Abstract
Previous studies showed that the serine/threonine kinase Unc51.1 is one of the earliest genes in neuronal differentiation and is required for granule cell axon formation. To examine the mechanism of Unc51.1 regulation of axon extension, we have identified two direct binding partners. The first, SynGAP, a negative regulator of Ras, is expressed within axons and growth cones of developing granule cells. Overexpression of SynGAP blocks neurite outgrowth by a mechanism that involves Ras-like GTPase cascade. The second binding partner is a PDZ domain-containing scaffolding protein, Syntenin, that binds Rab5 GTPase, the activity of which is attenuated by SynGAP. Thus, our results demonstrate that the Unc51.1-containing protein complex governs axon formation via Ras-like GTPase signaling and through regulation of the Rab5-mediated endocytic pathways within developing axons.
Keywords: Unc51.1, SynGAP, Syntenin, axon formation, vesicular membrane
The synaptic circuitry of the cerebellar cortex develops through the coordinated outgrowth of the axons and dendrites of the two principal classes of neurons, the granule cell and the Purkinje cell, and the subsequent formation of synaptic junctions among them. Parallel fibers of the granule cells form just after the granule cell progenitors exit the cell cycle, as an early step in the program of neuronal differentiation (Ramon y Cajal 1995). The cerebellar cortex provides an unique system for studying neurite outgrowth and connectivity. The granule cell, the most abundant neuron of the central nervous system (CNS), has facilitated analyses of the molecular and cellular mechanisms that underlie the basic steps in neuronal differentiation. The development of the cerebellar granule cell has been studied extensively, including mechanisms of proliferation, neuronal migration, axon formation, and synaptogenesis in vivo and in vitro.
To gain insight into initial steps of parallel fiber formation/axon outgrowth, we previously identified Unc51.1/Unc51.2, two murine homologs of the Caenorhabditis elegans unc-51 gene (Tomoda et al. 1999). In the worm, unc-51 is important for axon elongation (Hedgecock et al. 1985; Desai et al. 1988; Siddiqui 1990; McIntire et al. 1992; Ogura et al. 1994). The unc-51/Unc51.1/Unc51.2 genes belong to a subfamily of protein serine/threonine kinases. Unc51.1 is expressed in a number of neuronal populations, including the cerebellar granule cell, during development. The protein is localized to both axonal shafts and growth cones of extending axons. Retroviral infection of granule cell precursors with a kinase-deficient form of Unc51.1 demonstrates that Unc51.1 is essential for neurite extension/parallel fiber formation in cerebellar granule neurons (Tomoda et al. 1999). Others have recently reported that ULK1, a human ortholog of Unc51.1, interacts with the Golgi-associated ATPase enhancer of 16 kD (GATE-16) and with the γ2 subunit of GABA-A receptor associated protein (GABARAP; Okazaki et al. 2000). The GATE-16 is an essential factor for intra-Golgi transport (Sagiv et al. 2000), and the GABARAP has a possible role in the regulation of receptor trafficking (Wang et al. 1999). However, it is not yet known how these Unc51.1 binding partners function with Unc51.1 to regulate neurite formation.
To further investigate the molecular basis of Unc51.1/Unc51.2 function in neurite extension, we searched for additional proteins that bind Unc51.1/Unc51.2 in yeast two-hybrid screens. Interacting molecules identified included SynGAP, a synaptic GTPase-activating protein (GAP; Chen et al. 1998; Kim et al. 1998), and Syntenin, an endocytic vesicular membrane protein with PDZ domains (Grootjans et al. 1997; Fialka et al. 1999). SynGAP interacts with the PDZ domains of the PSD-95/SAP90 family of proteins via its C′-terminal amino acids, T/SXV (Kim et al. 1998), and is a major component of postsynaptic density (PSD), a dense cytoskeletal matrix found beneath the postsynaptic membrane that is notably prominent in excitatory synapses (Chen et al. 1998). SynGAP functions as a brain-specific Ras GTPase-activating protein and is associated in a large complex with PSD-95/SAP90, SAP102, and the NMDA receptors at excitatory synapses in cortex and hippocampus. Recent studies have shown that SynGAP contributes to neural plasticity by modulating Ras activity in brain (Komiyama et al. 2002; Kim et al. 2003). Syntenin contains two PDZ domains that bind cytoplasmic domains of various proteins, including syndecan, neurofascin, glutamate receptors, and ephrin-B2 (Grootjans et al. 1997; Lin et al. 1999; Koroll et al. 2001; Hirbec et al. 2003). Syntenin often serves as a scaffolding molecule for synaptic proteins (Hirbec et al. 2003).
In this report, we present evidence that SynGAP is expressed during early stages of granule neuron development and that Unc51.1 and SynGAP function in concert to regulate axon outgrowth of developing granule neurons. By using bioassays of primary granule neurons expressing either Unc51.1 or SynGAP or both, we show that Unc51.1 and SynGAP function cooperatively in axon formation during brain development. We also show that Unc51.1 affects the extent of SynGAP modulation of Ras activity, suggesting a functional link between Unc51.1 and SynGAP that could lead to long-term changes in gene expression responsible for axon formation. Syntenin provides a scaffold for Unc51.1 and for Rab5 GTPase, an endocytic membrane regulator, and SynGAP stimulates Rab5 GTPase activity in vitro. In addition, we show that the SynGAP-induced axon truncation is restored by overexpressing Ras or Rab5 GTPases, and we provide evidence that the dominant-negative Unc51.1 and SynGAP expression disorganizes vesicular membranes within extending axons of primary granule neuron culture. These studies suggest that Unc51.1 and its binding partners influence axon formation via the neuronal endocytic pathway.
Results
Unc51.1/Unc51.2 binds SynGAP
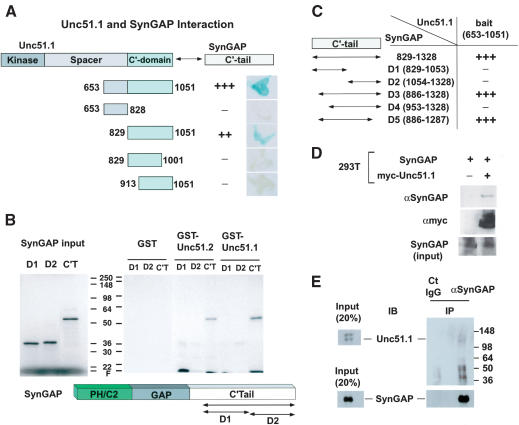
Previous studies suggested the importance of the C′-terminal domains of unc-51/Unc51.1/Unc51.2 in their biological function (Ogura et al. 1994; Tomoda et al. 1999). Thus, we used the C′-terminal region of Unc51.1 (amino acids 653–1051) and Unc51.2 (amino acids 531–1037) as baits to screen a yeast two-hybrid library prepared from postnatal day 6 (P6) mouse cerebellum. Screening of 3 million clones with Unc51.2 (amino acids 531–1037) and subsequent sequencing analyses revealed that one clone spanning ∼1.7 kb encoded the C′-terminal 500 amino acids of SynGAP (amino acids 829–1328). To examine whether SynGAP could also bind Unc51.1, the C′-terminal region of Unc51.1 (amino acids 653–1051) was tested in a yeast two-hybrid assay, and its association with Syn-GAP was confirmed (Fig. 1). This suggests that Unc51.1 and Unc51.2 may share similar structural features within their C′-terminal domains, as is expected from their primary sequences (70.8% similarity), and that they may function in overlapping molecular pathways.
Figure 1.
Unc51.1 binds SynGAP. (A) Summary of the yeast two-hybrid assay. The C′-terminal half of Unc51.1 (amino acids 653–1051) and its deletion mutants were tested for their ability to bind SynGAP α2 C′-tail (amino acids 829–1328) in a β-galactosidase filter assay. Relative binding strength, as judged by darkness of the blue color of representative colonies, is indicated: (+++) very strong; (++) strong; (+) weak; (–) no binding detected. (B) GST pull-down assay. The C′-tail of SynGAP (amino acids 829–1328) and its deletion mutants D1 (amino acids 829–1053) and D2 (1054–1328) were in vitro translated, labeled with [35S]-methionine, and applied to the columns made with GST fusion proteins (GST alone, GST–Unc51.1 C′-tail [amino acids 653–1051], or GST–Unc51.2C′-tail [amino acids 531–1037]). Bound proteins were eluted from the columns and analyzed with SDS-PAGE, followed by autoradiography. (C) Determination of the minimum region of SynGAP that binds Unc51.1. Various deletion mutants (D1–D5) of the C′-tail of SynGAP were tested for their ability to bind Unc51.1 C′-terminal domain (amino acids 653–1051) in a β-galactosidase assay. (D) Coprecipitation assay. A full-length SynGAP expression construct was transfected into HEK293T cells with or without the myc-tagged full-length Unc51.1 construct. Cell extracts were immunoprecipitated with a myc antibody, and the immune complex was analyzed by immunoblot using an anti-SynGAP antibody. (E) Membrane-enriched fractions extracted from P6 cerebellum were immunoprecipitated with either the anti-SynGAP antibody or the normal rabbit IgG, and the resulting immune complexes were analyzed with SDS-PAGE followed by immunoblot with an anti-Unc51.1 antibody and an anti-SynGAP antibody.
SynGAP exists as multiple alternatively spliced forms. There are at least seven variants in the 3′ portion of the SynGAP mRNA (α1, α2, β1, β2, β3, β4, and γ), and they encode five different protein isoforms (Li et al. 2001). The SynGAP clone isolated in our screen corresponded to the α2 isoform that encodes a protein 35 amino acids longer than that encoded by the α1 isoform, an isoform originally reported (Kim et al. 1998; Chen et al. 1998). The α1 isoform ends with TRV, through which it binds a PDZ domain of PSD-95. The α2 isoform has one base insertion within the C′-terminal tail region of the protein compared with the α1 isoform (Li et al. 2001), ends with ADH, and does not bind PSD-95 (data not shown). Both the α1 and α2 isoforms were expressed in P6 cerebellum as confirmed by RT–PCR, and both bound the Unc51.1/Unc51.2 baits, although the α2 isoform consistently displayed slightly higher affinity to the baits than did the α1 isoform in a yeast two-hybrid assay (data not shown). We reasoned that the C′-terminal sequence of SynGAP functions as a tag that determines subcellular localizations. We therefore generated GFP fusion proteins with the C′-tail domain of SynGAP α1 or α2, introduced them retrovirally into granule cells in culture, and found that the GFP-C′-tail (α2) was localized to extending axons but the GFP-C′-tail (α1) was only localized to cell soma (data not shown). This demonstrates that the α1 isoform with TRV motif could be the isoform that partitions into postsynaptic compartments, whereas the α2 isoform is the one that preferentially goes to axons. Therefore, we used only the α2 isoform in subsequent experiments.
To determine the minimum region of Unc51.1 that binds SynGAP, we generated a series of deletion constructs of Unc51.1 and tested them with SynGAP (amino acids 829–1328) in a β-galactosidase assay in yeast (Fig. 1A). Although the C′-terminal 399 amino acids of Unc51.1 (amino acids 653–1051), as well as the shorter C′-terminal domain of Unc51.1 (amino acids 829–1051), strongly bound SynGAP, the spacer region (amino acids 653–828) and subregions of the C′-terminal domain (amino acids 829–1001 and amino acids 913–1051) did not bind SynGAP. This result demonstrates that the C′-terminal domain of Unc51.1 (amino acids 829–1051) is the minimum requirement for the binding to SynGAP, and suggests an important role of this domain as a functional unit. It is noteworthy that this domain is highly conserved between the C. elegans unc-51 and the two murine homologs, Unc51.1 and Unc51.2 (Tomoda et al. 1999). To further confirm the binding of Unc51.1 to Syn-GAP, a glutathione S-transferase (GST) pull-down assay was performed. The bait regions of Unc51.1 (amino acids 653–1051) and Unc51.2 (amino acids 531–1037) were fused in-frame with a GST tag and tested for binding to the C′-terminal region of SynGAP (amino acids 829–1328) and its deletion mutants, D1 and D2 (amino acids 829–1053 and amino acids 1054–1328, respectively). In these assays, GST–Unc51.1 and GST–Unc51.2 specifically bound SynGAP (Fig. 1B). Smaller deletions within the C′ terminus of SynGAP did not bind to the GST fusion proteins, suggesting that the bulk of the C′-terminal domain of SynGAP is involved in binding to Unc51.1/Unc51.2. To determine the minimum region of SynGAP that binds Unc51.1, a series of deletion constructs of SynGAP (D1-D5) were generated and tested with the bait region of Unc51.1 in a yeast two-hybrid assay. Although deletion mutants, D1 and D2, did not bind Unc51.1 as was the case in the GST pull-down assay, the central region of SynGAP C′-tail (D5, amino acids 886–1287) was necessary for the binding (Fig. 1C).
To examine whether the full-length Unc51.1 binds the full-length SynGAP, a coimmunoprecipitation assay was done in a mammalian heterologous expression system. HEK293T cells were transfected with the full-length SynGAP expression construct with or without a construct expressing myc-tagged Unc51.1. The cell extracts were immunoprecipitated with a myc antibody, and the resulting immune complexes were tested for the presence of SynGAP. The antibody we used recognizes the GAP domain of SynGAP (Kim et al. 2003). SynGAP specifically interacted with Unc51.1 in this assay (Fig. 1D).
To confirm interaction of Unc51.1 with SynGAP in vivo, membrane-enriched fractions were prepared from P6 cerebellum and immunoprecipitated with an anti-SynGAP antibody to test for the presence of Unc51.1 in the resulting immune complex. Unc51.1 was specifically detected in the immunoprecipitates with SynGAP but not in the complex immunoprecipitated with the control normal rabbit IgG protein (Fig. 1E). Taken together, the results indicate that the Unc51.1 and SynGAP interact via their C′-terminal domains.
SynGAP expression during cerebellar development
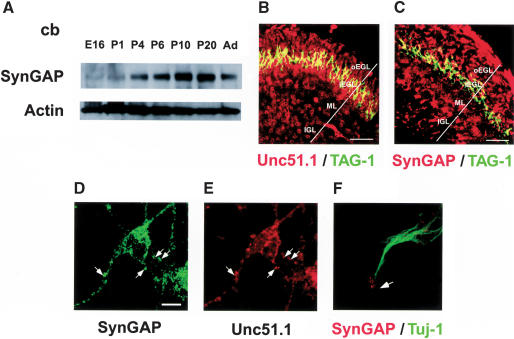
To further characterize SynGAP expression during cerebellar development, we prepared protein extracts from varying developmental stages of the cerebellum, starting from embryonic day 16 (E16) through an adult stage (P40), and analyzed them with Western blot using the anti-SynGAP antibody. SynGAP was detected as early as E16, and the levels of expression increased as the development proceeds (Fig. 2A). Expression reached maximum around P20, when synaptogenesis is actively ongoing in cerebellum, and expression was maintained into adulthood. Based on previous reports, SynGAP is thought to have a role for synaptic transmission or postsynaptic membrane organization (Komiyama et al. 2002; Kim et al. 2003). Therefore, it is notable that SynGAP is expressed before P4, when there are very few postsynaptic structures present. To further confirm the cellular distribution of SynGAP, we immunostained sagittal sections of the cerebellum prepared from P6 with the anti-SynGAP antibody. Granule cells were brightly stained in all stages of development (Fig. 2C). A similar pattern of staining was observed with an Unc51.1-specific antibody (Fig. 2B).
Figure 2.
Unc51.1 and SynGAP expression in developing cerebellum. (A) SynGAP protein is expressed throughout cerebellar development. Protein extracts prepared from varying stages of developing cerebellum (E16, P1, P4, P6, P10, P20, and adult) were analyzed by immunoblot. (B,C) Cellular distribution of Unc51.1 (B) and SynGAP (C) revealed by the antibody staining over P6 cerebellar sagittal sections (60 μm). TAG-1 antibody was used to mark differentiating granule cells in deeper external granular layer (iEGL). (oEGL) Outer external germinal layer that contains proliferating granule cells; (ML) molecular layer; (IGL) internal granular layer. (D,E) Subcellular distribution of SynGAP (D) and Unc51.1 (E) in granule neurons in culture. Arrows indicate punctae double-positive for Unc51.1 and SynGAP. (F) SynGAP localization in a growth cone (arrow). Tuj-1 was used to stain granule cell axons. Bar: D–F, 5 μm.
SynGAP is localized to axons
To provide a detailed view of the SynGAP protein localization within developing granule neurons, granule cells were purified from P6 mouse cerebellum, a time period when granule cell precursors are exiting the cell cycle and commencing differentiation. Purified granule neurons were cultured in vitro for 36 h and stained with the SynGAP antibody (GAP domain antibody). SynGAP immunoreactivity was seen in the cell soma and neurites of the granule cells and, at the growing tip of axon, the growth cone (Fig. 2D,F). Tuj-1, an antibody against the neuron-specific class III β-tubulin, was used to mark the axonal shafts of the granule cells. When double-stained with an Unc51.1 antibody, many SynGAP-positive domains of cells were also positive for Unc51.1 expression, suggesting possible colocalization of Unc51.1 and SynGAP proteins in granule cells (Fig. 2D,E).
Membrane localization of Unc51.1 and SynGAP
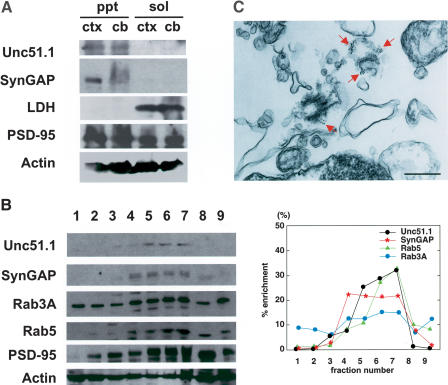
As a first step toward addressing the biological or biochemical processes that Unc51.1 and SynGAP proteins regulate, we examined the subcellular distribution of the proteins in intact neurons. After high-speed centrifugation of extracts from E16 cerebral cortex and P6 cerebellum, Unc51.1 and SynGAP were detected in the membrane fraction (Fig. 3A). To further analyze the subcellular localization of Unc51.1 and SynGAP, a step-wise sucrose gradient experiment was performed on the protein homogenates prepared from P6 cerebellum. Unc51.1 and SynGAP were enriched in fractions that were positive for the endocytic vesicular membrane marker Rab5 (Fig. 3B). In contrast, PSD-95, actin, and another vesicular membrane marker, Rab3A, were present in all fractions. To confirm the subcellular localization of Unc51.1, immunoelectron microscopy was carried out on cultures of purified P6 granule neurons by using an Unc51.1-specific antibody. Unc51.1-positive gold beads were specifically found within the cytoplasm bound to a subset of intracellular vesicles of single neurites (Fig. 3C). These experiments demonstrate that Unc51.1 is a vesicular membrane-associated protein within developing granule neuron processes.
Figure 3.
Membrane localization of Unc51.1 and SynGAP. (A) Both Unc51.1 and SynGAP are detected in crude membrane fractions, not in soluble fraction, of protein extracts prepared from E16 cerebral cortex (ctx) and P6 cerebellum (cb). LDH was exclusively identified in soluble fraction, and PSD-95 and actin were fractionated equally in membrane and soluble fractions. (B) Step-wise sucrose gradient fractionation of proteins extracted from P6 cerebellum. Each fraction (the top fraction, 1, toward the bottom fraction, 9) was analyzed with immunoblot with antibodies indicated; “% enrichment” of each protein (= [densitometry of a band from each fraction/total densitometry of bands from all fractions] × 100) was plotted in the graph. (C) An immunoelectron micrograph showing vesicular localization of Unc51.1 within an extending neurite of granule cells in culture. Bar, 300 nm.
SynGAP overexpression inhibits granule cell axon formation
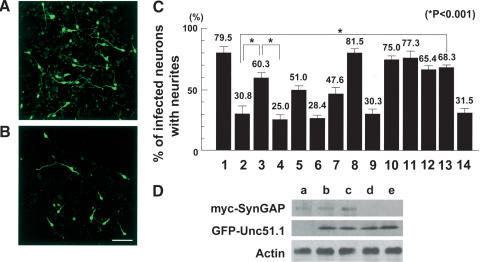
We previously showed that Unc51.1 is expressed in growth cones and functions in axon formation of the granule cells (Tomoda et al. 1999). The finding that SynGAP is expressed in growth cones prompted us to examine the role of the protein in the regulation of axon outgrowth. First, we analyzed the effect of SynGAP overexpression in granule cells in culture. We infected dissociated granule cells with a recombinant retrovirus expressing the full-length SynGAP. Our retroviral constructs contained a green fluorescent protein (GFP) driven by an internal β-actin promoter to serve as a marker for virally infected cells. Whereas ∼80% of cells infected with a control virus extended long neurites (50–200 μm), only ∼30% of cells expressing the virally encoded SynGAP extended neurites. (The rest of 70% cells infected extended short processes of only 5–20 μm in length; Fig. 4A–C, columns 1 and 2.) This finding indicates that SynGAP functions as a negative regulator of axon outgrowth. We have previously shown that overexpression of a kinase-deficient form of the Unc51.1 serine/threonine kinase in granule cells resulted in severe truncation of axons (Tomoda et al. 1999). Because we found that SynGAP and Unc51.1 interact and that SynGAP overexpression also results in axon truncation, we explored the interaction of both proteins. To assess the effect of Unc51.1 kinase activity on axon truncation caused by SynGAP, we coexpressed the SynGAP and Unc51.1 (either the wild-type or the kinase-deficient form) in granule cells and analyzed axon extension.
Figure 4.
SynGAP inhibition of cerebellar granule cell axon outgrowth and its rescue by Unc51.1. (A,B) Granule cells were purified from P6 cerebellum and infected with retroviruses expressing either GFP alone (A) or SynGAP + GFP (B), and axon outgrowth was visualized by a GFP antibody staining. Bar, 50 μm. (C) Quantitation of granule cells with neurites. Granule cells were infected with retroviruses expressing GFP alone (column 1), SynGAP + GFP (column 2), SynGAP + GFP–Unc51.1.wt (column 3), SynGAP + GFP–Unc51.1.KR (column 4), SynGAP + GFP–Unc51.1.wt/ΔC′-domain (column 5), SynGAP/ΔC′-tail + GFP (column 6), SynGAP/ΔC′-tail + GFP–Unc51.1.wt (column 7), GFP–Unc51.1.wt (column 8), GFP–Unc51.1.KR (column 9), SynGAP/Δ414-652+ GFP (column 10), GFP–H-Ras.wt (column 11), GFP–H-Ras17 (column 12), SynGAP + GFP–H-Ras.wt (column 13), or SynGAP + GFP–H-Ras17 (column 14), and the percentage of cells harboring neurites (>20 μm) were scored. A hundred cells per experiment were scored for the presence or absence of neurites and were counted from at least three individual experiments: 79.5% ± 6.2% (column 1), 30.8% ± 7.0% (column 2), 60.3% ± 4.5% (column 3), 25.0% ± 4.5% (column 4), 51.0% ± 2.7% (column 5), 28.4% ± 2.0% (column 6), 47.6% ± 6.1% (column 7), 81.5% ± 5.0% (column 8), 30.3% ± 5.4% (column 9), 75.0% ± 3.2% (column 10), 77.3% ± 5.5% (column 11), 65.4% ± 3.4% (column 12), 68.3% ± 1.8% (column 13) or 31.5% ± 4.8% (column 14) of cells expressing the indicated proteins extended neurites. Statistically significant differences (p < 0.001) were observed between columns 2 and 3, 3 and 4, and 2 and 13, as indicated by asterisks. (D) Expression levels of the proteins from the retroviruses used were evaluated. Equivalent titer (∼5 × 106 cfu) of each virus was used to infect NIH3T3 cells (∼1 × 106), and 48 h after the start of infection, cells were harvested and subjected for Western blotting by using myc and GFP antibodies. An actin antibody was used to ensure equal protein loading. Retroviruses used were myc-SynGAP + GFP (lane a), myc-SynGAP + GFP–Unc51.1.wt (lane b), myc-SynGAP + GFP–Unc51.1.KR (lane c), GFP–Unc51.1.wt (lane d), and GFP–Unc51.1.KR (lane e).
To achieve coexpression of the two proteins, SynGAP was placed in a multicloning site of the retroviral vector and its expression was driven by 5′-LTR promoter, whereas Unc51.1 was expressed as a fusion protein with GFP (GFP–Unc51.1), the expression of which was driven by an internal β-actin promoter. This expression scheme ensured that the two proteins were coexpressed within single neurons and allowed us to use GFP as a morphological marker. We evaluated the protein expression from these retroviral vectors by infecting NIH3T3 cells and confirmed that the expression from the 5′-LTR was not affected by the internal expression unit driven by the β-actin promoter (Fig. 4D). As shown previously, expression of the kinase-deficient form of Unc51.1 alone resulted in truncated neurites, whereas ∼80% of cells that express wild-type Unc51.1 generated long neurites (Fig. 4C, columns 8 and 9), consistent with our previous report (Tomoda et al. 1999). When SynGAP was expressed alone, only ∼30% of granule cell extended axons. However, when SynGAP and wild-type Unc51.1 were coexpressed, ∼60% of cells extended neurites (Fig. 4C, columns 2 and 3). In contrast, only 25% of the cells overexpressing SynGAP together with the kinase-deficient form of Unc51.1 extended neurites (Fig. 4C, column 4). Thus, Unc51.1 abrogated the effect of SynGAP on neurite extension, an effect that was dependent on Unc51.1 kinase activity. To address significance of Unc51.1–SynGAP interaction in axon formation, we coexpressed SynGAP and a form of Unc51.1 that lacks the SynGAP binding domain (ΔC′-domain, animo acids 829–1051). Approximately 50% of cells extended neurites (Fig. 4C, column 5), suggesting the importance of Unc51.1 binding to SynGAP in its rescuing activity. To further assess the significance of interaction of the two proteins, we coexpressed SynGAP/ΔC′-tail, that lacks Unc51.1 binding domain (animo acids 829–1328), with the full-length wild-type Unc51.1. Although expression of SynGAP/ΔC′-tail alone resulted in ∼28% neurite extension, a level comparable to that of the full-length SynGAP expression (Fig. 4C, columns 2 and 6), expression of Syn-GAP/ΔC′-tail with wild-type Unc51.1 resulted in 47% neurite extension, a lower level of recovery compared with the coexpression of full-length SynGAP and Unc51.1 (Fig. 4C, columns 3 and 7). These results demonstrate the significance of interaction of SynGAP and Unc51.1 in granule cell neurite extension.
Previous studies have addressed a role of Ras signaling in neurite extension in PC12 cells and in neuroblastoma cell lines (Bar-Sagi and Feramisco 1985; Noda et al. 1985; Eggert et al. 2000). In these reports, Ras has been reported to be a positive regulator of neurite extension. Because SynGAP is a Ras GTPase-activating protein that attenuates Ras signaling, SynGAP may affect axon extension via its GAP activity on Ras or Ras-like proteins. To investigate whether the GAP domain of SynGAP was responsible for neurite shortening, we constructed a mutant protein that deleted most of the GAP domain (SynGAP/Δ414–652), and overexpressed it retrovirally in granule cells. As shown in Figure 4C (column 10), ∼75% of the granule cells expressing SynGAP/Δ414–652 showed normal neurite lengths in vitro (50–200 μm), underscoring a role for GAP activity of SynGAP in axon extension. To determine whether Ras functions as an effecter of SynGAP in neurite extension, we coexpressed SynGAP with either GFP-fused wild-type H-Ras or a dominant-negative H-Ras17 in granule cells. When wild-type Ras or Ras17 was expressed alone, ∼77% or ∼65% of granule cells extended axons, respectively (Fig. 4C, columns 11 and 12). When SynGAP was coexpressed with wild-type Ras, ∼68% of granule cells extended axons (Fig. 4C, column 13). In contrast, when SynGAP and Ras17 were coexpressed, only 31% of cells extended neurites (Fig. 4C, column 14). This suggests that SynGAP activity in granule cell neurite extension may be through regulation of the Ras pathway.
Unc51.1 inhibits SynGAP activity
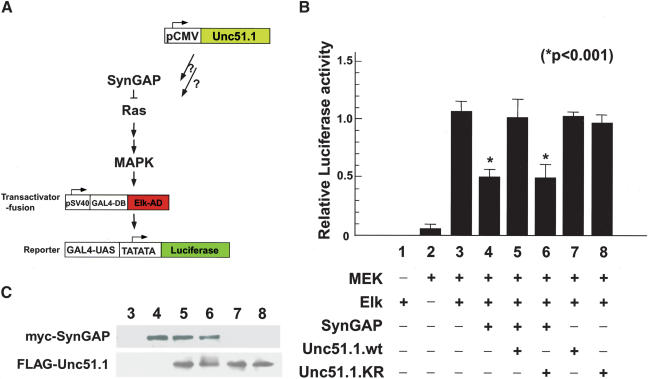
To explore the potential influence of SynGAP and Unc51.1 in Ras signaling, we used the PathDetect trans-reporting system (Stratagene), in which the luciferase reporter gene activity reflects the activation of Elk1 by MAP kinases in a mammalian heterologous expression system (Fig. 5A). Elk1 is a known substrate of the MAP kinase ERK, and functions as a transactivator that responds to the upstream Ras-MAP kinase signaling (Chang and Karin 2001). When HEK293 cells were transfected with MEK1 (MAP kinase kinase) and the GAL4-Elk-AD construct, a strong induction of luciferase activity was observed (Fig. 5B, column 3). When SynGAP was cotransfected, the reporter induction was attenuated by 50% (Fig. 5B, column 4). When wild-type Unc51.1 was cotransfected with SynGAP, the luciferase activity recovered to the control level (Fig. 5B, column 5). In contrast, cotransfection of the kinase-deficient form of Unc51.1 did not result in the recovery of the activity (Fig. 5B, column 6), suggesting that the Unc51.1 kinase activity inhibits SynGAP activity. To rule out the possibility that Unc51.1 activates Ras-MAP kinase signaling by itself, either the wild-type or the kinase-deficient form of Unc51.1 was transfected in the absence of SynGAP. Neither construct affected the luciferase activity (Fig. 5B, columns 7 and 8), indicating that Unc51.1 functions through SynGAP and does not directly affect Ras-MAP kinase signaling pathway in the assay conditions we tested. To control for the variation of transfection efficiency in this assay system, we used the second luciferase reporter (Renilla luciferase) to normalize the data. We also evaluated the level of protein expression from the Unc51.1 and SynGAP expression constructs and confirmed equivalent level of the protein expression in each condition (Fig. 5C).
Figure 5.
Influence of Unc51.1 and SynGAP on Ras-MAP kinase activity in a reporter assay and in primary granule cells in culture. (A) A reporter assay scheme. Effects of overexpression of SynGAP with or without Unc51.1 (either wild-type or the kinase-deficient form, KR) in HEK293 cells were evaluated by measuring the Ras-MAP kinase-Elk pathway-induced luciferase reporter activation. (B) HEK293 cells were transfected with various combinations of constructs as indicated, and the relative luciferase activity was expressed as a ratio of the relative light unit (RLU) normalized against the control RLU level. Total amount of DNA used for transfection was normalized with an empty vector (pcDNA3). The data are the mean ± S.E. of reporter activities from three independent experiments. Asterisks show significant decrease in the reporter activity (p < 0.001). (C) Expression levels of the proteins were evaluated by Western blotting using myc and Flag antibodies to confirm that equivalent amount of the proteins were used in the assays. Lane numbers (lanes 3–8) correspond to column numbers in B.
Previous reports showed that SynGAP is a Ras GTPase-activating protein that negatively regulates Ras signaling in vitro (Kim et al. 1998). Recent analyses of SynGAP knock-out mice confirm that Ras-MAP kinase signaling pathway is altered in mutant neurons (Komiyama 2002). Because SynGAP overexpression truncates axons of granule cells, we tested whether this effect resulted from attenuated MAP kinase signaling within affected neurons. We addressed this question by isolating granule cells and infecting them with high-titer retroviruses expressing either SynGAP or control protein (GFP). After 36 h of infection, the time period when we observed axon truncation, cells were harvested and assayed for MAP kinase signaling by Western blotting using antibodies against the whole MAP kinases (anti-p42/p44) as well as the phospho-specific (active form-specific) antibodies (anti-phospho-p42/p44). We observed no significant difference in the rate of active MAP kinase between the two populations, suggesting that the basal level of MAP kinase activities were not affected by SynGAP overexpression in granule cells (data not shown). This suggests that the SynGAP-Ras effect may be mediated through pathways other than that involving MAP kinase, or that SynGAP may have a downstream effecter other than Ras-MAP kinase pathway within granule cells, although it is possible that our measurement was not sensitive enough to detect transient or local changes in the MAP kinase activity within granule cells.
SynGAP stimulates Rab5 GTPase activity
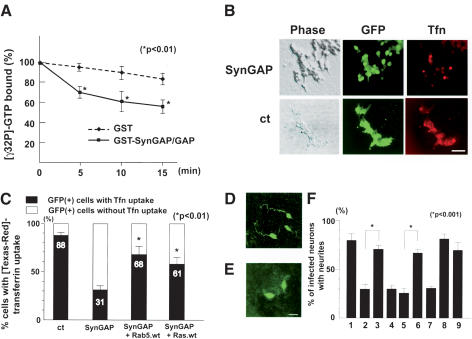
We have shown that SynGAP attenuates the Ras signaling pathway and that SynGAP-induced inhibition of neurite extension is restored by Ras coexpression in granule cells. Others have shown that Ras regulates endocytic pathways (Tall et al. 2001), and that p120 Ras GAP stimulates the GTPase activity of Rab5 in vitro (Liu and Li 1998). Rab5 is a member of the Ras-like small GTPases known to be a marker for early endosomes and is essential for endocytic membrane fusion and trafficking. To test whether SynGAP could modulate Rab5 GTPase activity in vitro, we generated a GST fusion protein of the GAP domain of SynGAP and incubated it with a purified Rab5 GST fusion protein in the presence of [γ-32P] GTP. GST–Rab5 fusion proteins exhibited a low intrinsic GTPase activity in the absence of a GAP domain of SynGAP. In contrast, addition of the GST–SynGAP/GAP domain fusion protein stimulated the GTPase activity of Rab5 (Fig. 6A). The functional interaction of SynGAP and Rab5 was consistent with our sucrose gradient fractionation experiments above, in which Rab5 cofractionated with Unc51.1 and SynGAP (Fig. 3B). Thus, SynGAP might regulate Rab5-mediated endocytosis in developing neurons.
Figure 6.
SynGAP functions as a Rab5 GAP. (A) The GAP domain of SynGAP stimulates Rab5 GTPase activity in vitro. Time course of the GTPase activity of the GST–Rab5 fusion protein in the presence or absence of the GAP domain of SynGAP. Incubation of a GST–Rab5 with a control GST protein (diamonds) had minimum effect on the slow intrinsic GTPase activity of GST–Rab5. In contrast, addition of the GST–SynGAP/GAP fusion protein accelerated the GTPase activity of GST–Rab5 (squares). Asterisks show significant decrease in the amount of bound [γ-32P]-GTP at each time point (p < 0.01). (B) HEK293T cells were transiently transfected with either SynGAP + GFP or GFP alone, and assayed for their ability to uptake [Texas-red]-conjugated transferrin. Transfected cells were identified by GFP epifluorescence. Although a majority of control cells showed transferrin uptake, fewer cells displayed uptake after SynGAP overexpression. Bar, 50 μm. (C) Quantitation of cells uptaking transferrin after SynGAP or SynGAP + Rab5/Ras overexpression. GFP-positive cells with or without transferrin were scored and counted from three independent experiments. The data are mean ± S.E. of percentages of transferrin-positive cells (closed column) or transferrin-negative cells (open column); 88.2% ± 3.6%, 31.5% ± 4.1%, 68.0% ± 8.0%, or 61.3% ± 4.9% of GFP-positive cells were transferrin-positive when transfected with vector + GFP, SynGAP + GFP, SynGAP + Rab5.wt + GFP, or SynGAP + Ras.wt + GFP plasmids, respectively. Asterisks show significant increase in transferrin uptake (p < 0.01) compared with SynGAP-induced down-regulation of the uptake. (D) Cerebellar granule cells infected with retroviruses expressing both SynGAP and GFP–Rab5 restored axon extension. (E) Cerebellar granule cells infected with retroviruses expressing SynGAP and GFP showed truncated morphology. Infected cells and morphology were determined by anti-GFP antibody staining. Bar, 10 μm. (F) Quantitation of granule cells harboring neurites. Granule cells were infected with retroviruses expressing GFP alone (column 1), SynGAP + GFP (column 2), SynGAP + GFP–Rab5.wt (column 3), SynGAP + GFP–Rab5.S34N (column 4), Unc51.1.KR + GFP (column 5), Unc51.1.KR + GFP–Rab5.wt (column 6), Unc51.1.KR + GFP–Rab5.S34N (column 7), GFP–Rab5.wt (column 8), or GFP–Rab5.S34N (column 9), and the percentage of cells harboring neurites (>20 μm) was scored. A hundred cells per experiment were scored for the presence or absence of neurites and were counted from at least three individual experiments. Data for columns 1 and 2 were carried over from Figure 4C: 79.5% ± 6.2% (column 1), 30.8% ± 7.0% (column 2), 72.0% ± 3.2% (column 3), 32.3% ± 3.0% (column 4), 25.8% ± 4.5% (column 5), 66.2% ± 3.2% (column 6), 33.1% ± 1.6% (column 7), 82.0% ± 6.1% (column 8), or 69.4% ± 8.3% (column 9) of cells expressing the indicated proteins extended neurites. Statistically significant differences (p < 0.001) were observed between columns 2 and 3, and 5 and 6, as indicated by asterisks.
To further investigate whether SynGAP functions as a Rab5 GAP, we overexpressed SynGAP and performed a transferrin uptake assay. Transferrin is a conventionally used marker for endocytosis, and its uptake by cells is known to reflect their endocytic membrane activity (Hopkins et al. 1990; Lamaze et al. 1996). Although 88% of the control cells expressing GFP alone took up Texas-red conjugated transferrin, only 31% of cells expressing SynGAP and GFP took up transferrin (Fig. 6B,C). When Rab5 or Ras was cotransfected with SynGAP, the percentage of cells that took up transferrin recovered to 68% or 61%, respectively. Thus, these results show that SynGAP functions as a Rab5 GAP and also as a RasGAP. Immunoblot was done to ensure that equivalent amounts of SynGAP proteins were expressed in this experiment (data not shown).
To study whether SynGAP-induced neurite truncation is mediated via attenuation of Rab5 pathway, we coexpressed Rab5 with SynGAP in granule cells and examined neurite extension. Although ∼30% of cells infected with the SynGAP expressing retrovirus extended axons (Fig. 6E,F, column 2), ∼72% of cells infected with a virus coexpressing SynGAP and wild-type Rab5 extended axons (Fig. 6D,F, column 3), and ∼32% of cells infected with a virus coexpressing SynGAP and a dominant-negative Rab5 (Rab5.S34N) extended axons (Fig. 6F, column 4). In control experiments, ∼82% of cells infected with a virus expressing wild-type Rab5 alone extended neurites (Fig. 6F, column 8), and ∼69% of cells infected with the Rab5.S34N expressing virus extended neurites (Fig. 6F, column 9). This demonstrates that the SynGAP effect on axon formation was mediated not only by the Ras signaling pathway but also by the Rab5 pathway in granule cells. To examine if Rab5 pathway can also reverse the neurite inhibition caused by Unc51.1.KR expression, we coexpressed Unc51.1.KR with Rab5. When Unc51.1.KR was coexpressed with wild-type Rab5, ∼66% of cells extended neurites (Fig. 6F, column 6). In contrast, when Unc51.1.KR was coexpressed with Rab5.S34N, ∼33% of cells extended neurites (Fig. 6F, column 7). Expression levels of the proteins from the retroviruses used were also evaluated as we previously did (Fig. 4D and its legend), and we confirmed that the equivalent amount of SynGAP, Unc51.1.KR, or Rab5 proteins was expressed in columns 2–4; 5–7; or 3, 4, 6–9, respectively, in Figure 6F (data not shown).
Membrane organization by Unc51.1 and SynGAP
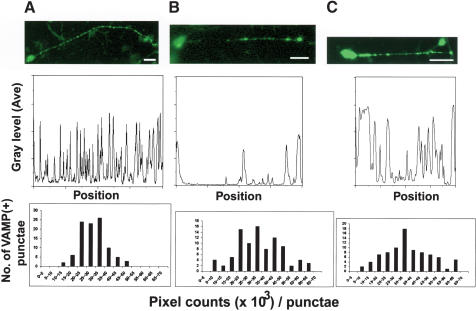
Our electron-microscopic observation of the membrane localization of Unc51.1, as well as the fact that SynGAP functions as a Rab5 GAP, prompted us to investigate a role of Unc51.1 and SynGAP in vesicular membrane regulation within extending axons. To directly address whether Unc51.1 or SynGAP overexpression could alter vesicular membrane structure and/or localization within extending axons, we infected granule cells with retroviruses designed to coexpress either SynGAP or the kinase-deficient Unc51.1 together with VAMP2–GFP fusion protein, a marker for subcellular membrane vesicles. In this assay, granule cells were induced to extend axons by plating them on a coated dish at the same time as we applied recombinant viruses, allowing granule cells to start extending axons prior to any sign of protein production from the infected viruses, thus enabling observation of vesicular structures within extending axons. At 24–36 h after the start of infection, we fixed and stained the culture with an antibody against GFP. Although cells in control culture had regularly aligned vesicular structures of uniform sizes along axonal shafts (Fig. 7A), a majority of cells overexpressing either SynGAP (∼54%) or the kinase-deficient Unc51.1 (∼60%) had irregularly aligned vesicles of various sizes within axons (Fig. 7B,C). They occasionally displayed abnormally large varicosities at the growth cones (∼10% of cells expressing SynGAP, and ∼8% of cells expressing the kinase-deficient Unc51.1), which were rarely seen in control cultures (data not shown). To measure the number and interval of punctate vesicular structures along the axon, line-scans were done for the neurons shown in the top panels of Figure 7, and the results were plotted in the middle graphs under each panel. To quantify the size of each VAMP2-positive vesicle, ∼100 vesicular structures along several axons were randomly chosen from each infection experiment, and the pixel size for each vesicle was measured. As shown in the bottom graphs of Figure 7, a majority of VAMP2-positive vesicles in the control culture were intermediate in size (20 ∼ 35 × 103 pixels/punctae). In contrast, the pixel size of vesicles in neurons infected with SynGAP or Unc51.1.KR viruses ranged from smaller to larger size compared with that of the control. The results demonstrate that the vesicular structure along the extending axons is disorganized after SynGAP or Unc51.1.KR overexpression.
Figure 7.
Disorganized vesicular membrane structures caused by Unc51.1.KR or SynGAP expression in cultured granule neurons. (Top) Although control cells expressing VAMP2–GFP fusion protein alone displayed regularly aligned VAMP-positive vesicles along axons (A), cells overexpressing SynGAP plus VAMP2–GFP (B) or Unc51.1.KR + VAMP2–GFP (C) had irregularly positioned vesicles. Bar, 10 μm. (Middle) Line-scan was done on the neurons shown above, and average gray levels were plotted along the entire length of the axons by using Metamorph analysis software (Universal Imaging Corporation). A peak within graphs corresponds to each VAMP2–GFP-positive punctae. (Bottom) Pixel counts per each punctae were done over ∼100 punctae for each infection experiment (VAMP2–GFP, left; SynGAP + VAMP2–GFP, center; Unc51.1.KR + VAMP2–GFP, right) by using Metamorph software, and each punctae was classified according to its pixel size. The X-axis shows the range of pixel sizes, and the Y-axis is the number of punctae that falls into each pixel size category shown in the X-axis.
Syntenin binds Unc51.1 and Rab5
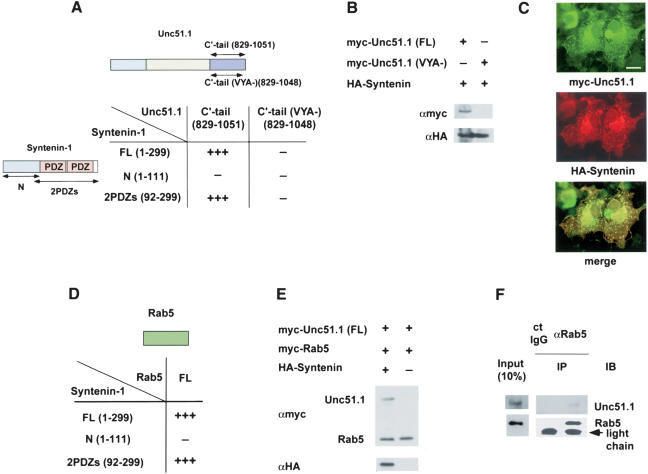
Our experiments on Unc51.1–SynGAP interactions as well as the finding that SynGAP functions as a regulator for Rab5-mediated endocytosis suggest the possibility that Unc51.1–SynGAP protein complex may function as a general regulator for endocytic pathway in neurons. In an effort to find a direct link between Unc51.1 and an endocytic pathway, we identified Syntenin as a binding partner for Unc51.1 in a yeast two-hybrid assay. Syntenin is an endocytic vesicular membrane protein with two PDZ domains (Grootjans et al. 1997; Fialka et al. 1999). Syntenin was found to bind Unc51.1 C′-tail domain (amino acids 653–1051; Fig. 8A). Because the last three amino acids of Unc51.1 are VYA, which is one of the typical PDZ binding consensus motifs (Harris and Lim 2001), we tested whether this VYA motif is responsible for Syntenin binding in a β-galactosidase assay in yeast as well as in a heterologous expression system in COS7 cells. In a two-hybrid assay, Unc51.1 C′-tail domain interacted specifically with the two PDZ domains of Syntenin. This interaction was abolished when the VYA motif was deleted (Fig. 8A). In the heterologous expression system, the HA-tagged full-length Syntenin bound the myc-tagged full-length Unc51.1 but not with the Unc51.1 lacking VYA (Fig. 8B). Thus, Unc51.1 specifically binds a scaffolding protein, Syntenin. In contrast, although SynGAP α1 isoform (with TRV motif at the C′ termini) bound the PDZ domains of Syntenin, SynGAP α2 isoform (with ADH at the C′ termini) did not bind Syntenin in a yeast two-hybrid assay (data not shown). Interaction of Unc51.1 and Syntenin was further confirmed by immunostaining of COS7 cells cotransfected with myc-Unc51.1 and HA-Syntenin. Almost all Unc51.1-positive signal (green) overlapped with Syntenin-positive signal (Fig. 8C, red). When myc-Unc51.1 lacking VYA and HA-Syntenin were coexpressed in COS7 cells, essentially the same pattern of staining was obtained as in Figure 8C (data not shown), although Unc51.1 (VYA–) and Syntenin did not bind in the yeast two-hybrid assay and in the coimmunoprecipitation assay. This suggests that the subcellular localization of Unc51.1 is not solely determined by Syntenin.
Figure 8.
Syntenin binds Unc51.1 and Rab5. (A) The C′-tail domain of Unc51.1 (amino acids 829–1051) and that deleting the last three amino acids (VYA–; amino acids 829–1048) were tested for their ability to bind the full-length Syntenin as well as its deletion mutants (N that spans the N′-terminal half of Syntenin, and 2PDZs that spans the two PDZ domains of Syntenin) in a β-galactosidase filter assay. Unc51.1 C′-tail displayed very strong binding (+++) toward the full-length Syntenin and the two PDZ domains of Syntenin. (B) COS7 cells were cotransfected with the HA-tagged full-length Syntenin and the myc-tagged full-length Unc51.1 or the Unc51.1 with VYA deletion. Immunoprecipitation was done with an anti-HA antibody, and the immune complex was analyzed with SDS-PAGE followed by immunoblot with an anti-myc antibody. The HA blot indicates the presence of Syntenin in the immune complex. (C) COS7 cells were cotransfected with the myc-Unc51.1 and HA-Syntenin constructs, fixed, and double-stained with a monoclonal myc (green) and a polyclonal HA antibodies (red). Bar, 10 μm. (D) The full-length Rab5 was tested for its ability to bind the full-length Syntenin as well as deletion mutants (N and 2PDZs) in a β-galactosidase filter assay. Rab5 displayed very strong binding (+++) toward the full-length Syntenin and the two PDZ domains of Syntenin. (E) COS7 cells were transfected with the myc-tagged full-length Unc51.1 and the myc-tagged Rab5 with or without the HA-tagged Syntenin. Immunoprecipitation was done with an anti-Rab5 antibody, and the immune complex was analyzed with SDS-PAGE followed by immunoblot with an anti-myc and an anti-HA antibodies. COS7 cells were cotransfected with the HA-tagged full-length Syntenin and the myc-tagged full-length Unc51.1 or the Unc51.1 with VYA deletion. Immunoprecipitation was done with an anti-HA antibody, and immune complex was analyzed with SDS-PAGE followed by immunoblot with an anti-myc antibody. The HA blot indicates the presence of Syntenin in the immune complex in a transfection-dependent manner. (F) Membrane-enriched fractions extracted from P6 cerebellum were immunoprecipitated with either the anti-Rab5 antibody or the normal rabbit IgG, and the resulting immune complexes were analyzed with SDS-PAGE followed by immunoblot with an anti-Rab5 antibody and an anti-Unc51.1 antibody.
Because both Rab5 and Syntenin are endocytic proteins, we tested whether Syntenin can link to several endocytic proteins. In an additional two-hybrid assay, we found that Syntenin binds Rab5, an early endosomal marker, with high affinity (Fig. 8D) and binds Rab7, a late endosomal marker, with lesser affinity. In contrast, Syntenin did not bind other small GTPases, including H-Ras, Rab3A, Rab6, RhoA, Rac1, and Cdc42, in yeast two-hybrid assays (data not shown). To determine the region of Syntenin that binds Rab5, either the N′-terminal domain or two PDZ domains of Syntenin were tested for interaction with Rab5 in a yeast two-hybrid assay. Two PDZ domains of Syntenin bound Rab5 specifically (Fig. 8D).
As Syntenin has been reported to act as a scaffolding protein that recruits several proteins, we next examined whether Syntenin could form a multiprotein complex with Unc51.1 and Rab5. When a coimmunoprecipitation experiment was done with an anti-Rab5 antibody from the extracts of COS7 cells transfected with HA-tagged Syntenin, myc-tagged Unc51.1 and myc-tagged Rab5, we found that Rab5 brought down both Unc51.1 and Syntenin (Fig. 8E). Rab5 did not bring down Unc51.1 in the absence of Syntenin. This suggests that Syntenin functions as a scaffolding protein that recruits both Rab5 and Unc51.1. To study whether these proteins form protein complex in vivo, proteins extracted from membrane-enriched fractions prepared from P6 mouse cerebellum were immunoprecipitated with an anti-Rab5 antibody, and the resulting immune complex was analyzed by SDS-PAGE. Unc51.1 was specifically identified within this immune complex (Fig. 8F), consistent with the sucrose gradient experiment (Fig. 3B). Thus, Unc51.1 appears to regulate axon extension, at least in part, through SynGAP, which attenuates both Ras and Rab5 signaling, and Syntenin provides a scaffold for Unc51.1 and endocytic machinery, which regulates endocytic pathway necessary for axon formation.
Discussion
Previous studies have established that members of the unc-51/Unc51.1/Unc51.2 gene family have an important role in axon formation of various neuronal cell types (Ogura et al. 1994; Tomoda et al. 1999). However, the signaling pathways in which the unc-51/Unc51.1/Unc51.2 genes function have, until now, remained poorly understood. Here we report the identification of SynGAP and Syntenin as binding partners for Unc51.1 and show that Unc51.1 and SynGAP colocalize within developing axons and are likely to work cooperatively to regulate axon length by modulating the Ras-like GTPase signaling pathway. The present results reveal a key linkage between the Unc51.1 function and the Ras-like GTPase signaling, which has previously been shown to act in axon extension of some neuronal cell lines (Bar-Sagi and Feramisco 1985; Noda et al. 1985; Eggert et al. 2000). In addition, our work suggests a possibility that the protein complex Unc51.1/SynGAP/Syntenin/Rab5 provides a scaffolding complex that regulates the endocytic membrane organization necessary during axon formation.
Binding of Unc51.1 with SynGAP
In the current experiments, we provided evidence that Unc51.1 binds SynGAP in a series of binding assays. In particular, the C′-terminal domain of Unc51.1 binds the C′-terminal half of SynGAP in the following assay systems: (1) yeast two-hybrid assay, (2) GST pull-down assay, and (3) coprecipitation assay in a mammalian heterologous expression system. Moreover, immunocytochemical experiments demonstrated that Unc51.1 and SynGAP colocalize within cell bodies, axonal shafts, and growth cones of extending granule cells in culture. Previous studies on SynGAP localization have been restricted to analyses of synaptic specialization of dendrites of mature cortical and hippocampal pyramidal neurons (Chen et al. 1998; Kim et al. 1998). Our studies underscore the potential role of these proteins at earlier stages of development.
Role of Unc51.1 and SynGAP in axon outgrowth
Ras has been reported to have a role for neurite formation in PC12 cells and neuroblastoma cell lines (Bar-Sagi and Feramisco 1985; Noda et al. 1985; Eggert et al. 2000). It has recently been shown that SynGAP has a Ras GAP activity toward H-Ras in a GTPase stimulating assay in vitro (Kim et al. 1998). In the present study, we showed that overexpression of SynGAP inhibited axon outgrowth of granule cells in culture, and that Ras coexpression with SynGAP reversed the axon inhibition effect. Therefore, one of the primary functions of SynGAP in the growing axons appears to be the attenuation of Ras signaling. This conclusion is supported by our reporter assay in a mammalian heterologous expression system, demonstrating the down-regulation of Ras-MAP kinase signaling by SynGAP. Moreover, overexpression of Syn-GAP lacking the GAP domain (SynGAP/Δ414–652) did not result in axon truncation, suggesting an important role of the GAP activity in the regulation of axon outgrowth. In addition, Unc51.1 partially rescued axon outgrowth defects in granule cells infected with SynGAP, likely through the ability of the Unc51.1 kinase to down-regulate the SynGAP activity. This is supported by the results from the reporter assay, in which Unc51.1 kinase activity decreased the effect of down-regulation of Ras signaling by SynGAP without directly affecting the Ras signaling in our reporter assay. The importance of physical and functional interaction of Unc51.1 and SynGAP was further supported by the neurite extension assay, in which SynGAP lacking the Unc51.1-binding domain (SynGAP/ΔC′-tail), or Unc51.1 lacking the SynGAP-binding domain (Unc51.1/ΔC′-domain.Δaa.829–1051) resulted in diminished capacity of Unc51.1 to rescue Syn-GAP-induced neurite inhibition. In our previous study, a dominant-negative version of Unc51.1/ΔC′-domain displayed diminished capacity to inhibit axon outgrowth (Tomoda et al. 1999). Taking our present data into account, it is likely that the C′-terminal domain of Unc51.1 is important in coordinating interaction with SynGAP and thereby regulating axon formation in concert with SynGAP. Taken together, the results suggest that Unc51.1 kinase activity counteracts the GAP activity of SynGAP, allowing the Ras signaling pathway to function in axon formation. At present, it is unclear how Unc51.1 functions to lower the SynGAP activity in the luciferase reporter assay and in the neurite extension assay. To date, we have not observed a direct phosphorylation of SynGAP by Unc51.1 in an in vitro kinase assay (data not shown). It remains possible that other cellular factor(s) facilitate Unc51.1 kinase activity in cells, as has been reported for Apg1 kinase, a yeast counterpart of Unc51.1 protein (Kamada et al. 2000). It also remains to be studied whether the MAP kinase pathway acts downstream of Unc51.1 and SynGAP to regulate axon formation in granule cells. In our experiments, a basal level of MAP kinase activity appears to be unaltered in granule cells overexpressing SynGAP. However, the MAP kinase pathway may have a role in a growth factor-induced or a guidance cue-dependent axon outgrowth or growth cone turning event, as has been reported recently in other neuronal cell types (Perron and Bixby 1999; Forcet et al. 2002).
In summary, SynGAP and Unc51.1 activities appear to be integrated to control neurite outgrowth through regulation of Ras-like small GTPases in developing granule neurons. Although SynGAP negatively regulates axon formation, Unc51.1 positively regulates axon extension by counteracting SynGAP via direct interaction, and the balance between Unc51.1 and SynGAP activities may ultimately determine axon length of the developing granule neurons.
Membrane localization of Unc51.1 and SynGAP and their possible role in membrane regulation
Immunoelectron microscopy of granule cells provided evidence that Unc51.1 associates with membrane vesicles. This interpretation is supported by our finding that Unc51.1 and SynGAP cofractionate in membranes in sucrose gradient fractionation. In addition, we found that SynGAP stimulates the GTPase activity of Rab5, a major regulator of endocytic vesicular organization. Among many Rab proteins, a role for Rab3A in synaptic vesicle recycling has been extensively studied (Lin and Scheller 2000). Rab5 is also localized to synaptic vesicles within neurons (Fisher von Mollard et al. 1994), and its role in endocytosis has been well documented in various non-neuronal cell types (Somsel Rodman and Wandinger-Ness 2000). However, its role in neurons has been less clear compared with that for Rab3A. In this report, we found that SynGAP functions as a Rab5 GAP in vitro and in a transferrin uptake assay. In addition, we found that overexpression of SynGAP or the kinase-deficient Unc51.1 resulted in abnormal vesicular structures within extending axons of granule cells. Moreover, we showed that SynGAP, as well as Unc51.1.KR-induced attenuation of axon extension, is restored by coexpression of wild-type Rab5, but not by a dominant-negative Rab5 in granule cells. Taken together, the results suggest that Unc51.1 and SynGAP are the regulators of membrane vesicles and organize membranes through the Rab GTPase pathway, thereby contributing to axon formation. Although Ras is considered to be important for a long-term maintenance of gene expression necessary for axon formation, the Rab GTPase pathway could be important for the development of endocytic processes during axon extension. The fact that both Ras and Rab5 restored the SynGAP-induced inhibition of axon outgrowth may suggest that SynGAP signals through multiple downstream effecters, or that Ras and Rab5 pathways functionally interact downstream of SynGAP. In this regard, it is interesting to note that Ras has been reported to regulate endocytic pathways through Rab5 GEF (Tall et al. 2001). At present, it is not clear whether the pathway downstream of SynGAP is mediated through Ras or Rab5 or both in developing granule cells. The exact mechanisms of how Ras or Rab5 functions in axon extension also remain elusive, as neither dominant-negative Ras nor Rab5 alone significantly affected axon formation. It is possible that SynGAP to Ras-like small GTPase pathway may not be a simple linear mode but rather a parallel signaling pathway, or it may be too complex to analyze because the exact physiological influence of Ras/Rab5 dominant-negative mutants on granule cells is unknown.
In support of our hypothesis, electron microscopic abnormalities of axons have been reported, such as enlargement of the diameter, atypical membranous vesicles, large varicosities at nerve termini, and cisternae-like structures in unc-51 mutant worm (McIntire et al. 1992). In addition, a recent report describes an abnormal positioning of presynaptic vesicles, as revealed by EGFP–VAMP marker, within axons of unc-51 mutants in C. elegans (Crump et al. 2001). Previously, the nematode Unc-14 was identified to bind Unc-51, and it was suggested that both unc-51 and unc-14 function cooperatively in axon formation (Ogura et al. 1997). The unc-14 mutant worms display phenotypes closely similar to those of unc-51 in C. elegans (McIntire et al. 1992). There has been no homolog of unc-14 gene found in mammals yet, but it is recently reported that the Unc-14 protein has a domain called RUN (Callebaut et al. 2001) that is a novel motif speculated to bind to several types of Ras-like GTPases, including Rab. It may therefore be interesting to see whether the unc-51/unc-14 pathway also regulates a Rab GTPase signaling pathway in C. elegans.
Recent evidence from yeast genetic screens searching for genes responsible for regulation of autophagy identified a gene, Apg1, a yeast gene structurally related to unc-51/Unc51.1/Unc51.2 (Matsuura et al. 1997). Autophagy is characterized by dynamic rearrangement of subcellular membranes and plays an important role for survival during nutrient deprivation in yeast (for review, see Klionsky and Emr 2000; Ohsumi 2001). Therefore, it would be interesting to see whether a role of Unc51.1 and its binding partners in axonal membrane organization may mechanistically resemble that of Apg1 during autophagy in yeast.
Role of Unc51.1 and synaptic protein complex in axon formation
Taken together, the binding assays and gene expression assays suggest that Unc51.1 binding to SynGAP regulates axon outgrowth, through the regulation of Ras or Rab5 signaling pathway. Although Ras is known to be a master switch that coordinates multiple signal transduction cascades (Marshall 1996; Olson and Marais 2000), Rab is considered to be a rather local switch that controls specific membrane fusion events during endocytosis and exocytosis (Ferro-Novick and Novick 1993; Pfeffer 1994; Zerial and McBride 2001). In addition, we found that Syntenin binds Rab5, but not with other small GTPases such as Ras, Rho family of GTPases, and Rab3A. This provides direct evidence that Syntenin is enriched in endocytic membranes (Fialka et al. 1999). Syntenin contains PDZ domains and is therefore thought to act as a scaffolding protein (Harris and Lim, 2001). Our results that Syntenin binds both Rab5 and Unc51.1 and that SynGAP functions as a Rab5 GAP argue for the existence of protein complexes that function in axon formation through regulation of endocytic membranes. Signals upstream of Unc51.1 and SynGAP may determine composition (both number and kind) of other protein members included in the complex/scaffold in a time- and location-specific manner, which might ultimately function in fine-tuning neuronal morphology. Antibodies against both Unc51.1 and SynGAP show a punctate pattern of staining, and in some area of the growing neurites, the two proteins are colocalized. Understanding the dynamics of protein localization within extending axons will likely yield more information on the trafficking of other synaptic components that may function during early steps of axon formation. Recent studies by others suggest a potential role of endocytosis in axon guidance, in which chemorepulsive guidance cues such as semaphorin 3A and ephrin 2A appear to induce endocytosis of axonal membranes during growth cone navigation (Castellani and Rougon 2002; Jurney et al. 2002; Zimmer et al. 2003). Others have also reported that endocytic machinery operates downstream of several axon guidance receptors that includes Robo and L1 (Kamiguchi and Lemmon 2000; Georgiou and Tear 2002; Keleman et al. 2002). In this regard, it will be necessary to investigate whether the scaffolding protein complex that contains Unc51.1 would function downstream of these guidance cues through regulation of endocytic membrane organization. In addition, a recent report suggests a new role of Rab5 for the regulation of synaptic vesicle size in Drosophila photoreceptor cells (Shimizu et al. 2003). Considering that Rab5 could regulate axon formation with Unc51.1 (Fig. 6) and that Unc51.1.KR induced disorganized membrane vesicles within extending granule cells (Fig. 7), it is interesting to study whether Unc51.1/Rab5-containing scaffolding complex would function in axon formation via a mechanism that involves synaptic vesicle size regulation.
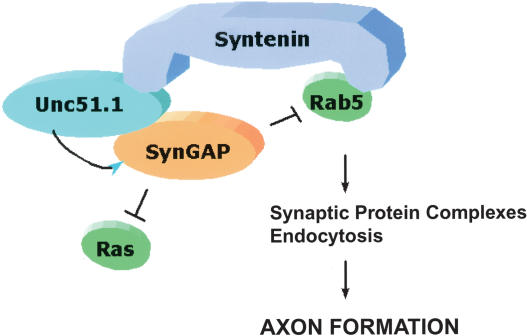
The central findings of this study are summarized in Figure 9. Within growing axons or at the growth cones, Unc51.1 down-regulates the GAP activity of SynGAP, which normally attenuates Ras and Rab5 signaling, thereby allowing Ras and Rab5 signaling to function in axon formation. In addition, Syntenin serves as a scaffold for Unc51.1 and Rab5. Together with our previous studies, these findings indicate a unique role for Unc51.1 in the early steps of cerebellar granule cell axon formation through a Rab5-mediated endocytic pathway. The protein complex that we described here, Unc51.1/SynGAP/Rab5/Syntenin, is the first to be shown to function in controling membrane organization essential during axon extension. Whereas previous attention has focused on guidance molecules and regulators of the cytoskeleton such as Rho family of small GTPases (Tessier-Lavigne and Goodman 1996; Mueller, 1999; Dickson 2002), our studies highlight the importance of protein assemblies with membrane vesicles that control endocytic pathways essential for axon outgrowth.
Figure 9.
Diagrammatic representation of the proposed mechanism by which Unc51.1 and its binding partners function in axon formation. Unc51.1 down-regulates the GAP activity of SynGAP, which allows Ras/Rab5 GTPases to function in axon formation, in part through endocytic pathways.
Materials and methods
Yeast two-hybrid assays
The yeast two-hybrid system was used to find proteins that interact with the C′-terminal domains of Unc51.1 and Unc51.2, according to the manufacture's procedures (ProQuest Two-Hybrid System, GIBCO-BRL/Life Technologies). The C′-terminal domains of Unc51.1 (amino acids 653–1051) and Unc51.2 (amino acids 531–1037) were generated by PCR by using a pair of oligonucleotides with restriction sites for Sal1 and Not1 (for Unc51.1: sense, 5′-GAGGTCGACACCACCTCGGAACC GTAC-3′; antisense, 5′-CTCGCGGCCGCTCAGGCATAG ACACCACTCAG-3′; for Unc51.2: sense, 5′-GAGGTCGACCCTCACCGATATCTATCAG-3′, antisense, 5′-CTCGCGGCCGCTCACACAGTTGCAGTGCTAC-3′) and were subcloned into the pPC97 yeast vector containing the GAL4 DNA-binding domain. The bait plasmid was transformed into MaV203 yeast cells together with a random-primed cDNA library from P6 mouse cerebellum subcloned into the Sal1/Not1 site of the pPC86 vector containing the GAL4 transcription activation domain. Positive clones were selected on plates lacking leucine, tryptophan, and histidine with 30 mM 3-aminotriazole and confirmed by filter assay for β-galactosidase activity.
Yeast interaction studies
The yeast two-hybrid system was used to check for interaction of SynGAP (amino acids 829–1328) with various deletion mutants of the Unc51.1 C′-terminal domain as well as for interaction of the full-length or deletion mutants of Syntenin with the C′-terminal tail of Unc51.1 or with Rab5. The Unc51.1 C′-terminal deletion mutants (amino acids 653–1051, 653–828, 829–1051, 829–1001, 913–1051, and 829–1048) and Rab5 (amino acids 1–215) were amplified by PCR and subcloned into pPC97. The C′-terminal domain of SynGAP (amino acids 829–1328) and the full-length (amino acids 1–299) or the deletion mutants (amino acids 1–111, amino acids 92–299) of Syntenin were subcloned into pPC86. The yeast vectors were cotransformed into MaV203 cells, and the resulting colonies were cultured without leucine and tryptophan and were tested for β-galactosidase activity by filter assays.
GST pull-down assay
Direct association of the C′-terminal domain of Unc51.1 and Unc51.2with the C′-terminal domain of SynGAP was analyzed by the pull-down assay using GST–Unc51.1/Unc51.2 fusion proteins. The C′-terminal domains of Unc51.1 (amino acids 653–1051) and Unc51.2 (amino acids 531–1037) were excised from pPC97 and subcloned in-flame into Sal1/Not1 site of pGEX4T-2 (Amersham-Pharmacia) to yield the GST–Unc51.1/Unc51.2fusion proteins. The construct was then introduced into BL21 bacteria (Stratagene), and following an induction of protein expression with isopropyl-β-D-thiogalactopyranoside (IPTG), the protein was purified on glutathione-Sepharose 4B beads. Deletion mutants of SynGAP (amino acids 829–1328 for the C′-terminal domain, 829–1053 for D1, and 1054–1328 for D2) were subcloned into the pRK5 vector, and the deletion mutant proteins were generated as [35S]-methionine-labeled proteins by in vitro transcription/translation reaction and applied to the GST column loaded with Unc51.1 or Unc51.2C′-terminal domains. The column was washed extensively, and the protein bound was analyzed by SDS-PAGE followed by autoradiography.
Coimmunoprecipitation assay
Coimmunoprecipitation assays in heterologous expression systems were done as follows. In brief, constructs for Unc51.1, SynGAP, Syntenin, or Rab5 were subcloned into pRK5 vector and transfected into HEK293T or COS7 cells with various combination indicated. Cells were homogenized in a TNE150 buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA) containing 1% Triton X-100, 0.1% deoxycholate (DOC), and 1 mM phenylmethylsulfonyl fluoride (PMSF) and centrifuged at 10,000 × g for 10 min to remove insoluble materials. The resulting lysate was mixed with a monoclonal antibody against myc (9E10), followed by the addition of Protein G plus/Protein A agarose (Oncogene Science) for precipitation of protein complex. The agarose pellet was washed five times with TNE150 supplemented with 0.5% Triton X-100. For in vivo coimmunoprecipitation experiments, the Protein A-antibody complex was prepared by preincubating 2μg of the affinity-purified SynGAP antibody (a gift from Richard Huganir, Johns Hopkins University, Baltimore, MD) or Rab5 antibody (Santa Cruz) with 50 μL of 1:1 slurry of Protein A-Sepharose for 1 h, and Protein A-antibody complex was centrifuged down at 2000 rpm for 2 min. The clarified supernatant of solubilized P2 fraction of a P6 cerebellar homogenate was then added to the Sepharose beads and the mixture incubated for 2h at 4°C. The mixture was washed once with immunoprecipitation buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 5 mM EDTA, 5 mM EGTA, and 1 mM PMSF) supplemented with 1% Triton X-100, twice with immunoprecipitation buffer with high salt (300 mM NaCl) plus 1% Triton X-100, and then three times with immunoprecipitation buffer. The proteins were eluted by Laemmli sample buffer (Laemmli 1970) and separated by SDS-PAGE. The gel was transferred to a polyvinylidine difluoride (PVDF, Millipore) membrane, and the membrane was blocked and immunoblotted. The antibodies used in the immunoblotting were anti-HA antibody (Covance, polyclonal HA.11, 1:500), anti-myc antibody (Oncogene Science, monoclonal, 9E10, 1:100), anti-SynGAP GAP domain antibody (Kim et al. 2003; polyclonal, 1:200), anti-Unc51.1 antibody (guinea pig polyclonal, 1:100), anti-Syntenin antibody (Synaptic Systems, polyclonal, 1:200), and anti-Rab5 antibody (Santa Cruz, polyclonal, 1:200).
GAP assay
The GAP assay was performed as described (Kim et al. 1998). In brief, 0.5–1 μM of GST–Rab5 was incubated with 20 nM [γ-32P]GTP (6000 Ci/mmole, DuPont-NEN) in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 1 mg/mL bovine serum albumin, and 1 mM dithiothreitol (DTT) for 10 min at room temperature. GST–SynGAP/GAP or control GST protein (200 ng) was added in 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 1 mM DTT and incubated for the indicated times. After stopping the reaction by adding five volumes of ice-cold 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 1 mM DTT, the mixture was filtered through hemagglutinin (HA) membranes (Millipore), and the filters were counted for 32P in scintillation cocktail.
Transferrin-uptake assay
The SynGAP or Rab5 cDNAs were subcloned into pRK5-myc vector and transiently transfected into 293T cells, together with EGFP expressing plasmid (pcDNA3-EGFP), using Lipofect-AMINE 2000 (Invitrogen). Twenty-four hours posttransfection, cells were incubated in serum-free medium for 1 h before the addition of 1 mg/mL bovine serum albumin and 20 μg/mL Texas Red-conjugated transferrin (Molecular Probes) for 20 min. After a 10-min acid incubation (50 mM glycine at pH 3.0, 100 mM NaCl) at 4°C, the cells were washed three times in PBS and fixed with 4% paraformaldehyde. The percentage of dye positive cells was calculated for a set of three experiments. In each experiment, >100 GFP-positive cells were scored.
Electron microscopy
Cerebellar granule cells prepared from P6 mouse were cultured at high density (10 million/35-mm plastic dish) for 24 h, fixed in 4% paraformaldehyde/0.25% glutaraldehyde/100 mM cacodylate (pH 7.4) for 30 min on ice, and then permeabilized in 0.05% Triton X-100/PBS for 5 min on ice. The cells were incubated with the anti-Unc51.1 antibody (rabbit polyclonal, 1:200; Tomoda et al. 1999) in PBS for 16 h at 4°C, washed three times with PBS, probed with a gold-conjugated anti-rabbit IgG (EM Science), and processed for electron microscopy as described (Shio and Lazarow 1981).
Subcellular fractionation and sucrose gradient experiments
E16 cerebral cortex and P6 cerebellum were homogenized by using a glass-Teflon homogenizer in homogenization buffer (0.32M sucrose, 50 mM HEPES, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 1× protease inhibitor cocktail [Sigma]) and the homogenate was spun at 700 × g to remove tissue debris. The unbroken debris was homogenized again, spun at 700 × g, and the combined supernatant was ultracentrifuged at 100,000 × g to separate crude membrane fraction (pellet) from soluble fraction (supernatant). For step-wise sucrose gradient fractionation experiments, the 700 × g supernatant (0.5 mL) was loaded on a 10%–40% sucrose gradient (10%, 20%, 30%, and 40% sucrose, 1 mL each), and centrifuged at 37,000 × g in SW55i rotor (Beckman) for 3 h at 4°C. After centrifugation, nine fractions (0.5 mL each) were recovered from the top of the gradient, and the protein concentration in each fraction was measured by Bradford method according to the protocol provided (Pierce). Ten micrograms of proteins per each fraction were applied to SDS-PAGE and analyzed by Western blotting using the following antibodies: anti-Unc51.1 (guinea pig polyclonal, 1:100), anti-SynGAP/GAP domain (rabbit polyclonal, 1:200), anti-Rab3A (rabbit polyclonal, Santa Cruz, 1:200), anti-Rab5 (rabbit polyclonal, Santa Cruz, 1:200), anti-PSD-95 (mouse monoclonal IgG2a, Upstate Biotechnology, 1:10,000), anti-LDH (Chemicon, goat polyclonal, 1:500), and anti-actin (monoclonal, Amersham-Pharmacia, 1:2000).
DNA constructs
Plasmids for Unc51.1 were as described (Tomoda et al. 1999). Expression constructs for SynGAP were provided from Richard Huganir (Kim et al. 1998). The yeast two-hybrid library was custom-made by GIBCO-BRL by using 15 μg of mRNA extracted from P6 mouse cerebellum. The cDNAs for Rab3A, Rab6, and Rab7 were amplified by PCR from P6 mouse cerebellum. Retroviral constructs were made by subcloning full-length coding regions of cDNAs for mouse Unc51.1, rat SynGAP, and human Rab5A into EcoR1/Not1 site of the pMSCX–β-actin–EGFP retroviral vector (a gift from David Solecki, The Rockefeller University, New York). For bicistronic expression of cDNAs with the retroviral expression system, one cDNA was inserted into the multicloning site of the pMSCX vector followed by the human β-actin promoter plus the second cDNA coding region N′-terminally tagged with EGFP. The EGFP was used as a morphological marker for the infected neurons.
Production of recombinant retroviruses
Recombinant ecotropic replication-incompetent retroviruses were produced as described (Tomoda et al. 1999). In brief, 293 cells were cotransfected with the recombinant retroviral constructs together with a pCL-Eco packaging construct (IMGEN). Twenty-four hours posttransfection, media was replaced with the granule cell media (Hatten 1985), and the culture supernatant containing the retroviruses was harvested 24 and 48 h later. Virus titers were estimated by infecting NIH3T3 cells with virus supernatants with serial dilution. Typically, the titers ranged from 4 × 105 to 1 × 107 cfu/mL.
Preparation and infection of primary granule cells
Cerebellar granule cells were purified from early postnatal C57BL/6J mice as described previously (Hatten 1985); 1 to 2 × 106 cells were cultured as cellular reaggregates in one well of a uncoated 16-well Lab-Tek glass chamber slides (Nunc), and infected with recombinant viruses overnight. After 18–24 h, the cultures were gently resuspended at 1 × 106 cells per milliliter and plated in Lab-Tek slides coated with poly-L-lysine (Sigma) and Matrigel (Becton Dickinson).
Immunocytochemistry
Antibody staining was as described (Gao et al. 1991). For immunoelectron microscopy, the affinity-purified rabbit anti-Unc51.1 antibody was used (Tomoda et al. 1999). For detection of Unc51.1 and SynGAP protein expression with cerebellum, P6 mice were perfused with 4% paraformaldehyde/PBS and postfixed in the same fixative containing 30% sucrose overnight at 4°C. Sagittal sections (60 μm) were made with a cryotome (Leica, SM2000R). To detect endogenous Unc51.1 protein, anti-Unc51.1 serum was generated by immunizing guinea pigs with the C′-terminal domain of Unc51.1 protein (amino acids 653–1051) prepared as a GST fusion protein in bacteria, and the resulting antiserum was affinity-purified by using the same domain of Unc51.1 as has been used for immunization. The specificity of the antibody was confirmed by Western blot using protein extracts prepared from P6 and adult mouse brain (data not shown). The dilution of other antibodies used was anti-TAG-1 antibody (monoclonal, IgM; 4D7; 1:2), anti-class III β-tubulin antibody (monoclonal, IgG2aκ; TUJ1; 1:1000, Covance). Specimens were viewed either with an epifluorescence illumination (Axiophot 100 microscope, Zeiss) or with a confocal scanning system Radiance 2000 (Bio-Rad). For detection of infected granule cells in dissociated culture, cells were fixed in 4% paraformaldehyde and probed with the anti-GFP antibody (Molecular Probes, polyclonal, 1:2000).
Acknowledgments
We are indebted to Helen Shio for electron microscopy, Yin Fang and Hirofumi Toda for expert technical assistance, Tomoyo Okada and Akihisa Mino for advice on GAP assay, Niels Adams for help with imaging, and Akira Sawa for advice on yeast two-hybrid. We also thank the following people for providing reagents: Richard Huganir for a SynGAP GAP domain antibody and SynGAP plasmids; Anat Ikin for Rab5 cDNA; Haruhiko Bito for RhoA, Rac1, and Cdc42 cDNAs; and David Solecki for a retroviral vector. We are grateful to Nat Heintz, Richard Huganir, Angus Nairn, Kathy Zimmerman, and David Solecki for critically reading the manuscript. Supported by grants from March of Dimes (1-FY99-510) and NIH (NS39991) to M.E.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1151204.
References
- Bar-Sagi D. and Feramisco, J.R. 1985. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell 42: 841–848. [DOI] [PubMed] [Google Scholar]
- Callebaut I., de Gunzburg, J., Goud, B., and Mornon, J.P. 2001. RUN domains: A new family of domains involved in Ras-like GTPase signaling. Trends Biochem. Sci. 26: 79–83. [DOI] [PubMed] [Google Scholar]
- Castellani V. and Rougon, G. 2002. Control of semaphorin signaling. Curr. Opin. Neurobiol. 12: 532–541. [DOI] [PubMed] [Google Scholar]
- Chang L. and Karin, M. 2001. Mammalian MAP kinase signalling cascades. Nature 410: 37–40. [DOI] [PubMed] [Google Scholar]
- Chen H.J., Rojas-Soto, M., Oguni, A., and Kennedy, M.B. 1998. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20: 895–904. [DOI] [PubMed] [Google Scholar]
- Crump J.G., Zhen, M., Jin, Y., and Bargmann, C.I. 2001. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 29: 115–129. [DOI] [PubMed] [Google Scholar]
- Desai C., Garriga, G., McIntire, S.L., and Horvitz, H.R. 1988. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature 336: 638–646. [DOI] [PubMed] [Google Scholar]
- Dickson B.J. 2002. Molecular mechanisms of axon guidance. Science 298: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Eggert A., Ikegaki, N., Liu, X., Chou, T.T., Lee, V.M., Trojanowski, J.Q., and Brodeur, G.M. 2000. Molecular dissection of TrkA signal transduction pathways mediating differentiation in human neuroblastoma cells. Oncogene 19: 2043–2051. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S. and Novick, P. 1993. The role of GTP-binding proteins in transport along the exocytic pathway. Annu. Rev. Cell Biol. 9: 575–599. [DOI] [PubMed] [Google Scholar]
- Fialka I., Steinlein, P., Ahorn, H., Bock, G., Burbelo, P.D., Haberfellner, M., Lottspeich, F., Paiha, K., Pasquali, C., and Huber, L.A. 1999. Identification of syntenin as a protein of the apical early endocytic compartment in Madin-Darby canine kidney cells. J. Biol. Chem. 274: 26233–26239. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G., Stahl, B., Walch-Solimena, C., Takei, K., Daniels, L., Khoklatchev, A., De Camilli, P., Sudhof, T.C., and Jahn, R. 1994. Localization of Rab5 to synaptic vesicles identifies endosomal intermediate in synaptic vesicle recycling pathway. Eur. J. Cell Biol. 65: 319–326. [PubMed] [Google Scholar]
- Forcet C., Stein, E., Pays, L., Corset, V., Llambi, F., Tessier-Lavigne, M., and Mehlen, P. 2002. Netrin-1-mediated axon outgrowth requires deleted in colorectal cancer-dependent MAPK activation. Nature 417: 443–447. [DOI] [PubMed] [Google Scholar]
- Gao W.-Q., Heintz, N., and Hatten, M.E. 1991. Cerebellar granule cell neurogenesis is regulated by cell–cell interactions in vitro. Neuron 6: 705–715. [DOI] [PubMed] [Google Scholar]
- Georgiou M. and Tear, G. 2002. Commissureless is required both in commissural neurones and midline cells for axon guidance across the midline. Development 129: 2947–2956. [DOI] [PubMed] [Google Scholar]
- Grootjans J.J., Zimmermann, P., Reekmans, G., Smets, A., Degeest, G., Durr, J., and David, G. 1997. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc. Natl. Acad. Sci. 94: 13683–13688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B.Z. and Lim, W.A. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114: 3219–3231. [DOI] [PubMed] [Google Scholar]
- Hatten M.E. 1985. Neuronal regulation of astroglial morphology and proliferation in vitro. J. Cell Biol. 100: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E.M., Culotti, J.G., Thomson, J.N., and Perkins, L.A. 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111: 158–170. [DOI] [PubMed] [Google Scholar]
- Hirbec H., Francis, J.C., Lauri, S.E., Braithwaite, S.P., Coussen, F., Mulle, C., Dev, K.K., Coutinho, V., Meyer, G., Isaac, J.T., et al. 2003. Rapid and differential regulation of AMPA and kainate receptors at hippocampal mossy fibre synapses by PICK1 and GRIP. Neuron 37: 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C.R., Gibson, A., Shipman, M., and Miller, K. 1990. Movement of internalized ligand-receptor complexes along a continuous endosomal reticulum. Nature 346: 335–339. [DOI] [PubMed] [Google Scholar]
- Jurney W.M., Gallo, G., Letourneau, P.C., and McLoon, S.C. 2002. Rac1-mediated endocytosis during ephrin-A2- and semaphorin 3A-induced growth cone collapse. J. Neurosci. 22: 6019–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150: 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H. and Lemmon, V. 2000. Recycling of the cell adhesion molecule L1 in axonal growth cones. J. Neurosci. 20: 3676–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K., Rajagopalan, S., Cleppien, D., Teis, D., Paiha, K., Huber, L.A., Technau, G.M., and Dickson, B.J. 2002. Comm sorts robo to control axon guidance at the Drosophila midline. Cell 110: 415–427. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Liao, D., Lau, L.F., and Huganir, R.L. 1998. SynGAP: A synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20: 683–691. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Lee, H.K., Takamiya, K., and Huganir, R.L. 2003. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J. Neurosci. 23: 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. and Emr, S.D. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama N.H., Watabe, A.M., Carlisle, H.J., Porter, K., Charlesworth, P., Monti, J., Strathdee, D.J., O'Carroll, C.M., Martin, S.J., Morris, R.G., et al. 2002. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J. Neurosci. 22: 9721–9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroll M., Rathjen, F.G., and Volkmer, H. 2001. The neural cell recognition molecule neurofascin interacts with syntenin-1 but not with syntenin-2, both of which reveal self-associating activity. J. Biol. Chem. 276: 10646–10654. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lamaze C., Chuang, T.H., Terlecky, L.J., Bokoch, G.M., and Schmid, S.L. 1996. Regulation of receptor-mediated endocytosis by Rho and Rac. Nature 382: 177–179. [DOI] [PubMed] [Google Scholar]
- Li W., Okano, A., Tian, Q.B., Nakayama, K., Furihata, T., Nawa, H., and Suzuki, T. 2001. Characterization of a novel synGAP isoform, synGAP-β. J. Biol. Chem. 276: 21417–21424. [DOI] [PubMed] [Google Scholar]
- Lin R.C. and Scheller, R.H. 2000. Mechanisms of synaptic vesicle exocytosis. Annu. Rev. Cell Dev. Biol. 16: 19–49. [DOI] [PubMed] [Google Scholar]
- Lin D., Gish, G.D., Songyang, Z., and Pawson, T. 1999. The carboxyl terminus of B class ephrins constitutes a PDZ domain binding motif. J. Biol. Chem. 274: 3726–3733. [DOI] [PubMed] [Google Scholar]
- Liu K. and Li, G. 1998. Catalytic domain of the p120 Ras GAP binds to RAb5 and stimulates its GTPase activity. J. Biol. Chem. 273: 10087–10090. [DOI] [PubMed] [Google Scholar]
- Marshall C.J. 1996. Ras effectors. Curr. Opin. Cell Biol. 8: 197–204. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada, M., Wada, Y., and Ohsumi, Y. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192: 245–250. [DOI] [PubMed] [Google Scholar]
- McIntire S.L., Garriga, G., White, J., Jacobson, D., and Horvitz, H.R. 1992. Genes necessary for directed axonal elongation or fasciculation in C. elegans. Neuron 8: 307–322. [DOI] [PubMed] [Google Scholar]
- Mueller B.K. 1999. Growth cone guidance: First steps towards a deeper understanding. Annu. Rev. Neurosci. 22: 351–388. [DOI] [PubMed] [Google Scholar]
- Noda M., Ko, M., Ogura, A., Liu, D.G., Amano, T., Takano, T., and Ikawa, Y. 1985. Sarcoma viruses carrying ras oncogenes induce differentiation-associated properties in a neuronal cell line. Nature 318: 73–75. [DOI] [PubMed] [Google Scholar]
- Ogura K., Wicky, C., Magnenat, L., Tobler, H., Mori, I., Muller, F., and Ohshima, Y. 1994. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes & Dev. 8: 2389–2400. [DOI] [PubMed] [Google Scholar]
- Ogura K., Shirakawa, M., Barnes, T.M., Hekimi, S., and Ohshima, Y. 1997. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes & Dev. 11: 1801–1811. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. 2001. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2: 211–216. [DOI] [PubMed] [Google Scholar]
- Okazaki N., Yan, J., Yuasa, S., Ueno, T., Kominami, E., Masuho, Y., Koga, H., and Muramatsu, M. 2000. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: Possible role of vesicular transport in axonal elongation. Brain Res. Mol. Brain Res. 85: 1–12. [DOI] [PubMed] [Google Scholar]
- Olson M.F. and Marais, R. 2000. Ras protein signalling. Semin. Immunol. 12: 63–73. [DOI] [PubMed] [Google Scholar]
- Perron J.C. and Bixby, J.L. 1999. Distinct neurite outgrowth signaling pathways converge on ERK activation. Mol. Cell Neurosci. 13: 362–378. [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. 1994. Rab GTPases: Master regulators of membrane trafficking. Curr. Opin. Cell Biol. 6: 522–526. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S. 1995. The cerebellum. In Histology of the nervous system of man and vertebrates. (ed. L.W. Swanson; trans. N. Swanson), pp. 3–123. Oxford University Press, New York, NY.
- Sagiv Y., Legesse-Miller, A., Porat, A., and Elazar, Z. 2000. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Kawamura, S., and Ozaki, K. 2003. An essential role of Rab5 in uniformity of synaptic vesicle size. J. Cell Sci. 116: 3583–3590. [DOI] [PubMed] [Google Scholar]
- Shio H. and Lazarow, P.B. 1981. Relationship between peroxisomes and endoplasmic reticulum investigated by combined catalase and glucose-6-phosphatase cytochemistry. J. Histochem. Cytochem. 29: 1263–1272. [DOI] [PubMed] [Google Scholar]
- Siddiqui S.S. 1990. Mutations affecting axonal growth and guidance of motor neurons and mechanosensory neurons in the nematode Caenorhabditis elegans. Neurosci. Res. 13 (Suppl): 171–190. [DOI] [PubMed] [Google Scholar]
- Somsel Rodman J. and Wandinger-Ness, A. 2000. Rab GTPases coordinate endocytosis. J. Cell Sci. 113: Pt 2, 183–192. [DOI] [PubMed] [Google Scholar]
- Tall G.G., Barbieri, M.A., Stahl, P.D., and Horazdovsky, B.F. 2001. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell 1: 73–82. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M. and Goodman, C.S. 1996. The molecular biology of axon guidance. Science 274: 1123–1133. [DOI] [PubMed] [Google Scholar]
- Tomoda T., Bhatt, R.S., Kuroyanagi, H., Shirasawa, T., and Hatten, M.E. 1999. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24: 833–846. [DOI] [PubMed] [Google Scholar]
- Wang H., Bedford, F.K., Brandon, N.J., Moss, S.J., and Olsen, R.W. 1999. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 397: 69–72. [DOI] [PubMed] [Google Scholar]
- Zerial M. and McBride, H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2: 107–117. [DOI] [PubMed] [Google Scholar]
- Zimmer M., Palmer, A., Kohler, J., and Klein, R. 2003. EphB–ephrinB bi-directional endocytosis terminates adhesion allowing contact mediated repulsion. Nat. Cell Biol. 5: 869–878. [DOI] [PubMed] [Google Scholar]