Abstract
Lack of the yeast Rrm3p DNA helicase causes replication defects at multiple sites within ribosomal DNA (rDNA), includingat the replication fork barrier (RFB). These defects were unaltered in rrm3 sir2 cells. When the RFB binding Fob1p was deleted, rrm3-generated defects at the RFB were eliminated, but defects at other rDNA sites were not affected. Thus, specific protein–DNA complexes make replication Rrm3p-dependent. Because rrm3-induced increases in recombination and cell cycle length were only partially suppressed in rrm3 fob1 cells, which still required checkpoint and fork restart activities for viability, non-RFB rrm3-induced defects contribute to rDNA fragility and genome instability.
Keywords: rDNA, helicase, replication, Sir2p, Rrm3p, Fob1p
Completion of DNA replication is critical for cell viability, yet many factors such as DNA damage, transcribing RNA polymerases, and protein complexes can impede the progress of replication forks. The yeast Rrm3p, a 5′-to-3′ DNA helicase (Ivessa et al. 2002), is needed for efficient fork progression at ∼1400 discrete sites throughout the genome. In its absence, replication forks pause at multiple sites in the ribosomal DNA (rDNA; Ivessa et al. 2000) as well as at telomeres (Ivessa et al. 2002), tRNA genes, centromeres, inactive replication origins, and the silent mating type loci (Ivessa et al. 2003). These paused replication forks have a propensity to break, which probably accounts for the increased recombination (Keil and McWilliams 1993; Ivessa et al. 2000, 2002, 2003) and elevated Ty1 transposition (Scholes et al. 2001) seen in rrm3 cells. Because of their widespread replication defects, rrm3 cells require the intra-S-phase checkpoint and fork repair activities for viability (Ivessa et al. 2003; Schmidt and Kolodner 2004; Torres et al. 2004).
Yeast rDNA exists as a single locus of ∼150 tandem repeats. The tandem nature of the rDNA array makes possible several modes of intrachromosomal recombination, including the Rad52p-dependent liberation of rDNA circles (Kim and Wang 1989; Park et al. 1999). Each rDNA repeat contains the Polymerase (Pol) I transcribed 35S rRNA gene and the Pol III transcribed 5S rRNA gene (Fig. 1A). The activity of Sir2p, a histone deacetylase that is rDNA-associated, makes rDNA chromatin structure more compact, which reduces Pol II transcription and rDNA recombination, including the generation of rDNA circles (for review, see Ivessa and Zakian 2002; Rusche et al. 2003).
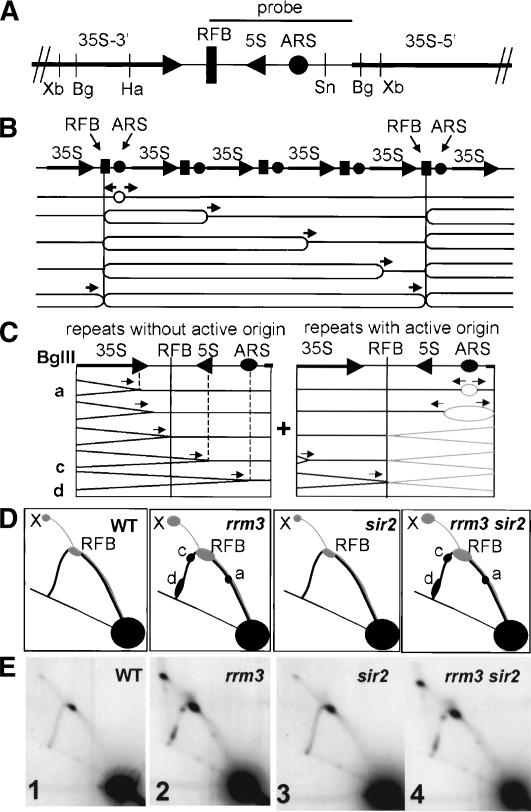
Figure 1.
rDNA replication in Saccharomyces cerevisiae. (A) Schematic of a single repeat of yeast rDNA. Shown are recognition sites for the restriction enzymes used [(Bg) BglII; (Xb) XbaI; (Ha) HaeII; (Sn) SnaBI] and the position of the probe used for all 2D gels. (B) Schematic of yeast rDNA replication (Brewer and Fangman 1988; Linskens and Huberman 1988). For simplicity the 5S rDNA gene is not shown. The large arrows indicate 35S rRNA genes, the rectangles indicate the RFB, and the circles represent the ARSs. (C) Schematics of BglII-digested rDNA. The left panel indicates the replication intermediates from forks initiated in upstream repeats. Vertical lines and lowercase letters indicate sites of replication fork pausing in rrm3 cells. The right panel shows a repeat with an active origin of DNA replication. Intermediates in gray are from leftward-moving forks and intermediates in black are from rightward-moving forks. (D) The 2D gel schematics show the pattern of replication intermediates in the indicated strains. The simple Y replication arc is a composite of rightward-moving forks from repeats without an active origin (black) and forks from repeats with an active origin (gray). Forks arrested (RFB) and converged (labeled X) at the RFB are indicated. (E) Sir2p does not contribute to rrm3-dependent pauses. 2D gels of BglII-digested rDNA from the indicated strains. (Panel 1) Wild type. (Panel 2) rrm3. (Panel 3) sir2. (Panel 4) rrm3 sir2. For these and subsequent 2D replication gels, digested DNA was Southern blot transferred and hybridized with the probe indicated in A.
Each rDNA repeat also contains a potential origin of DNA replication (ARS), although only ∼20% of the ARSs are active in a given S phase (Fig. 1B; Brewer and Fangman 1988; Linskens and Huberman 1988). Like rDNA recombination and Pol II transcription, origin activation is suppressed by Sir2p (Pasero et al. 2002). Replication of repeats with active origins is initially bidirectional (Brewer and Fangman 1988; Linskens and Huberman 1988). However, when the leftward-moving fork encounters the replication fork barrier (RFB), a cis-acting sequence near the 3′-end of the 35S transcription unit, it arrests. Fork arrest at the RFB is dependent on the RFB binding Fob1p (Kobayashi and Horiuchi 1996; Huang and Moazed 2003). At the end of rDNA replication, rightward-moving forks converge on forks arrested at the RFB, completing rDNA replication. Because of the RFB, rDNA is the only region of the yeast genome where replication is largely unidirectional. Fob1p is also required for most rDNA recombination, including generation of rDNA circles (Defossez et al. 1999; Johzuka and Horiuchi 2002).
rDNA replication is markedly altered in rrm3 cells (Ivessa et al. 2000). The abundance of forks converged at the RFB is elevated 10-fold without an increase in replication initiation, and forks arrested or converged at the RFB are more likely to break. In addition, rightward-moving forks pause near the beginning and end of the 35S gene, at the 5S rRNA gene, and at inactive ARSs. These replication defects and DNA breakage are associated with elevated rDNA recombination, including a large increase in rDNA circles (Keil and McWilliams 1993; Ivessa et al. 2000). Rrm3p, which is rDNA-associated in vivo, acts catalytically and probably directly to promote rDNA replication (Ivessa et al. 2000).
Sites whose replication depends on Rrm3p are assembled into stable, nonnucleosomal protein–DNA complexes. One model to explain the effects of Rrm3p on DNA replication is that the Rrm3p DNA helicase promotes movement of replication forks past protein–DNA complexes. This model predicts that disruption of the protein complex at a given site will render replication of that site Rrm3p-independent. In this paper, we test this hypothesis using rDNA. We report that Fob1p is required to cause rrm3-induced replication defects at the RFB. However, pausing at other sites in the rDNA, was unaffected by loss of Fob1p. Additionally, deletion of FOB1 did not eliminate the increased recombination, cell cycle progression defects, or synthetic lethal interactions seen in rrm3 cells. These data suggest that local protein–DNA complexes make rDNA replication-dependent on Rrm3p and show that Rrm3p-dependent pauses at sites other than the RFB contribute to the rDNA fragility and genome instability of rrm3 cells.
Results and Discussion
The histone deacetylase Sir2p represses transcription in three regions—rDNA, telomeres, and the silent mating type loci—where replication is Rrm3p-dependent (Rusche et al. 2003). To determine if Sir2p-mediated chromatin modifications render rDNA replication dependent on Rrm3p, we used two-dimensional (2D) gel electrophoresis to examine rDNA replication in rrm3 sir2 cells. If silent chromatin is responsible for rrm3-dependent pausing, then replication fork progression should be unimpeded in rrm3 sir2 cells. However, rDNA replication was indistinguishable in rrm3 and rrm3 sir2 cells (Fig. 1C–E). Therefore, the deacetylase Sir2p and its associated regional chromatin changes did not make the replication of rDNA Rrm3p-dependent.
Next we asked if site-specific protein–DNA complexes confer Rrm3p-dependent replication. FOB1 encodes a 65-kD protein that binds to the RFB and is required for RFB activity and most rDNA recombination (Kobayashi and Horiuchi 1996; Kobayashi et al. 1998; Huang and Moazed 2003). Fob1p is also required for the RFB binding of Sir2p and Net1p, two proteins that affect transcriptional repression in the rDNA (Huang and Moazed 2003). Thus, Fob1p is needed for the formation of a multiprotein complex at the RFB.
In wild-type cells, the RFB is a polar block to fork progression as only leftward-moving forks stop at the RFB (Brewer and Fangman 1988; Linskens and Huberman 1988). In rrm3 cells, the fraction of replication intermediates at the RFB in BglII-digested DNA is twice that of wild-type cells (Ivessa et al. 2000). As ∼90% of leftward-moving replication forks in wild-type cells arrest at the RFB (Brewer et al. 1992), the twofold increase in rrm3 cells cannot be explained by increased numbers of leftward-moving forks arresting at the RFB. Because leftward- and rightward-moving forks stopped at the RFB migrate to the same position in BglII-digested DNA (Fig. 1C), this increase is likely due to rrm3-dependent pausing of rightward-moving forks at the RFB.
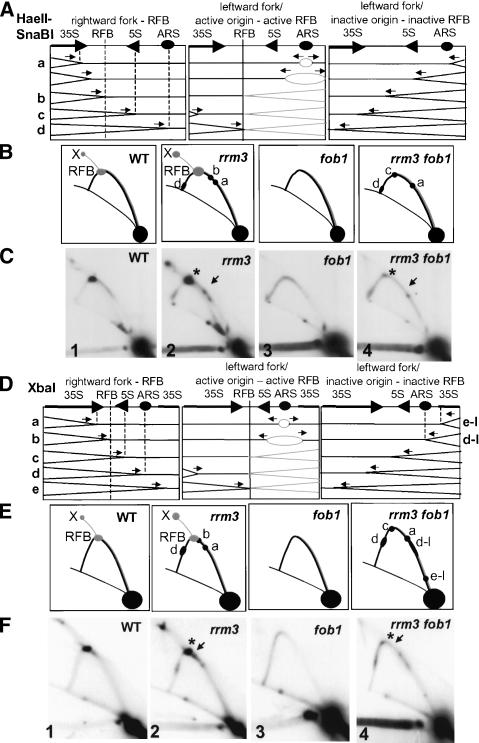
To establish that rightward-moving forks pause at the RFB in rrm3 cells, we examined DNA digested with both HaeII and SnaBI (Fig. 2A–C). In this double digest, the positions of rightward- and leftward-moving forks stopped at the RFB are easily distinguished. In wild-type cells, forks arrested and converged at the RFB (Fig. 2B,C, panel 1), and these structures were absent in fob1 cells (Fig. 2B,C, panel 3). DNA from rrm3 cells showed the expected increase in converged forks (labeled X) and pauses at the ARS (pause d) and near the 35S terminator (pause a; Fig. 2B,C, panel 2). In addition, a pause between the 35S terminator and the RFB was evident (Fig. 2C, panel 2, pause b indicated by arrow). This pause was ∼40% of the way through the fragment, the expected position for rightward-moving forks paused at the RFB (Fig. 2A, left panel). Leftward-moving forks arrested at the RFB, expected at ∼60% of the way through the fragment, generated the strong signal at the structure labeled RFB (Fig. 2C, panel 2, marked with an asterisk). Rightward-moving forks paused at the 5S gene were not visible in this digest because they comigrate with leftward-moving forks arrested at the RFB.
Figure 2.
Fob1p is required for rrm3-dependent pausing at the RFB but not at other rDNA sites. (A) Schematics of HaeII–SnaBI-digested rDNA. The first panel indicates replication forks initiated in upstream repeats. The middle pane indicates replication forks in repeats with an active origin and an active RFB. The right panel ndicates forks initiated in downstream repeats in an fob1 strain. These forks are only seen when there is no active RFB, as in fob1 cells. Vertical lines and lowercase letters indicate where replication forks pause in an rrm3 strain. (B) The 2D gel schematics show the pattern of replication intermediates in the indicated strains. Intermediates in gray are from forks initiated within the repeat, and intermediates in black are rightward-moving forks. When Fob1p is deleted, leftward-moving forks are also shown in black. Lowercase letters indicate the specific pause sites illustrated in A. (C) 2D gels of HaeII–SnaBI-digested rDNA. (Panel 1) Wild type. (Panel 2) rrm3. (Panel 3) fob1. (Panel 4)rrm3 fob1. The arrows in panels 2 and 4 indicate the expected position of the rightward-moving replication forks pausing at the RFB. The asterisks indicate the location of leftward-moving forks arrested at the RFB. (D–F) The same as A, B, and C except XbaI-digested DNA was examined. Pauses d-l and e-l are at the same sites as pauses d and e but are generated by leftward-moving forks moving through these sites.
If the presence of Fob1p at the RFB makes replication through this site Rrm3p-dependent, then rightward- and leftward-moving forks should not stop at the RFB in rrm3 fob1 cells. Consistent with this possibility, in rrm3 fob1 cells, the strong signal caused by leftward-moving forks arrested at the RFB was not detectable in HaeII–SnaBI-digested rDNA (Fig. 2C, panel 4, position marked by asterisk). The minor pause visible at this site can be attributed to rightward-moving forks pausing at the 5S rRNA gene. Likewise, rightward-moving forks no longer paused at the RFB (Fig. 2C, panel 4; arrow indicates site of paused rightward-moving fork in rrm3 cells). However, forks still paused at the inactive ARS (pause d) and near the end of the 35S rRNA gene (pause a) as well as at the 5S gene (pause c) in rrm3 fob1 cells. The same conclusions are reached by examining XbaI-digested DNA (Fig. 2D–F, panel 4; asterisk marks site of arrest of leftward-moving forks at the RFB; arrow marks site of pause of rightward-moving forks at the RFB). Although leftward-moving forks did not arrest and rightward-moving forks did not pause at the RFB in rrm3 fob1 DNA, the pauses at the end of the 35S rRNA gene (pause a), at the 5S gene (pause c), and at the inactive ARS (pause d) were not Fob1p-dependent (Fig. 2E,F, panel 4). Additionally, two new pauses were detected (d-l and e-l). These pauses can be attributed to leftward-moving forks paused at the start of the 35S transcription unit (pause e-l) and at the inactive ARS (pause d-l). These pauses are not seen in FOB1 rrm3 cells, because leftward-moving forks arrest at the RFB when Fob1p is present and thus do not reach these sites. Therefore, when Fob1p is absent such that replication of the rDNA is no longer unidirectional, the RFB, the promoter region for the 35S transcript and inactive ARSs impeded movement of both rightward- and leftward-moving forks in an Rrm3p-dependent way. We conclude that loss of Fob1p binding at the RFB in rrm3 cells eliminates pausing and arrest at the RFB but does not reduce pausing at other sites.
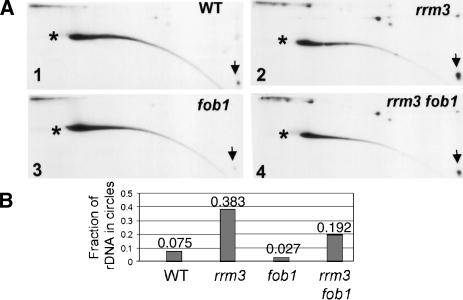
Because the number of rDNA circles is reduced in fob1 cells, circles are thought to arise from breakage of forks arrested at the RFB (Defossez et al. 1999; Versini et al. 2003). Paused and broken replication intermediates, as well as rDNA circles, are much more abundant in rrm3 than in wild-type cells (Ivessa et al. 2000). If rDNA circles arise solely from forks stalled at the RFB, then circles will be equally abundant in fob1 and rrm3 fob1 cells. Alternatively, if breakage also occurs at non-RFB sites of rrm3-induced pausing, then the circle content of rrm3 fob1 cells will be higher than that of an fob1 strain but lower than in an rrm3 strain. We used 2D gels to determine the fraction of rDNA in circles (Fig. 3A,B). In these gels, linear chromosomal rDNA is indicated by an asterisk, and supercoiled monomer circles are indicated by an arrow. Other circular forms, such as relaxed circles and multimers, run in an arc above linear DNA. The fraction of rDNA in circles was determined by dividing the amount of hybridization in all circular forms by the total rDNA hybridization in the gel. As expected, wild-type cells had more rDNA circles (7.5%, Fig. 3, panel 1) than fob1 cells (3%, Fig. 3, panel 3) and less than rrm3 cells (38%, Fig. 3, panel 2). The rDNA circle content of the rrm3 fob1 strain (19%, Fig. 3, panel 4) was intermediate between that of the fob1 and the rrm3 strains (Fig. 3). Although the absolute fraction of circles varied from experiment to experiment, in four of four experiments the fraction of rDNA in circles in the fob1 rrm3 strain was lower than in the rrm3 strain and higher than in either the fob1 or wild-type strain. Thus, forks paused at sites other than the RFB must also break and recombine to generate rDNA circles.
Figure 3.
Deletion of FOB1 only partially suppresses the generation of rDNA circles in rrm3 cells. (A) Undigested DNA was separated using 2D gels. The DNA was transferred to a membrane and hybridized simultaneously with two rDNA probes. One probe is indicated in Figure 1A; the other is a fragment of 35S rDNA. Asterisks indicate the position of linear chromosomal rDNA, and arrows indicate the presence of rDNA supercoiled monomer circles. Additional rDNA circle species form an arc above the chromosomal DNA. (B) Quantitation of a representative experiment. The number above each column is the amount of hybridization in circles divided by the total rDNA hybridization.
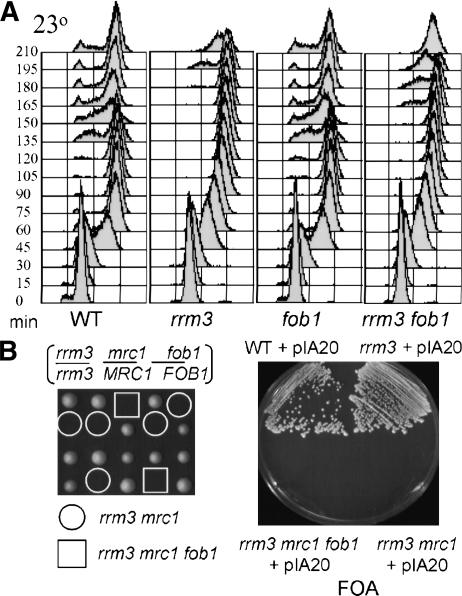
When G1-arrested cells are allowed to progress through the cell cycle, rrm3 and wild-type strains enter S phase with similar timing, but the rrm3 strain is delayed in moving from late S phase into the next cell cycle (Ivessa et al. 2002). There are ∼150 rDNA repeats, and forks arrested or converged at the RFB show a high level of breakage in the absence of Rrm3p (Ivessa et al. 2000). Therefore, the cell cycle delay might reflect a checkpoint response to the increase in forks pausing, converging, and breaking at the RFB. If this model is correct, the cell cycle defect should be mitigated in rrm3 fob1 cells. To test this idea, we used release from α-factor arrest to obtain synchronous rrm3 and rrm3 fob1 cultures and examined their progress through the cell cycle with fluorescent-activated cell sorting (FACS; Fig. 4A). The 23°C-grown wild-type and rrm3 cells were in S phase within 30 min after release. However, whereas many wild-type cells had a G1 DNA content by 135 min, G1-phase cells were not detected in the rrm3 culture until ∼195 min. The fob1 culture was indistinguishable from wild type. Thus, the absence of Fob1p alone had no evident effect on cell cycle progression. However, the rrm3 fob1 strain progressed through the cell cycle more slowly than either the wild-type or fob1 strain although not as slowly as rrm3 cells. G1-phase cells were evident in the rrm3 fob1 culture by 165 min, which was 30 min later than wild-type but 30 min earlier than rrm3 cells. Similar results were obtained for 30°C-grown cells (data not shown). Thus, the S-phase delay in rrm3 cells is probably caused in part by difficulties in resolving replication forks converged at the RFB. However, replication pausing at other sites must also contribute to the S-phase delay.
Figure 4.
Deletion of FOB1 only partially suppresses the rrm3 extended cell cycle. (A) G1-phase (α-factor-arrested) wild-type, rrm3, fob1, and rrm3 fob1 cells were released and allowed to progress through the cell cycle at 23°C. Aliquots were removed at indicated times in minutes and analyzed for DNA content by FACS. (B) Deletion of FOB1 does not rescue the rrm3 mrc1 synthetic lethality. Tetrad dissection of diploids homozygous for rrm3 and heterozygous for mrc1 and fob1. The four spores from a given tetrad are in a vertical line; dissected spores were grown on complete medium for 3 d at 30°C, and genotyped by replica plating. Assuming 2:2 segregation of the genetic markers allows one to identify rrm3 mrc1 double (circled) and rrm3 mrc1 fob1 triple mutants (boxed). Wild-type, rrm3, rrm3 mrc1, and rrm3 mrc1 fob1 spore products carrying pIA20 were streaked onto FOA and assessed for growth after 3 d at 30°C. Lack of growth on FOA indicates that the strain is inviable in the absence of pIA20.
The viability of rrm3 cells depends on several genes that act in the intra-S-phase checkpoint or that have roles in restarting replication forks (Tong et al. 2001; Ivessa et al. 2003; Ooi et al. 2003; Schmidt and Kolodner 2004; Torres et al. 2004). For example, Mrc1p, a transducer for the intra-S-phase checkpoint (Alcasabas et al. 2001), and the Sgs1p DNA helicase, which senses, stabilizes, and restarts stalled replication forks (for review, see Khakhar et al. 2003), are essential in cells lacking Rrm3p. Given the large effects of Rrm3p on fork stalling and breakage at the RFB, the lethality of these doubly mutant strains might be due to damage incurred at the RFB. To test this idea, we constructed rrm3 fob1 mrc1 or rrm3 fob1 sgs1 strains carrying the plasmid pIA20, which has the URA3 and RRM3 genes. Because URA3 cells die in the presence of 5-fluoroorotic acid (FOA), only cells that are viable in the absence of RRM3 will grow on FOA plates. Although wild-type and rrm3 cells grew well on FOA plates, rrm3 mrc1 fob1 and rrm3 sgs1 fob1 cells were dead (Fig. 4B; data not shown). Because deleting FOB1 did not suppress the lethality of rrm3 mrc1 or rrm3 sgs1 cells, replication defects at sites other than the RFB must contribute to making rrm3 viability dependent on the intra-S-phase checkpoint and fork restart activities.
Sites that show Rrm3p-dependent replication are assembled into stable nonnucleosomal protein–DNA complexes. Here we show that deleting the Sir2p histone deacetylase in an rrm3 strain did not alter the rrm3-dependent rDNA replication pauses. However, removal of the Fob1p complex eliminated fork stalling at the RFB but did not affect pausing at other rrm3-sensitive sites. Likewise, a mutation that prevents formation of the transcription preinitiation complex at a tRNA gene eliminates the rrm3-exacerbated replication pause at this gene, and mutations that prevent ORC or Rap1p binding to the HMR-E silent mating type silencer allow replication to proceed unimpeded through this site in rrm3 cells (Ivessa et al. 2003). Together, these data support a model in which Rrm3p promotes fork movement past nonnucleosomal protein–DNA complexes.
There are multiple examples of DNA helicases that are able to remove proteins from DNA in vitro (Bedinger et al. 1983; Kaplan and O'Donnell 2002; Krejci et al. 2003; Veaute et al. 2003). Thus, Rrm3p might act by removing protein complexes from DNA. Alternatively, Rrm3p might be part of a chromatin-remodeling activity or Rrm3p might assist the replicative helicase. Whatever its mechanism of action, proteins like Rrm3p are likely to be critical for genome stability as sequences whose replication is Rrm3p-dependent become fragile sites in its absence.
Materials and methods
Haploid derivatives of the diploid strain VPS107 (homozygous for ade2 ade3 leu2-3,112 ura3Δ trp1Δ lys2-801 can1) were used for 2D gel and FACS analyses. Null mutations of RRM3 were made as described (Ivessa et al. 2000), except that in the strains used for FACS, RRM3 was replaced with a hygromycin marker. Null mutations of SIR2, FOB1, SGS1, and MRC1 were made using a PCR-based strategy that precisely deleted the open reading frames, replacing SIR2 with LEU2, FOB1 with HA-URA3-HA, SGS1 with LEU2, and MRC1 with the hygromycin marker. To determine synthetic lethalities, derivatives of the diploid YPH501(homozygous for ura3-52 lys2-801 ade2-101 trp1Δ63 his3–Δ200 leu2–Δ1) carrying pIA20 were dissected and spore clones of the appropriate genotypes were identified by replica plating, then streaked onto FOA and assessed for growth after incubation for 3 d at 30°C. Conditions for 2D replication and circle gels are as described (Ivessa et al. 2000). For FACS analysis, spore clones of the appropriate genotypes were grown to log phase, arrested for 2 h with 2 μg/mL α-factor (Sigma) at 30°C, washed with YEPD + 20 μg/mL Pronase, released into fresh YEPD + 20 μg/mL Pronase at 23°C or 30°C, and samples collected at indicated time points. Cells were analyzed by FACS as described (Taggart et al. 2002).
Acknowledgments
We thank A. Ivessa and B. Lenzmeier for reagents and discussions and M. Mondoux, M. Sabourin, and L. Vega for comments on the manuscript. This work was supported by grant R37 GM26938 from the National Institutes of Health and a New Jersey Commission on Cancer Fellowship to J.B.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1154704.
References
- Alcasabas A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. [DOI] [PubMed] [Google Scholar]
- Bedinger P., Hochstrasser, M., Jongeneel, C.V., and Alberts, B.M. 1983. Properties of the T4 bacteriophage DNA replication apparatus: The T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell 34: 115–123. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. [DOI] [PubMed] [Google Scholar]
- Brewer B.J., Lockshon, D., and Fangman, W.L. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71: 267–276. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Prusty, R., Kaeberlein, M., Lin, S.J., Ferrigno, P., Silver, P.A., Keil, R.L., and Guarente, L. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3: 447–455. [DOI] [PubMed] [Google Scholar]
- Huang J. and Moazed, D. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes & Dev. 17: 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S. and Zakian, V.A. 2002. To fire or not to fire: Origin activation in Saccharomyces cerevisiae ribosomal DNA. Genes & Dev. 16: 2459–2464. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou, J.-Q., and Zakian, V.A. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou, J.-Q., Schulz, V.P., Monson, E.M., and Zakian, V.A. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes & Dev. 16: 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S., Lenzmeier, B.A., Bessler, J.B., Goudsouzian, L.K., Schnakenberg, S.L., and Zakian, V.A. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol. Cell 12: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Johzuka K. and Horiuchi, T. 2002. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7: 99–113. [DOI] [PubMed] [Google Scholar]
- Kaplan D.L. and O'Donnell, M. 2002. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell 10: 647–657. [DOI] [PubMed] [Google Scholar]
- Keil R.L. and McWilliams, A.D. 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135: 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar R.R., Cobb, J.A., Bjergbaek, L., Hickson, I.D., and Gasser, S.M. 2003. RecQ helicases: Multiple roles in genome maintenance. Trends Cell Biol. 13: 493–501. [DOI] [PubMed] [Google Scholar]
- Kim R. and Wang, J. 1989. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57: 975–985. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Horiuchi, T. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1: 465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck, D.J., Nomura, M., and Horiuchi, T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes & Dev. 12: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Van Komen, S., Li, Y., Villemain, J., Reddy, M.S., Klein, H., Ellenberger, T., and Sung, P. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Linskens M.H.K. and Huberman, J.A. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.L., Shoemaker, D.D., and Boeke, J.D. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35: 277–286. [DOI] [PubMed] [Google Scholar]
- Park P.U., Defossez, P.A., and Guarente, L. 1999. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P., Bensimon, A., and Schwob, E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & Dev. 16: 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L.N., Kirchmaier, A.L., and Rine, J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 27: 27. [DOI] [PubMed] [Google Scholar]
- Schmidt K.H. and Kolodner, R.D. 2004. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Scholes D.T., Banerjee, M., Bowen, B., and Curcio, M.J. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart A.K.P., Teng, S.-C., and Zakian, V.A. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista, M., Parsons, A.B., Xu, H., Bader, G.D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C.W., Bussey, H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Torres J.Z., Schnakenberg, S.L., and Zakian, V.A. 2004. The Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra S phase checkpoint and fork restart activities. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Veaute X., Jeusset, J., Soustelle, C., Kowalczykowski, S.C., Le Cam, E., and Fabre, F. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Versini G., Comet, I., Wu, M., Hoopes, L., Schwob, E., and Pasero, P. 2003. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22: 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]