Abstract
Recombinational repair of replication forks can occur either to a crossover (XO) or noncrossover (non-XO) depending on Holliday junction resolution. Once the fork is repaired by recombination, PriA is important for restarting these forks in Escherichia coli. PriA mutants are Rec− and UV sensitive and have poor viability and 10-fold elevated basal levels of SOS expression. PriA sulB mutant cells and their nucleoids were studied by differential interference contrast and fluorescence microscopy of 4′,6-diamidino-2-phenylindole-stained log phase cells. Two populations of cells were seen. Eighty four percent appeared like wild type, and 16% of the cells were filamented and had poorly partitioned chromosomes (Par−). To probe potential mechanisms leading to the two populations of cells, mutations were added to the priA sulB mutant. Mutating sulA or introducing lexA3 decreased, but did not eliminate filamentation or defects in partitioning. Mutating either recA or recB virtually eliminated the Par− phenotype. Filamentation in the recB mutant decreased to 3%, but increased to 28% in the recA mutant. The ability to resolve and/or branch migrate Holliday junctions also appeared crucial in the priA mutant because removing either recG or ruvC was lethal. Lastly, it was tested whether the ability to resolve chromosome dimers caused by XOs was important in a priA mutant by mutating dif and the C-terminal portion of ftsK. Mutation of dif showed no change in phenotype whereas ftsK1∷cat was lethal with priA2∷kan. A model is proposed where the PriA-independent pathway of replication restart functions at forks that have been repaired to non-XOs.
In Escherichia coli, accumulated evidence favors a model whereby replication forks routinely stop on their way from oriC to the terminus for a variety of reasons that include loss of helicase activity (arrest), encounter with a nick, or other type of DNA damage (collapse) (reviewed in refs. 1–3). Fork stoppage, repair, restart, and mitigation of the consequences of these events are therefore a common set of processes that occurs in most cells as they replicate their chromosomes. Repair of replication forks is accomplished by homologous recombination. Depending on the damage, different presynaptic enzymes such as RecBCD or RecFOR will aid RecA in the repair process and form a Holliday junction (reviewed in ref. 4). Postsynaptic enzymes for branch migration (RecG and RuvAB) and Holliday junction resolution (RuvC) will move and resolve the Holliday junction (5). Depending on resolution, exchange (crossing over) may occur between the sister chromosomes. If a crossover (XO) occurs then after replication has been restarted and completed, a chromosome dimer will be formed. The dif/XerCD/FtsK system then is needed to resolve these dimeric chromosomes to monomers (6, 7).
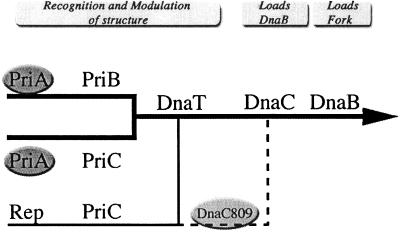
Replication restart proteins (RRPs) (3, 8) form multiple pathways (as defined genetically) to restart repaired forks as shown in Fig. 1 and ref. 9. Theoretically, the RRPs function equivalently to DnaA in that they form a protein-DNA complex so that the DnaB protein can be loaded onto the DNA (3, 10). However, unlike the loading of DnaB in a cell cycle, DNA sequence-dependent fashion at oriC, the RRPs can load DnaB anywhere on the chromosome by an oriC/dnaA-independent mechanism. Functionally, this is a key issue because the reasons for stopping can arise anywhere along the chromosome. Presumably, the RRPs load a single replication fork in the same direction the fork was traveling whereas DnaA loads two forks that replicate bidirectionally from oriC.
Figure 1.
Multiple biochemical pathways for replication restart from recombination intermediates. Three types of pathways: PriA-dependent (thick black lines), PriA-independent (thin black line), and PriA-independent using the suppressor dnaC809 (dashed line). Possible function(s) of the genes are suggested by the headings at the top. Full explanation of genes and pathways can be found elsewhere (9).
It is thought that PriA initiates the restart process by identifying and binding a recombinational intermediate (11, 12). In the absence of priA, cells are Rec− and UV sensitive and have poor viability, high basal levels of SOS expression, and poor plating of Mu phage. Extragenic suppressors of priA2∷kan have been found in dnaC (13). In vitro, DnaC810 can load DnaB onto single-stranded DNA in the absence of PriA or the other RRPs (dnaC810 has a different nucleotide change as compared with dnaC809 but produces the identical amino acid change) (14). In vivo, however, dnaC809 requires priC and rep to suppress priA2∷kan phenotypes (9).
To learn more about the PriA-independent pathway involving priC and rep, priA2∷kan mutant cells and their nucleoids were studied by differential interference contrast (DIC) and fluorescence microscopy of 4′,6-diamidino-2-phenylindole-stained log phase cells. Unexpectedly two populations of cells were found: wild-type-appearing cells and filaments with abnormal nucleoids. Factors contributing to the formation of these two populations were studied by mutating genes involved with cell division, recombination, and resolution of chromosome dimers. A model is proposed where the PriA-independent pathway can restart only replication when repair processes have resolved the Holliday junctions to noncrossovers (non-XOs).
Materials and Methods
Bacterial Strains.
All bacterial strains used in this work are derivatives of E. coli K-12 and are described in Table 1. Due to priA1∷kan strain's sensitivity to rich media (15), cultures for all experiments described, unless otherwise indicated, were grown in 56/2 minimal media (16) at 37°C. The protocol for P1 transduction has been described (16).
Table 1.
Strains of bacteria used in this work
| Strain | priAc | Relevant genotype and/or plasmid | Source or derivation |
|---|---|---|---|
| AD11 | + | ftsK1∷cat | (31) |
| CAG5052 | + | metB1 btuB3191∷Tn10 | (46) |
| DM4000a | + | M. Volkert | |
| GS1481 | + | ruvC64∷kan | B. Michel |
| JC10289 | + | del(srl-recA) 306∷Tn10 | (47) |
| JC13503 | + | del(srl-recA) 306∷Tn10 | JC10289→SK362e |
| JC13509b | + | (13) | |
| JC15716 | + | recO1504∷Tn5 | (48) |
| JC18983a | 2 | (13) | |
| JC19021b | 2 | (13) | |
| JC19008a | 2 | dnaC809 | (13) |
| JC19018a | 2 | dnaC809 zjj-202∷Tn10 | (13) |
| JC19098a | + | lexA3 malE∷Tn10 | (13) |
| JC19238a | + | dnaC809 metB1 btuB3191∷Tn10 | (9) |
| JC19250a | 2 | metB1 btuB3191∷Tn10 | (9) |
| JC19328b | + | del(srl-recA) 306∷Tn10 | JC13503→JC13509e |
| JJC315 | + | recB268∷Tn10 | B. Michel |
| N3695 | + | recG258∷kan | B. Lloyd |
| PK3881 | + | dif∷tet | P. Kuempel |
| PN105 | 2 | (18) | |
| SK362b | + | sulB+ | (49) |
| SS167b | + | del(srl-recA) 306∷Tn10 pSJS1222 (recA+) | pSJS1222→JC19328e |
| SS186b | 2 | del(srl-recA) 306∷Tn10 | JC19008→SS167e |
| SS324b | 2 | sulB+ | PN105→SK362e |
| SS328b | + | dif∷tet | PK3881→JC13509f |
| SS329b | 2 | dif∷tet | PN105→SS328e |
| SS380b | 2 | recO1504∷Tn5 | JC19008→SS481i |
| SS382b | + | sulA6209∷tet | TP606→JC13509e |
| SS394b | 2 | sulA6209∷tet | JC19008→SS382e |
| SS446b | + | recB268∷Tn10 | JJC315→JC13509g |
| SS447 | 2 | metB+ argE+ | Laboratory stock |
| SS459b | + | recB268∷Tn10 pDSW2 (recB+) | pDSW2→SS446e |
| SS460b | 2 | recB268∷Tn10 | PN105→SS459e |
| SS471b | + | recO1504∷Tn5 | JC15716→JC13509 |
| SS481b | + | recO1504∷Tn5 metB1 btuB3191∷Tn10 | CAG5052→SS471d |
| SS490b | + | metB1 btuB3191∷Tn10 | CAG5052→JC13509d |
| SS493a | + | recG258∷kan metB1 btuB3191∷Tn10 | N3695→JC19250i |
| SS702a | + | recG258∷kan dnaC809 metB1 btuB3191∷Tn10 | N3695→JC19238i |
| SS712a | 2 | recG258∷kan dnaC809 | JC19008→SS702h |
| SS725a | + | ruvC64∷kan metB1 btuB3191∷Tn10 | GS1481→JC19250i |
| SS728a | + | ruvC64∷kan dnaC809 metB1 btuB3191∷Tn10 | GS1481→JC19238i |
| SS735a | 2 | ruvC64∷kan dnaC809 | JC19018→SS728h |
| SS767b | + | lexA3 malE∷Tn10 | JC19098→JC13509g |
| SS768b | 2 | lexA3 malE∷Tn10 | SS447→SS767e |
| SS805b | + | ftsK1∷cat metB1 btuB3191∷Tn10 | AD11→SS490e |
| SS818b | + | ftsK1∷cat | AD11→JC13509e |
| SS822b | 2 | ftsK1∷cat dnaC809 zjj-202∷Tn10 | SS447→SS823e |
| SS823b | + | ftsK1∷cat dnaC809 zjj-202∷Tn10 | JC19018→SS818f |
| TP606 | + | sulA6209∷tet | (50) |
These strains have the following partial genotype: sulA∷Mu-d(Ap, lac, B∷Tn9) Δ(lac-pro)XIII hisG4 argE3 ara-14 xyl-5 mtl-1 rpsL31.
These strains have the following partial genotype: sulB103− lacMS286 Φ80dIIlacBK1 argE3 his-4 thi-1 xyl-5 mtl-1 SmRT6R.
The only allele of priA used in this study is priA2∷kan (18).
Select for tetracycline resistance and screen Met−.
Select for either kanamycin, tetracycline, ampicillin, or chloramphenicol resistance.
Select for tetracycline resistance and then screen for the presence of dnaC809 by PCR.
Select for tetracycline resistance and screen UV sensitive.
Select for Met+ and screen for the presence of priA2∷kan by PCR.
Select for kanamycin resistance and screen UV sensitive.
Microscopy Techniques.
Cultures were grown on a shaking water bath at 100 rpm to an optical density of 0.3 (A600) at 37°C. A drop (10 μl) of culture was applied to each well of a poly-l-lysine-coated microscope slide (8-well multitest slides, ICN) and incubated at room temperature for 10 min. The wells were washed 10 times with standard PBS and allowed to dry. Cells were fixed with 80% methanol for 5 min and stained with 0.5 μg/ml solution of 4′,6-diamidino-2-phenylindoledihydrochloride. After 2 min, the wells were washed five times with PBS, and a coverslip was applied. A Nikon Eclipse 600 light microscope with a SPOT digital camera was used to collect DIC and fluorescence micrographs. NIH imaging software, scion image (version 1.62a), was used to measure cell length. At least 500 cells from at least two individual trials were measured and used to calculate the percent filamentation.
Results
PriA2∷kan Cultures Have Two Populations of Cells.
We used DIC and fluorescence microscopy to visualize priA mutant cells and their nucleoids. Previous work revealed that priA mutant cells grown in rich media filament have poorly partitioned nucleoids and that sulA mutations suppressed the filamentation to a large degree (17, 18). Because it has since been shown that priA mutants are sensitive to rich media (15), a more in-depth study of priA2∷kan sulB103 mutant cells grown to midlog phase in minimal media at 37°C was done. SulB103 is an allele of ftsZ that causes FtsZ to be insensitive to the action of the SulA cell division inhibitor (19).
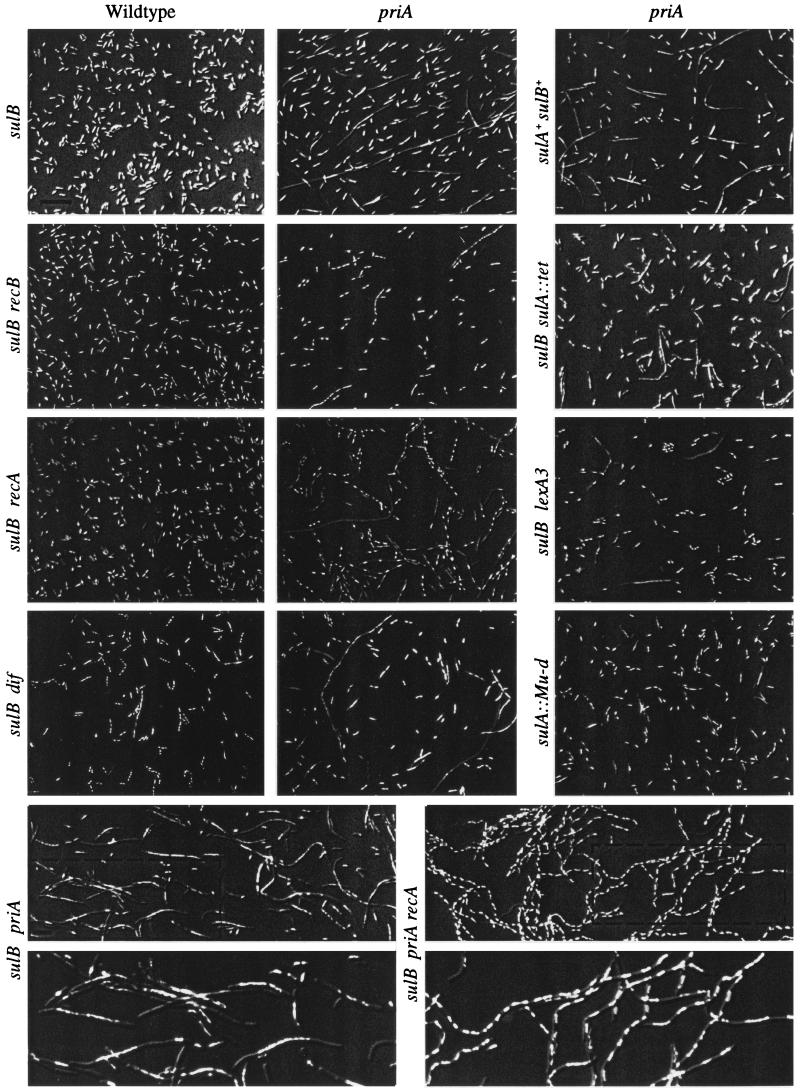
Based on counting and measuring the lengths of 2,710 cells, two distinct populations of cells were seen in a priA sulB culture (Fig. 2). Eighty four percent of the cells examined appeared like wild type [less than three cell lengths (6 μm) in size and showing one or two nucleoids] (Table 2). The other 16% of the population show cells that are larger than 6 μm in size, and their nucleoids show a variety of conformations. Some nucleoids occupy the length of the cell and have a “knobby” appearance, whereas others show a single large nucleoid in the middle of the filament [filaments are defined as cells larger than three cell lengths (6 μm) in size regardless of the number of nucleoids]. Both nucleoid types suggest that these cells had difficulty in properly partitioning their chromosomes (Par− is defined as cells containing multiple poorly condensed nucleoids). Some cells with no 4′,6-diamidino-2-phenylindole staining also are observed. These filaments were seen in early and late log phase growth (same proportion of the population) and with wet-mount preparations of live cells (data not shown).
Figure 2.
(On the opposite page.) Pictures of overlays of individual DIC microscopy and fluorescence microscopy images using Abode photoshop (version 5.5) are shown. Cells were prepared as noted in Materials and Methods. Genes mutated are indicated on the top and left sides. Wild type means priA+. See Table 1 for specific allele numbers and other genotypes of each strain. Strains are named starting from the top left and moving right. (Upper) Row 1: JC13509, JC19021, SS324. Row 2: SS446, SS460, SS392. Row 3: JC19328, SS186, SS768. Row 4: SS328, SS329, JC18983. (Lower) Row 1: JC19021, SS186. Row 2 shows enlargements of the regions in the dotted boxes of the pictures seen in the row above. The bars in the pictures indicate 10 μm.
Table 2.
Percentage of cells showing filamentation
| Strain | priA | recA | recB | recO | sulA | sulB | lexA | dif | Cells counted | Total filamentation | Distribution of cell sizes
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6–10 | 10–20 | 20–50 | >50 | |||||||||||
| SK362 | + | + | + | + | + | + | + | + | 500 | 0 | 0 | 0 | 0 | 0 |
| SS324 | 2 | + | + | + | + | + | + | + | 723 | 18.9 | 9.3 | 4.2 | 4.3 | 1.1 |
| JC13509 | + | + | + | + | + | 103 | + | + | 500 | 0 | 0 | 0 | 0 | 0 |
| JC19021 | 2 | + | + | + | + | 103 | + | + | 2,710 | 16.3 | 9.7 | 4.4 | 2.1 | 0.1 |
| SS382 | + | + | + | + | 6209 | 103 | + | + | 500 | 0 | 0 | 0 | 0 | 0 |
| SS394 | 2 | + | + | + | 6209 | 103 | + | + | 665 | 7.3 | 5.1 | 1.6 | 0.6 | 0 |
| DM4000 | + | + | + | + | Mu-d | + | + | + | 500 | 0 | 0 | 0 | 0 | 0 |
| JC18983 | 2 | + | + | + | Mu-d | + | + | + | 1,873 | 4.8 | 3.6 | 0.9 | 0.3 | 0 |
| SS481 | + | + | + | 1504 | + | 103 | + | + | 500 | 1.4 | 1.4 | 0 | 0 | 0 |
| SS380 | 2 | + | + | 1504 | + | 103 | + | + | 3,014 | 14.9 | 10.8 | 3.0 | 1.1 | 0 |
| SS446 | + | + | 268 | + | + | 103 | + | + | 3,698 | 1.5 | 1.3 | 0.1 | 0.1 | 0 |
| SS460 | 2 | + | 268 | + | + | 103 | + | + | 4,099 | 3.5 | 3.0 | 0.3 | 0.2 | 0 |
| JC19328 | + | 306 | + | + | + | 103 | + | + | 973 | 2.1 | 2.0 | 0.1 | 0 | 0 |
| SS186 | 2 | 306 | + | + | + | 103 | + | + | 2,200 | 28.1 | 17.6 | 6.6 | 3.0 | 0.9 |
| SS767 | + | + | + | + | + | 103 | 3 | + | 500 | 0 | 0 | 0 | 0 | 0 |
| SS768 | 2 | + | + | + | + | 103 | 3 | + | 2,020 | 7.0 | 3.8 | 1.9 | 1.0 | 0.3 |
| SS328 | + | + | + | + | + | 103 | + | tet | 501 | 0.8 | 0.6 | 0.2 | 0 | 0 |
| SS329 | 2 | + | + | + | + | 103 | + | tet | 1,683 | 18.7 | 11.5 | 4.0 | 3.0 | 0.2 |
The numbers for total filamentation and the distributions in the different size classes have been rounded off to the nearest tenth (0.1).
We confirmed previous studies that cultures of priA cells contained two populations of cells: one that appears as wild type and one that appears as partitioning-defective filaments. For log phase cells grown in minimal media, the proportions of cells in these two groups were quantitated to be 84% and 16%. In the remainder of this report, the cellular processes that give rise to these two population cells will be investigated.
High Basal Levels of SOS Expression and sulA Contribute to the Filamentation Seen in a priA sulB Strain.
It was interesting that both sulB (in minimal media) and sulA [in rich media (18)] mutations did not fully suppress the filamentation of a priA mutant as they do for lexA-defective mutants (20). Investigations as to why sulA and sulB mutations did not fully suppress the filamentation in a priA mutant (and did so differentially) were pursued. First, the level of filamentation of sulA priA strain grown in minimal media was determined and found to be 5% (Fig. 2, Table 2). The strain used, however, was of a different genetic background than the priA sulB strain (DM4000 vs. JC13509). Therefore a sulA mutation was introduced into the standard sulB strain. This sulA sulB priA mutant showed 7% filamentation. Previously it had been shown that the introduction of lexA3 to a priA sulA mutant decreased the expression of the sulA promoter 10-fold to just below levels seen in the priA+ control (13). To test whether lowering levels of sulA (in addition to other lexA-regulated genes) was important for the filamentation occurring in priA mutants, lexA3 was combined with the priA sulB mutations. Fig. 2 and Table 2 show that lexA3 decreased filamentation from 16% to 7%. Finally, it was still unclear why priA sulA and priA sulB mutants had different levels of filamentation. To test whether sulB suppressed any priA-induced filamentation, a priA sulB+ was constructed. It showed 18% filamentation.
We conclude that sulA is an important determinant for filamentation in a priA mutant. Either removing sulA or lowering its concentration significantly reduces filamentation. The effect of a sulB mutation is much smaller. It remains unclear, however, why the two effects are not equal and why they do not fully suppress the priA-induced filamentation as they do for SOS-induced filamentation.
RecA and recB Contribute to the Partitioning Defect Observed in the Filamenting Subpopulation of priA Mutant Cells.
Kuempel and colleagues (21) have shown that 15% of cells in a wild-type population undergo a XerCD-mediated recombination event at dif to produce hybrid dif sites. This number provides a minimum measure of the frequency of homologous recombination between sister chromosomes that result in XOs. The actual number could be greater if additional compensatory XOs took place after XO formation (6, 21). Because the percentage of cells showing a hybrid dif site was similar to the percentage of partitioning defective cells seen in a culture of a priA mutant, it was hypothesized that the partitioning defect was a result of recombinationally repaired replication forks where a XO had occurred. Then as in the dif system (6), removing presynaptic and synaptic recombination functions from a priA mutant should decrease the number of Par− nucleoids. To test this, del(recA)306, recB258∷Tn10, and recO1504∷Tn5 mutations were combined with a priA2∷kan mutation.
The recA priA and recB priA double mutant strains were viable and grew more slowly than priA, recA, or recB single mutants (data not shown). Several tests were performed to verify the genotypes of the double mutants and to check for suppressor mutations. These included reconstructing the strains, testing several different transductants, checking the alleles by PCR analysis, and sequencing the dnaC gene.
Removal of either recA or recB caused a dramatic change in the Par− filaments seen in the priA mutant (Fig. 2 and Table 2). In both cases the Par− phenotype nearly disappeared. This was more complete for the recB priA strain than for the recA priA strain. In some recA priA filaments, both Par+ and Par− nucleoids were observed. The most dramatic differences, however, between the strains were in the length and number of filaments. Removing recB from the priA mutant reduced filamentation from 16% to 3% and removing recA increased filamentation from 16% to 28%. These priA recA filaments observed were much longer as well (Table 2). Removal of recO from the priA mutant had little effect on the appearance or number of the Par− cells (Table 2).
We conclude that under these conditions the recombination processes contributed by recB and recA, but not recO, help produce the Par− phenotype seen in priA mutants. The difference in filamentation of the recB priA and recA priA mutant strains may be attributable to the removal of RecA-mediated SOS induction.
RecG258∷kan and ruvC64∷kan Are Synthetically Lethal with priA2∷kan.
Above, we hypothesized that the 16% of the priA mutant cells (the Par− filaments) are the product of XOs. Two possibilities can explain the wild-type appearance of the remaining 84%: either they did not experience a repair event or the repair event was resolved to a non-XO. Michel et al. (22) have suggested that resolution of Holliday structures created during the recombinational repair of replication forks in rep mutants are biased to the production of non-XOs [other bias resolution observed in palindrome-induced DNA damage (23)]. If this bias also occurs in priA mutants, then removing functions that cause this biased resolution may result in fewer viable cells. This assumes that at least the same number of forks need repair and that priA cells in which forks are repaired to XOs are inviable. Candidate genes for providing this biased resolution include genes involved with branch migration (recG and ruvAB) and Holliday junction resolution (ruvC) (24). Therefore, recG258∷kan and ruvC64∷kan were added to a priA mutant. Because initial results indicated that both of these mutations were synthetically lethal with priA2∷kan, this was tested definitively by using a previously described P1 transduction assay (9). Briefly, priA2∷kan is introduced to a metB1 recipient (also containing the other mutation to be tested) by selecting Met+ transductants. These are scored for the inheritance of priA2∷kan by PCR analysis. Normal cotransduction frequencies can be monitored by testing for the conversion of the nearby btuB locus (from TetR to TetS). If no priA2∷kan transductants are found in 32 independent events, then there is a 3 in 107 chance this could have occurred by random chance. Table 3 shows that in a recG and ruvC mutant, respectively, none of 34 or 24 independent Met+ transductants tested inherited priA2∷kan. Conversion of the btuB locus occurred with a frequency similar to wild type.
Table 3.
Number of cotransduction events of priA2∷kan, btu+ with metB+ into strains harboring mutations in either ruvC, recG, or ftsK
| Strain | Genotype of recipient
|
Met+ transductants
|
|||||
|---|---|---|---|---|---|---|---|
| ruvC | recG | ftsK | PriA+ TetR | PriA+ TetS | PriA− TetR | PriA− TetS | |
| JC19250* | + | + | + | 5/56 | 7/56 | 20/56 | 24/56 |
| SS725 | 64 | + | + | 11/24 | 13/24 | 0/24 | 0/24 |
| SS493 | + | 258 | + | 18/34 | 16/34 | 0/34 | 0/34 |
| SS805 | + | + | 1 | 19/30 | 11/30 | 0/30 | 0/30 |
The data from this control strain is taken from ref. 9 for the convenience of the reader.
It is concluded that ruvC and recG are essential genes in a priA mutant. This is consistent with a model where the priA-independent pathway can only restart recombinationally repaired replication forks to non-XOs that are produced through biased resolution of Holliday junctions.
Dif-Mediated Resolution of Dimers Is Not Essential in priA Mutants, but the Presence of ftsK+ Is Required.
The above data are compatible with a model where recombinational repair of replication forks in a priA mutant are preferentially resolved to the non-XO state 84% of the time. Because dif and the machinery (xerCD and ftsK) used to resolve dimers are only needed when XOs occur, this locus and these gene products should not be needed in a priA mutant. To test this, dif∷tet was first introduced into our test strain (JC13509). It was expected that this mutation by itself would cause a moderate amount of filamentation and defects in chromosome partitioning (25). We found, however, that dif∷tet produced only a small amount (0.8%) of filamentation (Fig. 2 and Table 2). This smaller than expected amount may be due to the sulB103 mutation and would make it unlikely that we should see any change in filamentation upon mutating dif in a priA2∷kan sulB103 mutant. This was tested and only a small difference was observed (Fig. 2 and Table 2). Similar results were seen with xerC and xerC priA mutants (data not shown). Although we cannot be sure whether there is an effect of dif∷tet in a priA mutant, we can conclude that dif is not essential in a priA mutant. This is consistent with the model.
FtsK mutations are pleiotrophic, causing deficiencies in cell division (26), dif-mediated segregation of resolved dimers (7, 27–30), and regulation of uspA gene expression (31). FtsK1∷cat is an insertion mutation that allows expression and function of the amino-terminal domain essential for cell division (32). Because ftsK also is required for dif-mediated recombination, we attempted to combine ftsK1∷cat with priA2∷kan. Table 3 shows that this was not possible and that ftsK1∷cat and priA2∷kan are synthetically lethal.
It is concluded that the absence of dif/xerCD-mediated resolution of dimers does not alter significantly the number or appearance of the Par− filamentous priA cells. The ftsK gene product, however, is essential for the viability under these conditions. Roles of ftsK in cell division, segregation of chromosomes (other than XerCD/dif-mediated), and/or gene regulation may be responsible for the observed synthetic lethality between ftsK1∷cat and priA2∷kan.
Synthetic Lethality Between priA and ftsK, recG, and ruvC Is Rescued by dnaC809.
The observations presented here that mutations in ftsK, recG, and ruvC are synthetically lethal with priA2∷kan mutations represent the third report of mutations that are synthetically lethal with priA. Two previous reports showed that rep and priC mutations also were synthetically lethal with a priA mutation (9, 33). The synthetic lethality between mutations in those two cases (rep/priC with priA) could not be rescued by dnaC809. This is noteworthy because in all other cases tested, dnaC809 suppresses all other known priA-mutant phenotypes (9, 13). For this reason, it was hypothesized that rep and priC were required for dnaC809 suppression of priA2∷kan and for the PriA-independent pathway (Fig. 1) (9). To test whether ruvC, recG, and ftsK were also part of this pathway, dnaC809 was first combined with each of these mutations and then we attempted to introduce priA2∷kan into these double mutants. The ability to construct all three triple mutants is shown in Table 1 (see strains SS822, SS735, and SS712). Thus, in all three cases, dnaC809 could rescue the synthetic lethality seen between priA and ftsK, recG and ruvC mutations. It is therefore concluded that although recG, ruvC, and ftsK are essential in a priA mutant, they are different from priC and rep and are not needed for the PriA-independent pathway.
Discussion
In this paper the appearance of log phase 4′,6-diamidino-2-phenylindole-stained priA2∷kan mutant cells were visualized by DIC and fluorescence microscopy. Two populations of cells were seen: 84% of the cells appeared like wild type whereas 16% were Par− filaments. Mutations that affect cell division (sulA, ftsK), SOS regulation (lexA, recA), recombination (recA, recB, recO), the ability to resolve dimer chromosomes (dif, ftsK), and the ability to resolve and/or branch migrate Holliday junctions (ruvC and recG) were used to investigate how the two populations are produced. To explain these results, we formulated a model (see Fig. 3 and below for specifics) based on the consequences of forming XO products during the recombinational repair and the restart of forks in the absence of PriA. Aspects of SOS regulation and cell division (see below) also are invoked to explain the extent of filamentation seen in some of the double mutants.
Figure 3.
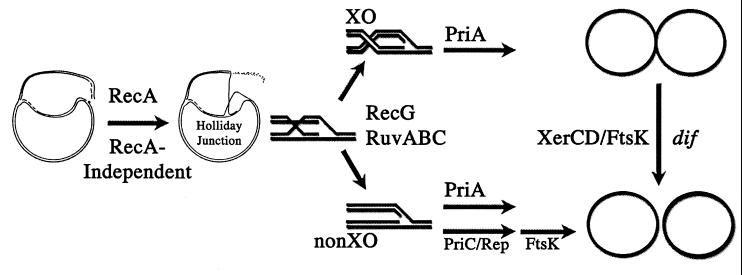
A model for the reactivation of collapsed replication forks emphasizing a differentiation in the usage of replication restart pathways specified by Holliday junction resolution products. The flow of the diagram is from left to right. A broken chromosome is depicted. This is converted by RecA-dependent pathway to a Holliday junction. In the absence of RecA, RecA-independent pathways become available. See text for explanation. Other proteins such as RecBCD and RecFOR also may aid in these reactions. Resolution of the Holliday junction to either a XO or non-XOs is catalyzed by RecG and RuvABC. If available, PriA can restart replication forks from both resolution products. The PriA-independent pathway (PriC and Rep), however, can only restart from non-XO products. dif-mediated xerCD site-specific recombination can resolve chromosome dimers formed by XOs to monomeric circles. FtsK is important for partitioning chromosome dimers resolved at dif. It also has an additional role in the viability of priA2∷kan strains. This role could be in either chromosome segregation or cell division.
The Model.
Several lines of evidence lead to the hypothesis that recombinational repair of replication forks results in either a XO or non-XO (reviewed in ref. 1). It is presumed that depending on the reason for the fork stoppage, the cell will use appropriate recombination functions to repair the fork. Many models call for production and resolution of a Holliday junction that leads to either the XO or non-XO structure. When replication has been completed, the XO will produce a dimeric chromosome and non-XO will produce two monomers. If a XO has occurred and a chromosome dimer is formed, this must be resolved to monomers before cell division can take place. Resolution of the dimer depends on at least three factors: dif, XerCD, and FtsK (7, 28, 30).
Our working model hypothesizes that the formation of partitioning-defective filaments in a priA mutant is a consequence of the inability to restart DNA replication at XOs produced by recombinational repair of replication forks (Fig. 3). This idea was adapted from models proposed for dif-mediated recombination (i.e. refs. 21 and 23). Several assumptions were made to more fully consider the role of replication restart in a priA mutant cell. The first assumption is that commitment to Holliday junction resolution in a particular orientation and/or resolution itself occurs before restart of the replication forks by the PriA-independent pathway. This is necessary because of the second assumption: the PriA-independent pathway can only restart replication forks when repair has or will lead to the non-XO state. Presumably priA+ cells do not have this limitation because restart and resolution of the dimers occurs without incident. The third assumption is that the Par− filaments lead to inviability. Fourth, all rounds of chromosome replication experience at least one event where repair/restart of the fork is necessary.
Other models also may be possible. For instance, in a priA mutant, failure to correctly restart any repaired fork, regardless of XO formation, could lead to the Par− filaments. This model, however, does not explain the effects of the rec mutants on the Par− filaments or the synthetic lethality of priA and recG/ruvC mutations.
Comparisons with Existing dif/xerCD Data.
Although the percentage of Par− filaments seen in priA mutants is very similar to the percentage of XerCD-mediated recombination at dif seen in a wild-type strain (6, 21), this could be purely coincidental. The observation that recA mutant cells show hardly any XerCD-mediated recombination at dif sites (6) and that the priA recA double mutants show very little of the Par− nucleoids suggests, however, that this comparison may not be purely coincidental. This becomes less general, however, when the recB data are reviewed. RecB mutants show only a 50% decrease in dif-mediated recombination in priA+ cells whereas they show almost no Par− nucleoids in a priA mutant. It is possible that the differences observed between the sets of experiments are due to the types of media used (rich vs. minimal) or the pleiotropic effects of having a priA mutation (high basal levels of SOS expression in the recB mutant and not the recA mutant). No measurement for XerCD-mediated recombination event at dif sites has been published for a recO mutant.
Can Bias in Holliday Junction Resolution Explain the Data?
It is noteworthy that the priA recB mutant showed essentially no Par− nucleoids (even though there was still 3% filamentation). This suggests that the processing of Holliday junctions in this mutant was almost entirely to the non-XO state. Because recB is mutated, it is plausible that the presynaptic recombination functions used to repair these forks are accomplished by RecFOR (4). This suggests that repair by RecFOR (in the absence of RecBCD) favors non-XO. This agrees with Cromie and Leach (23) who observed that repair of a gap produced by DNA replication of a palindrome (a RecFOR substrate) leads to non-XOs.
In the above discussion, it is inferred that the key enzyme complex to catalyze the production of a XO or non-XO is RuvABC. It has been shown in vitro that the way in which RuvAB binds to a Holliday junction dictates how RuvC will resolve the junction (34). Because Holliday junctions are symmetric, one can reasonably ask how the asymmetry is initiated so that resolution can be bias in a particular situation. One idea is that the replication fork itself confers the asymmetry for loading of RuvAB. This could be determined by either the DNA directly [structure of the DNA at the fork or combination of new and parental strand (methylation)] or the presence of replication proteins or both. Depending on the type of damage, a different set of proteins or DNA structures could exist to dictate RuvAB loading.
A New Relationship Between RuvC and RecG?
The synthetically lethal combinations of ruvC priA and recG priA mutations could be a consequence of that both of these genes must conspire to help resolve Holliday junctions to the non-XO state when priA is not available. Alternately, it is possible, that in a priA mutant, it is the balance of RuvABC and RecG activities that is important for viability. Currently we favor the former explanation, but we have no reason to reject the latter. If in a priA mutant, ruvC and recG do conspire to yield viability, then this would indicate a different relationship between ruvC and recG than has been seen for Hfr-mediated recombination. In this process, these mutants show additive effects (35). One reason a different relationship between ruvC and recG may exist could be that the DNA substrates produced by conjugation are different from those generated during repair of replication forks.
McGlynn and Lloyd (36) reported construction of a ΔruvAC65 priA2∷kan strain and referred to unpublished data of a recG priA double mutant. One possible explanation for why McGlynn and Lloyd may be able to construct the ΔruvAC65 priA2∷kan double mutant and we could not construct a ruvC64∷kan priA2∷kan strain is that the ΔruvAC65 mutant is more viable. It has been shown that in a recBC sbcBC background, a ruvA60 ruvC64∷kan double mutant has a 15-fold greater viability than the ruvC64∷kan single mutant (37). Another possibility is that ΔruvAC65 deletes ruvA, yebB (a gene of unknown function), and ruvC (38) whereas ruvC64∷kan affects only ruvC. It is possible that in a priA mutant either the absence of yebB allows viability or RuvAB without RuvC causes inviability. Because McGlynn and Lloyd give no details as to the genotype, construction, or characterization of their recG priA double mutant, we are unable to comment on the differences between their results and ours.
DnaC809 Provides a Test for Different Types of Synthetic Lethality that Mutations May Have with priA2∷kan.
From the accumulated data presented in this paper and others (9, 33), it is clear that mutations in many genes are synthetically lethal with priA2∷kan. This is most easily understood in that these genes do something essential in a priA mutant. One, however, can distinguish these synthetic lethalities into at least two groups by an experimental criterion: whether or not the synthetic lethality is suppressed by dnaC809. Combinations of mutations in priA and recG, ruvC, ftsK, or recF (39) are suppressed with dnaC809. PriA with priC or rep mutations are not. One therefore can hypothesize that priC and rep have an essential role in the dnaC809 suppression of priA2∷kan where the other genes do not.
Repair of Replication Forks in a recA Mutant.
Currently, the major proposed mechanisms for repair of replication forks necessitate the process of healing the forks using homologous recombination. If so, then how can recA mutants survive? Most models also hypothesize that formation and resolution of the Holliday junction is essential for the healing process. The question then becomes how can one make Holliday junctions in a recA mutant? At least three recA-independent pathways for Holliday junction formation at a replication fork can be hypothesized. All three are presumably very inefficient and only occur to an appreciable degree in a recA mutant cell. In any given recA mutant cell, any combination of these could occur. The first is replication fork reversal (33). This is where the replication fork “backpedals,” the two newly synthesized strands are allowed to hybridize, and this in turn forms a Holliday junction. Second, in vivo overproduction of RecO protein can indirectly suppress the UV sensitivity of a recA mutant and in vitro RecO protein can catalyze D-loop formation (40). If RecO activity is concentrated in a recA mutant and if the two arms of a broken fork are in close proximity, then RecO could catalyze the strand invasion that could lead to a Holliday junction. Such a mechanism is compatible with a factory model for DNA replication (41). Lastly, Cao and Kogoma (42) showed that strains defective in recA lexA polA have significantly higher levels of UV resistance and recombination proficiency than a recA single mutant and this depended on recF.
Unusual Effects on Cell Division.
Analysis of priA and recA mutant cells is difficult because these mutations are pleiotrophic, affecting replication restart, recombination, and regulation of SOS- or LexA-regulated genes. The LexA regulon comprises at least 31 known genes involved in DNA repair, mutagenesis, and cell division through the SOS response (43). The absence of each of these genes affects SOS expression in important ways. In priA mutants, SOS levels are increased (18); whereas in the absence of recA, SOS expression cannot be induced and remain at basal levels (44). Thus to explain the combined effects of the two mutants together one has to consider not only the absence of recombination and poorer replication restart in a cell, but also changes in SOS expression. It is interesting that sulA is SOS-regulated and induced by priA2∷kan. In the recA priA double mutant, SOS induction should not occur and yet levels and sizes of the filaments actually increase. Therefore we propose that the reason filaments are more abundant and longer in a culture of the priA recA strain is that FtsK, a critical cell division protein that is induced by the SOS response, may be rate limiting (31, 45). Consistent with this proposal is that the appearance of these priA recA mutant cells are similar to the appearance of ftsK mutant cells at the nonpermissive temperature (26). Other explanations also may be possible.
Other Implications of the Data.
In this work it is postulated that the PriA-independent pathway can only restart replication forks at structures where the repair process has made non-XO. One question that arises from this way of thinking is what is the difference in structure (or what the structures are) at XOs and non-XOs that limits the PriA-independent pathway to function at the latter? A related question is what is the mechanism of PriA function that allows it to work at both structures? These remain important issues to address. Another issue raised is whether the 8- to 10-fold higher basal SOS expression in culture of priA mutant cells is due to all of the cells in culture equally having 8- to 10-fold higher levels of expression, or whether only a portion of the cells, in this case the Par− filaments, are fully induced for SOS expression and the remaining 84% are not induced at all. Lastly, this work suggests that the reason priA mutants are Rec− is that these cells cannot restart replication forks at XOs needed to form recombinants.
Acknowledgments
We thank Benedicte Michel, Peter Kuempel, Tony Poteete, Bob Lloyd, and Thomas Nystrom for supplying strains, Patrick Higgins for insight into the interpretation of these results, and Petra Levin, Joseph Chen, and William Rosche for advice in performing the microscopy. This work was supported by Grant RPG-99–194-01-GMC from the American Cancer Society. The microscope facility used in this work is supported by National Science Foundation Grant BBS 8714235.
Abbreviations
- XO
crossover
- non-XO
noncrossover
- DIC
differential interference contrast
- RRP
replication restart protein
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 3.Marians K J. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 4.Clark A J, Sandler S J. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 5.Stahl F W. Genetics. 1994;138:241–246. doi: 10.1093/genetics/138.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiner W W, Kuempel P L. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner W, Liu G, Donachie W D, Kuempel P. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 8.Sandler S J, Marians K J. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler S J. Genetics. 2000;155:487–497. doi: 10.1093/genetics/155.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bramhill D, Kornberg A. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Marians K J. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 12.McGlynn P, Al-Deib A, Liu J, Marians K, Lloyd R. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 13.Sandler S J, Samra H S, Clark A J. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Xu L, Sandler S J, Marians K J. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asai T, Kogoma T. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willetts N S, Clark A J, Low B. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee E H, Kornberg A. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurse P, Zavitz K H, Marians K J. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi E, Lutkenhaus J. J Bacteriol. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huisman O, D'Ari R, George J. J Bacteriol. 1980;144:185–191. doi: 10.1128/jb.144.1.185-191.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steiner W W, Kuempel P L. Mol Microbiol. 1998;27:257–268. doi: 10.1046/j.1365-2958.1998.00651.x. [DOI] [PubMed] [Google Scholar]
- 22.Michel B, Recchia G D, Penel-Colin M, Ehrlich S D, Sherratt D J. Mol Microbiol. 2000;37:180–191. doi: 10.1046/j.1365-2958.2000.01989.x. [DOI] [PubMed] [Google Scholar]
- 23.Cromie G A, Leach D R F. Mol Cell. 2000;6:815–826. doi: 10.1016/s1097-2765(05)00095-x. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd R G, Low K B. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2236–2255. [Google Scholar]
- 25.Kuempel P L, Henson J M, Dircks L, Tecklenburg M, Lim D F. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- 26.Begg K J, Dewar S J, Donachie W D. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X C, Weihe E K, Margolin W. J Bacteriol. 1998;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Recchia G D, Aroyo M, Wolf D, Blakely G, Sherratt D J. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle D S, Grant D, Draper G C, Donachie W D. J Bacteriol. 2000;182:4124–4127. doi: 10.1128/jb.182.14.4124-4127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Draper G C, Donachie W D. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 31.Diez A, Gustavsson N, Nystrom T. Mol Microbiol. 2000;36:1494–1503. doi: 10.1046/j.1365-2958.2000.01979.x. [DOI] [PubMed] [Google Scholar]
- 32.Diez A A, Farewell A, Nannmark U, Nystrom T. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 34.van Gool A J, Hajibagheri N M, Stasiak A, West S C. Genes Dev. 1999;13:1861–1870. doi: 10.1101/gad.13.14.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd R G. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 37.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudd K E. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandler S J. Mol Microbiol. 1996;19:871–880. doi: 10.1046/j.1365-2958.1996.429959.x. [DOI] [PubMed] [Google Scholar]
- 40.Luisi-DeLuca C. J Bacteriol. 1995;177:566–572. doi: 10.1128/jb.177.3.566-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemon K P, Grossman A D. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y, Kogoma T. Genetics. 1995;139:1483–1494. doi: 10.1093/genetics/139.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez De Henestrosa A R, Ogi T, Aoyagi S, Chafin D, Hayes J J, Ohmori H, Woodgate R. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 44.Walker G. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1400–1416. [Google Scholar]
- 45.Wang L, Lutkenhaus J. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 46.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Csonka L N, Clark A J. Genetics. 1979;93:321–343. doi: 10.1093/genetics/93.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandler S J, Clark A J. J Bacteriol. 1994;176:3661–3672. doi: 10.1128/jb.176.12.3661-3672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zieg J, Kushner S R. J Bacteriol. 1977;131:123–132. doi: 10.1128/jb.131.1.123-132.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy K C, Campellone K G, Poteete A R. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]