Abstract
The production of small RNAs (sRNAs) from phased positions set by microRNA-directed cleavage of trans-acting-siRNA-producing locus (TAS) transcript has been characterized extensively; however, the production of sRNAs from non-phased positions remains unknown. We report three cis-cleavages that occurred in TAS3 transcripts in Vitis vinifera, by combining high-throughput sRNA deep sequencing information with evolutional conservation and genome-wide RNA degradome analysis. The three cis-cleavages can be deciphered to generate an orderly cleavage cascade, and can also produce distinct phasing patterns. Each of the patterns, either upstream or downstream of the cis-cleaved position, had a set of sRNAs arranged in 21-nucleotide increments. Part of the cascading cis-cleavages was also conserved in Arabidopsis thaliana. Our results will enhance the understanding of the production of sRNAs from non-phased positions that are not set by microRNA-directed cleavage.
Keywords: trans-acting siRNA, cis-cleavage, ta-siRNA-producing locus, miRNA
1. Introduction
In plants, many endogenous small RNAs (sRNAs), including microRNAs (miRNAs), heterochromatic small interfering RNAs (siRNAs), natural antisense siRNAs, and trans-acting siRNAs (ta-siRNAs), play important roles in regulating gene expression networks [1,2]. The sRNAs are also valuable tools for functional genomics studies. Usually, the sRNAs silence gene expression by either degrading mRNA or repressing translation but, in a few cases, they also generate a population of secondary siRNAs. The ta-siRNAs are secondary siRNAs that are produced by a miRNA-targeted trigger that bridges the pathways of miRNA and siRNA regulation. ta-siRNAs can regulate plant development, metabolism, and responses to biotic and abiotic stresses, and thus have received more attention in the recent decade [3–5].
During the biogenesis of ta-siRNA, a single-stranded RNA is transcribed from a ta-siRNA-producing locus (TAS) and then cleaved by a phase-initiator (a miRNA or, in some cases, a ta-siRNA). Then, RNA-dependent RNA polymerase 6 (RDR6)-dependent conversion of the resulting fragments into double-stranded RNA and its subsequent cleavage by dicer-like 4 (DCL4) at every ~21 nucleotide (nt) relative to the phase-initiator cleavage site generates ~21-nt phased sRNAs. Some of the phased sRNAs become ta-siRNAs by binding argonaute (AGO) proteins to direct a trans-cleavage of targeted mRNAs [5–7]. Plant TASs can be classified into at least eight families, based on initiator-dependence, sequence similarity, and target gene identity. TAS1 and TAS2 are targets of miR173 and their ta-siRNAs can target the pentatricopeptide repeat genes [8]. TAS3 is a target of miR390 and its ta-siRNA can target the auxin response factor gene family [8]. The initiator of TAS4 is miR828, and the TAS4 ta-siRNA can target the MYB transcription factor gene family [9]. TAS5 is triggered by miR482 and its ta-siRNA can target the Bs4 resistance gene [10]. miR156 and miR529 initiate TAS6, which targets an mRNA that encodes a zinc finger protein [11]. miR828 initiates TAS7, which can target 13 genes, including genes that encode the leucine-rich receptor protein kinase-like protein and a calcium-transporting ATPase [12]. At1g63130 is a pentatricopeptide repeat gene that was reported to be cleaved by TAS2-derived ta-siR2140 [13]. TAS3 is flanked by two miR390 binding sites; one of which can be cleaved by the interaction of miR390 and AGO7, and another that is non-cleavable. Both binding sites are critical for the biogenesis of the TAS3 ta-siRNAs. In contrast, other TASs have only a single miRNA binding site and are cleaved by the interaction of miRNA/ta-siRNA and AGO1. Recently, AGO2 has been reported to mediate cis-cleavage of TAS1c, although its slicer activity has not been demonstrated so far [14]. Taken together, it might be expected that many of the sequenced sRNAs could be mapped onto the phased positions set by the phase-initiator. Yet, while many of the sRNAs were successfully mapped, unexpectedly, many were mapped onto non-phased positions; that is, the intervals between the phased positions [12,13]. These sRNAs have been called “non-phased sRNAs” and how they are produced remains unclear.
The recent publication of the degradome library generated from cleaved mRNA fragments and sRNA libraries generated by high-throughput deep sequencing has enabled the study of all the cleavages that occur in a TAS transcript [15,16]. Here, we studied cis-cleavage of grapevine TAS3 and found a cascading cis-cleavage, which produce sRNAs from the so-called non-phased positions and broaden the known scope of non-phased sRNA production.
2. Results and Discussion
2.1. Overview of Small RNA Distribution on TAS3 from Vitis vinifera
Previously we reported that the TAS3 from Vitis vinifera (vviTAS3) can be targeted by vvi-miR390 to trigger ta-siRNA production in grapevine [12]. Here, to determine the distribution of sRNAs on the vviTAS3 transcript, a sRNA library from grapevine leaves (GEO: GSM458927) was used. To improve mapping confidence, only the sRNAs that mapped to a single site on the whole V. vinifera genome were used, because they could be attributed with certainty to a particular locus.
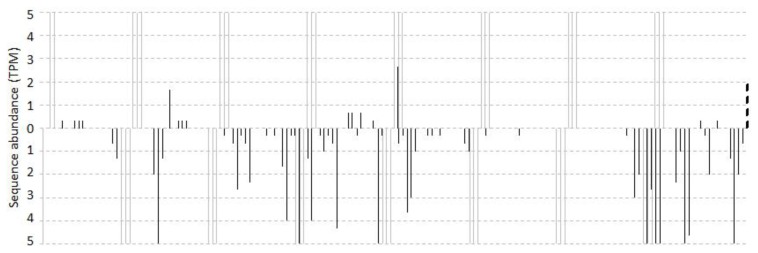
As a result of the mapping, we detected 131 unique sRNAs, representing 3969 reads, which matched perfectly to vviTAS3 (Figure 1). The 5′ ends of the reads occupied 79 positions on the transcript. Only 14 (18%) of the positions were found to belong to phased positions set by vvi-miR390 when a 1-nt offset from the phased positions was allowed. After filtering out sRNAs that had TPM (tags per million) values of five or less, some unique sRNAs that mapped to non-phased positions remained (Figure 1). The percentage of phased sRNA positions increased from 18% to 40%. Together, these results showed that some sRNAs are really generated from non-phased positions, and might even be functional because of the relatively high levels at which they are often expressed.
Figure 1.
Abundance distribution of sRNAs mapped to vviTAS3. The number of reads with a 5′ end at each position is plotted. Bars above the sequence represent sense reads; those below represent antisense reads. The cleavage site of vvi-miR390 is marked by a vertical dotted line. Vertical gray lines indicate the miRNA-set 21-nt phased cleavage, allowing a 1-nt offset. Regions with TPMs greater than five are not shown.
2.2. Computational Prediction and Validation of cis-Cleavages
Recent reports have shown that many functional siRNAs belong to a class of 21–22-nt 5′U/A sRNAs [14]. Therefore, we filtered out the potential siRNAs from the mapped sRNAs by limiting the length of the reads to 21 nt and the 5′ end to U/A. As a result, we detected 35 sRNAs that were mapped to the antisense strand that passed the rule.
Additionally, it has been reported that many functional cleaved positions tend to be conserved through evolution, and they have been found to be highly conserved in alignments of genomic sequences from different species [8]. To identify conserved positions in the vviTAS3 transcript, we compiled a dataset of TAS3 sequences from eight dicotyledonous plants; namely, V. vinifera, Ricinus communis, Populus trichocarpa, Arabidopsis thaliana, Malus domestica, Fragaria vesca, Prunus persica, and Glycine max, and aligned them using ClustalX2 [17] with the default parameters (Figure 2). The multiple sequence alignment showed that there are no insertions or deletions among the TAS3 sequences, except for a one-nucleotide deletion in vviTAS3 between position 116 and 117. The conserved positions were filtered by requiring each position to be conserved in at least six of the species, and to correspond to the 10th position of the 35 candidate cis-acting sRNAs. We found that 19 of the 35 sRNAs passed these rules.
Figure 2.
Alignment of TAS3 DNA sequences from eight dicotyledonous plant species. The numbers in the last row indicate the number of species with the same nucleotide as vviTAS3 at each of the marked positions. Only numbers over six are shown. vvi, Vitis vinifera; rco, Ricinus communis; ptr, Populus trichocarpa; ath, Arabidopsis thaliana; mdo, Malus domestica; fve, Fragaria vesca; ppe, Prunus persica; gma, Glycine max.
Finally, using a parallel sequencing sRNA library that contained degradome tags from grapevine leaves [18], we validated the predicted cis-cleaved positions on the vviTAS3 transcript by requiring that the cis-cleaved positions overlapped with the 5′ end of the RNA degradation fragment mapped onto the TAS. As a result, three of the positions, 63, 85, and 138, were validated. The corresponding cis-acting siRNAs (ca-siRNAs) were three 21-nt 5′U ca-siRNAs and one 22-nt 5′U ca-siRNA (Table 1). It has been reported that the size of the sRNAs and the 5′-terminal nucleotide are critical for the sorting of AGO. AGO1 binds 21-nt 5′U sRNAs, but in some cases, it also binds 22-nt 5′U sRNAs [14]. In Arabidopsis, the 22-nt 5′U 3′D10(−) from TAS1c has been reported to mediate its cis-cleavage by binding to AGO1, and the 21-nt 5′U 3′D6(−) from TAS1c has also been shown to mediate TAS1c cleavage by binding to AGO1; however, in this case, the cleaved site is not its original site [14]. In this study, we found three ca-siRNAs that were 21-nt 5′U sRNAs and one that was a 22-nt 5′U sRNA. These results implied that the four ca-siRNAs were all loaded to AGO1.
Table 1.
Validated cis-cleaved positions on vviTAS3 transcript and their ca-siRNAs.
| cis-cleaved position | ca-siRNA | |
|---|---|---|
| Name | Sequence | |
| 63 | ca-siRM72 | UGGAAAACGAAGAAGAAAUGC |
| 85 | ca-siRM94 | UGGUAUAGAGUUCAUGACAAG |
| 138 | ca-siRM147 | UGAGUUGGGCGGAAACGGGGA UGAGUUGGGCGGAAACGGGGAA |
It has been demonstrated that two miR390 binding sites located on each side of TAS3 are critical for TAS3 ta-siRNAs biogenesis [8]. Therefore, we looked for ca-siRNA targeting sites on vviTAS3 and the flanking 300 bp upstream and downstream of the gene where the two miR390 binding sites were located. We found one targeted site on vviTAS3 for each of the ca-siRNAs. This finding suggested that cis-cleavage might use a different mechanism from the mechanism used by miR390 to initiate the cleavage of vviTAS3.
We used the same method and criteria to examine the antisense strand that was not targeted by miR390. Surprisingly, no ca-siRNA targeting sites were detected on the antisense strand of vviTAS3. The asymmetrical distribution between the targeted and non-targeted strand might imply that the non-targeted strand is readily in a double-stranded RNA form and is constantly processed by DCL4 and, therefore, protected from cis-cleavage. In addition, it might support the hypothetically biological function of cis-cleavage, i.e., inactivating TAS transcription to feedback control ta-siRNA’s production, as the sense strand was a template strand synthesizing antisense strand, so it would be more effective when the cis-cleavages preferred to occur in sense strand rather than in antisense strand.
2.3. Cascading cis-Cleavages
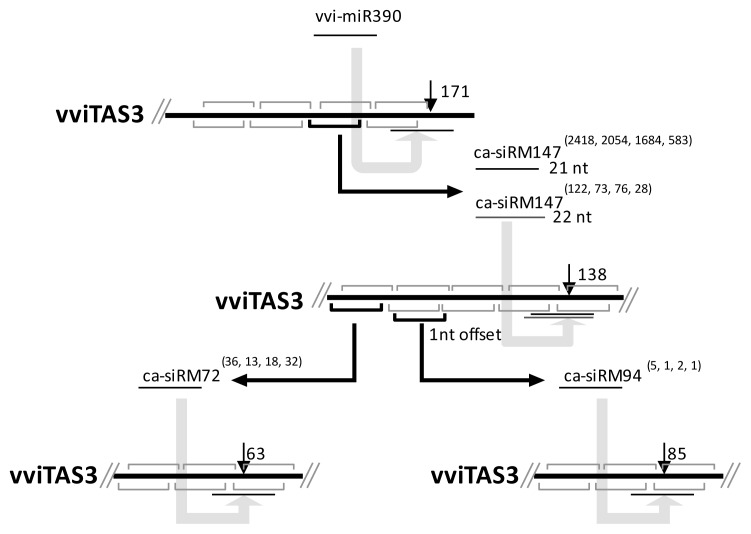
After identifying the cis-cleaved positions and their ca-siRNAs, we investigated how ca-siRNA production is triggered. It has been shown that ca-siRM147 can be triggered by miR390 [12], but for other ca-siRNAs the triggers remain unclear because they are out of the register set by miR390. To identify possible initiators, we investigated the distribution of cis-cleaved position and the locations of ca-siRNA 5′ ends on the vviTAS3 transcript. We found that the 5′ end of ca-siRM72 occurred precisely at the register set by ca-siRM147 in which the cleaved site of ca-siRM147 located on sense strand was 65 nt away from the 5′ end of ca-siRM72 on antisense strand. The 5′ end of ca-siRM94 was offset by 1 nt from the register set by ca-siRM147. Because the cleavage of phasing sRNAs often occurs within 1–2 nt of the phased position [8,13], we propose that the production of ca-siRM147, ca-siRM72, and ca-siRM94 is triggered by miR390, ca-siRM147, and ca-siRM147, respectively. The cascading cis-cleavage that we have proposed is shown schematically in Figure 3.
Figure 3.
Cleavage cascades generated by cis-cleavages of siRNA on the sense strand of vviTAS3. The four numbers in the brackets indicate the raw abundance of ca-siRNA in grapevine berries, leaves, inflorescences, and tendrils, respectively.
2.4. The Accumulated Levels of ca-siRNAs
When we analyzed the accumulated levels of the ca-siRNAs in the grapevine leaf, berry (GEO: GSM458930), inflorescence (GEO: GSM458929), and tendril (GEO: GSM458928) libraries, we found that the levels were in agreement with the cleavage cascades. The abundance of the ca-siRNAs that were located upstream of the cascade was always higher than the abundance of the ca-siRNAs that were downstream. For example, in the grapevine leaf library, the 21-nt ca-siRM147 located upstream had 2054 sequenced reads, while ca-siRM72, located downstream of ca-siRM147 cleavage, had only 13 sequenced reads. Moreover, the ca-siRNAs that occurred precisely at the register were always more abundant than those that occurred out of the register. For example, in the grapevine leaf library, ca-siRM72 had 13 sequenced reads, while ca-siRM94, which was shifted by 1 nt from the phased positions set by ca-siRM147, had only one sequenced read. Similar results were obtained for the other three tissues (Figure 3).
2.5. cis-Cleavages Produced sRNAs in Increments of Approximately 21 nt
To test whether or not cis-cleavages can also produce phased sRNAs in increments of approximately 21 nt, we searched for sRNAs with 5′ ends that overlapped the predicted phased and non-phased positions by allowing an offset of 1 nt. As expected, each cis-cleavage had a set of corresponding phased sRNAs arranged in ~21-nt increments upstream and downstream of the cleavage position (Table 2).
Table 2.
Phased patterns set by different cis-cleavages.
| cis-cleaved position | Upstream of cis-cleaved position | Downstream of cis-cleaved position | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Number of phased sRNAs | Number of non-phased sRNAs | p value * | Number of phased sRNAs | Number of non-phased sRNAs | p-value | |
| 63 | 2 | 12 | 1.11 × 10–1 | 3 | 47 | 4.92 × 10–1 |
| 85 | 5 | 27 | 7.54 × 10–3 | 4 | 30 | 1.00 × 10–1 |
| 138 | 8 | 41 | 4.71 × 10–4 | 3 | 16 | 7.43 × 10–2 |
p-values less than 0.01 are in bold font.
To test whether or not the phased patterns produced from cis-cleavages were statistically significant, we developed an improved equation (see Experimental Section) by modifying previous algorithms [12,13,19] to evaluate the phasing pattern set by cis-cleavage. First, the new equation is not constrained by the previous 231-bp length requirement [12,13,19], but requires only a multiple of 21 nt, which provides a more accurate TAS evaluation, especially for TASs that are longer or shorter than 231 bp. Second, our equation uses a variable s to reflect the maximum offset from a phase position [12,19], making the evaluation more flexible. This equation could be applied to TAS identification in the future. Using our improved algorithm [12,13,19], we determined that two cis-cleavages had significant phasing patterns (p-value < 0.01). When the number of sRNAs located in the phased positions set by ca-siRNAs was counted, we found that 54 unique sRNA located in non-phased positions set by miRNA390 were included in the phasing patterns. These results suggested that some common processes might be used for both miRNA-mediated ta-siRNA production and the cis-cleavage of siRNA.
2.6. The Conservation of ca-siRNAs and Cascading cis-Cleavages
To examine the conservation of cis-cleavages further, we first looked for the presence of ca-siRNAs in the sRNA datasets of V. vinifera, A. thaliana, M. domestica, and P. persica downloaded from the Gene Expression Omnibus (GEO) or the plant MPSS databases. We found that although the accumulation levels of the four ca-siRNAs varied in the different species, they were expressed in all four species, except for 22-nt ca-siRNA147, which was not detected in P. persica (peach) (Table 3).
Table 3.
Conservation of ca-siRNAs in the sRNA datasets of four species.
| ca-siRNAs | Average of normalized abundance (TPM) | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Name | Length | Grapevine | Apple | Peach | Arabidopsis |
| ca-siRM72 | 21 nt | 6 | 2 | 1 | 1 |
| ca-siRM94 | 21 nt | 1 | 3 | 14 | 1 |
| ca-siRM147 | 2 1nt | 419 | 32 | 9 | 2 |
| 22 nt | 18 | 1 | 0 | 2 | |
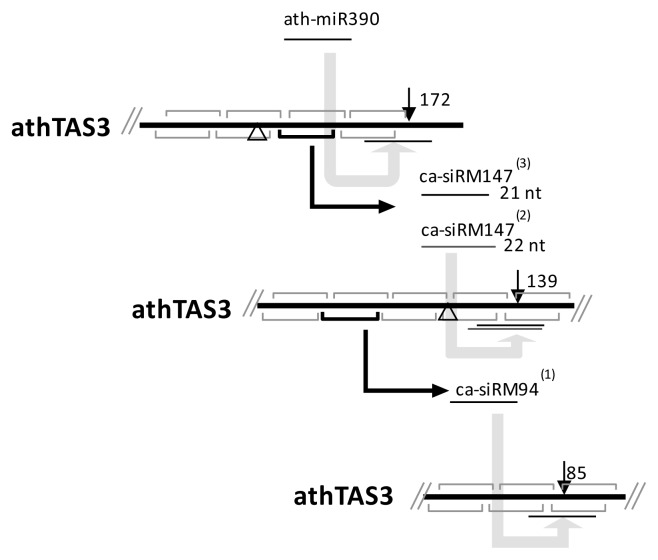
We then evaluated the corresponding cis-cleavages based on the Col7d samples (GEO: GSE20197) and the degradome library (GEO: GSM280227) from Arabidopsis, and found that, two cis-cleavages occurred in positions 85 and 139 (equivalent to position 138 in vviTAS3 because of the nucleotide deletion in the vviTAS3 sequence) were also validated on athTAS3 (Figure 4). The accumulated levels of ca-siRNAs in Col7d sample were also in agreement with the cleavage cascades. In which, the 21-nt and 22-nt ca-siRM147 located upstream had, respectively, three and two sequenced reads, while ca-siRM94, located downstream of ca-siRM147 cleavage, had one sequenced reads. In a previous study [5], it was suggested that cis-cleavage occurred on position 139 in athTAS3, although the ca-siRNA and functional sRNAs were not found. Here, we detected the ca-siRNA and a secondary ca-siRNA product (ca-siRNA94), which we believe provides enough evidence to establish the cis-cleavage on TAS3. The findings reported here for athTAS3 indicate that cascading cis-cleavage is conserved.
Figure 4.
Cleavage cascades generated by cis-cleavages of siRNA on the sense strand of athTAS3. The triangle indicates the position of the nucleotide deletion in vviTAS3. The number in the bracket indicates the raw abundance of ca-siRNA in Col7d (GEO: GSE20197).
3. Experimental Section
3.1. Sources of sRNA Libraries
In this study, we used four deep sequencing sRNA datasets; namely, two degradome library and two sRNA libraries. All the datasets were downloaded from the Gene Expression Omnibus (GEO). The GEO accession numbers for these libraries are given in the Results section.
3.2. Evaluation of Phasing Patterns Set by cis-Cleavage
Once a cis-cleaved position was determined, the numbers of phased and non-phased positions were counted upstream and downstream of the cleavage sites respectively. Phased positions refer to positions arranged in 21-nt increments relative to the cleavage position as well as to positions shifted by s nt relative to the positions of 21-nt increments. Non-phased positions are all the other positions. The p-value of each detected phasing pattern was calculated based on a random hyper-geometric distribution using an improved equation based on previously used algorithms [12,13,19,20]:
| (1) |
Where L is the length of the detected pattern and is a multiple of 21, K1 is the number of phased positions having sRNA hits, K2 is the number of non-phased positions having sRNA hits, and s is the maximum allowed offset from the phase position.
3.3. Expressional Conservation of ca-siRNAs
The expressional conservation of the grapevine ca-siRNAs was investigated by performing a search against 84 sRNA libraries from grapevine, apple, Arabidopsis, and peach. The sRNA libraries of grapevine (GEO: GSE18405) and apple (GEO: GSE36065) were downloaded from the GEO and the sRNA libraries of Arabidopsis and peach were used from the MPSS databases [21]. The normalized abundance is the raw expression value divided by the total number of signatures and multiplied by 1,000,000.
4. Conclusions
In this work, we reexamined the distribution of sRNAs on vviTAS3 using a stringent threshold that used only the sRNAs that mapped to a single site on the whole V. vinifera genome and that had normalized abundant values of one or more TPM. Our results showed that the non-phased positions were indeed located by some of the uniquely mapped sRNAs. We identified three cis-cleavages that directed by four ca-siRNAs at positions 63, 85, and 138 on vviTAS3 by combining computational predictions and validation. We found that three cis-cleavages, together with their ca-siRNAs, formed a cascading cis-cleavage. The accumulated levels of four ca-siRNAs in the berry, leaf, inflorescence, and tendril libraries of V. vinifera also agreed with the cascade. A comparative analysis showed that the expression levels of the four ca-siRNAs were conserved among grapevine, apple, peach, and Arabidopsis, and part of the cis-cascade was also identified in Arabidopsis. We also found that sRNAs were located at the phased positions set by ca-siRNA. These results broaden the known scope of non-phased sRNA production. We also developed an improved equation by modifying previous algorithms to evaluate the phasing pattern set by cis-cleavage. It could be applied to TAS identification in the future.
Acknowledgments
This work was supported by the National Science Foundation of China (Grant No. 31171273) and the Qing Lan Project of Jiangsu Province, China.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Axtell M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 2.Chen X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Allen E., Xie Z., Gustafson A.M., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F., Orban R., Baker B. SoMART: A web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 2012;70:891–901. doi: 10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- 11.Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant. 2012;5:1281–1294. doi: 10.1093/mp/sss036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C., Li G., Wang J., Fang J. Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics. 2012;28:2561–2568. doi: 10.1093/bioinformatics/bts500. [DOI] [PubMed] [Google Scholar]
- 13.Chen H.M., Li Y.H., Wu S.H. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajeswaran R., Aregger M., Zvereva A.S., Borah B.K., Gubaeva E.G., Pooggin M.M. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 2012;40:6241–6254. doi: 10.1093/nar/gks242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.German M.A., Pillay M., Jeong D.H., Hetawal A., Luo S., Janardhanan P., Kannan V., Rymarquis L.A., Nobuta K., German R., et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 17.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 18.Pantaleo V., Szittya G., Moxon S., Miozzi L., Moulton V., Dalmay T., Burgyan J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. Cell Mol. Biol. 2010;62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 19.Dai X., Zhao P.X. pssRNAMiner: A plant short small RNA regulatory cascade analysis server. Nucleic Acids Res. 2008;36:W114–W118. doi: 10.1093/nar/gkn297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Wang J., Hua X., Fang J., Zhu H., Gao X. A mutation degree model for the identification of transcriptional regulatory elements. BMC Bioinforma. 2011;12 doi: 10.1186/1471-2105-12-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano M., Nobuta K., Vemaraju K., Tej S.S., Skogen J.W., Meyers B.C. Plant MPSS databases: Signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]