SUMMARY
Lipid droplets (LDs) are the major fat storage organelles in eukaryotic cells, but how their size is regulated is unknown. Using genetic screens in C. elegans for LD morphology defects in intestinal cells, we found that mutations in atlastin, a GTPase required for homotypic fusion of endoplasmic reticulum (ER) membranes, cause not only ER morphology defects, but also a reduction in LD size. Similar results were obtained after depletion of atlastin or expression of a dominant-negative mutant, whereas overexpression of atlastin had the opposite effect. Atlastin depletion in Drosophila fat bodies also reduced LD size and decreased triglycerides in whole animals, sensitizing them to starvation. In mammalian cells, co-overexpression of atlastin-1 and REEP1, a paralog of the ER tubule-shaping protein DP1/REEP5, generates large LDs. The effect of atlastin-1 on LD size correlates with its activity to promote membrane fusion in vitro. Our results indicate that atlastin-mediated fusion of ER membranes is important for LD size regulation.
INTRODUCTION
Lipid droplets (LDs) are the main organelle for fat storage in eukaryotic cells (Walther and Farese, 2012). LDs consist of a core of neutral lipids, comprising triglycerides (TAG) and sterol esters (SE), and a surrounding phospholipid monolayer. The size of LDs varies in response to changes in nutrient availability, increasing when nutrients are amply available, and decreasing during starvation. Although the enzymes involved in synthesis and degradation of neutral lipids have been identified, the mechanism of this regulation remains poorly understood.
The endoplasmic reticulum (ER) membrane likely plays a major role in the generation and expansion of LDs. Electron microscopy studies show that the ER is tightly associated with LDs, and a physical coupling of the two organelles is a prerequisite for LD expansion (Blanchette-Mackie et al., 1995; Robenek et al., 2009; Wilfling et al., 2013). In Caenorhabditis elegans, the binding of LDs to the ER can be facilitated by an interaction between the ER protein FATP1, a fatty acyl-coenzyme A synthetase, and the lipid-droplet-associated protein DGAT2, an enzyme that converts diglycerides into triglycerides (Xu et al., 2012). In addition, there is evidence in yeast that the cytoplasmic leaflet of the phospholipid bilayer of the ER membrane is continuous with the phospholipid monolayer of LDs (Jacquier et al., 2011). The physical link between the ER and LDs, as well as the ER localization of many enzymes involved in neutral lipid synthesis (Coleman and Lee, 2004; Yen et al., 2008), suggests that the ER plays a major role in LD size regulation.
The ER is a single, continuous membrane system comprising the nuclear envelope and a peripheral network of membrane tubules and sheets (Shibata et al., 2006). The tubules have high membrane curvature in cross-section, which is stabilized by members of two distinct, evolutionarily conserved ER protein families, one called the reticulons (or Rtns in yeast), and the other DP1/REEP5 in mammals and Yop1p in yeast (Voeltz et al., 2006). All of these proteins are anchored in the ER membrane by two hydrophobic hairpin structures. The reticulons and DP1/Yop1p probably stabilize the high membrane curvature by forming a wedge-like structure in the lipid bilayer as well as arc-like scaffolds around the tubules (Hu et al., 2008; Shibata et al., 2009; Voeltz et al., 2006). In vertebrates, DP1/REEP5 is one of six related gene products. While REEP6 is highly similar to DP1/REEP5, REEP1–4 are distinguished by having very short N-terminal hydrophilic domains preceding the first hydrophobic segment and by containing C-terminal microtubule-binding domains (Park et al., 2010).
The ER tubules are connected into a network by membrane fusion, a process mediated by the metazoan atlastin proteins and their yeast functional ortholog, Sey1p (Hu et al., 2009, 2011; Orso et al., 2009; Zhu et al., 2003). The atlastins belong to the dynamin family of GTPases. They consist of a GTPase domain, a three-helix bundle, two closely spaced transmembrane segments, and a cytoplasmic C-terminal tail (Bian et al., 2011; Byrnes and Sondermann, 2011). A role for the atlastins in ER fusion is suggested by the observation that the depletion of atlastins or overexpression of dominant-negative atlastin mutants leads to long, nonbranched ER tubules in tissue culture cells (Hu et al., 2009). Atlastin depletion also causes ER morphology defects in Drosophila melanogaster neurons and muscles (Orso et al., 2009). In addition, antibodies to atlastin inhibit ER network formation in Xenopus egg extracts (Hu et al., 2009). Finally, proteoliposomes containing purified Drosophila atlastin or yeast Sey1p undergo GTP-dependent fusion in vitro (Anwar et al., 2012; Bian et al., 2011; Orso et al., 2009). Both the atlastins and Sey1p physically and genetically interact with the tubule-shaping proteins (Hu et al., 2009; Park et al., 2010), suggesting a functional interplay between these two protein classes. Significantly, mutations in a neuronally expressed isoform of atlastin (atlastin-1) or in REEP1 cause hereditary spastic paraplegia in humans, a neurodegenerative disease that affects corticospinal axons (Blackstone, 2012).
In this paper, we present evidence that proteins determining ER morphology play a role in LD size regulation. Specifically, we report that atlastin affects LD size in C. elegans, Drosophila, and mammalian cells. We present evidence that atlastin regulates LD size through its membrane fusion activity. Our results also suggest that regulation of LD size by atlastin has significant consequences for the organism with respect to fat storage and tolerance to nutrient deprivation.
RESULTS
Atlastin Regulates LD Size in C. elegans
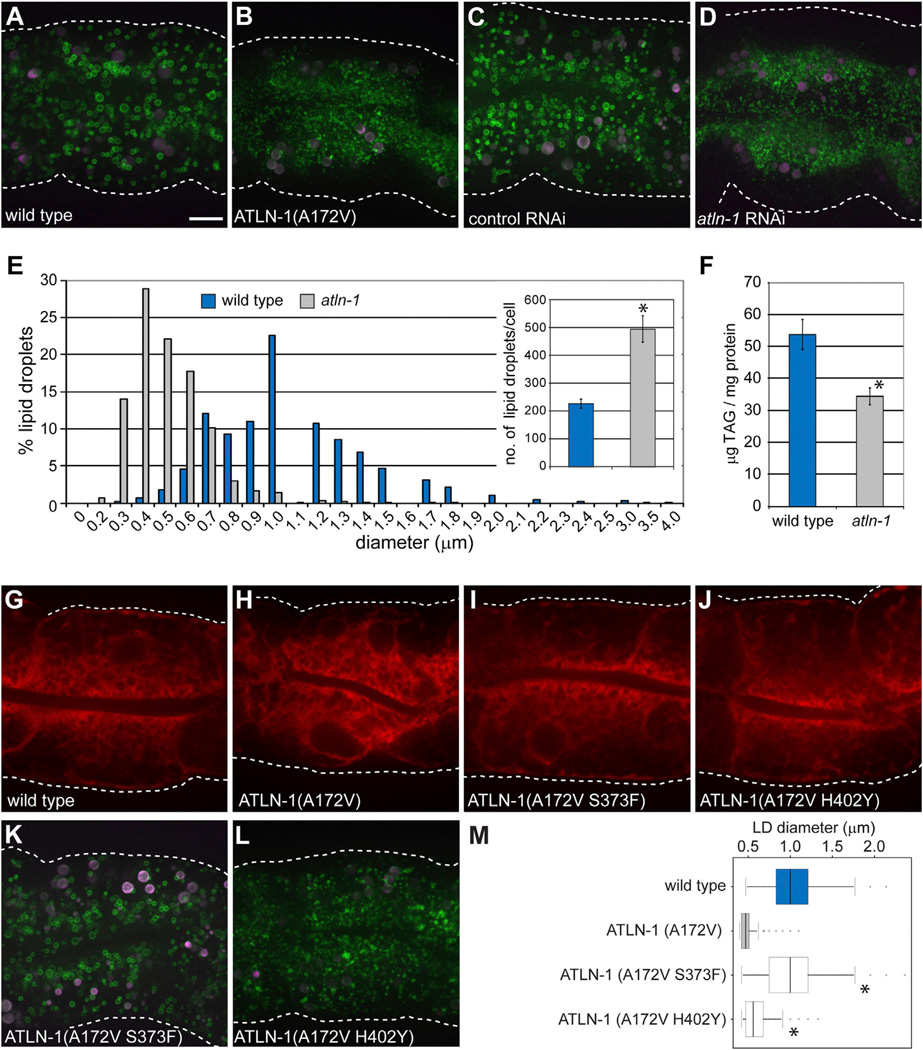
To identify genes that regulate LD size and distribution in C. elegans, we performed an unbiased forward genetic screen. Animals overexpressing the R01B10.6∷GFP fusion protein, which allowed visualization of a subset of LDs in C. elegans (H.Y.M., unpublished data), were mutagenized with ethyl methanesulfonate. Mutant animals with LD morphology changes in intestinal cells, the major site of fat storage in worms (Mak, 2012), were selected with a microfluidic sorting device (Chung et al., 2008; Crane et al., 2012). We identified two recessive mutant alleles, a013 and a015, of the gene Y54G2A.2. Sequence analysis indicated that Y54G2A.2 is a homolog of the mammalian atlastins. We therefore named it atln-1 for atlastin-1. The a013 and a015 alleles encode the mutations A363V and A172V, respectively. We focused our analysis on atln-1(a015) because it causes a stronger phenotype. Similar to atlastins in other species, the mRuby∷ATLN-1 fusion protein localizes to the ER when expressed at physiological levels (Figures S1A–S1C).
To analyze in more detail the effect of mutant ATLN-1 on LDs in intestinal cells, we used a GFP fusion of DGAT-2 (GFP∷DGAT-2), an established LD marker (Xu et al., 2012). In wild-type animals, the diameter of the LDs ranged from 0.3 to 4 µ.m (mode ~1 µm) (Figures 1A and 1E). In addition, the LDs were uniformly distributed throughout the cell (Figure 1A). In contrast, mutant animals expressing ATLN-1(A172V) had significantly smaller LDs, ranging in size from 0.2 to 1.8 µm (mode ~0.4 µm) (Figures 1B and 1E), and the LDs were largely excluded from the basolateral cell cortex. Similar changes in LD size and distribution were observed when ATLN-1 was depleted by RNA interference (RNAi) (Figures 1C and 1D). Consistent with the morphological changes, lipid analysis by gas chromatography and mass spectrometry showed that mutant animals have 36% lower triglyceride levels compared with wild-type animals (Figure 1F). As expected from the established role of atlastin in mammals and Drosophila, mutant worms expressing ATLN-1(A172V) also showed changes in ER morphology, determined by visualizing an ER targeted fluorescent protein (signal sequence∷mCherry∷HDEL; Figure 1H versus Figure 1G). Quantification of ATLN-1 RNAi-treated animals showed that the ER is depleted from the lateral regions of the cells (Figure S1D). The simultaneous visualization of ER and LDs showed that the altered distribution of LDs in mutant animals is correlated with the contraction of the ER (Figures S1E–S1J). This is consistent with the notion that LDs are closely associated with the ER.
Figure 1. ATLN-1 Regulates LD Size and Distribution in C. elegans.
(A) Visualization of lipid droplets (LDs) using the LD marker GFP∷DGAT-2 (hjSi56) in a larval L4 stage animal grown at 25°C. The image shows the second intestinal segment. White dotted lines indicate the cell boundaries on the basal side. GFP is in green and autofluorescence in magenta. A projection of 8 µm z stacks is shown. Scale bar= 10 µm (applies to all other panels).
(B) As in (A), but with a atln-1(a015) mutant worm expressing the ATLN-1(A172V) protein.
(C) As in (A), but worms were treated with a control RNAi.
(D) As in (A), but worms were depleted of ATLN-1 by RNAi.
(E) Distribution of LD size in wild-type and ATLN-1(A172V) animals grown at 20°C. Ten animals of each group were analyzed. The inset shows the number of LDs in wild-type and atln-1(a015) mutant animals. Results are given as means ± SEM derived from ten animals of each genotype. *p < 0.05.
(F) Quantification of triglycerides by gas chromatography-mass spectrometry. Results are given as means ± SEM derived from six independent populations of animals of each genotype grown at 20°C. *p < 0.05.
(G) Image from a single focal plane of a wild-type animal at larval L4 stage expressing a luminal ER mCherry marker (hjSi158).
(H) As in (G), but with an animal expressing ATLN-1(A172V).
(I) As in (G), but with an animal expressing ATLN-1 carrying the original A172V mutation and the suppressor mutation S373F.
(J) As in (G), but with an animal expressing ATLN-1 carrying the original A172V mutation and the suppressor mutation H402Y.
(K) As in (A), but with an animal expressing ATLN-1(A172V S373F).
(L) As in (A), but with an animal expressing ATLN-1(A172V H402Y).
(M) Quantification of LD size. The number of LDs with a diameter ≥0.4 µm in the second intestinal segment was determined in ten cells of five animals for each strain, all grown at 25°C. Total number of LDs measured: wild-type = 2,060, ATLN-1(A172V) = 2,761, ATLN-1(A172V S373F) = 2,215, ATLN-1(A172V H402Y) = 5,503. The median is indicated by a vertical line in the box. The upper and lower quartiles are indicated by horizontal lines and outliers by circles. *p < 0.05, t test against ATLN-1(A172V).
See also Figures S1 and S2.
Given that the expression levels of wild-type ATLN-1 and ATLN-1(A172V) are similar (Figure S2A), it appears that the function of ATLN-1 is compromised by the A172V mutation. We note that residue A172 is conserved in human atlastin-1 and its mutation to proline causes hereditary spastic paraplegia (SPG3A) (Sauter et al., 2004). Consistent with the notion that atlastin activity correlates with LD size, ATLN-1 overexpression resulted in enlarged LDs in C. elegans intestinal cells (Figures S2B–S2D). However, ATLN-1 is not essential for LD biogenesis. In fact, mutant animals expressing ATLN-1(A172V) contain more LDs than wild-type animals (Figure 1E inset). Taken together, these results indicate that ATLN-1 has an effect on the size and distribution of LDs and on the cellular lipid homeostasis in C. elegans.
Effect of Suppressor Mutations in ATLN-1 on LDs and ER Morphology
To confirm that ATLN-1 has a role in LD and ER morphology, we searched for intragenic suppressors of the LD phenotype seen with mutant animals expressing ATLN-1(A172V). Mutant worms expressing GFP∷DGAT-2 were mutagenized and screened for restored LD morphology. We identified seven intragenic suppressor alleles, two of which (S373F and H402Y) encode amino acid substitutions in the three-helix bundle domain. In wild-type atlastin, the three-helix bundle domain undergoes a major conformational change relative to the GTPase domain upon GTP hydrolysis, which ultimately drives membrane fusion (Bian et al., 2011; Byrnes and Sondermann, 2011). The S373F and H402Y mutations did not affect the expression level of ATLN-1 (Figure S2A). Animals expressing ATLN-1(A172V S373F) or ATLN-1(A172V H402Y) showed a normal distribution of LDs and the ER (Figures 1I and J and Figures 1K and 1L). However, while the S373F suppressor mutation rescued LD size to about that of wild-type animals (Figures 1K and 1M), the H402Y suppressor restored LD size only partially (Figures 1L and 1M). It therefore appears that different atln-1 alleles have distinct effects on ER morphology, LD size, and LD distribution.
Sustained Requirement for ATLN-1 in Maintaining LD Size in C. elegans
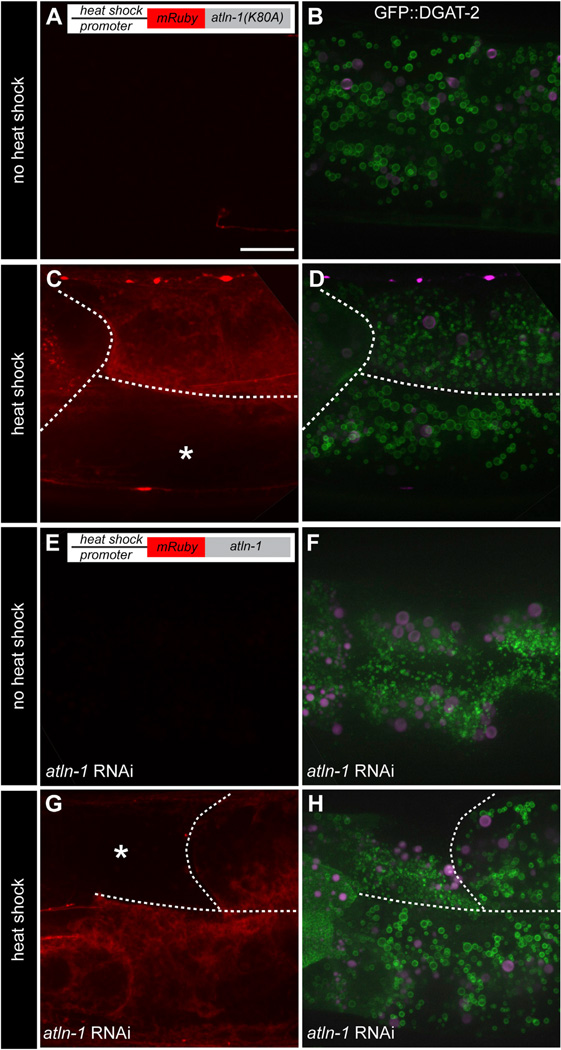
Next, we asked whether atlastin function is required to maintain LD size. We overexpressed a dominant-negative form of ATLN-1 (ATLN-1[K80A]) in wild-type larvae, using a transgene under the control of a heat-shock promoter. The analogous mutation in mammalian atlastin is GTPase defective and causes ER morphology defects (Bian et al., 2011; Hu et al., 2009). Expression of ATLN-1(K80A) upon heat shock caused a reduction in LD size (Figures 2C and 2D), whereas no ATLN-1(K80A) was synthesized and no changes in LD size were observed without heat shock (Figures 2A and 2B). Heat shock per se did not cause changes in LD size, as neighboring cells not expressing ATLN-1(K80A) had normal LDs (Figure 2D). These results indicate that atlastin function is continuously required to maintain the normal size of LDs.
Figure 2. Sustained Requirement for ATLN-1 in Maintaining LD Size in C. elegans.
(A) A transgenic animal was analyzed, which carried a heat-shock-inducible transgene for expression of a dominant-negative ATLN-1 mutant (hjEx17[mRuby∷ATLN-1(K80A)]) and a single-copy transgene expressing an LD marker (hjSi56[GFP∷DGAT-2]). Shown is a control for the absence of mRuby∷ATLN-1(K80A) expression without heat-shock treatment. The second intestinal segment of a larval L4 animal was imaged. A projection of 8 µm z stacks is shown. Scale bar = 10 µm.
(B) As in (A), but the cells were imaged for LDs by analyzing GFP fluorescence (in green). Auto-fluorescence is shown in magenta.
(C) As in (A), but with an animal subjected to heat shock. Expression of ATLN-1 is visualized by mRuby fluorescence (in red). The white dotted lines indicate the cell boundaries. The asterisk indicates a cell not expressing ATLN-1(K80A).
(D) As in (B), but with an animal subjected to heat shock.
(E) A transgenic animal was analyzed, which carried a heat-shock-inducible transgene for expression of wild-type ATLN-1 (hjEx15[mRuby∷ATLN-1]) and a single-copy transgene expressing an LD marker (hjSi56 [GFP∷DGAT-2]). The endogenous ATLN-1 was depleted by RNAi. Shown is a control for the absence of mRuby∷ATLN-1 expression without heat-shock treatment. The second intestinal segment of a larval L4 animal was imaged.
(F) As in (E), but the cells were imaged for LDs by analyzing GFP fluorescence.
(G) As in (E), but with an animal subjected to heat shock.
(H) As in (F), but with an animal subjected to heat shock. The same observations as in (D) and (H) were made with animals carrying independent extrachromosomal arrays for ATLN-1(K80A) and ATLN-1 expression, respectively. All animals were grown at 25°C.
In a complementary approach, we asked whether the small size of LDs, caused by ATLN-1 depletion, could be reversed by the subsequent expression of ATLN-1. To this end, the level of ATLN-1 was lowered by RNAi, starting from the embryonic stage, and wild-type ATLN-1 was ectopically expressed in larval animals under the heat-shock promoter from an RNAi-resistant transgene. Heat-shock-induced expression of ATLN-1 restored normal LD size (Figures 2G and 2H), whereas LDs remained small in animals not subjected to heat shock (Figures 2E and 2F). These results confirm that ATLN-1 is required to maintain LD size in all developmental stages of C. elegans.
Atlastin Affects LD Size in Drosophila Fat Bodies
We next investigated if atlastin plays a role in regulating LD size in Drosophila. The Drosophila genome encodes a single atlastin gene (atl/CG6668) (Lee et al., 2008; Orso et al., 2009). We focused our analysis on the fat body, the major organ of fat storage in flies (Kühnlein, 2012). Atl was depleted by expressing an UAS-RNAi construct under control of the fat-body-specific Gal4 driver CgGal4 (Asha et al., 2003). As a control, we separately knocked down Brummer (Bmm), a lipase involved in neutral lipid mobilization (Grönke et al., 2005), using the same fat-body-specific Gal4 driver. Label-free imaging of the fat bodies by coherent anti-Stokes Raman scattering (CARS) indicated that atl depletion leads to smaller LDs (Figure 3A). While LDs in wild-type fat bodies had a maximum diameter of ~12 µm with 12.4% above 4 µm (1,494 LDs measured; Figure 3A), LDs in atl-depleted fat bodies had a maximum diameter of ~7 µm with only 3.3% above 4 µm (1,467 LDs measured). As expected, depletion of the Bmm lipase had the reverse effect, significantly increasing the size of LDs (Figure 3A). Similar observations were made when LDs were stained with BODIPY493/503 (Y.G., unpublished data). The morphological changes of the LDs were also confirmed by transmission electron microscopy (TEM) (Figure 3B, upper panels). TEM analysis of serial sections, followed by three-dimensional reconstruction, showed that the ER network is significantly less dense in atl knockdown animals (Figure 3B, lower panels and Figure S3), probably indicative of less branching of ER tubules, as observed in HeLa cells depleted of atlastins (Hu et al., 2009). Interestingly, atl knockdown not only had an effect on LD size, but also reduced the total triglyceride content in the whole organism; adult female and male flies had 80% and 90% lower triglyceride levels, respectively (Figure 3C). As a consequence, these flies were more sensitive to starvation challenge; they died much faster than wild-type flies when placed on 1% agar without nutrients (Figure 3D). Taken together, these results show that, like in C. elegans, loss of Drosophila atlastin function has a pronounced effect on LD size. Furthermore, atlastin’s effect on LDs influences organismic triglyceride levels and energy homeostasis in both C. elegans and Drosophila.
Figure 3. Atlastin Regulates LD Size in Drosophila Fat Bodies.
(A) LDs were imaged infresh third-instar larval fat bodies by CARS. Analyzed were fat bodies from wild-type animals (w1118), or flies depleted of the lipase Bmm or of atlastin (Atl) by expression of RNAi constructs under the fat-body-specific CgGal4 driver. The white dotted lines indicate the cell boundaries. Scale bar= 10µm. The right panel shows the quantification of LD size determined from five independent fields for each condition. A total of 1,494 LDs were counted for the control and 1,497 LDs for the Atl knockdown. LD sizes were grouped as indicated. *p < 0.05, t test.
(B) Transmission electron micrographs of fat bodies of wild-type and ATL knockdown animals. The upper panels show LDs and the lower panels show the ER. Scale bar = 10 µm (upper panel) and 0.5 mm (lower panel).
(C) The triglyceride levels of 1 day virgin female and male flies were determined. Controls for the ATL knockdown were performed with wild-type animals (CgGal4/w1118) and animals not expressing the Gal4 driver (UAS-AtlRNAi/w1118). Values are mean ± SD from three experiments. *p < 0.001, determined by a oneway ANOVA F-test, followed by a Bonferroni’s post hoc test.
(D) Female and male flies were starved and the number of surviving animals was determined. Values are average ± SD of three experiments.
See also Figure S3.
Atlastin’s Fusion Activity Is Required to Form Giant LDs in Mammalian Cells
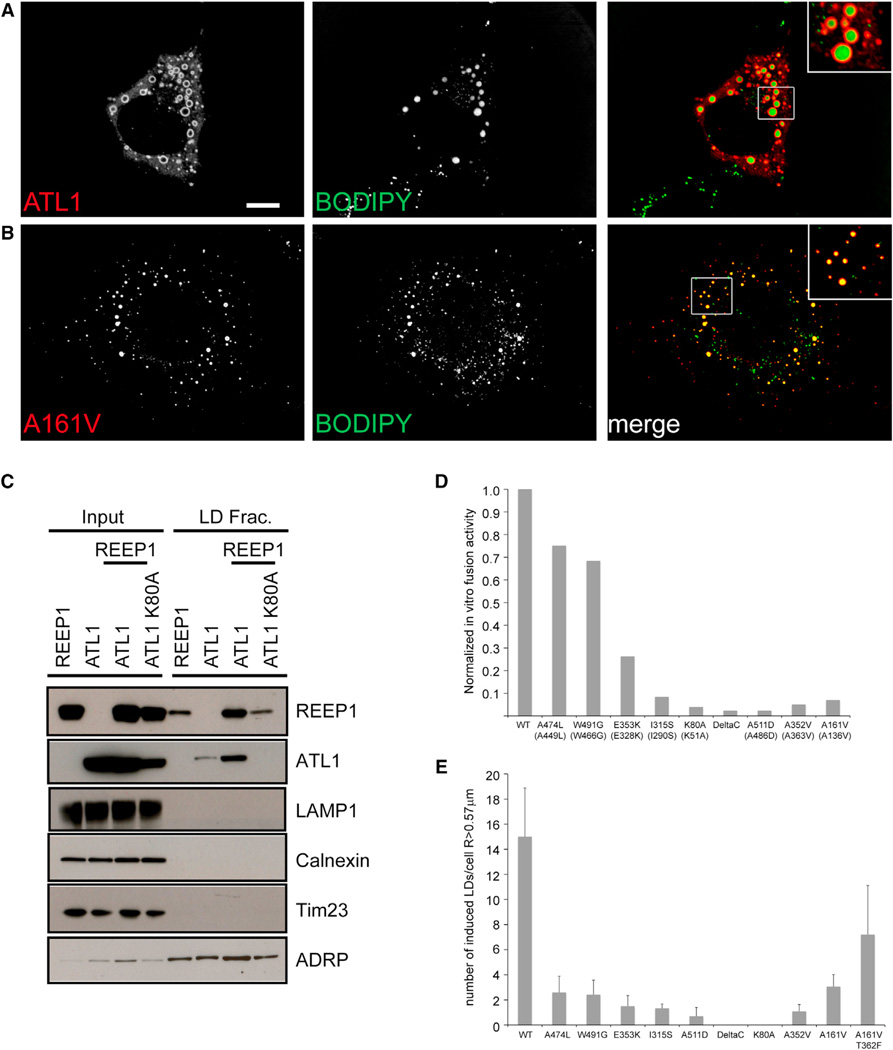
To further explore how atlastin affects LD size, we performed experiments in mammalian tissue culture cells. When human atlastin-1 (ATL1) was expressed alone in COS7 cells, no obvious changes in LD size were observed (Figures S4A–S4C), although the ER morphology was visibly altered at higher expression levels, as previously reported (Hu et al., 2009). By contrast, when human ATL1 was coexpressed with human REEP1, a member of the DP1/REEP5 protein family, large BODIPY-positive spherical structures were seen (Figure 4A). ATL1 and REEP1 both localized to the rim of these structures (shown for ATL1 in the inset of Figure 4A). In addition, a population of much smaller endogenous LDs was still seen and did not stain for ATL1. Expression of REEP1 alone resulted in its localization on microtubules (Figure S4C), as previously reported (Park et al., 2010), consistent with the existence of a microtubule-binding domain in REEP1. Large BODIPY-stained structures were also induced upon coexpression of REEP1 with either ATL2 or ATL3 (Figure S4D), although the LDs were somewhat smaller with ATL2 and were not always uniformly stained with BODIPY in the case of ATL3. Similar observations were made when ATL1 was coexpressed with REEP2, 3, or 4 (Figure S4D). In contrast, no large LDs were generated upon coexpression of ATL1 with REEP5 (Figure S4D) or REEP6 (data not shown), consistent with the fact that REEP1–4 are structurally distinct from REEP5/6. Cell-fractionation experiments showed that some ATL1 and REEP1 molecules localized to LDs, as shown by copurification with the LD marker adipocyte differentiation-related protein (ADRP) and separation from markers of other organelles (Figure 4C). REEP1 expression alone resulted in a small amount of the protein cofractionating with LDs (Figure 4C), although giant LDs were not visibly induced (Figure S4C). Taken together, these data indicate that the simultaneous overexpression of atlastins and REEP1–4 leads to the appearance of large LDs in mammalian cells. Because overexpression of ATLN-1 alone is sufficient to induce large LDs in C. elegans intestinal cells (Figures S2B–S2D), it appears that REEP proteins may not be limiting in these cells.
Figure 4. The Generation of Large LDs in COS7 Cells Is Dependent on the Fusion Activity of Atlastin.
(A) Atlastin-1 (ATL1) tagged at its N terminus with myc was expressed together with REEP1 tagged at its C terminus with hemagglutinin (HA). The cells were stained for ATL1 with myc antibodies (and for REEP1 with HA antibodies, data not shown), as well as for LDs with BODIPY 493. The right panel shows a merged image, and the inset shows an enlarged view of the boxed area.
(B) As in (A), but with ATL1(A161V), corresponding to C. elegans ATLN-1 A172V, instead of wild-type ATL.
(C) REEP1 and myc-tagged atlastin (ATL) were expressed alone or together in HEK293 cells, as indicated. As a control, an ATL1 mutant was used (ATL1[K80A]). The cells were lysed and the extracts were subjected to centrifugation. Ten micrograms of protein of the total extract (input) and of the floated fraction containing LDs were analyzed by SDS-PAGE and immunoblotting with different antibodies. LAMP1, calnexin, Tim23, and ADRP are markers for lysosomes, ER, mitochondria, and LDs, respectively.
(D) Relative fusion activity of wild-type and mutant Drosophila atlastin. The fusion efficiency of the purified proteins was determined by an in vitro lipid mixing assay using reconstituted proteoliposomes (Bian et al., 2011; Liu et al., 2012). The fusion efficiency of each mutant was normalized with respect to the activity of the wild-type protein, determined in the same experiment. The mutations of the Drosophila protein are indicated in brackets below the corresponding mutations in human atlastin.
(E) The number of large LDs in COS7 cells coexpressing REEP1 and either wild-type or mutant ATL1 was quantitated. Images were analyzed with the software ImageJ, and LDs with a diameter larger than 1 µm were counted. The average number of large LDs per cell is plotted for each mutant (± SEM; n = 10 cells; total LDs counted 1,500–2,000).
See also Figure S4.
Next, we asked whether the induction of giant LDs requires the membrane fusion activity of ATL1. We generated a Drosophila atlastin mutant of ATL1 (A136V), analogous to the A172V C. elegans mutant found in the screen. Drosophila ATL (A136V) was purified and reconstituted into proteoliposomes, and fusion activity was tested by lipid mixing using a fluorescent-lipid dequenching assay (Bian et al., 2011; Orso et al., 2009). The activity of the mutant was severely reduced compared with that of wild-type Drosophila ATL1 (Figure 4D). When the equivalent mutant of human ATL1 (A161V) was coexpressed with REEP1 in COS7 cells, only a few large LDs were generated (Figures 4B and 4E), suggesting that the fusion activity of ATL1 is required to generate large LDs. This assumption was confirmed by testing the generation of large LDs with several other mutants of human ATL1, corresponding to Drosophila ATL1 mutants that had previously been shown to be defective in in vitro fusion assays (Bian et al., 2011; Liu et al., 2012). Indeed, there was a reasonable correlation between the fusion activity of ATL1 mutants and the number of large LDs generated upon coexpression with REEP1 (Figures 4D and 4E). For example, mutant K80A (K51A in Drosophila ATL) had a strong defect in both assays. Consistent with the absence of giant LDs, the coexpression of ATL1(K80A) with REEP1 resulted in smaller amounts of REEP1 floating with LDs (Figure 4C). The suppressor mutation T362F found in the screen in C. elegans (S373F in C. elegans) partially restored the ability of the A161V mutant to induce large LDs (Figure 4E), although the in vitro fusion activity was low, likely because the Drosophila protein was not stable after purification (R.W.K., unpublished data). Taken together, our results indicate that the membrane fusion activity of atlastin is required to support the generation of large LDs.
DISCUSSION
Our results provide an unexpected, evolutionarily conserved link between the atlastins and LD size regulation. Gain- and loss-of-function studies in C. elegans, Drosophila, and mammalian cells suggest that the atlastins have a significant effect on LD size, which strongly correlates with their in vitro membrane fusion activity. We propose that atlastin-mediated ER fusion is required for LD expansion. Nevertheless, atlastin does not seem to be required for the initial steps of LD formation, i.e., the local accumulation of neutral lipids between the two leaflets of the phospholipid bilayer that results in their separation to form a hydrophobic inclusion surrounded by a phospholipid monolayer. This is because LDs persist upon atlastin depletion by RNAi in C. elegans, Drosophila, and several mammalian cell lines (Y.G. and Y.S., unpublished data). In yeast, it has been reported that LDs remain associated with the ER during their subsequent growth phase (Jacquier et al., 2011). It seems possible that LDs are associated with a specific ER domain. This ER domain could be a dense ER network or a sheet structure around the LDs, and its formation may depend on atlastin-mediated fusion of ER membranes. In principle, atlastin could also move into LDs and fuse them directly, but atlastin does not seem to localize to LDs in C. elegans or mammalian cells. We therefore favor the idea that atlastin affects LD size indirectly through its role in ER membrane fusion. Consistent with this assumption, LD size reduction correlates with ER morphology defects, although LD size appears to be more sensitive to defects in atlastin function based on our observation in C. elegans (Figures 1K–1M).
Regardless of the exact mechanism by which atlastin affects LD size, it is clear that atlastin’s function is important for fat storage in worms and flies. It seems possible that ER morphology defects may also cause defects in lipid metabolism in humans. This has important implications for the pathogenesis of hereditary spastic paraplegias, because many other forms are caused by mutations in proteins directly involved in lipid biosynthesis (Blackstone, 2012).
EXPERIMENTAL PROCEDURES
Strains and Transgenes
The wild-type C. elegans strain was Bristol N2. All animals were maintained at either 20°C or 25°C. The following alleles were used: LG IV, atln-1(a013), atln-1(a015), atln-1(a015 hj60), and atln-1(a015 hj62). The following transgenes were used:
hjSi56[vha-6p∷3xFLAG-TEV-GFP∷dgat-2∷let-858 3′UTR] IV
hjSi158[vha-6p∷SEL-1(1–79)∷mCherry∷HDEL∷let-858 3′UTR ] I
hjSi185[vha-6p∷3xFlag∷mRuby∷atln-1 (RNAi resistant) cDNA∷let-858 3′ UTR] II
hjEx15, hjEx16[hsp∷3xFlag∷mRuby∷atln-1 (RNAi Resistant) cDNA∷let-858 3′ UTR (10ng/µl); tub-1∷gfp (20ng/µl); pBluescript (70ng/µl)] injected into hjSi56.
hjEx17, hjEx18[hsp∷3xFlag∷mRuby∷atln-1(K80A) (RNAi Resistant) cDNA∷let-858 3′ UTR (10ng/µl); tub-1∷gfp (20ng/µl); pBluescript (70ng/µl)] injected into hjSi56.
hjEx19, hjEx20[vha-6p∷3xFlag∷mRuby∷Atlastin cDNA∷let-858 3′ UTR (30ng/µl); pBluescript (70ng/µl)] injected into hjSi56.
C. elegans Genetic Screens
The atln-1 (a013) and atln-1 (a015) alleles were isolated as second-generation progeny of ethyl-methanesulfonate (EMS)-mutagenized hjIs60[vha-6p∷R01B10.6∷gfp] animals, which expressed the R01B10.6∷GFP fusion protein in intestinal cells. Mutant animals with altered GFP signal were monitored and selected using a microfluidic device mounted on a wide-field microscope (Leica DM4500) equipped with a 63×, numerical aperture (NA) 1.4 oil objective, and a charge-coupled device (CCD) camera (Infinity 3–1; Lumenera). The a013 and a015 alleles were genetically linked to hjIs60, which placed them on LG IV. The genome of mutant animals carrying the a013 and a015 alleles were sequenced (Illumina), and atln-1 was the only gene on LG IV that harbored independent missense mutations. Molecular cloning of the atln-1 gene was confirmed by transgenic rescue of atln-1 (a015) with a vha-6p∷gfp∷atln-1 transgene. The atln-1 (a015) allele was separated from hjIs60 by genetic recombination and outcrossed twice with wild-type N2 animals.
In addition to phenotypes associated with LD size and distribution, the atln-1(a015) mutant animals also have a low brood size at 25°C. We mutagenized atln-1(a015) hjSi56[gfp∷dgat-2] animals with EMS and selected for animals that had an elevated brood size over ten generations at 25°C. Mutant animals were then examined for their LD size and distribution using the GFP∷DGAT-2 marker. We identified molecular lesions of the atln-1 gene in animals carrying the hj60 and hj62 alleles by Sanger sequencing. In both cases, we detected the mutation associated with the parental a015 allele and an additional missense mutation in the atln-1 gene. The atln-1(a015 hj60) and atln-1(a015 hj62) mutants were outcrossed twice with wild-type N2 animals to confirm that the suppression of the atln-1 (a015) phenotypes was not due to unlinked extragenic mutations.
Live Imaging of C. elegans
Confocal images of L4 larval animals were acquired on a spinning disk confocal microscope (Ultraview, Perkin Elmer) using a 100×, NA 1.45 oil Plan-Apochromat objective, on a CCD camera (Orca-R2, Hamamatsu) controlled by the Volocity software (PerkinElmer). For GFP, a 488 nm laser was used for excitation and signals were collected with a 500–555 nm emission filter. For mRuby or mCherry, a 561 nm laser was used for excitation and signals were collected with a 415–475 nm and 580–650 nm dual-pass emission filter. For autofluorescence from lysosome related organelles, a 488 nm laser was used for excitation and signals were collected with a (415– 475 nm) (580–650 nm) dual-pass emission filter. Optical sections were taken at 0.25 µm intervals and z stacks of 8.0 µm were exported from Volocity to Imaris 6.2.1 (Bitplane) for identification, annotation, and measurement of LD diameter. Maximum projection of the same z stacks were also exported from Volocity and processed in Photoshop (Adobe).
Ectopic Expression of ATLN-1 in C. elegans
Heat-shock experiments were performed on L2 larval animals at 33°C for 30 min in a water bath. Animals were allowed to recover and develop into L4 stage at 20°C for 24 hr before imaging. The strain hjSi56[vha-6p∷3xFLAG-TEV-GFP∷dgat-2∷let-858 3’UTR]; hjEx15[hsp∷3xFlag∷mRuby∷atln-1 (RNAi Resistant) cDNA∷let-858 3’ UTR] was used for ectopic overexpression of ATLN-1. The strain hjSi56[vha-6p∷3xFLAG-TEV-GFP∷dgat-2∷let-858 3′UTR]; hjEx17[hsp∷3xFlag∷mRuby∷atln-1 (K80A)(RNAi Resistant) cDNA∷let-858 3′ UTR] was used for ectopic overexpression of ATLN-1(K80A).
Lipid Analysis of C. elegans
Lipid extraction and quantification of triglyceride levels by thin-layer chromatography and gas chromatography from synchronized L4 larval animals were performed as previously described (Zhang et al., 2010).
Antibodies and Western Blotting
Antibodies against ATLN-1 were raised by simultaneous injection of two peptides into rabbits (1METTPQNEHNEHQQQQHAGHVED, 555RTANATAAP GAADGLRKRH) (YZ3315, Yenzym). Antibodies were purified with Melon gel (45206, Pierce) and used in western blotting at a dilution of 1:100. Anti-histone H3 antibodies (ab1791, Abcam) were used at 1:2,000. Images were captured with a GeneGnome5 bio imager (Syngene) and quantified with the GeneTools analysis software (Syngene). Antibodies used for analysis of the cellular fractions by western blot were against REEP1: rabbit polyclonal against residues 174–191 of human REEP1 (Park et al., 2010); Calnexin: mouse monoclonal (IgG1, clone 37; BD Biosciences Transduction Laboratories); ADRP: rabbit polyclonal (LifeSpan BioScience).
Drosophila Experiments
Fly stocks were raised on standard corn meal-agar medium (Nutri-Fly BF formula, Genesee Scientific, San Diego, CA). The CgGal4 (Asha et al., 2003) driver line was backcrossed to the white1118 line (ID 60000) from the Vienna Drosophila RNAi center (VDRC) for more than ten generations. Male UAS-RNAi Atl transgenic flies (ID 6719, VDRC) were crossed with CgGal4 virgin flies. Fat bodies from third-instar larvae of the progeny were dissected in PBS and imaged immediately on a FluoView FV1000 MPE microscope (Olympus America, Center Valley, PA) using the CARS application. A XLPlanN 25×/1.05w MP objective lens was used, and the Mai Tai DeepSea laser (Spectra-physics) was tuned to 800 nm. For transmission electron microscopy, fat bodies were processed following the Malachite Green staining protocol (Cocchiaro et al., 2008). Serial sections were cut at 0.09 µm thickness, and individual micrographs were collected and stacked. Micrographs were acquired using a JEOL 1400 Transmission Electron Microscope (JEOL, Peabody, MA) at 80 kV. Image stacks were aligned, analyzed, and visualized using freeware Reconstruct Software (SynapseWeb, written by John C. Fiala, Ph.D.). For triglyceride measurement, 1 day virgin flies were processed according to a published protocol (Hildebrandt et al., 2011) using the Infinity Triglycerides reagent (TR22421, Thermo Fisher Scientific). For starvation assays, 1 day virgin flies were starved in vials with 1% agar, and surviving flies were scored every 8 hr.
Protein Expression in COS Cells
Myc tagged wild-type human atlastin-1 (mycATL1), mycATL1(K80A), and hemagglutinin (HA)-tagged REEP1 (REEP1-HA) were described previously (Park et al., 2010). Point mutants in ATL1 corresponding to Drosophila mutants defective in fusion were generated by site-directed mutagenesis using the Quikchange kit (Stratagene) and verified by sequencing.
COS7 cells were transfected using 1 µg DNA per construct and the transfection reagent lipofectamine (Invitrogen). The cells were incubated for 12 hr at 37°C in DMEM, 10% fetal bovine serum, and 1 mM sodium pyruvate in the presence of 5% CO2. For immunofluorescence and fluorescence microscopy, cells were trypsinized and transferred onto acid-washed glass coverslips and incubated for 24 hr in a 12-well tissue culture dish (BD Biosciences). Cells were fixed by replacing the medium with PBS containing 4% paraformaldehyde and incubated for 20–30 min at room temperature. The cells were washed with PBS and permeablized by incubation for 10 min in PBS containing 0.1% Triton X-100 (Thermo). Immunocytochemistry was carried out by incubating the permeabilized cells with antibodies against the Myc or HA epitope (Roche, mouse monoclonal 9E10 and rat 3F10, respectively), diluted in PBS containing 1% calf serum (GIBCO). The cells were washed three times in PBS and then incubated with Alexa-Fluor-labeled secondary antibodies and 0.1 µg/ml BODIPY 493 (Invitrogen).
Images were captured using a Yokogawa spinning-disk confocal on a Nikon TE2000U inverted microscope with a 100× Plan Apo NA 1.4 objective lens, a Hamamatsu ORCA ER-cooled CCD camera, and MetaMorph software. For quantification of the number of large LDs, the Myc-stained images were converted to 8 bit grayscale images and thresholded using the software ImageJ. The binary images were processed by the ‘‘fill holes’’ and ‘‘watershed’’ functions. LDs were counted by using the ‘‘analyze particle’’ function, and the diameters were calculated from the measured area of each LD. LDs with a diameter >1 µm were considered to be large. For image quantification, at least ten cells per experimental condition were analyzed. The bar diagrams show the average number of giant lipid droplets per cell ± SEM. For image presentation, brightness levels and bit depth were adjusted using Adobe Photoshop.
Isolation of Lipid Droplets by Subcellular Fractionation
HEK293 cells were treated with 200 µM oleic acid for 24 hr. The cells were harvested and lipid droplets were isolated by subcellular fractionation on a sucrose gradient as described previously (Cermelli et al., 2006; Martin and Parton, 2008). After centrifugation at 150,500 × g (Beckman TLS55 rotor for 60 min), 0.1 ml was taken from the top of the gradient, precipitated by trichloroacetic acid, and washed with acetone. Aliquots of the cell lysate (10 µg protein; ~4% of the starting material) and ~5% of LD fraction were resolved by SDS-PAGE and immunoblotted with the indicated markers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sabrina Caldwell for the initial mapping of altn-1 alleles, and the Stowers Institute Microscopy Center for assistance with imaging. We thank the Mayo Clinic Microscopy and Cell Analysis Core Facility for assistance with TEM and CARS applications. We thank the Nikon Imaging Center at Harvard Medical School for help. This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH (C.B.), a Mayo Clinic New Investigator Startup Fund and Richard F. Emslander Career Development Award (Y.G.), the National Institutes of Health grants R01-GM099844 (R.V.F.), R01AG035317 (H.L.), R01GM088333 (H.L.), the National Science Foundation grant CBET 0954578 (H.L.), and the Stowers Institute for Medical Research (H.Y.M.). T.A.R. is a Howard Hughes Medical Institute investigator.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.04.015.
LICENSING INFORMATION
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
REFERENCES
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 2012;197:209–217. doi: 10.1083/jcb.201111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H, Nagy I, Kovacs G, Stetson D, Ando I, Dearolf CR. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics. 2003;163:203–215. doi: 10.1093/genetics/163.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. USA. 2011;108:2216–2221. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat. Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. USA. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Crane MM, Stirman JN, Ou CY, Kurshan PT, Rehg JM, Shen K, Lu H. Autonomous screening of C. elegans identifies genes implicated in synaptogenesis. Nat. Methods. 2012;9:977–980. doi: 10.1038/nmeth.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Bickmeyer I, Kühnlein RP. Reliable Drosophila body fat quantification by a coupled colorimetric assay. PLoS ONE. 2011;6:e23796. doi: 10.1371/journal.pone.0023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011;147:1226–1231. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Kühnlein RP. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J. Lipid Res. 2012;53:1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Paik D, Bang S, Kang J, Chun B, Lee S, Bae E, Chung J, Kim J. Loss of spastic paraplegia gene atlastin induces age-dependent death of dopaminergic neurons in Drosophila. Neurobiol. Aging. 2008;29:84–94. doi: 10.1016/j.neurobiolaging.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA. 2012;109:E2146–E2154. doi: 10.1073/pnas.1208385109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak HY. Lipid droplets as fat storage organelles in Caenorhabditis elegans: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. J. Lipid Res. 2012;53:28–33. doi: 10.1194/jlr.R021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Characterization of Rab18, a lipid droplet-associated small GTPase. Methods Enzymol. 2008;438:109–129. doi: 10.1016/S0076-6879(07)38008-7. [DOI] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robenek H, Buers I, Hofnagel O, Robenek MJ, Troyer D, Severs NJ. Compartmentalization of proteins in lipid droplet biogenesis. Biochim. Biophys. Acta. 2009;1791:408–418. doi: 10.1016/j.bbalip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Sauter SM, Engel W, Neumann LM, Kunze J, Neesen J. Novel mutations in the Atlastin gene (SPG3A) in families with autosomal dominant hereditary spastic paraplegia and evidence for late onset forms of HSP linked to the SPG3A locus. Hum. Mutat. 2004;23:98. doi: 10.1002/humu.9205. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV, Jr, Mak HY. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010;107:4640–4645. doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu PP, Patterson A, Lavoie B, Stadler J, Shoeb M, Patel R, Blackstone C. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 2003;278:49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.