Abstract
MDM2 Binding Protein (MTBP) has been implicated in cancer progression. Here we demonstrate one mechanism by which MTBP inhibits cancer metastasis. Overexpression of MTBP in human osteosarcoma cell lines lacking wild-type p53 did not alter primary tumor growth in mice but significantly inhibited metastases. MTBP downregulation increased the migratory potential of MDM2−/−p53−/− mouse embryonic fibroblasts, suggesting that MTBP inhibited cell migration independently of the Mdm2-p53 pathway. Co-immunoprecipitation and mass spectrometric analysis identified alpha-actinin-4 (ACTN4) as a MTBP-interacting protein. Endogenous MTBP interacted with and partially colocalized with ACTN4. MTBP overexpression inhibited cell migration and filopodia formation mediated by ACTN4. Increased cell migration by MTBP downregulation was inhibited by concomitant downregulation of ACTN4. MTBP also inhibited ACTN4-mediated F-actin bundling. We furthermore demonstrated that nuclear localization of MTBP was dispensable for inhibiting ACTN4-mediated cell migration and filopodia formation. Thus, MTBP suppresses cell migration, at least partially, by inhibiting ACTN4 function. Our study not only provides a mechanism of metastasis suppression by MTBP, but also suggests MTBP as a potential biomarker for cancer progression.
Keywords: MDM2, MTBP, ACTN4, filopodia, metastasis
Introduction
Acquisition of metastatic potential by cancer cells is a significant event, often leading to clinically incurable disease. However, the exact mechanism of gaining metastatic potential remains incompletely understood at the molecular and biochemical levels (1). Metastasis is a multi-step process, including detachment of tumor cells from the primary tumor, invasion, migration, intravasation, survival in the vasculature, extravasation, and colonization of a secondary site (2). Increased migratory potential clearly plays a crucial role in the metastatic cascade. Cell polarization marks the beginning of migration, followed by the extension of processes such as lamellipodia and filopodia in the direction of migration (3, 4). Filopodia are transient, thin, hairlike protrusions containing parallel actin bundles, which requires cross-linking of actin filaments (4). Several proteins, including fascin, α-actinin, espin, plastin, and villin, participate in filopodia formation (4–7).
MDM2 Binding Protein (MTBP) was originally identified as a protein that interacts with the oncoprotein MDM2, using a yeast two-hybrid screen (8). MDM2 is a major negative regulator of the tumor suppressor p53 (9). Overexpression of MTBP inhibits proliferation of several human cancer cell lines regardless of p53 status. This MTBP function is nullified by simultaneous overexpression of MDM2 (8). These results support a potential role for MTBP in the suppression of tumorigenesis. To better understand the physiological function of MTBP, we previously generated mice with disruption of the MTBP gene (10). Homozygous deletion of MTBP results in early embryonic lethality not rescued by a concomitant deletion of p53, suggesting that the lethality is independent of p53. MTBP heterozygous mice (MTBP+/−) do not present with any obvious phenotypes (10). However, in mice predisposed to tumorigenesis, MTBP haploinsufficiency significantly increases tumor metastasis (10). Mouse embryonic fibroblasts (MEFs) from MTBP+/− mice show a higher migratory potential than wild-type MEFs (10), further supporting the involvement of MTBP in regulation of cell migration and metastasis. Clinically, Vlatković et al. (11) reports that loss of MTBP expression is associated with reduced survival of patients with head and neck carcinoma, and serves as an independent prognostic factor when p53 is mutated in tumors. Thus, MTBP plays a definitive role in inhibiting cell migration and tumor progression. However, the mechanism through which MTBP inhibits metastasis remains unknown.
We hypothesized that MTBP inhibits cancer cell migration by interacting with a protein involved in cell motility. Our co-immunoprecipitation and mass spectrometric analysis identified alpha-actinin-4 (ACTN4) as a MTBP-interacting protein, one involved with cell motility and cancer metastasis (12–16). We determined that endogenous MTBP and ACTN4 interacted intracellularly, and that MTBP inhibited ACTN4-mediated cell migration, filopodia formation, and F-actin bundling. Thus, inhibition of ACTN4 function appears to be one mechanism through which MTBP suppresses tumor migratory potential, thereby attenuating cancer metastasis.
Results
MTBP suppresses osteosarcoma metastasis independently of p53
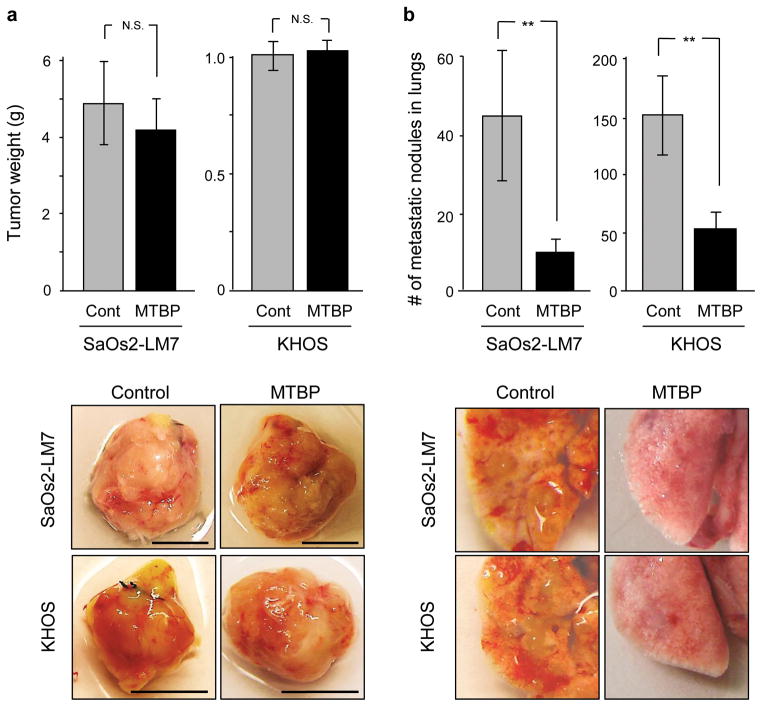
Our previous results indicate that MTBP haploinsufficiency in mice increases tumor metastasis (10). Loss of MTBP expression is also shown to be associated with reduced survival of head and neck carcinoma patients (11). To further our understanding of MTBP-mediated suppression of tumorigenesis and metastasis, we established an orthotopic tumor cell transplantation assay using the human osteosarcoma cell lines SaOs2-LM7 and KHOS. Both cell lines lack the functional p53 activity. We first generated stable SaOs2-LM7 and KHOS cell lines that constitutively overexpress MTBP. Mice were then intrafemorally injected with empty vector-infected cells (control) or MTBP-overexpressing cells. Mice in both groups were sacrificed at the same day after injections to examine for the weight of primary tumors at injected sites and the number of lung metastatic nodules. As compared to controls, MTBP overexpression did not alter the primary tumor weight (Figure 1a), but significantly reduced the quantity of metastatic pulmonary nodules in lungs in both cell lines (Figure 1b), illustrating suppression of tumor metastasis by MTBP independently of p53.
Figure 1.
MTBP suppresses osteosarcoma metastasis independently of p53. NOD-scid IL2Rγ-null mice were intrafemorally injected with SaOs2-LM7 and KHOS cells that were stably infected with either empty (grey, cont) or MTBP-encoding (black, MTBP) lentiviral vectors. Mice were monitored for the development of primary tumors and lung metastases. Approximately 4 and 6 weeks after injections of KHOS and SaOs2-LM7 cells, respectively, mice were sacrificed and examined for the weight of primary tumors at injected sites (a) and the number of lung metastatic nodules (b). Representative pictures of primary tumors and pulmonary metastases from SaOs2-LM7 and KHOS injected mice are shown below graphs. Error bars are means ± S.D. from four independent mice. **, P < 0.01; t test. N.S., not significant. Scale bar: 1 cm.
MTBP inhibits cell migratory potential independently of MDM2 and p53
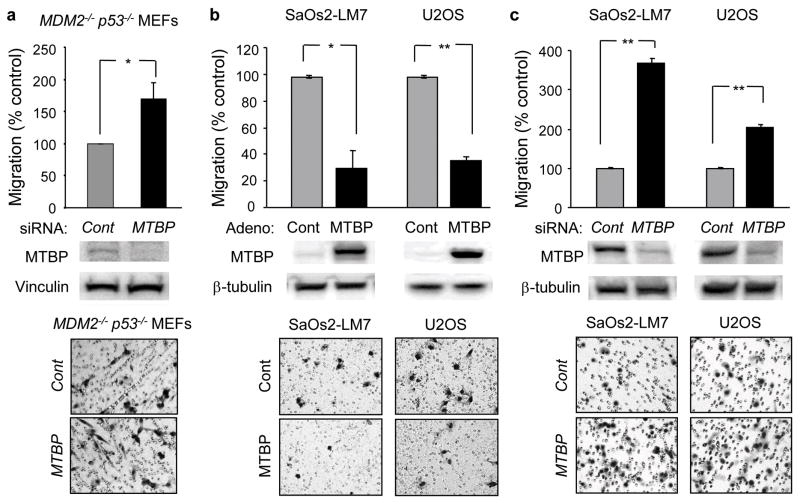
We previously demonstrated that MTBP+/− MEFs migrate faster than wild-type MEFs (10). Furthermore, MTBP overexpression in a mouse p53-null osteosarcoma cell line significantly decreases its invasive potential (10). To determine whether MTBP inhibits cell migration independently of the MDM2-p53 pathway, we examined the effect of MTBP downregulation on the migratory potential of MDM2−/−p53−/− MEFs. As expected, MTBP downregulation resulted in an increased cell migration (Figure 2a). We next examined the effect of modulating MTBP expression on the migratory potential of human osteosarcoma cells. We infected SaOs2-LM7 (p53 null) and U2OS (wild-type p53) cells with empty (control) or MTBP-encoding adenoviral vectors (20 multiplicity of infection: MOI) and performed transwell migration assays. MTBP overexpression reduced cell migration by 70% when compared with controls in both the cell lines (Figure 2b). The inhibition of migratory potential by MTBP was observed in a dose-dependent manner (Figure S1). Conversely, MTBP downregulation using MTBP-specific siRNA significantly enhanced the migratory potential of both cell lines compared to cells transfected with control siRNA (Figure 2c). These results indicate that MTBP suppresses cell migration independently of the MDM2-p53 pathway.
Figure 2.
MTBP inhibits cell migratory potential independently of MDM2 and p53. (a) MDM2−/−p53−/− MEFs were transfected with non-target (grey, cont) or MTBP-specific (black, MTBP) siRNAs. (b) SaOs2-LM7 and U2OS cells were infected with empty (grey, cont) or MTBP-encoding (black, MTBP) adenoviruses (Adeno). (c) SaOs2-LM7 and U2OS cells were transfected with non-target (grey, cont) or MTBP-specific (black, MTBP) siRNAs. Thirty six (36) hours after manipulation of MTBP expression, cells were examined for migratory potential. Migrating cells were stained and entire fields were counted. Representative images are shown below the graphs. Graphs showing relative cell migration (%) compared to the number of migrating cells in control. Western blotting results below the graphs showing successful manipulation of MTBP expression. Error bars are means ± S.D. from three independent experiments. *, P < 0.05 and **, P < 0.01; t test.
MTBP interacts with ACTN4 and inhibits ACTN4-induced cell migration
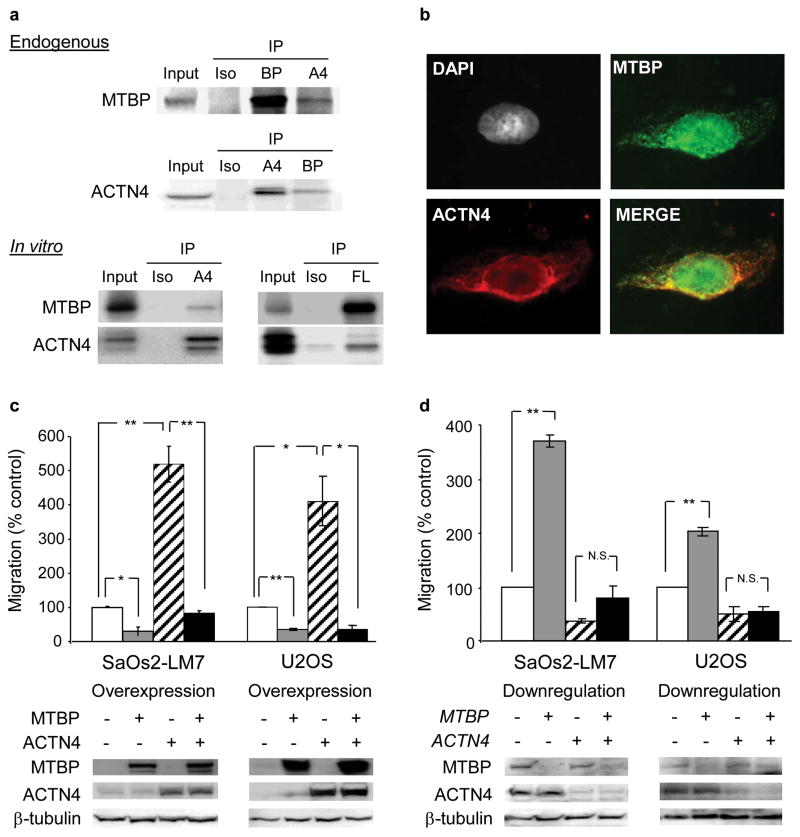
Given the MDM2-independent suppression of cell migration by MTBP, we hypothesized that MTBP interacts with proteins involved in cell motility. To test this hypothesis, we first performed co-immunoprecipitation and mass spectrometric analysis using MTBP-overexpressing U2OS cells, which identified 36 proteins potentially interacting with MTBP (Table 1). Four of the proteins identified, ACTN4 (17), vimentin (18), actin (19), and tropomyosin 3 (TPM3) (20) are related to cell motility. We then validated endogenous interactions of these proteins with MTBP. Under our experimental conditions using RIPA or Cell Lytic M buffers, the only confirmed endogenous interaction was between MTBP and ACTN4 (Figure 3a, endogenous), since we failed to detect interactions of MTBP with vimentin, actin, and TPM3 (Figure S2). We also confirmed that MTBP did not interact with vinculin, another cytoskeletal protein (21), suggesting that MTBP specifically interacted with ACTN4 (Figure S2). The MTBP-ACTN4 interactions were further supported by the studies using proteins synthesized with the TnT Quick Coupled Transcription/Translation Systems (Figure 3a, in vitro). Immunofluorescence studies using SaOs2-LM7 cells also revealed a partial colocalization of MTBP and ACTN4, mainly in the cytoplasm (Figure 3b).
Table 1.
MTBP-interacting proteins identified by co-immunoprecipitation and mass spectrometry
| Protein | Size (kDa) |
|---|---|
| Clathrin heavy chain 1 | 191.5 |
| Alpha-actinin-4 | 105.0 |
| Elongation factor 2 | 95.3 |
| Transitional endoplasmic reticulum ATPase | 89.3 |
| Heat shock protein 90 | 83.2 |
| Nucleolin | 76.7 |
| Glucose regulated protein 75 | 73.5 |
| Heat shock protein 70 | 71.0 |
| Alpha fetoglobulin | 67.3 |
| Pyruvate kinase | 57.8 |
| T complex protein 1 | 57.4 |
| Prolyl 4-hydroxylase | 57.1 |
| ATP synthase | 56.3 |
| Vimentin | 53.6 |
| Tubulin | 49.6 |
| Aspartate aminotransferase | 47.4 |
| Alpha enolase | 47.1 |
| Eukaryotic initiation factor 4A1 | 46.1 |
| Actin | 41.7 |
| Phosphoserine aminotransferase | 40.4 |
| Fructose bisphosphate aldolase A | 39.3 |
| Poly rC binding protein 1 | 37.5 |
| L lactate dehydrogenase | 36.7 |
| Glyceraldehyde 3 phosphate dehydrogenase | 36.0 |
| Annexin | 35.9 |
| Malate dehydrogenase | 35.5 |
| Heterogeneous nuclear ribonucleoprotein | 34.4 |
| ADP ATP translocase 2 | 32.9 |
| Tropomyosin 3 | 32.8 |
| 40S ribosomal protein | 32.8 |
| Nucleophosmin | 32.5 |
| Proliferating cell nuclear antigen | 28.8 |
| 14-3-3 | 27.7 |
| Chloride intracellular channel protein 1 | 26.9 |
| GTP binding nuclear protein Ran | 24.4 |
| Nascent polypeptide associated complex | 23.4 |
Figure 3.
MTBP interacts with ACTN4 and inhibits ACTN4-mediated cell migration. (a) Whole cell extracts from SaOs2-LM7 cells were immunoprecipitated (IP) with MTBP (BP) or ACTN4 (A4) antibodies (endogenous). Immunoprecipitants were subjected to western blotting for MTBP or ACTN4. Matched isotype antibodies (Iso) were used as negative controls. In vitro translated ACTN4 and FLAG-tagged MTBP were incubated in PBS and immunoprecipitated with ACTN4 (A4, left) or FLAG antibodies (FL, right), followed by immunoblotting for MTBP or ACTN4, respectively (in vitro). (b) Immunofluorescence was performed to examine endogenous colocalization of MTBP and ACTN4 in SaOs2-LM7 cells using indicated antibodies. DNA was stained using DAPI. Results were analyzed on a Leica epifluorescence microscope. A merge picture indicates that MTBP and ACTN4 partially colocalize mainly in the cytoplasm (yellow). (c, d) SaOs2-LM7 and U2OS cells stably infected with either empty (ACTN4−) or ACTN4-encoding (ACTN4+) lentiviral vectors were infected with empty (MTBP−) or MTBP-encoding (MTBP+) adenoviruses (c). Cells stably infected with empty (ACTN4−) or ACTN4 shRNA-encoding (ACTN4+) lentiviral vectors were transiently transfected with non-targeted (MTBP−) or MTBP-specific (MTBP+) siRNAs (d). Thirty six (36) hours after manipulation of MTBP expression, migration assays were performed. Migrating cells were stained and entire fields were counted. Graphs showing relative cell migration (%) compared to the number of migrating cells in control. Western blotting results below the graphs demonstrating successful manipulation of MTBP and ACTN4 expression. Error bars are means ± S.D. from three independent experiments. *, P < 0.05 and **, P < 0.01; t test. N.S., not significant. Control: white, MTBP alone manipulated: grey, ACTN4 alone manipulated: oblique, MTBP and ACTN4 manipulated: black.
We next tested whether MTBP attenuated ACTN4-mediated cell migration. To achieve this, we generated stable ACTN4-overexpressing SaOs2-LM7 and U2OS cell lines or those with empty lentiviral vectors as controls. ACTN4 overexpression increased the migratory potential of both SaOs2-LM7 and U2OS cell lines (Figure 3c). As expected, MTBP overexpression using an MTBP-encoding adenoviral vector in control cells significantly suppressed the migratory potential of both cell lines. Importantly, simultaneous overexpression of MTBP in ACTN4-overexpressing cells reduced cell migration at similar levels to that of controls in both cell lines, suggesting that MTBP inhibited ACTN4-mediated cell migration (Figure 3c). We also examined whether MTBP downregulation increased migratory potential of cells depleted for ACTN4. We transfected control or ACTN4-downregulated cells with MTBP-specific siRNA. MTBP downregulation enhanced cell migration only in control cells, whereas it failed to do so in ACTN4-downregulated cells (Figure 3d), suggesting that an increase in cell migration by MTBP downregulation is at least partially dependent on ACTN4 in these cell lines. Intriguingly, in KHOS cells where ACTN4 overexpression failed to enhance migratory potential, MTBP overexpression still suppressed cell migration (Figure S3). These results suggest that MTBP affects cell migratory potential, at least in part, by suppressing ACTN4 function, but it may be involved in other pathways that lead to suppression of cell migration.
MTBP inhibits filopodia formation and cross-linking of F-actin induced by ACTN4
ACTN4 is an actin-binding protein that cross-links actin filaments into bundles to form filopodia (22). Considering this, we assessed the effect of MTBP on ACTN4-induced filopodia formation and found that MTBP overexpression significantly inhibited the filopodia formation induced by ACTN4 overexpression in both SaOs2-LM7 and U2OS cells (Figure 4a). Suppression of filopodia formation by MTBP was observed in a dose-dependent manner in both the cell lines (Figure S4). To elucidate the mechanism by which MTBP suppressed ACTN4-mediated filopodia formation, we performed actin bundling assays using luciferase (Figure 4b) as a negative control. ACTN4 successfully cross-linked F-actin into bundles, as its addition to the reaction increased the amount of actin in the pellet fraction. The amount of actin and ACTN4 in the pellet fraction was significantly reduced when MTBP was added, suggesting that ACTN4-mediated F-actin cross-linking was inhibited by MTBP (Figure 4b). These results indicate that MTBP attenuates ACTN4-mediated filopodia formation through inhibition of actin cross-linking. However, in KHOS cells where ACTN4 overexpression did not enhance filopodia formation, MTBP overexpression still suppressed filopodia formation (Figure S5), suggesting the possibility that MTBP affects filopodia formation via multiple pathways.
Figure 4.
MTBP inhibits ACTN4-mediated filopodia formation and cross-linking of F-actin. (a) SaOs2-LM7 (left) and U2OS (right) cells with or without ACTN4 overexpression were infected with empty (control) or MTBP-encoding (MTBP) adenoviruses. Forty eight (48) hours later, phalloidin staining was performed. Cells (n=100) were examined for filopodia formation. The percentages of cells positive for filopodia formation (top) and representative phalloidin staining pictures (bottom). (b) Actin bundling assay. In vitro translated MTBP and/or ACTN4 proteins were incubated with purified F-actin in the assay reaction, followed by protein sedimentation. Luciferase (Luc) was used as a negative control. Supernatant (S) and pellet (P) fractions were subjected to western blotting for actin, ACTN4, and MTBP (left). Graph showing the percentage of sedimented bundled F-actin (pellet) as analyzed by densitometry (right). Error bars are means ± S.D. from three independent experiments. **, P < 0.01; t test. N.S., not significant.
Nuclear localization of MTBP is dispensable for suppression of cell migration and filopodia formation mediated by ACTN4
MTBP localizes to both the nucleus and cytoplasm, while ACTN4 is largely limited to the cytoplasm (Figure 3b). We therefore hypothesized that nuclear localization of MTBP would not be required for inhibiting cell migration and filopodia formation mediated by ACTN4. To test this hypothesis, we generated a mutant MTBP lacking the capacity for nuclear localization (MTBPnls) by substituting alanine for lysine and arginine in the nuclear localization domain of MTBP. MTBPnls was exclusively localized to the cytoplasm (Figure 5a). Overexpression of MTBPnls in SaOs2-LM7 cells significantly suppressed ACTN4-induced cell migration (Figure 5b) and filopodia formation (Figure 5c). These results indicated that nuclear localization of MTBP is dispensable for suppression of cell migration and filopodia formation mediated by ACTN4. To examine if MTBPnls still interacted with ACTN4 and to understand which region of MTBP was required for the MTBP-ACTN4 interaction, we performed co-immunoprecipitation studies following transfection of MTBPnls and a series of deletion mutants into 293T cells which had undetectable levels of endogenous MTBP (Figure 5d, 293T). We found that MTBPnls did interact with endogenous ACTN4 in 293T cells and that the C-terminal region from amino acids 760 to 904 of MTBP was required for the MTBP-ACTN4 interaction (Figure 5d). Using proteins synthesized with the in vitro coupled Transcription/Translation Systems, we further confirmed the crucial role of this C-terminal region of MTBP in interaction with ACTN4 (Figure 5d, in vitro). Taken together, our study strongly suggests that inhibition of ACTN4 function through interaction with C-terminal region of MTBP appears to be one of the mechanisms by which MTBP suppresses tumor migratory potential.
Figure 5.

Nuclear localization of MTBP is not required for inhibiting ACTN4 function. (a) Nuclear localization signal (NLS) domain in MTBP with mutated amino acids (underlined, left). Immunofluorescence images demonstrating the localization of FLAG-tagged full-length MTBP (MTBP) and mutant MTBP (MTBPnls, right). (b) SaOs2-LM7 cells with or without ACTN4 overexpression were infected with lentiviral vectors encoding empty (control) or MTBPnls mutant (MTBPnls), followed by cell migration assays. Migrating cells were stained, and entire fields were counted. Relative cell migration (%) compared to the number of migrating cells in control (top) and representative staining pictures (bottom). (c) Phalloidin staining was performed using the same cells as in Figure 5b. Cells (n= 100) were examined for filopodia formation. The percentages of cells positive for filopodia formation (top) and representative phalloidin staining pictures (bottom). Error bars are means ± S.D. from three independent experiments. *, P < 0.05 and **, P < 0.01; t test. (d) 293T cells which had undetectable levels of endogenous MTBP were transfected with Flag-tagged full-length MTBP (MTBP), NLS mutant (MTBPnls) and deletion mutants (F4 and ΔC). A diagram showing regions deleted for MTBP mutants. Forty-eight hours later, co-immunoprecipitation (IP) studies were performed using ACTN4 antibody (A4), followed by western blotting for MTBP. Matched isotype antibodies (Iso) were used as negative controls. In vitro synthesized ACTN4 and mutant MTBP (MTBPnls, F4, and ΔC) were co-incubated for co-IP studies using ACTN4 or FLAG (FL) antibodies, followed by immunoblotting for MTBP or ACTN4.
Discussion
In this study, we have demonstrated that MTBP suppresses osteosarcoma metastasis in vivo and attenuates cell migration and filopodia formation by inhibiting ACTN4 function. ACTN4, which was first cloned by Honda et al. (13), plays a crucial role in cytoskeletal organization of the actin network by cross-linking F-actin (23, 24). Increased levels of ACTN4 enhance the motility of colorectal cancer cells (14), while its downregulation dramatically reduces the migratory potential of glioblastoma cells (25). Thus, ACTN4 has been implicated in cancer cell migration and metastasis (13, 14, 25–28).
Our results also indicate that MTBP suppresses cell migration via multiple pathways, as it suppressed the migration and filopodia formation of KHOS cells, which failed to respond to ACTN4 overexpression. These results suggest that additional proteins involved in cell migration and filopodia formation may also interact with and are affected by MTBP. Alternatively, MTBP may suppress cancer metastasis through mechanisms other than migration that are essential to the metastatic cascade. Further investigations are required to address these issues.
We also demonstrate that nuclear localization of MTBP is not required for suppressing filopodia formation and cell migration. MTBP mainly localizes to the nucleus, increasing the likelihood that MTBP has other biological functions than inhibition of ACTN4 function. To this end, we recently reported that MTBP is involved with mitotic progression and chromosome segregation (29). Since chromosome instability has an established association with cancer progression, maintenance of chromosome integrity by MTBP may be another pathway through which it suppresses cancer progression.
The recent discovery of a novel class of proteins, termed metastasis suppressors, provides novel diagnostic and therapeutic targets for cancer. Metastasis suppressors are defined as proteins whose expression results in the inhibition of a neoplastic cell’s ability to metastasize with little effect on primary tumor growth (30, 31). With our results illustrating that MTBP has little effect on primary tumor growth but it suppresses osteosarcoma metastasis, MTBP meets the criteria for a metastasis suppressor. Most of these proteins play a role in inhibiting tumor growth at the secondary sites, in addition to inhibiting other aspects of the metastatic cascade (30, 31). Whether MTBP exclusively inhibits cell migration or if it also suppresses colonization at secondary sites remains to be determined. Nonetheless, a clearer understanding of the biology of metastasis suppressors will greatly facilitate the discovery of novel therapeutic strategies aimed at cancer metastasis.
This study and several other reports support the role of MTBP in suppressing tumor progression (8, 10, 11, 29). However, two reports suggest an oncogenic role of MTBP; One is by Bradley et al. (32) in which MTBP overexpression resulted in inhibited MDM2 self-ubiquitination and subsequent p53 degradation. The other is by Bouska et al. (33) where reduced MTBP expression decreased Myc-induced B-cell lymphomagenesis. This discrepancy might suggest that MTBP’s function is context dependent. In fact, the expression levels of MTBP vary among different types of cancer. In B-cell lymphomas, an increased MTBP expression is observed as compared to normal human lymphoid tissue controls (34). On the other hand, recent publication by Vlatković et al. (11) indicates that loss of MTBP is associated with a worse prognosis of head and neck carcinoma patients and it serves as an independent prognostic marker of p53. Further investigations are required to understand the association of MTBP expression with clinicopathological relevance in various types of cancer, as well as context-dependency of the MTBP function. Nonetheless, our study illustrates one mechanism by which MTBP inhibits cancer metastasis independently of the MDM2-p53 pathway, which provides new insights into cancer cell migration and metastasis and may help advance therapies for metastatic disease.
Materials and methods
Cell lines and mice
U2OS and KHOS/NP (R-970-5) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). SaOs2-LM7 cells were kindly provided by Eugenie S. Kleinerman at the UT MD Anderson Cancer Center (35). Non-obese diabetic/severe combined immunodeficiency (NOD-scid) IL2Rγ-null mice at 6 weeks old were purchased from Charles River laboratories (Wilmington, MA) and used for intrafemoral injection studies. Mice were maintained under specific pathogen free conditions, and experimental procedures were performed according to the protocol approved by Institutional Animal Care and Use Committee.
Antibodies
For western blotting, rabbit polyclonal anti-ACTN4 (210-356-C050, Enzo Lifesciences, Exeter, UK), rabbit polyclonal anti-ACTN4 (19096-1-AP, ProteinTech, Chicago, IL), goat polyclonal anti-MTBP (K20, Santa Cruz Biotech, Santa Cruz, CA), mouse monoclonal anti-RGS-His (34610, Qiagen, Valencia, CA), rabbit polyclonal anti-actin (AAN01, Cytoskeleton, Denver, CO), mouse monoclonal anti-vinculin (10R-C105a, Fitzgerald, Acton, MA), mouse monoclonal vimentin (ab8978, Abcam, Cambridge, MA), rabbit polyclonal tropomyosin 3 (10737-1-AP, ProteinTech), and mouse monoclonal anti-β-tubulin (T0198, Sigma, St. Louis, MO) were used. For immunoprecipitation, rabbit polyclonal anti-ACTN4 (210-356-C050, Enzo Lifesciences), rabbit monoclonal anti-ACTN4 (GTX62422, GeneTex, Irvine, CA), mouse monoclonal anti-FLAG (M2, Sigma), and rabbit polyclonal anti-MTBP (hC2) were used. The hC2 antibody was generated by Bethyl Laboratories (Montgomery, TX) using a 17 amino acid synthetic peptide at the C-terminus of human MTBP (NNAVQVIDWVLEKTSKK). For immunofluorescence studies, rabbit polyclonal anti-MTBP (H-40, Santa Cruz), goat polyclonal anti-ACTN4 (C20, Santa Cruz), and mouse monoclonal anti-FLAG were used.
Viral vectors, plasmids, and siRNAs transfection
An adenoviral vector encoding mouse MTBP was described previously (10). Empty adenoviral vector was purchased from Vector Biolabs (Philadelphia, PA). The pCDH-EF1-MCS-T2A-copGFP lentiviral vector was purchased from System Biosciences (Mountain View, CA), and RGS-His-tagged ACTN4 or Flag-tagged MTBP or MTBPnls were inserted into this vector. The pGIPZ lentiviral vector encoding ACTN4-specific shRNA (RHS3979-9622968) was purchased from Open Biosystems (Lafayette, CO). Human MTBP-specific siRNA (GCCACAUUGAUUCACUCAGUU) and non-target siRNA were purchased from Dharmacon RNAi Technologies (Lafayette, CO). Transfection of siRNA was performed using HiPerFect (QIAGEN) transfection reagent according to manufacturer’s instructions. N-terminal FLAG-tagged full-length MTBP, MTBPnls, and deletion mutants were inserted into pEF/FRT/V5-D-TOPO vector (Invitrogen, Grand Island, NY) and transfected using jetPrime transfection reagent (Polyplus Transfection, New York, NY) according to manufacturer’s instructions.
In vitro cell migration assays
MTBP expression was manipulated in cells using an MTBP-encoding adenoviral vector or MTBP-specific siRNA. ACTN4 expression was stably modulated with lentiviral vectors encoding either ACTN4 cDNA or its specific shRNA. Empty vectors were used as controls. Migration assays were performed 36 hours after transfection/infection using 24-well transwell chambers (6.5 mm diameter, 8 μm pore size; Corning Inc, Corning, NY). Cell suspension (10,000) in 100 μl of 0.2% FBS-containing medium was added onto upper compartments of the chamber and 10% FBS in DMEM was added to the lower compartments as chemoattractant. Cells were then allowed to migrate across the membrane by incubating at 37°C in CO2 incubator for 12–16 hours. The non-migrating cells were removed from the upper face of the filters using cotton swabs, while the cells migrated to the lower face of the filters were fixed and stained with Diff-Quik Stain Set (Dade Behring, Newark, DE). Stained cells in the entire fields were counted under an inverted microscope.
Western blotting
Western blotting was performed using a standard method as previously described (29). To make whole cell extracts, we used RIPA buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% sodium deoxycholate, 0.1% Triton X-100, 0.1% SDS) supplemented with protease inhibitor cocktail.
Co-immunoprecipitation
Cells were lysed with RIPA or Cell Lytic M (Sigma) buffers supplemented with protease inhibitor cocktail (Roche, Indianapolis, IN) consisting of 1 mM PMSF, 0.2 mM sodium orthovanadate, and 100 mM sodium fluoride. Approximately 400–600 μg of whole cell lysates were incubated with protein-specific antibodies overnight at 4°C, followed by precipitation of the antibody-protein complex using protein A/G plus-agarose beads (Santa Cruz). In each immunoprecipitation experiment, a matched isotype antibody was used as a negative control. After washing four times with lysis buffer, precipitated protein complexes were extracted with 2xSDS sample buffer for western blotting.
Immunofluorescence and filopodia detection
Cells were grown on poly-D-lysine/laminin-coated glass coverslips (BD Biosciences, Franklin Lakes, NJ) in a 24-well plate. Following fixation of cells with freshly prepared 1% paraformaldehyde in PBS for 20 minutes, cells were permeabilized with 0.2% Triton-X 100 in PBS (PBS-T) for 3 minutes and were blocked in 3% BSA in PBS-T for 30 minutes, followed by incubation with appropriate protein-specific antibodies at room temperature for one hour or 4°C overnight. After PBS wash, cells were incubated with the appropriate fluorophore-conjugated secondary antibodies for one hour. To visualize F-actin for filopodia formation assays, cells were incubated with rhodamine-phalloidin (Invitrogen). Samples were mounted in the ProLong Gold Antifade Reagent with DAPI (Invitrogen), and results were analyzed with a Leica epifluorescence microscope (Leica Microsystems Inc., Bannockburn, IL).
F-actin bundling assay
MTBP and ACTN4 proteins were synthesized in vitro using the TnT Quick Coupled Transcription/Translation Systems (Promega, Madison, WI). F-actin stock was prepared by incubating 200 μg of non-muscle actin (Cytoskeleton) in 200 μl of general actin buffer (5mM Tris-HCl, pH 8.0 and 0.2 mM CaCl2) for 30 minutes on ice, followed by the addition of 20 μl of actin polymerization buffer (50 mM KCl, 2 mM MgCl2, and 1 mM ATP) and incubation for 1 h at room temperature. Thirty (30) μl of F-actin was mixed with translated MTBP (3.0 μl) and/or ACTN4 (10.0 μl), and incubated at room temperature for 30 minutes. The mixture was then centrifuged at 14,000g at room temperature for 30 minutes. Supernatant (S) and pellet (P) fractions were proportionally loaded on a SDS-polyacrylamide gel and subjected to western blotting for actin. Band intensities were quantified using a Bio-Rad VersaDoc imaging system (Bio-Rad, Hercules, CA).
Intrafemoral injection
Cells (1,000,000 cells) in 15 μl of HBSS were injected into the femoral bone marrow space of anesthetized NOD-scid IL2Rγ-null immunocompromised mice as previously described (36). Following tumor cell injection, bone wax was used to seal the hole to avoid the leakage. Mice were monitored daily for the development of primary tumors and signs of labored breathing or lethargy. Mice injected with KHOS and SaOs2-LM7 cells were dissected approximately 4 and 6 weeks after injections, respectively. Mice injected with empty vector-infected cells (control) and those with MTBP-overexpressing cells were sacrificed at the same day, and the weight of primary tumors and the number of metastatic pulmonary nodules (>0.5 mm) were determined by gross examination at a full necropsy.
Statistical analysis
Each experiment was performed independently at least thrice. Statistical analysis was performed using GraphPad Prism software (GraphPad Software). Statistical significance was assessed by comparing mean values (± SD) using Student’s t test. p<0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Drs. Indu Kheterpal, Yuki Tochigi, and Louis Marrero for technical assistance. We also thank Ms. Kristy-Le T. Nguyen for technical assistance and editing the manuscript. This work was supported by grants from P20 RR020152-02 (P.L.D), RSG-09-169-01-CSM (T.I), LCRC start-up (T.I.), and National Foundation for Cancer Research-Center for Metastasis Research (D.R.W.).
Abbreviations List
- MDM2
murine double minute
- MTBP
MDM2 binding protein
- ACTN4
alpha-actinin-4
- MEFs
mouse embryonic fibroblasts
- siRNA
short interfering RNA
- shRNA
short hairpin RNA
- PBS
phosphate buffered saline
- DMSO
dimethyl sulfoxide
- DAPI
4′,6-diamidino-2-phenylindole
- SDS
sodium dodecyl sulfate
- S.D
standard deviation
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author contribution: Neeraj Agarwal, Amit S. Adhikari, Swathi V. Iyer, Kevon Hekmatdoost, and Tomoo Iwakuma all performed experiments. Danny R. Welch provided us with crucial materials and discussion. Neeraj Agarwal, Swathi V. Iyer, and Tomoo Iwakuma wrote the manuscript.
Supplementary information is available at the Oncogene website (http://www.nature.com/onc).
References
- 1.Mina LA, Sledge GW., Jr Rethinking the metastatic cascade as a therapeutic target. Nat Rev Clin Oncol. 2011;8(6):325–32. doi: 10.1038/nrclinonc.2011.59. Epub 2011/04/20. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Xu Z, Wang Y. Recent advances in breast cancer metastasis suppressor 1. Int J Biol Markers. 2011;26(1):1–8. doi: 10.5301/jbm.2011.6267. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005;17(5):559–64. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9(6):446–54. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 5.Lindberg U, Karlsson R, Lassing I, Schutt CE, Hoglund AS. The microfilament system and malignancy. Semin Cancer Biol. 2008;18(1):2–11. doi: 10.1016/j.semcancer.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Kirfel G, Rigort A, Borm B, Herzog V. Cell migration: mechanisms of rear detachment and the formation of migration tracks. Eur J Cell Biol. 2004;83(11–12):717–24. doi: 10.1078/0171-9335-00421. [DOI] [PubMed] [Google Scholar]
- 7.Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65(9):687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd MT, Vlatkovic N, Haines DS. A novel cellular protein (MTBP) binds to MDM2 and induces a G1 arrest that is suppressed by MDM2. J Biol Chem. 2000;275(41):31883–90. doi: 10.1074/jbc.M004252200. [DOI] [PubMed] [Google Scholar]
- 9.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1(14):993–1000. [PubMed] [Google Scholar]
- 10.Iwakuma T, Tochigi Y, Van Pelt CS, Caldwell LC, Terzian T, Parant JM, et al. Mtbp haploinsufficiency in mice increases tumor metastasis. Oncogene. 2008;27(13):1813–20. doi: 10.1038/sj.onc.1210827. [DOI] [PubMed] [Google Scholar]
- 11.Vlatkovic N, El-Fert A, Devling T, Ray-Sinha A, Gore DM, Rubbi CP, et al. Loss of MTBP expression is associated with reduced survival in a biomarker-defined subset of patients with squamous cell carcinoma of the head and neck. Cancer. 2011;117(13):2939–50. doi: 10.1002/cncr.25864. Epub 2011/06/22. [DOI] [PubMed] [Google Scholar]
- 12.Koizumi T, Nakatsuji H, Fukawa T, Avirmed S, Fukumori T, Takahashi M, et al. The role of actinin-4 in bladder cancer invasion. Urology. 2010;75(2):357–64. doi: 10.1016/j.urology.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 13.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140(6):1383–93. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F, et al. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology. 2005;128(1):51–62. doi: 10.1053/j.gastro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Menez J, Le Maux Chansac B, Dorothee G, Vergnon I, Jalil A, Carlier MF, et al. Mutant alpha-actinin-4 promotes tumorigenicity and regulates cell motility of a human lung carcinoma. Oncogene. 2004;23(15):2630–9. doi: 10.1038/sj.onc.1207347. [DOI] [PubMed] [Google Scholar]
- 16.Hayashida Y, Honda K, Idogawa M, Ino Y, Ono M, Tsuchida A, et al. E-cadherin regulates the association between beta-catenin and actinin-4. Cancer Res. 2005;65(19):8836–45. doi: 10.1158/0008-5472.CAN-05-0718. [DOI] [PubMed] [Google Scholar]
- 17.Barbolina MV, Adley BP, Kelly DL, Fought AJ, Scholtens DM, Shea LD, et al. Motility-related actinin alpha-4 is associated with advanced and metastatic ovarian carcinoma. Lab Invest. 2008;88(6):602–14. doi: 10.1038/labinvest.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrix MJ, Seftor EA, Chu YW, Trevor KT, Seftor RE. Role of intermediate filaments in migration, invasion and metastasis. Cancer Metastasis Rev. 1996;15(4):507–25. doi: 10.1007/BF00054016. Epub 1996/12/01. [DOI] [PubMed] [Google Scholar]
- 19.Vignjevic D, Montagnac G. Reorganisation of the dendritic actin network during cancer cell migration and invasion. Semin Cancer Biol. 2008;18(1):12–22. doi: 10.1016/j.semcancer.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Choi HS, Yim SH, Xu HD, Jung SH, Shin SH, Hu HJ, et al. Tropomyosin3 overexpression and a potential link to epithelial-mesenchymal transition in human hepatocellular carcinoma. BMC Cancer. 2010;10:122. doi: 10.1186/1471-2407-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mierke CT. The role of vinculin in the regulation of the mechanical properties of cells. Cell Biochem Biophys. 2009;53(3):115–26. doi: 10.1007/s12013-009-9047-6. [DOI] [PubMed] [Google Scholar]
- 22.Weins A, Schlondorff JS, Nakamura F, Denker BM, Hartwig JH, Stossel TP, et al. Disease-associated mutant alpha-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci U S A. 2007;104(41):16080–5. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhina S, Wang YL, Murata-Hori M. Alpha-actinin is required for tightly regulated remodeling of the actin cortical network during cytokinesis. Dev Cell. 2007;13(4):554–65. doi: 10.1016/j.devcel.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao H, Wu C, Wells A. Phosphorylation of alpha-actinin 4 upon epidermal growth factor exposure regulates its interaction with actin. J Biol Chem. 2010;285(4):2591–600. doi: 10.1074/jbc.M109.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen S, Dong M, Kumar S. Isoform-specific contributions of alpha-actinin to glioma cell mechanobiology. PLoS One. 2009;4(12):e8427. doi: 10.1371/journal.pone.0008427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quick Q, Skalli O. Alpha-actinin 1 and alpha-actinin 4: contrasting roles in the survival, motility, and RhoA signaling of astrocytoma cells. Exp Cell Res. 2010;316(7):1137–47. doi: 10.1016/j.yexcr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi S, Honda K, Tsuda H, Hiraoka N, Imoto I, Kosuge T, et al. Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin Cancer Res. 2008;14(17):5348–56. doi: 10.1158/1078-0432.CCR-08-0075. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S, Tsuda H, Honda K, Onozato K, Takano M, Tamai S, et al. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod Pathol. 2009;22(4):499–507. doi: 10.1038/modpathol.2008.234. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal N, Tochigi Y, Adhikari AS, Cui S, Cui Y, Iwakuma T. MTBP plays a crucial role in mitotic progression and chromosome segregation. Cell Death Differ. 2011;18(7):1208–19. doi: 10.1038/cdd.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steeg PS, Ouatas T, Halverson D, Palmieri D, Salerno M. Metastasis suppressor genes: basic biology and potential clinical use. Clin Breast Cancer. 2003;4(1):51–62. doi: 10.3816/cbc.2003.n.012. [DOI] [PubMed] [Google Scholar]
- 31.Hurst DR, Welch DR. Metastasis suppressor genes at the interface between the environment and tumor cell growth. Int Rev Cell Mol Biol. 2011;286:107–80. doi: 10.1016/B978-0-12-385859-7.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brady M, Vlatkovic N, Boyd MT. Regulation of p53 and MDM2 activity by MTBP. Mol Cell Biol. 2005;25(2):545–53. doi: 10.1128/MCB.25.2.545-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28(15):4862–74. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odvody J, Vincent T, Arrate MP, Grieb B, Wang S, Garriga J, et al. A deficiency in Mdm2 binding protein inhibits Myc-induced B-cell proliferation and lymphomagenesis. Oncogene. 2010;29(22):3287–96. doi: 10.1038/onc.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia SF, Worth LL, Turan M, Duan XP, Kleinerman ES. Eradication of osteosarcoma lung metastasis using intranasal gemcitabine. Anticancer Drugs. 2002;13(2):155–61. doi: 10.1097/00001813-200202000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Adhikari AS, Agarwal N, Wood BM, Porretta C, Ruiz B, Pochampally RR, et al. CD117 and Stro-1 Identify Osteosarcoma Tumor-Initiating Cells Associated with Metastasis and Drug Resistance. Cancer Res. 2010;70(11):4602–12. doi: 10.1158/0008-5472.CAN-09-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.