Zheng Y., Boutros M. C., Altshuller Y. M., Frohman M. A., Kraner S. D., Mandel G. (1995). REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 [DOI] [PubMed] [Google Scholar]
- Dallman J. E., Allopenna J., Bassett A., Travers A., Mandel G. (2004). A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J. Neurosci. 24, 7186–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino G. I., Schwartz Y. B., Farkas G., McCabe D., Elgin S. C., Pirrotta V. (2004). Polycomb silencing blocks transcription initiation. Mol. Cell 13, 887–893 [DOI] [PubMed] [Google Scholar]
- Deng W. M., Bownes M. (1997). Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development 124, 4639–4647 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Skeath J. B. (1996). Neurogenesis in the insect central nervous system. Curr. Opin. Neurobiol. 6, 18–24 [DOI] [PubMed] [Google Scholar]
- Dorsett D. (1993). Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics 134, 1135–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., Ward L. D., Brugman W., de Castro I. J., Kerkhoven R. M., Bussemaker H. J., et al. (2010). Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M., Morcillo P., Dorsett D. (2001). Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21, 4807–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P. G., Gerasimova T. I. (1989). Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220, 121–126 [DOI] [PubMed] [Google Scholar]
- Georgiev P., Kozycina M. (1996). Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T. I., Gdula D. A., Gerasimov D. V., Simonova O., Corces V. G. (1995). A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82, 587–597 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G. (1992). DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6, 1865–1873 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Spana C., Corces V. G. (1986). On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5, 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R., Giles K., Gowher H., Xiao T., Xu Z., Yao H., Felsenfeld G. (2012). Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta 1819, 644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurudatta B. V., Corces V. G. (2009). Chromatin insulators: lessons from the fly. Brief. Funct. Genomic. Proteomic. 8, 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., Mortin M. A., Corces V. G. (1992). The RNA polymerase II 15-kilodalton subunit is essential for viability in Drosophila melanogaster. Mol. Cell. Biol. 12, 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., Gdula D. A., Coyne R. S., Corces V. G. (1993). A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7, 1966–1978 [DOI] [PubMed] [Google Scholar]
- Heath H., Ribeiro de Almeida C., Sleutels F., Dingjan G., van de Nobelen S., Jonkers I., Ling K. W., Gribnau J., Renkawitz R., Grosveld F., et al. (2008). CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 27, 2839–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Lemke S. B., Levine M. (2012). ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 26, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Li L., Qin Z. S., Corces V. G. (2012). Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- Jeong K., Kim-Ha J. (2003). Expression of Rbp9 during mid-oogenesis induces apoptosis in egg chambers. Mol. Cells 16, 392–396 [PubMed] [Google Scholar]
- Kermekchiev M., Pettersson M., Matthias P., Schaffner W. (1991). Every enhancer works with every promoter for all the combinations tested: could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1, 71–81 [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Baker B. S. (1993). The Drosophila gene rbp9 encodes a protein that is a member of a conserved group of putative RNA binding proteins that are nervous system-specific in both flies and humans. J. Neurosci. 13, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J. (2000). Isolation of true Rbp9 null alleles by imprecise P element excisions. Mol. Cells 10, 61–64 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kim J., Kim Y. J. (1999). Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Mol. Cell. Biol. 19, 2505–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug W. S., Bodenstein D., King R. C. (1968). Oogenesis in the suppressor of hairy-wing mutant of Drosophila melanogaster. I. Phenotypic characterization and transplantation experiments. J. Exp. Zool. 167, 151–156 [DOI] [PubMed] [Google Scholar]
- Kuhn E. J., Geyer P. K. (2003). Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15, 259–265 [DOI] [PubMed] [Google Scholar]
- Kurshakova M., Maksimenko O., Golovnin A., Pulina M., Georgieva S., Georgiev P., Krasnov A. (2007a). Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell 27, 332–338 [DOI] [PubMed] [Google Scholar]
- Kurshakova M. M., Krasnov A. N., Kopytova D. V., Shidlovskii Y. V., Nikolenko J. V., Nabirochkina E. N., Spehner D., Schultz P., Tora L., Georgieva S. G. (2007b). SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 26, 4956–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B., Roelens I., Jacob S. (2006). CoREST-like complexes regulate chromatin modification and neuronal gene expression. J. Mol. Neurosci. 29, 227–239 [DOI] [PubMed] [Google Scholar]
- Lobanenkov V. V., Nicolas R. H., Adler V. V., Paterson H., Klenova E. M., Polotskaja A. V., Goodwin G. H. (1990). A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753 [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N. (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier K., Mathieu E. L., Finkernagel F., Reuter L. M., Scharfe M., Doehlemann G., Jarek M., Brehm A. (2012). LINT, a novel dL(3)mbt-containing complex, represses malignant brain tumour signature genes. PLoS Genet. 8, e1002676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., Henikoff J. G., Feng X., Ahmad K., Russell S., White R. A., et al. (2010). A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6, e1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L., Wood I. C. (2007). Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8, 544–554 [DOI] [PubMed] [Google Scholar]
- Pai C. Y., Lei E. P., Ghosh D., Corces V. G. (2004). The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16, 737–748 [DOI] [PubMed] [Google Scholar]
- Park S. J., Yang E. S., Kim-Ha J., Kim Y. J. (1998). Down regulation of extramacrochaetae mRNA by a Drosophila neural RNA binding protein Rbp9 which is homologous to human Hu proteins. Nucleic Acids Res. 26, 2989–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., Coyne R. S., Corces V. G. (1988). The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2, 1205–1215 [DOI] [PubMed] [Google Scholar]
- Parnell T. J., Viering M. M., Skjesol A., Helou C., Kuhn E. J., Geyer P. K. (2003). An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100, 13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A., Amadio M., Quattrone A. (2008). Defining a neuron: neuronal ELAV proteins. Cell. Mol. Life Sci. 65, 128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab J. R., Kamakaka R. T. (2010). Insulators and promoters: closer than we think. Nat. Rev. Genet. 11, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C., Heath H., Krpic S., Dingjan G. M., van Hamburg J. P., Bergen I., van de Nobelen S., Sleutels F., Grosveld F., Galjart N., et al. (2009). Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J. Immunol. 182, 999–1010 [DOI] [PubMed] [Google Scholar]
- Richter C., Oktaba K., Steinmann J., Müller J., Knoblich J. A. (2011). The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat. Cell Biol. 13, 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S., White K. (1991). Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443–461 [DOI] [PubMed] [Google Scholar]
- Roseman R. R., Pirrotta V., Geyer P. K. (1993). The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R. R., Johnson E. A., Rodesch C. K., Bjerke M., Nagoshi R. N., Geyer P. K. (1995a). A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141, 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R. R., Swan J. M., Geyer P. K. (1995b). A Drosophila insulator protein facilitates dosage compensation of the X chromosome min-white gene located at autosomal insertion sites. Development 121, 3573–3582 [DOI] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., et al. ; modENCODE Consortium (2010). Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C. J., Paquette A. J., Anderson D. J. (1996). Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA 93, 9881–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M., Li H. B., Gorchakov A. A., Minoda A., Shanower G., Alekseyenko A. A., et al. (2012). Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22, 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 [DOI] [PubMed] [Google Scholar]
- Soller M., Li M., Haussmann I. U. (2010). Determinants of ELAV gene-specific regulation. Biochem. Soc. Trans. 38, 1122–1124 [DOI] [PubMed] [Google Scholar]
- Soshnev A. A., Li X., Wehling M. D., Geyer P. K. (2008). Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 4, e1000159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., He B., Baxley R. M., Jiang N., Hart C. M., Tan K., Geyer P. K. (2012). Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 40, 5415–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N., Montavon T., Leleu M., Galjart N., Duboule D. (2010). Functional analysis of CTCF during mammalian limb development. Dev. Cell 19, 819–830 [DOI] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G. (1988). The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2, 1414–1423 [DOI] [PubMed] [Google Scholar]
- Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tastan O. Y., Maines J. Z., Li Y., McKearin D. M., Buszczak M. (2010). Drosophila ataxin 2-binding protein 1 marks an intermediate step in the molecular differentiation of female germline cysts. Development 137, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda L., Kaido M., Lim Y. M., Kato K., Aigaki T., Hayashi S. (2006). An NRSF/REST-like repressor downstream of Ebi/SMRTER/Su(H) regulates eye development in Drosophila. EMBO J. 25, 3191–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzolovsky G., Deng W. M., Schlitt T., Bownes M. (1999). The function of the broad-complex during Drosophila melanogaster oogenesis. Genetics 153, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bemmel J. G., Filion G. J., Rosado A., Talhout W., de Haas M., van Welsem T., van Leeuwen F., van Steensel B. (2013). A network model of the molecular organization of chromatin in Drosophila. Mol. Cell 49, 759–771 [DOI] [PubMed] [Google Scholar]
- Wan L. B., Pan H., Hannenhalli S., Cheng Y., Ma J., Fedoriw A., Lobanenkov V., Latham K. E., Schultz R. M., Bartolomei M. S. (2008). Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development 135, 2729–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E. J., Berg C. A. (2005). Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech. Dev. 122, 241–255 [DOI] [PubMed] [Google Scholar]
- Xiong W. C., Okano H., Patel N. H., Blendy J. A., Montell C. (1994). repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 8, 981–994 [DOI] [PubMed] [Google Scholar]
- Yamasaki Y., Lim Y. M., Niwa N., Hayashi S., Tsuda L. (2011). Robust specification of sensory neurons by dual functions of charlatan, a Drosophila NRSF/REST-like repressor of extramacrochaetae and hairy. Genes Cells 16, 896–909 [DOI] [PubMed] [Google Scholar]
- Yang J., Corces V. G. (2011). Chromatin insulators: a role in nuclear organization and gene expression. Adv. Cancer Res. 110, 43–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Corces V. G. (2012). Insulators, long-range interactions, and genome function. Curr. Opin. Genet. Dev. 22, 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.