Abstract
RNA polymerase II (Pol II) elongation in metazoans is thought to require phosphorylation of serine 2 (Ser2-P) of the Pol II C-terminal domain (CTD) by the P-TEFb complex, CDK-9/cyclin T. Another Ser2 kinase complex, CDK-12/cyclin K, which requires upstream CDK-9 activity has been identified in Drosophila and human cells. We show that regulation of Ser2-P in C. elegans soma is similar to other metazoan systems, but Ser2-P in the germline is independent of CDK-9, and largely requires only CDK-12. The observed differences are not due to differential tissue expression as both kinases and their cyclin partners are ubiquitously expressed. Surprisingly, loss of CDK-9 from germ cells has little effect on Ser2-P, yet CDK-9 is essential for germline development. By contrast, loss of CDK-12 and Ser2-P specifically from germ cells has little impact on germline development or function, although significant loss of co-transcriptional H3K36 trimethylation is observed. These results show a reduced requirement for Pol II Ser2-P in germline development and suggest that generating Ser2-P is not the essential role of CDK-9 in these cells. Transcriptional elongation in the C. elegans germline thus appears to be uniquely regulated, which may be a novel facet of germline identity.
Keywords: C. elegans, CDK-9, CDK-12
INTRODUCTION
The large subunit of RNA polymerase II (Pol II) contains a C-terminal domain (CTD) that is composed of repeats of a heptapeptide sequence with consensus Y1S2P3T4S5P6S7 (42 in C. elegans). The phosphorylation states of specific residues in the repeats are thought to be important for proper mRNA synthesis. Pol II initially recruited to genes is hypophosphorylated on the CTD. Serine 5 of the repeat (Ser5) becomes phosphorylated during initiation, and serine 2 (Ser2) is phosphorylated on elongating Pol II (Egloff and Murphy, 2008; Zhou et al., 2012). Dephosphorylation of both Ser2 and Ser5 is important for recycling of Pol II complexes for reinitiation (Cho et al., 1999). The different phosphorylation marks are thought to help recruit various Pol II transcription factors, RNA processing machinery and histone/nucleosome modifiers at the proper time during the transcription cycle (Meinhart et al., 2005; Egloff and Murphy, 2008).
The transition between initiation and elongation is a regulated process in many metazoans, controlled by positive and negative regulatory factors that interact with Pol II (Adelman and Lis, 2012). Pol II elongation is inhibited by the combined activity of two complexes, DSIF (DRB-sensitivity inducing factors Spt4 and Spt5) and NELF (negative elongation factor). Phosphorylation of these factors releases inhibition of Pol II elongation and this appears tightly linked to Pol II CTD Ser2 phosphorylation (Yamaguchi et al., 2013). Whereas DSIF is highly conserved among eukaryotes, NELF is absent in many eukaryotes, including C. elegans (Narita et al., 2003). It is unclear whether Pol II elongation is a regulated step of transcription in this system, as Pol II pausing is not commonly detected during normal development (Whittle et al., 2008; Baugh et al., 2009; Kruesi et al., 2013).
In budding yeast, Ser2 and DSIF phosphorylation requires two kinases, Bur1 and Ctk1. Bur1 phosphorylates both DSIF and Ser2, but Ctk1 is responsible for the majority of Ser2 phosphorylation (Ser2-P) (Qiu et al., 2009). In metazoans, the P-TEFb complex, which is composed of Cdk9 and cyclin T, has been considered the sole DSIF/NELF and Pol II Ser2 kinase because loss of Cdk9 activity leads to loss of Ser2-P (Wood and Shilatifard, 2006). However, the closest metazoan homolog of yeast Ctk1, Cdk12, was shown to be required for a large fraction (∼60%) of Ser2-P in Drosophila and humans (Bartkowiak et al., 2010). It was thus suggested that Ser2-P in metazoans may match the elongation process in budding yeast: Cdk9 phosphorylates the Pol II CTD and DSIF (and NELF when present), and these events precede and are required for Cdk12-mediated phosphorylation of Ser2 on elongating Pol II (Bartkowiak and Greenleaf, 2011). This model could explain the requirement for Cdk9 for all Ser2-P observed in metazoans.
Here, we show that Ser2-P is differentially regulated in the somatic and germ cell lineages in C. elegans. Ser2-P regulation in the soma requires CDK-9 acting upstream of CDK-12 for Ser2-P; however, Ser2-P in the germline is independent of CDK-9 and instead requires only CDK-12. Surprisingly, CDK-12-dependent Ser2-P is not required for germline development. These results show that Ser2-P is not an obligate component of Pol II-mediated gene expression in all tissues, and further suggest that fundamental differences in transcriptional regulation may be important features of soma and germline distinction.
MATERIALS AND METHODS
Strains used
Strains used: wild-type N2 (Bristol); cdk-9(tm2884) I/hT2 qIS48; cdk-12(tm3846) IV/qC1 qIS26; cdk-12(ok3664) IV/qC1 qIS26; cit-1.2(gk241) III; mes-4(bn85) V/DnT1-GFP; dpy-18(e364) pie-1(zu127) III/qC1; rrf-1(pk1417) I; rrf-3(1426) II; and met-1(n4337) I. Transgenic lines were created for this study (see supplementary material Tables S2, S3). Some strains were provided by the CGC (Minneapolis, MN, USA), funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and the National Bioresource Project for the Experimental Animal ‘Nematode C. elegans’ (Kawada-cho, Shinjuku-ku, Tokyo). Worms were grown at 20°C except where indicated.
RNA interference
RNAi knock down for all experiments, except where indicated (supplementary material Fig. S7), was performed by soaking and feeding as previously described (Furuhashi et al., 2010) with the following modifications: 1 μg/μl dsRNA was used and animals were grown on RNAi plates for 24 hours before analysis. RNAi was considered efficient if 100% embryonic lethality (for cdk-9, cyclin T or fcp-1) or larval arrest (for cdk-12 and ccnk-1) was observed.
Immunofluorescence
Immunofluorescence analyses on methanol/formaldehyde-fixed specimens was performed as previously described (Seydoux and Dunn, 1997) using the following antibodies: rat anti-Ser2-P (3E10, 1/10 dilution), rat anti-Ser5-P (3E8, 1/10), rat anti-Ser7-P (4E12, 1/10) (Chapman et al., 2007), goat anti-PIE-1 (cN-19, 1/100, Santa Cruz Biotech, Dallas, TX, USA, #sc9245), rabbit anti-PGL-1 [1/10,000 (Kawasaki et al., 1998)] and mouse anti-H3K36me3 [CMA333, 0.25 μg/ml (Furuhashi et al., 2010)]. Methanol/acetone fixation (Schaner et al., 2003) was used for anti-GFP (Millipore MAB3580, 1/200), anti-AMA-1 (AMA-1 N-terminal, 1/10,000; Novus Biological, Littleton, CO, USA, #38520002), anti-H3K4me2 [CMA303, 1/20 (Kimura et al., 2008)] and anti-P-granules [OIC1D4, 1/4 (Strome and Wood, 1983)]. Incubation with primary antibodies was for 12-16 hours at 4°C. Secondary antibodies included Alexa Fluor 594- and Alexa Fluor 488-conjugated donkey antibodies (1/500) (Molecular Probes, Eugene, OR, USA).
Specimens were examined on a Leica DMRXA microscope equipped with a Q-Imaging Retiga-SRV CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) using Simple PCI software (Hamamatsu Photonics). Fiji (Schindelin et al., 2012) was used for quantification of raw immunofluorescence data. Average nuclear signal intensity was measured for Z2/Z2 or three representative nuclei. Average background signal, identified as non-nuclear staining from three surrounding representative regions, was subtracted. Standard error was determined according to Ashcroft and Pereira (Ashcroft and Pereira, 2003). Embryos with ∼100-300 cells were analyzed. For embryo staging, the number of nuclei in an embryo was counted using Fiji (Schindelin et al., 2012). Non-overlapping z-stacks were taken of each embryo. The DAPI channel of each layer was transformed into a binary image, the Watershed protocol was applied to separate individual nuclei, and the number of particles (nuclei, 200-2000 pixel units) was counted.
Quantification of anti-Ser2-P was performed using the 3E10 antibody rather than H5 antibody because the H5 antibody recognizes an AMA-1-independent signal in mitotic nuclei that interferes with quantitative approaches (Walker et al., 2004); however, reported results were consistent between both antibodies.
Protein isolation and western blot
Seventy-five young adult worms following RNAi were transferred to 5 μl 1× PBS in a 1.5 ml microfuge tube. Lysis buffer (75 μl) [100 mM Tris HCl (pH 8.0), 3 mM MgCl2, 300 μM KCl, 0.1% NP40, 20% glycercol, 40 mM DTT] was added before snap freezing in liquid nitrogen. Samples were thawed and refrozen in liquid nitrogen twice before sonication for 10 minutes (Diagenode Bioruptor, high setting) in a 4°C water bath. Samples were centrifuged at 4°C for 1 minute at 13,000 g and supernatant removed to a new tube. Protein concentrations were analyzed by Bradford (Bio-Rad Laboratories, Hercules, CA, USA).
Equal amounts of protein were run on a 4-20% SDS-PAGE gel (Bio-Rad) and transferred for western analyses. Membranes were probed with anti-Ser2-P (H5, 1/5000, Covance, Princeton, NJ, USA), anti-AMA-1 N-terminal antibody (1/10,000) or anti-actin (1/10,000, Millipore, Billerica, MA, USA, MAB1501) and detected with HRP-conjugated secondaries using SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL, USA).
Transgenic lines
Constructs were generated by conventional cloning methods (supplementary material Tables S4, S5). Promoters, ORFs and 3′ UTRs were amplified from C. elegans N2 genomic DNA and construct tags were amplified from indicated plasmids (supplementary material Table S4) using Phusion Taq (Finnzymes, Waltham, MA, USA). Transgene components were cloned into pCR-Blunt II-TOPO Zero Blunt Cloning Plasmid (Invitrogen, Grand Island, NY, USA). Finished constructs were subcloned into MOS-SCI plasmids [cit-1.1, cit-1.2, ccnk-1 and spt-5 into pCFJ151; cdk-9 into pCFJ356; and cdk-12 into pCFJ352, from AddGene (Cambridge, MA, USA)]. Kinase-dead point mutants were created using QuikChange XL (Aglient, Santa Clara, CA, USA).
All transgenes made in this study were made by the standard MOS-SCI protocol targeting integration into either chromosome I (ttTi4348) or chromosome II (ttTi5605) loci (Frøkjær-Jensen et al., 2008). Insertion was confirmed by PCR and at least two independent expressing lines were assayed. All lines for each transgene showed identical expression patterns. Representative transgenic lines used for this study are indicated (supplementary material Table S2).
Fluorescence and DNA visualization in intact worms
Five to 10 adult worms were picked and quickly moved between the following solutions in a multi-well depression slide: ice-cold 80% methanol, 45 seconds; room temperature 2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), 30 seconds; room temperature deionized H2O, 2-10 minutes. Worms were mounted under a glass coverslip in 7 μl 5% glycerol on a slide. This maintained GFP and mCherry signal and allowed DAPI to stain all nuclei except those in embryos.
RNA isolation and qRT-PCR
Three-hundred animals washed with M9 were transferred to 100 μl M9. Trizol (220 μl) (Invitrogen) was added before snap freezing in liquid nitrogen. Samples were thawed and refrozen four times, and 45 μl chloroform added before spinning for 15 minutes at 4°C. Nucleic acids were precipitated with isopropanol and resuspended in 100 μl nuclease-free water. The RNeasy kit (Qiagen, Valencia, CA, USA) and DNase treatment was used. RNA (1 μg) was used for cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad) following manufacturer’s instructions. cDNA corresponding to 50 ng RNA was used for qPCR analysis using SsoFast (Bio-Rad) following manufacturer’s instructions on a CFX96 (Bio-Rad) machine. Transcript levels were normalized to either 18S or U6 levels for each sample. The average of two technical replicates from three biological samples was plotted with s.e.m.
Brood size analysis
Twenty worms of each genotype were transferred to individual plates at the L4 stage, moved every 7 hours and the number of embryos on each plate were counted until no offspring were produced. After 21 hours, unhatched embryos on plates were counted as dead. Adults that died during analysis were excluded from results. cdk-12(ok3664)/cdk-12(ok3664); cdk-12:pal-1 3′UTR (cdk-12 mutant) animals analyzed were the offspring of balanced cdk-12 mutants maintained to prevent suppressor selection.
RESULTS
Tissue-specific requirements for CDK-9 and CDK-12 in Ser2 phosphorylation
Previous studies have indicated that CDK-9 activity is required for all detectable Ser2-P in early C. elegans embryos (Shim et al., 2002), but Ser2-P is minimally affected by CDK-9 depletion in the two primordial germ cells (PGCs), Z2/Z3 (Furuhashi et al., 2010). Although these initial studies implicated TLK-1 as a Ser2 kinase in the germ cells, the cell-cycle defects in tlk-1(RNAi) embryos made it difficult to conclude this was a direct effect (Furuhashi et al., 2010). To further investigate embryonic Ser2 kinase requirements, we depleted other components of the CDK-9/cyclin T complex as well as homologs of the newly described CDK-12/cyclin K Ser2 kinase complex (supplementary material Fig. S1).
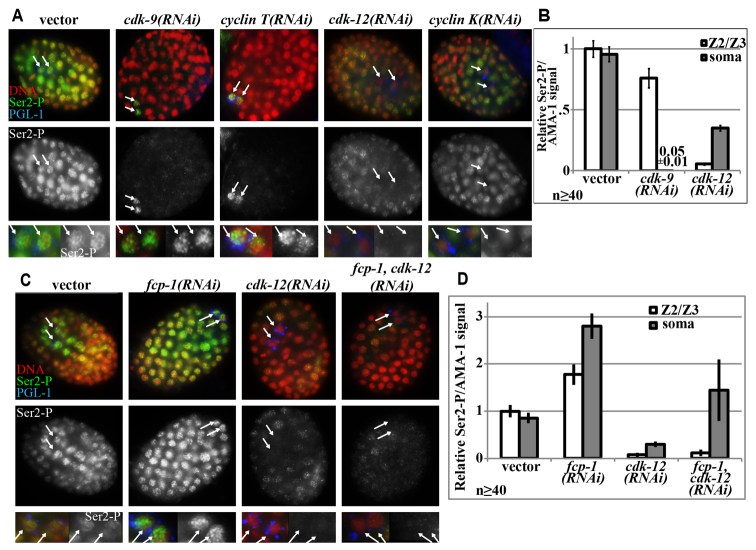
As previously observed, cdk-9(RNAi) resulted in loss of detectable Ser2-P in all embryonic somatic cells, but significant levels of Ser2-P remained unaffected by this treatment in Z2/Z3 (Fig. 1A,B; supplementary material Fig. S2A). Loss of the two cyclin T paralogs yielded identical results to cdk-9(RNAi) (Fig. 1A; supplementary material Fig. S2E). Knock down of either CDK-9 or cyclin T caused defective gastrulation and early embryonic arrest at ∼120 cells (supplementary material Fig. S3). These phenotypes correlate with defective somatic zygotic transcription and also occur with depletion of the large subunit of Pol II, AMA-1 (Powell-Coffman et al., 1996). Thus, although CDK-9 and cyclin T are important for both zygotic transcription and Ser2-P in somatic cells, neither is required for Ser2-P in Z2/Z3.
Fig. 1.
Kinase and cyclin dependency of Ser2-P in embryonic soma and germ cells. (A) Anti-Ser2-P immunofluorescence (green) analyses of embryos exposed to RNAi targeting Cdks and cyclins, as indicated, and counterstained with DAPI (red). Arrows indicate the two primordial germ cells, Z2 and Z3 (Z2/Z3), marked by anti-PGL-1 (blue). (B) Quantification of anti-Ser2-P immunofluorescence relative to that of anti-AMA-1 (total Pol II) in either Z2, Z3 or representative somatic nuclei, normalized to vector controls. Error bars indicate s.e.m. (C) Anti-Ser2-P immunofluorescence (green) analyses of embryos exposed to RNAi targeting cdk-12 and/or fcp-1, as in A. (D) Quantification of Ser2-P immunofluorescence, as in B.
CDK-12 is a CTD kinase responsible for significant Ser2-P in both Drosophila and human cells (Bartkowiak et al., 2010; Blazek et al., 2011; Kohoutek and Blazek, 2012). A previously uncharacterized gene, B0285.1, is the closest homolog to Drosophila CDK12 in C. elegans (supplementary material Fig. S1). In contrast to cdk-9(RNAi), cdk-12(RNAi) embryos had undetectable Ser2-P levels in the PGCs, and a ∼60% reduction of Ser2-P in embryonic somatic nuclei (Fig. 1A,B; supplementary material Fig. S2A). The RNAi conditions used were efficient and specific for each gene (supplementary material Fig. S4A).
Cyclin K is the cyclin partner of CDK-12 in other systems (Bartkowiak et al., 2010; Blazek et al., 2011; Kohoutek and Blazek, 2012). RNAi of the closest sequence homolog to Drosophila cyclin K, F43D2.1 (hereafter named ccnk-1), phenocopies cdk-12(RNAi), and is likely the cyclin partner of CDK-12 (Fig. 1A,B; supplementary material Fig. S2F). Knock down of either CDK-12 or cyclin K results in early larval arrest, but no specific developmental defects were observed other than what appeared to be arrested growth (supplementary material Fig. S3).
These results demonstrate that CDK-12, rather than CDK-9, is the predominant Ser2 kinase in embryonic germ cells. The Ser2-P antibody used in this study [3E10 antibody (Chapman et al., 2007)] is specific for Pol II in C. elegans, as ama-1(RNAi) depletes all antibody binding (Furuhashi et al., 2010). Effects observed with cdk-12(RNAi) are specific for Ser2 in the embryo, as another phosphoepitope, Ser5-P, was unaffected by cdk-12(RNAi). We observed decreases in Ser5-P with cdk-9(RNAi) (supplementary material Fig. S2B,D), which could be an indirect effect, but CDK-9 has been reported to phosphorylate Ser5 in human cells (Glover-Cutter et al., 2009).
Decreased Ser2-P after cdk-12 knockdown could be an indirect result of increased Pol II CTD phosphatase activity. Knockdown of the best characterized Pol II CTD Ser2 phosphatase, FCP-1, resulted in increases in Ser2-P levels in embryonic somatic and germline nuclei (Fig. 1C,D; supplementary material Fig. S2G). However, RNAi against fcp-1 in combination with cdk-12(RNAi) caused a Ser2-P reduction in Z2/Z3 similar to cdk-12(RNAi) alone, so reduction of Ser2-P in these cells is not due to an ectopic increase in FCP-1 activity (Fig. 1C,D; supplementary material Fig. S2G).
Embryonic germline-soma conversion yields switch in kinase dependency
The maternal protein PIE-1 inhibits Ser2 phosphorylation and Pol II transcription in the C. elegans embryonic germline precursor cells, called P-cells (Ghosh and Seydoux, 2008). PIE-1 is proposed to inhibit Ser2 phosphorylation by sequestering P-TEFb until PIE-1 is degraded at the birth of Z2/Z3, and degradation of PIE-1 in Z2/Z3 correlates with Ser2-P appearance in these cells [Fig. 2A (Seydoux and Dunn, 1997)]. However, our results suggest that the germline Ser2-P kinase inhibited by PIE-1 is likely to be CDK-12/cyclin K. Indeed, delaying PIE-1 degradation in Z2/Z3 by knocking down ZIF-1, a protein involved in PIE-1 degradation, caused a delay in appearance of CDK-12-dependent Ser2-P in Z2/Z3 (Fig. 2A-C).
Fig. 2.
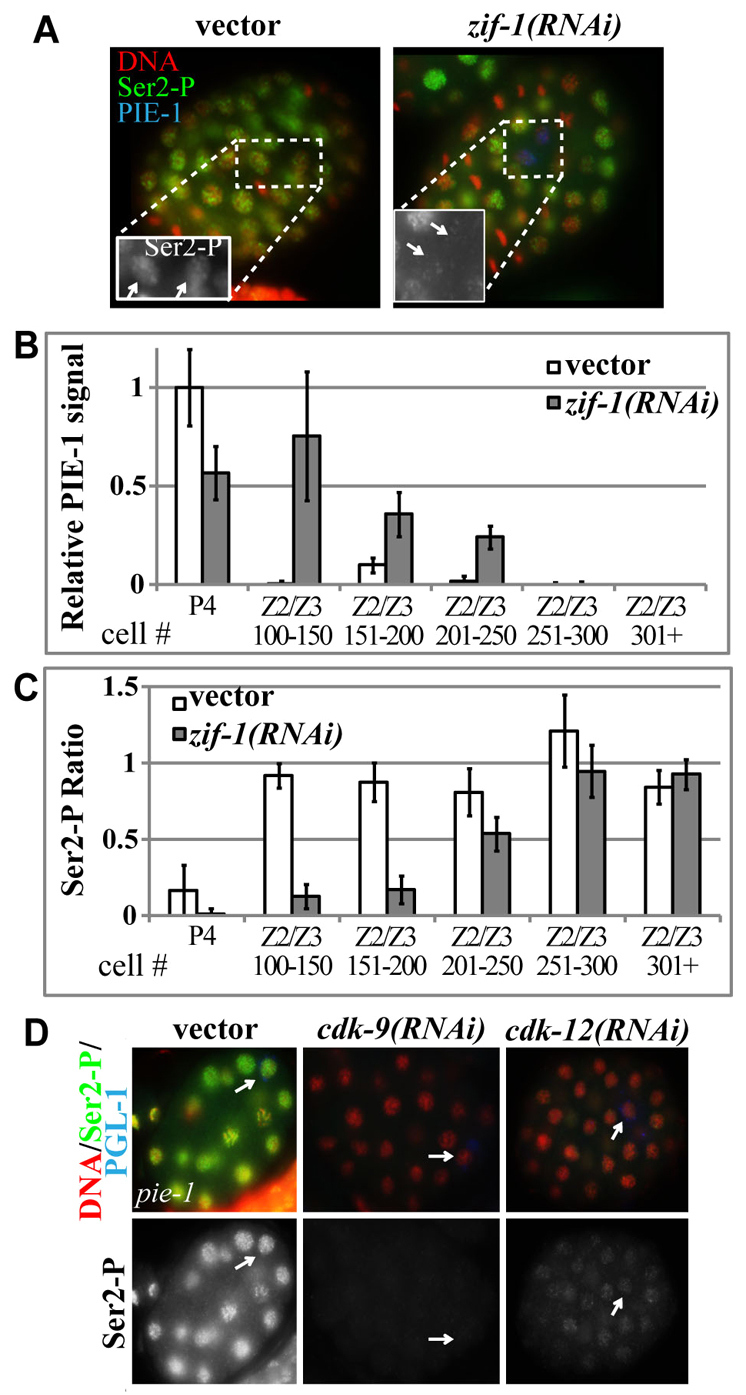
PIE-1 regulates CDK-12-dependent Ser2-P. (A) Anti-Ser2-P (green) and PIE-1 (blue) immunofluorescence analyses of embryos exposed to RNAi targeting zif-1 and counterstained with DAPI (red). No PIE-1 signal is detected in Z2/Z3 of vector-treated embryos because PIE-1 is degraded by this point; but PIE-1 remains in Z2/Z3 following zif-1(RNAi). Boxes outline region expanded in inset, with germ cells indicated by arrows. (B) Quantification of PIE-1 immunofluorescence in embryos with the Z2/Z3 precursor cell P4 or in embryos in which Z2/Z3 have been specified, staged by cell number and normalized to levels in P4 vector control. (C) Quantification of Ser2-P immunofluorescence ratio in PGCs relative to surrounding somatic nuclei in embryos staged by cell number as in B. Error bars indicate s.e.m. n≥10. (D) Anti-Ser2-P immunofluorescence (green) analyses of pie-1(zu127) embryos as in Fig. 1. Embryos at the 20- to 30-cell stage are displayed.
We next analyzed the germline correlation with CDK-12-dependent Ser2-P. Loss of PIE-1 in the P-cells results in a transformation of germline into soma and premature Ser2-P appearance. We tested whether the ectopic Ser2-P that appears in P-cells of pie-1(zu127) mutants is CDK-12 dependent. Transformation from germline to soma is accompanied by a switch to CDK-9-dependent Ser2-P in pie-1(zu127) mutant embryos (Fig. 2D). This further illustrates a tight correlation between germline identity and CDK-9-independent Ser2-P regulation.
CDK-12-dependent Ser2-P is not essential for early germ cell proliferation
We next examined the requirement for CDK-12 in post-embryonic germ cells. Z2/Z3 are born in the embryo after about 2 hours of development (∼100 cells), remain mitotically quiescent through the remaining 10 hours of embryogenesis and re-enter the cell cycle after hatching (Sulston et al., 1983). cdk-12(RNAi) embryos arrest after hatching, so we examined germ-line Ser2-P in L1 larval offspring of RNAi-treated animals. cdk-12(RNAi) larvae had decreased Ser2-P in somatic nuclei and no detectable Ser2-P in the dividing germ cells (Fig. 3A,B). Thus, CDK-12 is required for Ser2-P in post-embryonic germ cells. The Ser2-P observed in cdk-12(RNAi) soma is presumably due to CDK-9 activity, although this could not be verified because of early embryonic lethality from cdk-9(RNAi).
Fig. 3.
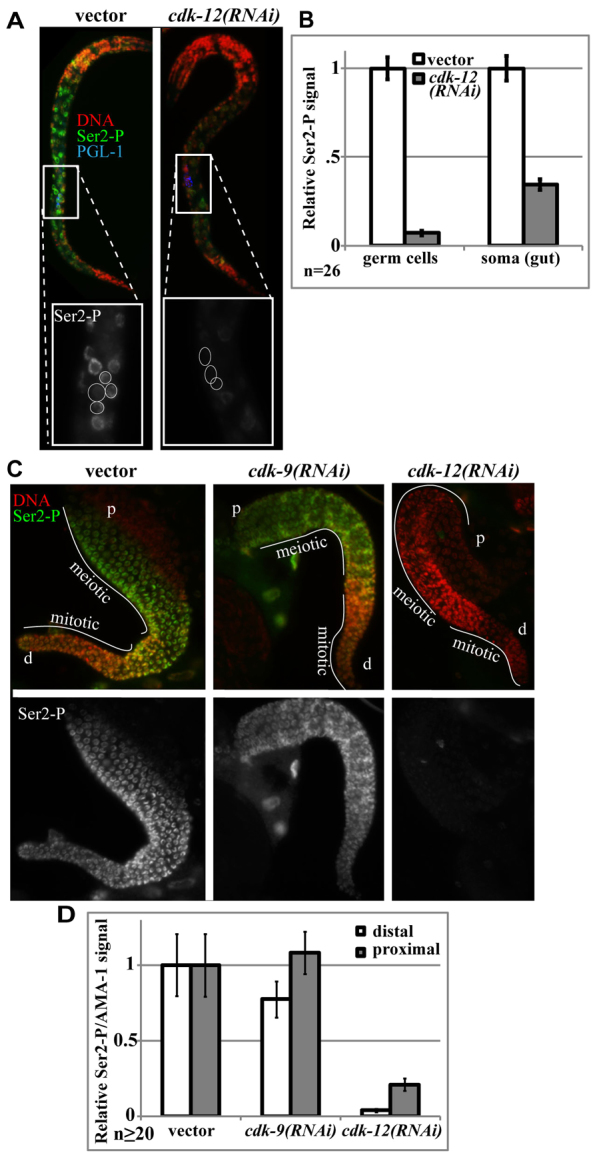
Ser2-P in proliferating germline cells is independent of P-TEFb and requires CDK-12. (A) Anti-Ser2-P immunofluorescence analyses of hatched larvae fed for 6 hours that developed from embryos exposed to cdk-12(RNAi) as in Fig. 1. Box indicates region expanded in inset with germ cells outlined. (B) Quantification of anti-Ser2-P immunofluorescence signal in germ cells relative to surrounding gut nuclei and normalized to RNAi vector controls. Error bars indicate s.e.m. (C) Anti-Ser2-P immunofluorescence analyses of dissected ovaries exposed to the indicated RNAi conditions (L4+48 hour RNAi treatment). Mitotic (stem cell pool) and meiotic regions of the gonad are indicated. Distal and proximal ends of the gonad are marked by ‘d’ and ‘p’, respectively. (D) Quantification of anti-Ser2-P immunofluorescence signal versus total AMA-1 protein in nuclei at the extreme distal or proximal end of the gonad. Error bars indicate s.e.m. Signal is normalized to vector-treated signal.
Importantly, the cdk-12(RNAi) larvae examined arise from embryos with no history of detectable Ser2-P in their germ cells. Despite absence of Ser2-P, the larval germ cells showed some proliferation before arresting with the rest of the somatic lineages in the larvae (supplementary material Fig. S3). The larval arrest appeared to be related to a lack of growth, as the animals moved and fed normally but did not progress into further larval stages and eventually died.
CDK-12 is the predominant germline Ser2 kinase
We next examined the requirements for CDK-9 and CDK-12 in adult germ cells. The adult gonad is a syncytium of germ cell nuclei produced from a pool of proliferating stem cells at the distal end of the gonad, and these cells progress sequentially through meiosis and gametogenesis as they migrate toward the proximal end. Thus, the adult gonad contains germ cells linearly arranged in discrete, sequential stages of development that are highly transcriptionally active. We targeted each kinase by RNAi and used CDK-9:mCherry and CDK-12:GFP transgenes (described below) to assess RNAi knockdown. By initiating RNAi treatment at the last larval (L4) stage, we obtained adults with loss of detectable kinase protein in younger cells in the distal end of the gonad, and near complete loss in older nuclei at the proximal end (supplementary material Fig. S4B). Kinase RNAi in presence or absence of transgenes yielded similar results.
Surprisingly, although cdk-9(RNAi) caused loss of CDK-9:mCherry below detection, this had little effect on Ser2-P (Fig. 3B,C; supplementary material Fig. S5G). By contrast, cdk-12(RNAi) caused near complete loss of Ser2-P in all germ cells (Fig. 3B,C). Some Ser2-P remained detectable in older proximal nuclei in cdk-12(RNAi) gonads, presumably owing to CDK-12 protein produced before RNAi treatment was maximal. Similar RNAi results were observed in male gonads (data not shown). A substantial decrease of Ser2-P in cdk-12(RNAi) germ cells, which represent roughly half of the total nuclei in an adult worm, was also apparent by western blot analyses (supplementary material Fig. S5A). Thus, Ser2-P in germ cells at all stages of the germ cell cycle is largely independent of CDK-9/P-TEFb and dependent on CDK-12.
Importantly, neither cdk-9(RNAi) nor cdk-12(RNAi) significantly affected Ser5-P levels in adult germ cells (supplementary material Fig. S5C). By contrast, cdk-12(RNAi) caused a significant reduction of Ser7-P levels (supplementary material Fig. S5D), but it is unclear whether this is a direct effect.
CDK-9, CDK-12 and cyclin partners are ubiquitously expressed
Tissue-specific differences in Ser2 kinase requirements could be due to differences in expression of the kinases and/or their cyclin partners. In the absence of available antibodies for these components in C. elegans, we analyzed their expression using low copy, integrated, fluorescently tagged transgenes under control of their endogenous promoters and 3′UTRs (Frøkjær-Jensen et al., 2008).
All components tested were ubiquitously expressed (Fig. 4A). The expression of the CDK-9, CDK-12 and cyclin T (CIT-1.2) transgenes recapitulate endogenous protein function because the transgenes rescue their respective mutant strains (a cyclin K mutation is unavailable; supplementary material Table S1). The kinase activity of both CDK-9 and CDK-12 is essential, as transgenes expressing kinase-inactivated point mutants did not rescue (data not shown). Thus, tissue-specific expression is unlikely to underlie the different kinase requirements observed.
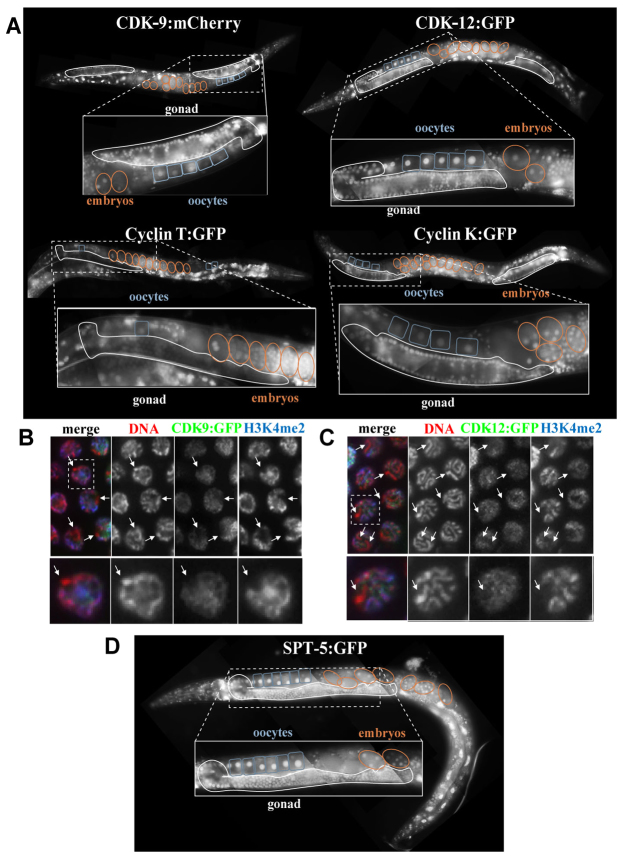
Fig. 4.
CDK-9, CDK-12, their cyclin partners, and SPT-5 are ubiquitously expressed. (A) Expression in young adult hermaphrodites of transgenic, single-copy insertions of fluorescently tagged CDK-9 (mCherry and GFP-tagged versions show the same pattern, data not shown), cyclin T (CIT-1.2 shown, CIT-1.1 has the same pattern; patterns identical with GFP or FLAG tags, not shown), CDK-12, and cyclin K. Outlines around the adult gonad (white), oocytes (blue) and embryos (orange) show these factors are expressed in the germline and maternally loaded into embryos. Boxes outline regions expanded. (B,C) Co-staining gonads with GFP and H3K4me2 show chromosomes that lack H3K4me2 (X chromosome) also lack CDK-9 and CDK-12. Representative nuclei (dotted boxes) are shown below. (D) Expression in young adult hermaphrodite of the fluorescently tagged DSIF component SPT-5, as in A.
We further analyzed the subnuclear distribution of the fusion proteins in adult germ cells. The X chromosome is transcriptionally suppressed in meiotic germ cells in C. elegans (Kelly et al., 2002). Both CDK-9 and CDK-12 and their component cyclins are associated with autosomal chromatin but largely excluded from the X chromosome until late oogenesis, when X-linked transcription activates (Fig. 4B,C; cyclin T/K data not shown). Thus, although only CDK-12 activity is required for Ser2-P in the germline, both the CDK-9/cyclin T and CDK-12/cyclin K complexes are distributed in low-resolution patterns that correlate with active transcription.
Both cyclin T paralogs are ubiquitously expressed; however, the transgenic expression of each appeared reduced in the adult gonad and embryonic germ cells relative to somatic expression (Fig. 4A; supplementary material Fig. S6A,B). We increased cyclin T expression in germ cells to test whether reduced expression correlates with the decreased requirement on CDK-9/cyclin T for Ser2-P. We observed no increase in CDK-9-dependent Ser2-P in the germline as a result of increased cyclin T expression (supplementary material Fig. S6C,D).
Tissue-specific differences in Ser2 kinase requirements could also be due to the presence or absence of other factors involved in transcription elongation. We examined the expression of SPT-5, the major functional component of DSIF in C. elegans and other systems (Swanson and Winston, 1992; Yamaguchi et al., 1999; Shim et al., 2002). As observed for the CTD kinases and cyclins, SPT-5:GFP is ubiquitously expressed and also excluded from the X chromosome in adult meiotic germ cells (Fig. 4D and data not shown). Thus, tissue-specific differences in Pol II Ser2 phosphorylation are not likely due to differential expression of the kinases, cyclins or the DSIF complex.
CDK-9, but not CDK-12, is required for proper germline development or transcription
We next examined whether either kinase is essential for germline development. The RNAi analyses described above were performed on late larval (L4) worms, after substantial germline development had occurred, so we performed RNAi starting at the L1 stage. cdk-9(RNAi) in L1 worms results in larval arrest so the effect on germline development could not be assessed. cdk-12(RNAi) in wild-type animals resulted in sterility of nearly all animals, but the sterility was not observed when cdk-12(RNAi) was performed in rrf-1(pk1417) mutant animals (supplementary material Fig. S7). RNAi is ineffective in rrf-1 mutant soma, resulting in largely germline-specific RNAi effects (Sijen et al., 2001; Kumsta and Hansen, 2012). The sterility observed in cdk-12(RNAi) worms thus appears to be an indirect effect due to CDK-12 knockdown in soma rather than a direct effect of knockdown in the germline. The converse experiment is not readily feasible [mutants defective in germline-specific RNAi have fertility defects on their own (Kumsta and Hansen, 2012)], but we also observed increased sterility after cdk12(RNAi) in rrf-3(pk1427) worms, a strain with enhanced somatic RNAi but not enhanced germline RNAi [supplementary material Fig. S7 (Sijen et al., 2001)].
To definitively determine whether CDK-9 and CDK-12 are essential for germline development, we developed a method to specifically remove CDK-9 and CDK-12 activity from only the germline. Homozygous CDK-9 and CDK-12 deletions mutants arrest early in larval development because of somatic requirements, so we provided kinase expression solely in somatic tissues to allow larval growth and adult development. To do this, the endogenous 3′UTRs of the rescuing kinase transgenes were replaced with the 3′UTR of the pal-1 gene, which prevents translation in the germline until late oogenesis in the adult (Merritt et al., 2008). The CDK-9 and CDK-12 transgenes with the pal-1 3′UTR show ubiquitous somatic expression, but no expression in post-embryonic germ cells until late in adult oogenesis (Fig. 5A). Expressing the kinase:pal-1 3′UTR transgenes in the respective mutants therefore allows rescue of kinase function in all somatic lineages, but no rescue in post-embryonic germline development.
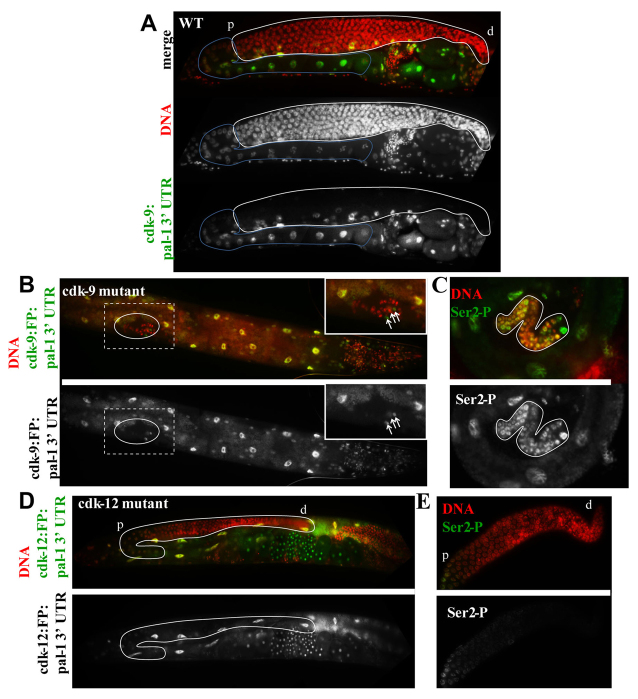
Fig. 5.
CDK-9, but not CDK-12 or Ser2-P, is essential for germline development. (A) Expression of both pal-1 3′UTR-tagged kinases (only cdk-9:pal-1 3’UTR is shown) is observed in all somatic cells, but not in germ cells until late oogenesis. White outline surrounds adult germline, blue outline surrounds start of transgene expression. (B) Expression of cdk-9:pal-1 3′UTR in homozygous cdk-9(tm2884) young adult hermaphrodite worms. Boxed region is expanded in inset. White oval surrounds germ cells. Arrows indicate somatic gonad nuclei. (C) Anti-Ser2-P immunofluorescence analyses of cdk-9(tm2884), cdk-9:pal-1 3′UTR hermaphrodite ovaries (outlined). (D) Expression of cdk-12:pal-1 3′UTR in homozygous cdk-12(ok3664) young adult hermaphrodite worms. White outline surrounds gonad. (E) Anti-Ser2-P immunofluorescence analysis of cdk-12(ok3664), cdk-12:pal-1 3′UTR hermaphrodite gonads. Distal and proximal ends of the gonad are marked by ‘d’ and ‘p’, respectively.
Expression of the kinase:pal-1 3′UTR transgenes fully rescued somatic development in the respective mutants: the transgene-rescued mutants grew to adults in parallel with their heterozygous siblings and showed no obvious somatic phenotypes. The absence of CDK-9 or CDK-12 activities in germ cells, however, had very different consequences.
The absence of CDK-9 in germ cells caused dramatic sterility, with animals producing only ∼50-100 germ cells per gonad (∼1/10th normal size, Fig. 5B). The germ cells produced, however, exhibited robust Ser2-P levels, further supporting the conclusion that Ser2-P in germ cells does not require CDK-9 (Fig. 5C). No CDK-9:mCherry is detectable in germ cells at any larval stage, suggesting that some proliferation can still occur in the absence of CDK-9 activity. Although persistence of trace amounts of maternally derived CDK-9 mRNA and/or protein cannot be ruled out, its activity would have been at very low levels and distributed among a significant number of nuclei.
Surprisingly, cdk-12(ok3664) animals expressing the cdk-12:pal-1 3′UTR transgene showed few germline defects. Neither CDK-12:GFP nor Ser2-P was detected at any post-embryonic germ cell stage until late oogenesis (Fig. 5D,E), yet these animals produced large numbers of functional germ cells and were fertile (Fig. 5D). Therefore, neither CDK-12 nor Ser2-P phosphorylation is essential for germ cell proliferation and development at normal temperatures. Interestingly, 100% of homozygotes expressing the cdk-12:pal-1 3′UTR transgene were sterile when raised at 25°C instead of 20°C (data not shown). The interpretation of this is unclear because these animals also showed misregulation of the pal-1 3′UTR transgene in the germline, which complicates the analysis.
Fertility of animals lacking CDK-12 at 20°C, however, was not completely normal. Homozygous cdk-12(ok3664) animals expressing the cdk-12:pal-1 3′UTR transgene produced fewer offspring than their heterozygous siblings and fully rescued cdk-12(ok3664) animals (Fig. 6A). However, no dramatic rise in embryonic lethality was observed in any strain (Fig. 6A). Therefore, neither CDK-12 nor Ser2-P is crucial for germline development.
Fig. 6.
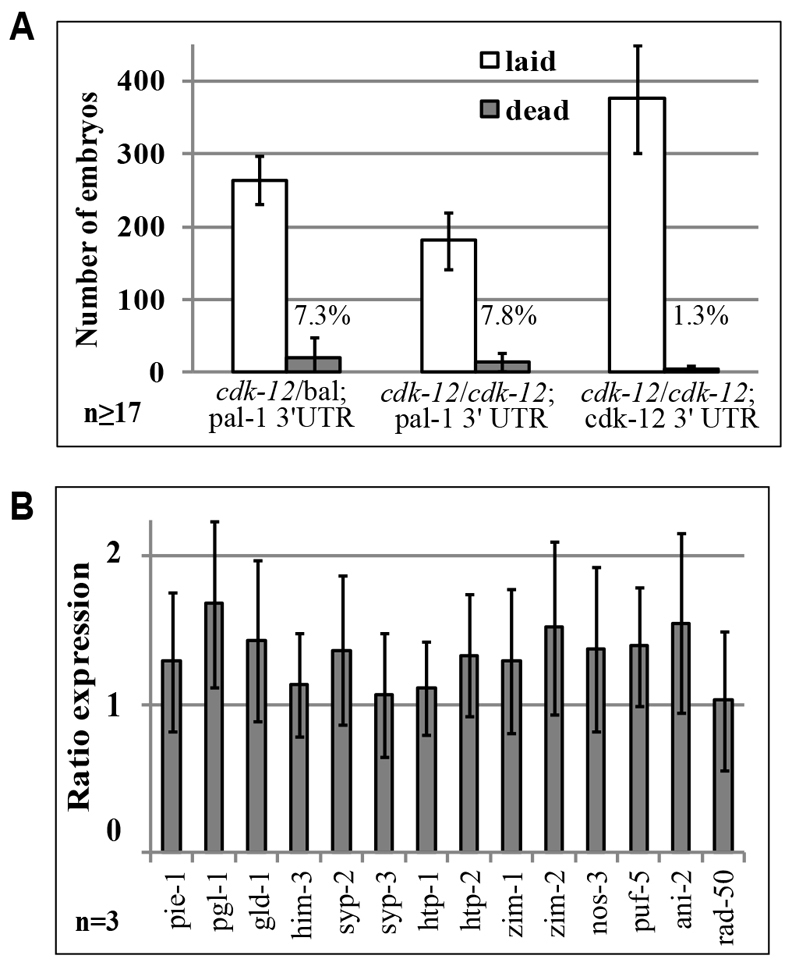
Loss of CDK-12 and Ser2-P from the germline does not dramatically affect germline function. (A) Brood size and embryonic lethality of cdk-12(ok3664)/+; cdk-12:pal-1 3′UTR (WT), cdk-12(ok3664)/cdk-12(ok3664); cdk-12:pal-1 3′UTR (cdk-12 mutant) or cdk-12(ok3664)/cdk-12(ok3664); cdk-12:cdk-12 3′UTR (cdk-12 mutant rescued with cdk-12 transgene) animals. The cdk-12 balancer results in both embryonic and larval arrest when homozygous. Error bars represent s.e.m. (B) qRT-PCR analysis of germline restricted transcripts in whole animals. RNA from each sample was normalized to 18S RNA (similar results were obtained when normalized to U6 RNA) and the ratio of expression in cdk-12(ok3664)/cdk-12(ok3664); cdk-12:pal-1 3′UTR (cdk-12 mutant) animals versus balanced cdk-12(ok3664)/+; cdk-12:pal-1 3′UTR (wild type) controls is displayed. The average of two technical replicates from three biological samples is plotted ± s.e.m. No sample is statistically different from control. For both analyses, cdk-12(ok3664)/cdk-12(ok3664); cdk-12:pal-1 3′UTR (cdk-12 mutant) animals analyzed were F1 offspring of balanced cdk-12 mutants in order to prevent suppressor selection.
We further examined transcript levels of a panel of germline-specific genes in cdk-12:pal-1 3′UTR transgene rescued mutants. Surprisingly, no substantial decrease in transcript was observed for any of the 14 genes assayed, most of which actually tended towards higher levels of expression (Fig. 6B). Therefore, neither CDK-12 nor Ser2-P is crucial for germline transcription.
CDK-12 is required for normal H3K36me3 levels in germ cells
We did not observe dramatic defects in RNA production in germlines lacking detectible Ser2-P, so we next examined whether loss of Ser2-P results in defects in another process linked to this mark. Ser2-P has been shown to regulate histone H3 lysine 36 trimethylation (H3K36me3) levels by recruiting the K36 methyltransferase Set2 during Pol II transcription elongation (Kizer et al., 2005). C. elegans has two H3K36 methyltransferases (HMT): MET-1, a transcription-dependent K36 HMT and MES-4, a transcription-independent H3K36 HMT (Bender et al., 2006; Furuhashi et al., 2010; Rechtsteiner et al., 2010). We tested whether transcription-dependent (i.e. MET-1 dependent) H3K36me3 in the germline is dependent on CDK-12.
Previous studies have shown that H3K36 dimethylation (H3K36me2) in the adult germline requires MES-4 (Bender et al., 2006). We found that mes-4(RNAi) or mes-4(bn85) mutants still exhibit substantial levels of H3K36 trimethylation (H3K36me3) in adult germ cells (Fig. 7A,C; supplementary material Fig. S8A,C). All the offspring of the mes-4(RNAi) animals were sterile, indicating efficient knockdown of MES-4, which is essential for fertility (Garvin et al., 1998). No H3K36me3 was detected in met-1(n4337); mes-4(RNAi) animals (data not shown). The H3K36me3 in mes-4(RNAi) germlines is thus due to transcription-dependent activities of MET-1, allowing us to assess the requirement for CDK-12 in transcription-dependent H3K36me3.
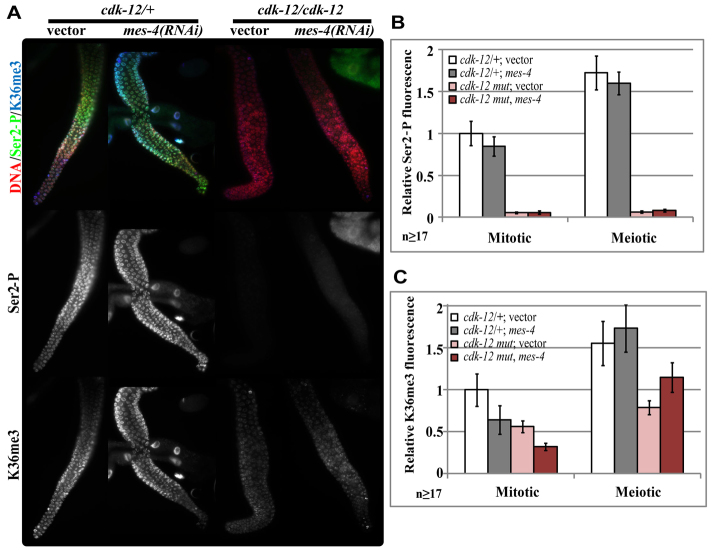
Fig. 7.
CDK-12 regulates H3K36me3 levels. (A) Ser2-P and H3K36me3 immunofluorescence images of hermaphrodite gonads in either cdk-12(ok3664)/+, cdk-12:pal-1 3′UTR (wild type) or cdk-12(ok3664)/cdk-12(ok3664), cdk-12:pal-1 3′UTR (cdk-12 mutant) young adults exposed to the indicated RNAi conditions. (B) Quantification of Ser2-P immunofluorescence signal ± s.e.m. (C) Quantification of H3K36me3 immunofluorescence signal ± s.e.m. Nuclei selected for analysis were from the most distal mitotic and meiotic regions of the gonad.
The decreased Ser2-P in cdk-12:pal-1 3′UTR germ cells correlated with a significant decrease in MET-1-dependent H3K36me3 (Fig. 7A,B). Both total H3K36me3 levels and MES-4-independent H3K36me3 levels showed significant decreases (Fig. 7A,C). Similar results were obtained with cdk-12(RNAi) in mes-4(bn85) mutants (supplementary material Fig. S8A,C). Thus, normal levels of H3K36me3 in the C. elegans germline require Ser2-P produced by CDK-12.
We also observed decreases in H3K36me3 levels after cdk-9(RNAi) in mes-4(bn85) mutant germ cells (supplementary material Fig. S8A,C). This was surprising, as loss of CDK-9 has little effect on Ser2-P levels in germ cells. Given the severe germ cell phenotypes caused by decreased CDK-9, this may be an indirect effect on H3K36me3 levels, although a similar decrease in H3K36me3 levels has been observed in yeast Bur1 mutants (Chu et al., 2006).
DISCUSSION
Tissue-specific Ser2-P regulation
Although phosphorylation of Ser2 of the Pol II CTD has been studied for over 20 years, the role of CDK-12 in this process has only recently been appreciated (Bartkowiak et al., 2010). This is understandable considering that the loss of P-TEFb (CDK-9/cyclin T) activity in most analyses caused loss of Ser2-P, thereby obviating an apparent requirement for additional kinases. The severe effect on Ser2-P observed with loss of CDK-9 indicates a role for P-TEFb upstream of CDK-12 (Bartkowiak and Greenleaf, 2011). Somatic tissues in C. elegans embryos match this model: knock down of P-TEFb results in complete loss of Ser2-P. By contrast, CDK-12/cyclin K knock down results in a 60% decrease in Ser2-P, a decrease remarkably consistent with that observed in other systems (Bartkowiak et al., 2010; Blazek et al., 2011).
Ser2-P regulation in soma also agrees with current models of Ser2-P regulation in the transcription cycle (Fig. 8A). CDK-9, cyclin T and FCP-1 knock downs cause defective gastrulation, a phenotype associated with defective zygotic transcription in C. elegans (Powell-Coffman et al., 1996). Although this phenotype could be due to other functions of these proteins, it seems likely that these effects are due to defective Pol II transcription, i.e. CDK-9 knock down prevents proper Pol II elongation, whereas FCP-1 prevents proper Pol II recycling, in agreement with the current models (Cho et al., 1999; Fuda et al., 2012).
Fig. 8.
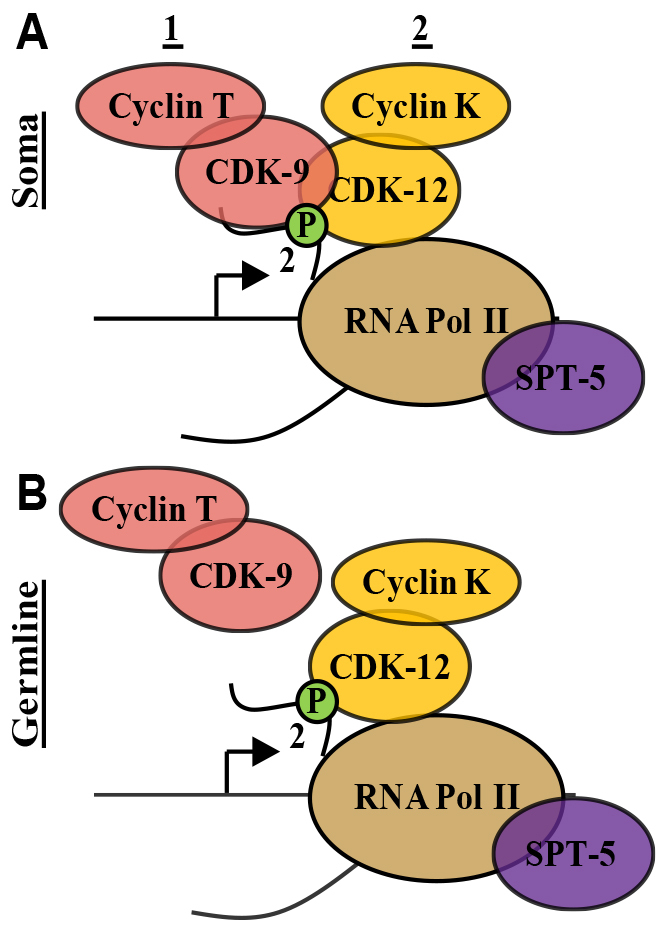
Schematic of Ser2-P-mediated regulation in C. elegans. (A) In somatic lineages, both CDK-9 and CDK-12 contribute to Ser2-P levels, but CDK-9 is required upstream of CDK-12 activity. (B) In the germline, Ser2-P is independent of CDK-9 but instead requires CDK-12. Kinase and cyclins are ubiquitously expressed, as is the major DSIF component SPT-5.
Our results show that transcription regulation in the C. elegans germline dramatically differs from current models of metazoan transcription elongation: CDK-9 does not play a major role in phosphorylation of Ser2, and Ser2-P added by CDK-12 does not require upstream CDK-9 activity (Fig. 8B). This is the first demonstration of Pol II Ser2-P not requiring, directly or indirectly, CDK-9/P-TEFb activity in a multicellular organism. Furthermore, the pie-1 mutant demonstrates that this is a fundamental difference between somatic and germline Pol II regulation, as transformation of germline to soma is accompanied by a switch from CDK-12- to CDK-9-dependent regulation of Ser2-P.
Unique regulation of Pol II kinases is a common feature of germ cell specification in many organisms (Nakamura and Seydoux, 2008), yet we were surprised to identify P-TEFb-independent Ser2-P in the C. elegans PGCs, Z2/Z3. Previous studies have suggested that inhibition of P-TEFb by PIE-1 is important for transcriptional repression in the germline precursors that give rise to the PGCs, Z2/Z3 (Zhang et al., 2003; Ghosh and Seydoux, 2008). PIE-1 degradation at the birth of Z2/Z3 was modeled to release P-TEFb inhibition. We find instead that PIE-1 activity correlates with CDK-12 repression in the P-cells, suggesting that PIE-1 may regulate Pol II by inhibiting multiple CTD kinases. Indeed previous studies have shown that PIE-1 also inhibits Ser5 phosphorylation by CDK-7, which may be more crucial for suppression of Pol II in the P-cells (Ghosh and Seydoux, 2008).
Kinase requirements in development
Although animals lacking CDK-9 and CDK-12 function arrest at different points, both kinases are essential for normal somatic development and growth. Their kinase activities are essential for somatic development, as kinase-dead point mutants do not rescue the lethality of deletion mutants. Although both of these kinases play roles in somatic Ser2-P levels, interpretation of this should be made with caution as it is unclear whether Pol II is the sole, or even the essential, target for either CDK-9 or CDK-12 in somatic tissues.
In the germline, CDK-9 is required for normal germline development, yet paradoxically is not significantly required for its most-studied activity: phosphorylation of Ser2 in the Pol II CTD. CDK-9 has other well-characterized roles in transcriptional regulation (Pirngruber et al., 2009), and the association of CDK-9 with transcriptionally active chromatin in the adult gonad indicates it has a role in germline transcription. However, whether the essential function of CDK-9 in the germline is restricted to transcription regulation is unclear.
Because Ser2-P in germ cells does not require CDK-9, we had the unique opportunity to assess the requirement for Ser2 phosphorylation in transcription apart from any other essential roles of CDK-9. Surprisingly, the high levels of Ser2-P in germ cells, which are dependent on CDK-12, were not required for germline development or fertility. Germlines lacking CDK-12 develop germline stem cells, progress successively through meiosis, and produce functional gametes and substantial numbers of viable offspring. Although brood size is reduced, transcription does not appear to be. Thus, neither CDK-12 nor Ser2-P is required for substantial transcriptional activity at any stage of germ cell development in C. elegans. Therefore, our results indicate that CTD phosphoepitopes may not always be accurate indicators of different stages of transcriptional regulation; e.g. absence of Ser2-P may not always reflect an absence of elongating Pol II.
CTD phosphorylation provides docking sites on elongating Pol II for complexes involved in transcript processing and nucleosome dynamics. Given the essential and predominant role of co-/post-transcriptional processing in C. elegans germ cells, it is unlikely these mechanisms are grossly affected in the cdk-12 mutant at optimal growth conditions. We observed a dramatic decrease in co-transcriptional H3K36me3 caused by loss of CDK-12. The role of H3K36 methylation, which is largely limited to gene bodies, decreases chromatin remodeling activity in the wake of Pol II elongation and helps restrict initiation to the correct promoter (Buratowski and Kim, 2010; Venkatesh et al., 2012). Co-transcriptional H3K36me3 by MET-1 is neither essential for viability nor for fertility under normal growth conditions in C. elegans (Andersen and Horvitz, 2007), but its absence could affect chromatin integrity and transcription fidelity, and more dramatically impact the transcriptome during post-embryonic growth or during stress. Pol II holoenzyme associations with RNA processing factors or other chromatin-modifying complexes could also be more dependent on Ser2-P during growth or stress. Germ cell development appears more reliant on CDK-12 activity at higher temperature, as the cdk-12(ok3664) mutant rescued with the cdk-12:pal-1 3′UTR construct is sterile at 25°C. This requirement under stress is similar to that observed in budding yeast, where deletion of Ctk1, the major Ser2 kinase, causes cold-sensitive growth (Lee and Greenleaf, 1991).
We observed very small but quantifiable amounts (<5% in germline cdk-12 mutants) of Ser2-P in cdk-12(RNAi) and mutant germ cells. This small signal could represent CDK-9-dependent Ser2-P that occurs in a subset of loci that are essential for germ cell development and viability. Indeed, this could explain the requirement of CDK-9 for fertility: the germline/soma difference in kinase requirements may reflect a difference in gene-specific requirements for Ser2-P regulation. It is even possible that the residual H3K36me3 observed in germ cells lacking CDK-12 could be due to low levels of CDK-9 dependent Ser2-P. Although this is possible, the loss of 95% of detectable Ser2-P does not dramatically affect germline gene expression, so the requirement for CDK-9 for fertility does not generally equate with a requirement for Ser2-P. Importantly, decreased correlation between transcription and Ser2-P may be a general property of pluripotent cells, as Ser2-P is undetectable in melanocyte stem cells and in other stem cells (Freter et al., 2010). In any case, regulation of Ser2-P in C. elegans germ cells is clearly unique from other described systems: the majority of Ser2-P is not downstream of CDK-9 activity and is not required for germ cell development.
Mechanism behind tissue-specific kinase requirements on Ser2-P
The regulation of Ser2-P in embryonic soma matches the current model of metazoan Ser2-P regulation: CDK-9 is required ‘upstream’ of CDK-12. Previous studies suggest that the upstream role of CDK-9 in metazoans is to alleviate negative regulation causing promoter-proximal ‘paused’ polymerase, i.e. Pol II that has initiated transcription but cannot progress to elongation. Pausing is largely mediated by the negative regulator, NELF (Yamaguchi et al., 2013). C. elegans does not have homologs to NELF components. Furthermore, paused polymerase signatures are also largely absent from C. elegans Pol II profiles, excluding a few hundred genes in starved C. elegans larvae (Baugh et al., 2009; Kruesi et al., 2013). Thus, although the upstream requirement of CDK-9 for somatic Ser2-P in embryos cannot be directly related to NELF-mediated pausing in other systems, it is intriguing to speculate that there may be another mechanism of Pol II elongation regulation in embryonic soma employing DSIF in combination with soma-specific factors.
By contrast, germline regulation of Ser2-P more closely matches that in yeast where the CDK-9 homolog, Bur1, is not required for Ser2 phosphorylation by the CDK-12 homolog Ctk1 (Qiu et al., 2009). The similarity between this tissue and yeast is particularly interesting as both organisms lack NELF-mediated pausing, and thus do not appear to require CDK-9/Bur1 for the progression of initiated Pol II into elongation and CDK-12/Ctk1-mediated Ser2-P. As in C. elegans germline, Bur1/CDK-9 is essential in yeast, yet the essential target is still unclear in both organisms.
The striking differences between these two modes of Ser2 regulation likely reflect inherent differences in regulating the transition between Pol II initiation and elongation in soma versus germline. Control of this transition may be more important in somatic development, in which the spatial and temporal regulation of tissue-specific gene expression is tightly regulated. The transition to elongation may not be as tightly regulated in C. elegans germ cells, which predominantly rely on post-transcriptional regulation for spatial and temporal protein patterns (Merritt et al., 2008). There is also substantial epigenetic contribution to the guidance of germ cell transcription in C. elegans that might decrease reliance on dynamic promoter regulation by kinases. For example, histone methylation contributes to an ‘epigenetic memory’ of transcription and chromatin organization produced in germ cells of the previous generation, and this memory is essential for normal germ line development (Furuhashi et al., 2010; Rechtsteiner et al., 2010; Gaydos et al., 2012). A structural memory of open chromatin, reinforced by active transcription in each generation that is stabilized in the germ line through successive generations, may obviate the need for the complications of multi-step transcriptional control seen in somatic development.
In summary, we have identified novel and tissue-specific regulation of Ser2-P in C. elegans. We believe that this demonstration of P-TEFb-independent Ser2-P in the C. elegans germline, the first to our knowledge in a multicellular organism, will provide a unique model for understanding the role of Ser2-P in transcription regulation during development. It is also intriguing to speculate that the different kinase requirements for Ser2-P and germline-specific modes of Pol II elongation regulation may be related to germline immortality.
Note added in proof
A recent publication (Cecere et al., 2013) has proposed that the ZFP-1(AF10)/DOT-1 complex may play a role in Pol II transcription elongation regulation and might be involved in Pol II pausing in C. elegans.
Supplementary Material
Acknowledgments
We thank Rebecca L. Adams, Dr Sujata Bhattacharyya, Dr Roger Deal, Dr Daniel Reines, Dr Jason Lieb and members of the Kelly lab for helpful discussions and comments on the manuscript. We also thank Dr Christian Frøkjær-Jensen for reagents and advice on the MOS-SCI method.
Footnotes
Funding
This work was supported by the National Institutes of Health [GM 077600 to W.G.K.] and by the National Science Foundation Graduate Research Fellowship Program (E.A.B.). Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
E.A.B. conceived the project, designed and performed most experiments, and prepared the manuscript. C.R.B. performed fcp-1 RNAi experiments and helped with quantification of RNAi experiments. J.H.A. contributed to the larval RNAi experiments. W.G.K. supervised the study and preparation of the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.095778/-/DC1
References
- Adelman K., Lis J. T. (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C., Horvitz H. R. (2007). Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134, 2991–2999 [DOI] [PubMed] [Google Scholar]
- Ashcroft S. J. H., Pereira C. (2003). Practical Statistics for the Biological Sciences: Simple Pathways to Statistical Analyses. Basingstoke, UK; New York, NY: Palgrave Macmillan; [Google Scholar]
- Bartkowiak B., Greenleaf A. L. (2011). Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription 2, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L. (2010). CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24, 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Demodena J., Sternberg P. W. (2009). RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324, 92–94 [DOI] [PubMed] [Google Scholar]
- Bender L. B., Suh J., Carroll C. R., Fong Y., Fingerman I. M., Briggs S. D., Cao R., Zhang Y., Reinke V., Strome S. (2006). MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133, 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D., Kohoutek J., Bartholomeeusen K., Johansen E., Hulinkova P., Luo Z., Cimermancic P., Ule J., Peterlin B. M. (2011). The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25, 2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Kim T. (2010). The role of cotranscriptional histone methylations. Cold Spring Harb. Symp. Quant. Biol. 75, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G., Hoersch S., Jensen M. B., Dixit S., Grishok A. (2013). The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol. Cell 50, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007). Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- Cho H., Kim T. K., Mancebo H., Lane W. S., Flores O., Reinberg D. (1999). A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13, 1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y., Sutton A., Sternglanz R., Prelich G. (2006). The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J. D., Barton G. J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Wu X., Riedel C. G., Ruvkun G. (2009). A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 459, 1079–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Murphy S. (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24, 280–288 [DOI] [PubMed] [Google Scholar]
- Freter R., Osawa M., Nishikawa S. (2010). Adult stem cells exhibit global suppression of RNA polymerase II serine-2 phosphorylation. Stem Cells 28, 1571–1580 [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S. P., Grunnet M., Jorgensen E. M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda N. J., Buckley M. S., Wei W., Core L. J., Waters C. T., Reinberg D., Lis J. T. (2012). Fcp1 dephosphorylation of the RNA polymerase II C-terminal domain is required for efficient transcription of heat shock genes. Mol. Cell. Biol. 32, 3428–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi H., Takasaki T., Rechtsteiner A., Li T., Kimura H., Checchi P. M., Strome S., Kelly W. G. (2010). Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin 3, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin C., Holdeman R., Strome S. (1998). The phenotype of mes-2, mes-3, mes-4 and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics 148, 167–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Rechtsteiner A., Yuen K. W., Muroyama A., Egelhofer T., Gaydos L., Barron F., Maddox P., Essex A., Monen J., et al. (2012). An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., Strome S. (2012). Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2, 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D., Seydoux G. (2008). Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation. Genetics 178, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R. P., Bentley D. L. (2009). TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29, 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y. H., Kirchner J., Kaminker J., Wood W. B., Strome S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K., Villeneuve A. M., Reinke V. (2002). X-chromosome silencing in the germline of C. elegans. Development 129, 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. (2008). The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 33, 61–73 [DOI] [PubMed] [Google Scholar]
- Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. (2005). A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohoutek J., Blazek D. (2012). Cyclin K goes with Cdk12 and Cdk13. Cell Div. 7, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi W. S., Core L. J., Waters C. T., Lis J. T., Meyer B. J. (2013). Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2, e00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C., Hansen M. (2012). C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS ONE 7, e35428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Greenleaf A. L. (1991). CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1, 149–167 [PMC free article] [PubMed] [Google Scholar]
- Liu J., Kipreos E. T. (2000). Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 17, 1061–1074 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P. (2005). A structural perspective of CTD function. Genes Dev. 19, 1401–1415 [DOI] [PubMed] [Google Scholar]
- Merritt C., Seydoux G. (2010). The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development 137, 1787–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G. (2008). 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18, 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Seydoux G. (2008). Less is more: specification of the germline by transcriptional repression. Development 135, 3817–3827 [DOI] [PubMed] [Google Scholar]
- Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D. K., Hasegawa J., Omori M., Inukai N., et al. (2003). Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirngruber J., Shchebet A., Johnsen S. A. (2009). Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle 8, 3636–3642 [DOI] [PubMed] [Google Scholar]
- Powell-Coffman J. A., Knight J., Wood W. B. (1996). Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev. Biol. 178, 472–483 [DOI] [PubMed] [Google Scholar]
- Qiu H., Hu C., Hinnebusch A. G. (2009). Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtsteiner A., Ercan S., Takasaki T., Phippen T. M., Egelhofer T. A., Wang W., Kimura H., Lieb J. D., Strome S. (2010). The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 6, e1001091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner C. E., Deshpande G., Schedl P. D., Kelly W. G. (2003). A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G., Dunn M. A. (1997). Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124, 2191–2201 [DOI] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I. (2011). OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6, e20085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Walker A. K., Shi Y., Blackwell T. K. (2002). CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16, 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R. H., Fire A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 [DOI] [PubMed] [Google Scholar]
- Strome S., Wood W. B. (1983). Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35, 15–25 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Winston F. (1992). SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., Kumar S., Natarajan K., Workman J. L. (2012). Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 [DOI] [PubMed] [Google Scholar]
- Walker A. K., Shi Y., Blackwell T. K. (2004). An extensive requirement for transcription factor IID-specific TAF-1 in Caenorhabditis elegans embryonic transcription. J. Biol. Chem. 279, 15339–15347 [DOI] [PubMed] [Google Scholar]
- Whittle C. M., McClinic K. N., Ercan S., Zhang X., Green R. D., Kelly W. G., Lieb J. D. (2008). The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 4, e1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A., Shilatifard A. (2006). Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle 5, 1066–1068 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Wada T., Watanabe D., Takagi T., Hasegawa J., Handa H. (1999). Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274, 8085–8092 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Shibata H., Handa H. (2013). Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta 1829, 98–104 [DOI] [PubMed] [Google Scholar]
- Zhang F., Barboric M., Blackwell T. K., Peterlin B. M. (2003). A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 17, 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Li T., Price D. H. (2012). RNA polymerase II elongation control. Annu. Rev. Biochem. 81, 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.