Abstract
In order to independently study the numerous variables that influence cell movement, it will be necessary to employ novel tools and materials that allow for exquisite control of the cellular microenvirenment. In this work, we have applied advanced 3D micropatterning technology, known as two-photon laser scanning lithography (TP-LSL), to poly(ethylene glycol) (PEG) hydrogels modified with bioactive peptides in order to fabricate precisely designed microenvirenments to guide and quantitatively investigate cell migration. Specifically, TP-LSL was used to fabricate cell adhesive PEG-RGDS micropatterns on the surface of non-degradable PEG-based hydrogels (2D) and in the interior of proteolytically degradable PEG-based hydrogels (3D). HT1080 cell migration was guided down these adhesive micropatterns in both 2D and 3D, as observed via time-lapse microscopy. Differences in cell speed, cell persistence, and cell shape were observed based on variation of adhesive ligand, hydrogel composition, and patterned area for both 2D and 3D migration. Results indicated that HT1080s migrate faster and with lower persistence on 2D surfaces, while HT1080s migrating in 3D were smaller and more elongated. Further, cell migration was shown to have a biphasic dependence on PEG-RGDS concentration and cells moving within PEG-RGDS micropatterns were seen to move faster and with more persistence over time. Importantly, the work presented here begins to elucidate the multiple complex factors involved in cell migration, with typical confounding factors being independently controlled. The development of this unique platform will allow researchers to probe how cells behave within increasingly complex 3D microenvironments that begin to mimic specifically chosen aspects of the in vivo landscape.
Introduction
Cell motility is critical for a variety of functions within healthy human physiology. For example, embryonic development, wound healing, angiogiogensis, and the regulation of the immune system are just some of the processes that rely on the ability of cells to migrate within the native extracellular matrix (ECM)1. Additionally, abnormal cell migratory behaviours are implicated in a number of devastating conditions including vascular disease, osteoporosis, multiple sclerosis, and metastatic cancer1. An in-depth understanding of cell motility is therefore critical for applications ranging from tissue engineering to disease pathology and drug discovery.
Much of what is known about cell migration is based upon observations of cells cultured on tissue culture plastic2. While 2D studies have served as a valuable first step to understanding cell motility, cells within the body do not migrate on flat, non-deformable surfaces. Rather, cells exist within a 3D microenvironment that regulates migration via a number of interconnected physical and biochemical mechanisms. In order to better mimic this complex microenvironment, scientists have moved towards the use of biomaterial systems to study cell motility. Researchers have used biomaterials to observe dramatic differences between cells migrating on 2D surfaces and those within 3D matrices. For example, Cukierman et al. demonstrated that human foreskin fibroblasts on a cell-derived 3D extracellular matrix migrated 1.5 times faster and with a less spread morphology when compared with cells cultured on 2D tissue culture plastic3. Additionally, Zaman et al. showed that as 3D Matrigel matrices were softened, human prostate carcinoma cells migrated at maximal speeds when fewer adhesion molecules were present4. This migration trend is in direct contrast with two-dimensional migration studies and is likely explained by the forces a 3D matrix places on all cell surfaces, with less stiff surfaces serving as a better environment for matrix degradation, remodelling, and subsequent motility4. Additionally, researchers have also studied cell migration on "1D" micropatterned lines as opposed to flat 2D surfaces and discerned increases in migration speed and pronounced differences in protein expression and migration mode5.
While previous research has clearly demonstrated a difference between 2D and 3D cell migration, the direct cause of these differences has been difficult to discern. Cell migration in the native extracellular matrix can be linked to at least 5 different factors, namely adhesion molecules, chemotactic growth factors, matrix stiffness, matrix porosity, and the proteolytic susceptibility of the matrix1. The natural ECM matrices typical of migration studies provide very little ability to control and tune matrix properties, making it difficult to parse out the factors responsible for differences in motility. In collagen or Matrigel environments, for example, one may attempt to probe the effect of increasing material stiffness on cell migration by increasing the protein concentration in the hydrogel material. In addition to increasing the stiffness of the material, however, such a hydrogel would also have significantly altered adhesive ligand concentration, pore size, diffusion of growth factors, and proteolytic susceptibility. Further, studies comparing 2D cell migration on one material (e.g. tissue culture plastic) and 3D cell migration within a different material (e.g. Matrigel) are confounded by differences in biochemical growth factors present within a naturally derived matrix.
The development of custom biomaterials in combination with novel micropatterning technologies will allow for increased control of cell migration under precisely designed conditions, and ultimately shed light on complex cell motility processes that have yet to be discovered or understood. Acrylate terminated, poly(ethylene glycol) (PEG) based hydrogels offer a highly-tunable, biomaterial environment that may allow for the elucidation of many of these confounding factors6. Such photochemically crosslinked hydrogels are cell non-adhesive and act as a blank slate into which custom bioactivity may be introduced7–9. This property of PEG-based hydrogels eliminates the unknown and uncontrollable cell-growth factor interactions that cloud many cell migration systems. The PEG-based hydrogel system also allows for the incorporation of modulatable quantities of adhesion molecules without altering the hydrogel material properties. Additionally, 2D migration may be studied on the surface of non-degradable PEG-based hydrogels while 3D migration may be studied within enzymatically degradable PEG-based networks, allowing for direct comparison of migration modes.
In previous work, acrylate-terminated PEG-based hydrogels have been modified via photochemical reactions to covalently link acrylate-modified bioactive molecules to the hydrogel backbone in a spatially controlled manner. For example, the fibronectin derived cell adhesion molecule acrylate-PEG-RGDS has previously been photochemically immobilized into micropatterns on and within PEG-based hydrogels6,7,9,10. Such light-initiated reactions offer opportunities for fabrication of specifically designed geometrical patterns of bioactive molecules to guide cell migration. Additionally, these reactions occur similarly using both non-degradable PEG-diacrylate hydrogels (PEG-DA) as well as enzymatically degradable acrylate-terminated PEG hydrogels (PEG-PQ), which will allow for a comparison of cell migration within bioactive micropatterns on both 2D and 3D materials.
We have previously developed and optimized a technique known as two-photon laser scanning lithography (TP-LSL) in order to crosslink PEG-RGDS into 3D forms within PEG-DA and PEG-PQ hydrogels 11–14. This technique involves permeating a hydrogel with PEG-RGDS (or any other acrylate-PEG-biomolecule), and then using a two-photon laser to covalently crosslink the PEG-RGDS into desired 3D forms. This procedure results in high-resolution, 3D volumes of patterned bioactivity within the hydrogel network11. TP-LSL has been utilized previously to guide some simple behaviours of cells. For example, early work from our laboratory has shown the direct guidance of fibroblast migration via 3D PEG-RGDS channels within enzymatically degradable PEG-based hydrogels12. Additionally, we have previously demonstrated the guidance of vascular cells to 3D PEG-RGDS patterns that mimicked the structure of endogenous vessels13. While these early works demonstrate the feasibility and promise of guiding cell behaviour in hydrogels using a 3D patterned biochemical cue, they have yet to utilize the potential of the TP-LSL technique to investigate cell motility over various micropatterned conditions.
In this work, TP-LSL was implemented to guide and quantitatively study cell motility. Specifically, PEG-RGDS micropatterns of controlled size and concentration were fabricated both on the surface of non-degradable PEG-DA hydrogels and embedded within enzymatically susceptible PEG-PQ hydrogels. HT1080 cells were then seeded onto micropatterned PEG-DA hydrogels (2D) or encapsulated directly adjacent to micropatterned PEG-PQ hydrogels (3D), and subsequent cell migration was observed. The PEG-RGDS patterns guided cell migration, allowing for the direct tracking of individual cells within a controlled microenvironment. The 2D and 3D experimental groups were directly compared in terms of cell speed, cell persistence and cell shape. Patterns of various size and volume were also utilized to manipulate cell persistence and guide cells on the microscale. Finally, the cell migratory effects of varying factors such as PEG-RGDS ligand concentration and PEG-PQ hydrogel composition were analyzed to probe the confounding factors of cell migration. Overall, the current study demonstrates that the modification of PEG-based hydrogels with TP-LSL can serve as a novel tool to investigate cell motility within precisely designed biomimetic microenvironments. This quantitative investigation of cell migration over various micropatterned conditions begins to elucidate many of the interwoven factors that affect how cells move within the extracellular matrix and leads the way toward a more complete understanding of cell motility.
Materials and Methods
Synthesis of Poly(ethylene glycol) Diacrylate
Poly(ethylene glycol) (PEG 6000 Da; Fluka) was reacted overnight with acryloyl chloride (Sigma) at a 1:4 molar ratio and with triethyl amine (TEA; Sigma) at a 1:2 molar ratio (PEG:TEA) all in anhydrous dichloromethane (DCM; Sigma). The resultant PEG-DA product was purified via washing with 2M K2CO3, and subsequent removal of the chloride salt via phase separation. Anhydrous MgSO4 was added to dry the PEG-DA. The product was then filtered, precipitated in diethyl ether, and filtered again. The final PEG-DA product was dried overnight under vacuum, and characterized by 1H-NMR.
Synthesis of Enzymatically Degradable Acrylate-Terminated PEG Macromer
To synthesize enzymatically degradable PEG hydrogel precursors, the peptide GGGPQGIWGQGK (abbreviated PQ) was first synthesized via solid phase peptide synthesis with standard F-Moc chemistry using an APEX396 peptide synthesizer (Aapptec). Successful peptide synthesis was confirmed with matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-ToF; Bruker Daltonics). The PQ peptide was then reacted overnight with acrylate-poly(ethylene glycol)-succinimidyl valerate (PEG-SVA, Laysan Bio) at a 1:2.1 (PQ:PEG-SVA) molar ratio in HEPBS conjugation buffer to form acrylate-PEG-PQ-PEG-acrylate (PEG-PQ). HEPBS conjugation buffer consisted of 20 mM n-(2-Hydroxyethyl)piperizine-N’-(4-butanesulfonic acid) (Santa Cruz Biotechnology), 100 mM sodium chloride, 2 mM calcium chloride, and 2 mM magnesium chloride at pH 8.5. The resultant PEG-PQ product was dialyzed against Millipore water for 12 h using a 3500 MWCO regenerated cellulose membrane (Spectrum Laboratories) and then lyophilized to dryness. PEG-PQ was characterized via gel permeation chromatography (GPC, Polymer laboratories) with detectors for UV–vis and evaporative light scattering.
Synthesis and Fluorescent Labelling of Acrylate-PEG-RGDS
The RGDS peptide (American Peptide) was reacted overnight with PEG-SVA at a 1.2:1 (RGDS:PEG-SVA) molar ratio in HEPBS conjugation buffer in order to form acrylate-PEG-RGDS (PEG-RGDS). The product was dialyzed against Millipore water for 12 h using a 3500 MWCO regenerated cellulose membrane and then lyophilized to dryness prior to characterization by GPC. To fluorescently label PEG-RGDS, Alexa Fluor® 633 carboxylic acid succinimidyl ester (AF633-SE; Invitrogen) in dimethylformamide was reacted with PEG-RGDS in 0.1 M sodium bicarbonate buffer (pH 8.3) at a 1:10 molar ratio (PEG-RGDS:AF4633-SE). This reaction was rapidly mixed for 2 hr and dialyzed for 12 h against Millipore water with a 3500 MWCO regenerated cellulose membrane. The resulting monoacrylate-PEG-RGDS-Alexa Fluor®633 was lyophilized to dryness and stored at −20 °C.
Quantification of RGDS Concentration in Patterned PEG-based Hydrogels
To quantify the concentration of RGDS crosslinked into PEG-based hydrogels, samples were degraded and an ninhydrin assay was used to measure amine content as compared to a known standard curve15. Hydrogels were soaked in 20 mM PEG-RGDS and a 720 nm laser (60 mW/µm2 intensity, scan speed 12.5 µs/pixel) was utilized to pattern a total volume of 0.75 mm3 within each hydrogel sample with a laser scan occurring every 3 µm in the Z direction. Samples were washed in HBS for 2 days with at least 4 buffer changes to remove unpatterned PEG-RGDS. A standard curve was created where each standard reference point contained a non-patterned PEG-DA hydrogel, as well as a known concentration of PEG-RGDS. Standards and samples were lyophilized overnight, dissolved in hydrochloric acid (HCl), and then placed on the rotovap to remove all HCl. Samples and standards were next dissolved in 0.1 M sodium citrate (pH 5), sonicated, centrifuged, and mixed with the Ninhydrin reagent(Sigma) at a 1:1 ratio. The solutions were then boiled for 20 min, and the resulting product was assayed for absorbance at 570 nm to determine the amount of RGDS in each patterned hydrogel. When 20 mM PEG-RGDS was soaked into PEG-DA hydrogels, the patterned concentration was determined to be 2.98 mM PEG-RGDS. This functional PEG-RGDS concentration indicates that approximately 15% of the original 20 mM PEG-RGDS that was soaked into the PEG-DA hydrogels was covalently conjugated using the TP-LSL patterning process. For the remainder of this work, a 15% conjugation efficiency was assumed for all samples.
Cell Maintenance
HT1080s (ATCC) were maintained in culture with Eagle's minimal essential medium (EMEM) (ATCC) supplemented with 10 % fetal bovine serum, 2 mM L-glutamine, 1 U/ml penicillin, and 1 µg/ml streptomycin. Cells were cultured at 37 °C with 5% CO2 and the media was replenished every 2 days. Cells were used from passages 2– 6.
Fabrication of PEG-RGDS Patterns to Guide 2D and 3D Cell Migration
Glass coverslips were incubated in a solution of 75% sulphuric acid (VWR) and 25% hydrogen peroxide (30% w/w, VWR) for 1 h with gentle agitation. Coverslips were then rinsed with Millipore water and incubated in 85 mM 3-(Trimethoxysilyl)propyl methacrylate (Fluka) in ethanol (pH 4.5) to introduce surface methacrylate groups to the glass16. A hydrogel precursor solution of 5% PEG-DA with 3.4 µl/ml N-vinyl pyrrolidone (NVP), 1.5 % v/v triethanolamine (Fluka BioChemika) and 10 µM eosin Y (Sigma) was prepared in Hepes Buffered Saline(HBS). For 3D migration studies, PEG-PQ was substituted for the PEG-DA. Two 380 µm polydimethylsiloxane (PDMS) spacers were pressed against a glass slide that had been incubated in Sigmacoat (Sigma). A methacylated coverslip was then sealed against the PDMS spacers to form a hydrogel mold. 6 µl of hydrogel precursor solution was injected into the mold and exposed to white light (Fiberlite, Dolan Jenner) for 25 s, yielding a crosslinked PEG-based hydrogel. Immobilized PEG-based hydrogels were incubated in 5 mM PEG-RGDS in HBS along with 50 µM PEG-RGDS-633 and 10 µl/ml of 300 mg/ml 2, 2-dimethoxy-2-phenylacetophenone in NVP for 15 min. (In experiments where variation of PEG-RGDS was tested, the soaking concentrations contained either 0.5 mM, 1 mM, 5 mM, or 20 mM PEG-RGDS). The hydrogels were then placed on the stage of a Zeiss LSM 710 Multi-photon microscope and patterned. Four rectangular regions of interest were designated, with lengths of 500 µm and widths of 10, 25, 50, and 100 µm, respectively. The microscope was then focused on the hydrogel, and a titanium/sapphire laser (Chameleon) was tuned to 720 nm and set with an intensity of 60 mW/µm2 and a laser scan speed of 12.5 µs/pixel. Scanning over the regions of interest resulted in exact shape specified by the regions of interest. For 2D studies, the laser was scanned every 3 µm for a distance 51 µm above and below the PEG-DA hydrogel surface to ensure PEG-RGDS was crosslinked to the surface of the hydrogel. For 3D studies, the laser was focused in the interior of the PEG-PQ hydrogels. The depth of each pattern was also varied for 3D studies. The 100 µm wide pattern was iteratively scanned with a 3 µm adjustment between each scan until the pattern depth reached 100 µm. The 50 µm, 25 µm and 10 µm patterns were fabricated in a similar way so that final pattern dimensions for 3D migration were: 100 × 100 × 500 µm, 50 × 50 × 500 µm, 25 × 25 × 500 µm, and 10 × 10 × 500 µm (Supplementary Figure 1). For both 2D and 3D migration, patterned hydrogels were washed in HBS under gentle rocking for 48 h with at least 4 buffer changes to remove precise excitation of photoinitiator molecules and subsequent crosslinking of PEG-RGDS to the PEG-based hydrogel in the unbound PEG-RGDS.
Guiding Cell Migration on 2D PEG-RGDS Surface Patterns
HT1080s at approximately 90% confluency were fluorescently labelled via a 1 h incubation with 1 µg/ml green CMFDA Cell Tracker® (Invitrogen) in EMEM. After incubation, cells were washed with PBS and fresh media was added. HT1080s were then harvested and seeded onto PEG-DA hydrogels whose surfaces had been patterned with PEG-RGDS. HT1080s were seeded at cell densities of 70,000 cells/cm2 for hydrogels patterned using 20 mM PEG-RGDS, 90,000 cells/cm2 for hydrogels pattered using 5 mM PEG-RGDS and 110,000 cells/cm2 for hydrogels patterned using 0.5 mM and 1 mM PEG-RGDS. After seeding, cells were incubated in fresh media and allowed to adhere. After 24 h, the hydrogels were washed gently with PBS to remove non-adherent cells and imaged (Figure 1a). The migration of HT1080s on PEG-DA hydrogel surfaces was monitored using a confocal microscope (Zeiss5 LIVE, Plan-Apochromat 20x objective with 0.8 numerical aperture: for green CMFDA Cell Tracker excitation = 489 nm, emission BP filter = 500–525 nm; for Alexafluor 633 excitation = 633 nm, emission long pass filter = 650 nm). Time-lapse microscopy was performed with an image acquired every 2 min for 6 h. The hydrogels were maintained at 37°C with 5 % CO2 at all times.
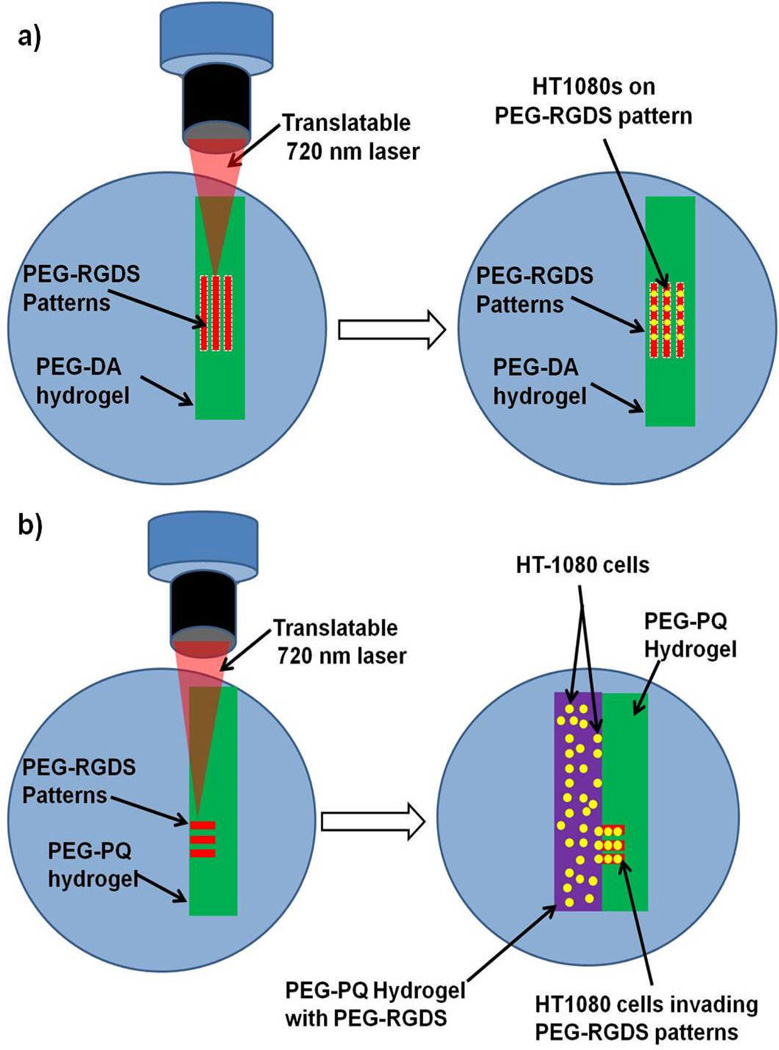
Fig. 1.
(a) PEG-RGDS micropatterns (red) were fabricated using two-photon laser scanning lithography on a PEG-DA hydrogel surface (green).Fluorescently labelled HT1080s (yellow) were seeded onto surfaces and cells were observed to adhere and migrate on the PEG-RGDS micropatterns. (b) PEG-RGDS micropatterns (red) were fabricated using two-photon laser scanning lithography within an immobilized PEG-PQ hydrogel (green).Fluorescently labelled HT1080s (yellow) were photoencapsulated within a different PEG-PQ hydrogel with ubiquitous PEG-RGDS (purple). The cellularized hydrogel was immobilized directly adjacent to the patterned hydrogel, and HT1080s were observed to migrate into PEG-RGDS patterns.
Guiding Cell Migration into 3D PEG-RGDS Micropatterns
HT1080s were fluorescently labelled as described above. A hydrogel precursor solution of 10 % PEG-PQ with 3.4 µl/ml NVP, 1.5 % v/v triethanolamine (Fluka BioChemika), 10 µM eosin Y (Sigma) and 3.5 mM PEG-RGDS was made in HBS. Fluorescently labelled HT1080s were harvested and suspended in the precursor solution at a cell density of 30,000 cells/µl. Hydrogel molds were fabricated using a 380 µm thick PDMS spacer and a glass slide exposed to Sigmcoat®. The coverslip with the attached patterned PEG-PQ hydrogel was placed face down onto the spacer so that a mold was formed between the spacer and the existing patterned hydrogel. 6 µl of the cell-containing precursor solution was injected into the mold and exposed to white light (Fiberlite, Dolan Jenner) for 25 s, yielding a crosslinked, cell-laden PEG-PQ hydrogel directly adjacent to the PEG-PQ hydrogel with patterned PEG-RGDS. The hydrogels were then immersed in EMEM and maintained at 37°C and 5% CO2 for 12 h to allow cells to adhere and begin migration into the hydrogel with patterned PEG-RGDS (Figure 1b). The 3D migration of HT1080s was monitored using a Zeiss 5 LIVE confocal microscope as described above.
Staining Nuclei and F-Actin to Visualize Cell Structure
After 18 h in culture, gels to be stained were fixed in 4% paraformaldehyde for 20 min and then washed with PBS. The gels were next incubated in 0.5% Triton X-100 for 30 min to permeabilize the cells and washed again with PBS prior to blocking with 1% bovine serum albumin (BSA) in PBS for 30 min to reduce non-specific binding. Finally, the gels were washed again in PBS and a staining solution of 2 µM DAPI with 1:100 rhodamine phalloidin (Invitrogen) in PBS was applied for 12 h. Cell structures within hydrogels were visualized on a confocal microscope (Zeiss5 LIVE, Plan-Apochromat 20x objective with 0.8 numerical aperture: for rhodamine phalloidin excitation = 532 nm, emission BP filter = 560–675 nm; for DAPI excitation = 405 nm, emission band pass filter = 415–580 nm).
Analysis of Cell Migration
The migration of HT1080s was analyzed with Imaris (Bitplane) software. Cells were tracked using an autoregressive motion gapclose 3 algorithm with all cells that were tracked for at least 2 h considered for analysis. With the exception of the experiment that varied pattern width, cells were analyzed on or within 100 µm wide patterns. Imaris was used to calculate average cell migration speed, average cell persistence, and average cell guidance ratio. Cell migration speed was defined as the total distance a cell travelled divided by the total time. Cell persistence was defined as the total cell displacement from its starting position divided by the total distance a cell travelled. Cell guidance ratio was defined as the total distance a cell moved in the direction parallel to the pattern divided by the total distance it travelled. In addition, cell area and circularity were calculated using ImageJ (National Institutes of Health) on images of HT1080s fixed and stained with DAPI and phalloidin. This analysis was performed by first tracing the perimeter of each cell and then calculating cell area as the total number of square micrometers within the cell tracing and cell circularity according to Equation 1 below.
| (1) |
HT1080s were evaluated for cell speed, cell persistence, cell guidance ratio, cell area, and cell circularity under a variety of conditions. Cells migrating on 2D surfaces were compared to those migrating within 3D hydrogels. Cells were also analyzed on or within various pattern widths, various PEG-RGDS concentrations, various PEG hydrogel compositions, and at various time points. When comparing more than 2 groups, one-way ANOVA and subsequent post hoc analysis via the Bonferonni method were used to statistically analyze differences. When comparing only 2 groups, a student's T test was applied to determine significance. In all cases, p < 0.05 was considered statistically significant.
Results
Comparing 2D and 3D Cell Migration using TP-LSL and PEG-Based Hydrogels
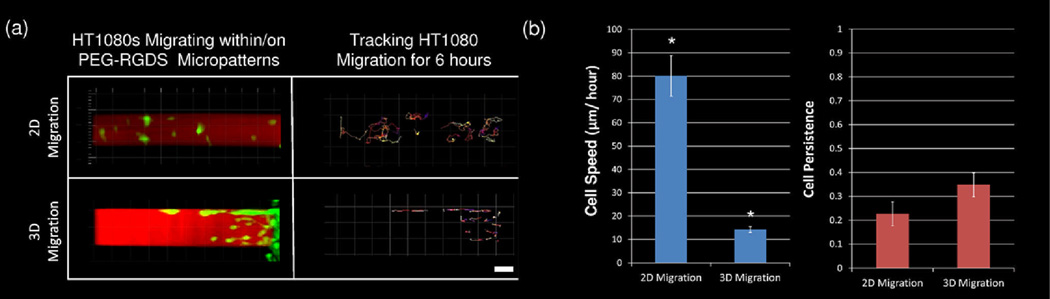
The overarching goal of this work was to develop two-photon laser scanning lithography (TP-LSL) as a tool to investigate many of the confounding factors that influence cell migration. We first investigated cell motility on and within micropatterned PEG-based hydrogels in order to elucidate the inherent differences between 2D and 3D cell migration. Representative images of labelled HT1080s migrating in 2D and 3D PEG-RGDS micropatterns are shown in Figure 2. In both cases, the movement of each cell was tracked over a 6 h period with the passage of time indicated by the changing colour of the tracking lines with early times indicated by blues and reds and later times indicated by yellows and whites (Figure 2a). Cells migrating on non-degradable PEG-DA surfaces changed their position in only the X and Y dimensions, while cells migrating within enzymatically degradable PEG-PQ hydrogels changed position in the X, Y, and Z dimensions. An analysis of cell speed indicated that HT1080s migrated over 5 times faster on 2D hydrogel surfaces than within 3D PEG-PQ hydrogels (80 µm/h-2D, 14 µm/h-3D) (Figure 2b).
Fig. 2.
(a) HT1080s (green) are depicted migrating on the surface of a PEG-DA hydrogel (2D) or within a PEG-PQ hydrogel (3D) 1 day after cell seeding. Migrating cells are confined to a 100 µm wide pattern of PEG-RGDS (red) and were tracked for 6 h with the passage of time indicated by colour changes from blue to red to yellow to white. All images are representative. Scale bar = 50 µm. (b) Cell tracking data was used to calculate cell speed and cell persistence for both 2D and 3D cell migration with cell speed significantly increased with 2D migration (*p<0.05), while cell persistence trended higher with 3D migration.
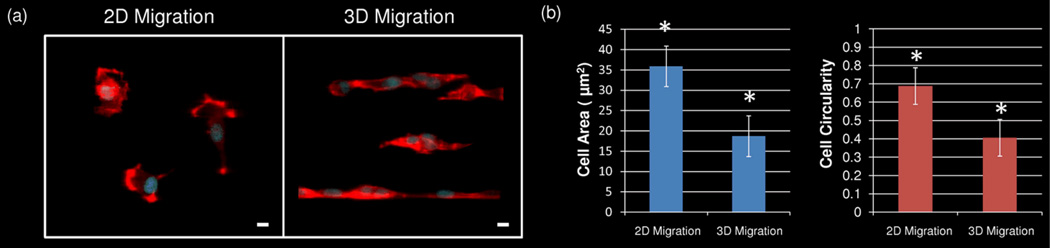
Analysis of how much cell movement contributed to total cell displacement, known as cell persistence, was also performed. Average cell persistence trended higher in 3D as opposed to on 2D surfaces (Figure 2b). This indicates that when cells are migrating within a matrix, changes in direction of migration occur only periodically and are relatively subdued. Contrastingly, when cells are migrating on a surface, changes in direction of migration occur rapidly and dramatically. A closer investigation of the size and shape of the migrating HT1080s was next conducted. Migrating cells on both 2D hydrogel surfaces and within 3D hydrogels were fixed and stained with DAPI and phalloidin to visualize the nuclei and actin cytoskeleton (Figure 3a). HT1080s were observed to be smaller and more elongated in 3D than on 2D surfaces. These differences were quantified in Figure 3b to show that average cell area decreased from 36 µm2 on 2D hydrogel surfaces to 18 µm2 within 3D hydrogels, and cell circularity, which measures how closely a cell approximates a circle, dropped from nearly 0.7 (2D) to 0.4 (3D), with 1 representing a perfect circle.
Fig. 3.
HT1080s were seeded onto PEG-DA surfaces (2D) with PEG-RGDS micropatterns and encapsulated adjacent to PEG-PQ hydrogels with PEG-RGDS micropatterns (3D). (a) Cells were allowed to migrate for 12 h on/within PEG-RGDS patterns before they were stained with DAPI and phalloidin to visualize the nuclei and actin cytoskeleton. All images are representative. Scale bar = 10 µm. (b) Cell area and cell circularity were quantified for both 2D and 3D cell migration. Total cell area and overall cell circularity were significantly greater for cells on 2D hydrogel surfaces as compared to cells migrating through micropatterned 3D hydrogels. (*p<0.05)
Controlling 2D and 3D Cell Migration with PEG-RGDS Patterns of Varying Size
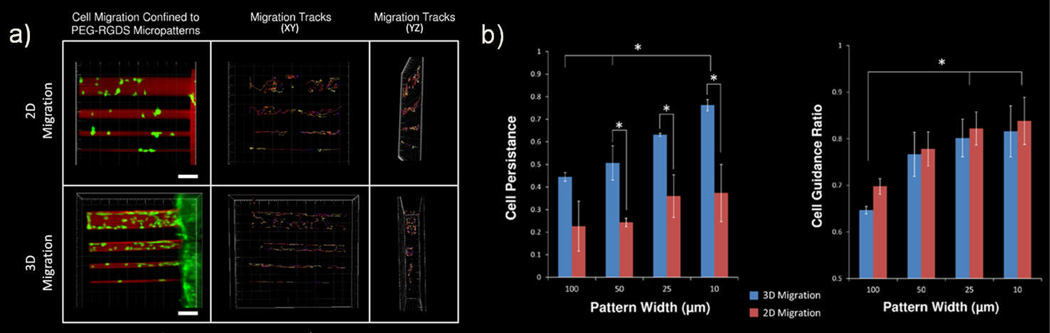
TP-LSL was utilized to micropattern PEG-RGDS tracks of various sizes in order to apply an increasing degree of control to the migratory path of mobile cells in both 2 and 3 dimensions. Representative images show that the PEG-RGDS micropatterns successfully applied an increasing degree of control on HT1080 cell migration in both 2D and 3D (Figure 4a). Cells were tracked for 6 h and their migration paths were confined to the area (2D) or volume (3D) of micropatterned PEG-RGDS. Migration tracks of cells are shown in the normal straight on view (XY) as well as a side view (YZ) (Figure 4a). The YZ view demonstrates how HT1080s migrating in 3D are confined to their PEG-RGDS micropattern in both the XY dimension and the YZ dimension. Cells migrating in the smallest 3D PEG-RGDS patterns (10 × 10 × 500 µm) are so limited in the YZ direction that their tracks begin to resemble straight lines, similar to those of cells migrating on the smallest 2D micropatterns.
Fig. 4.
(a) In 2D, HT1080s were guided on the surface of PEG-DA hydrogels by patterns of PEG-RGDS that vary in width. In 3D, HT1080s (green) were guided within PEG-PQ hydrogels by patterns of PEG-RGDS (red) that vary in width and height. HT1080 cells were tracked for 6 h with the passage of time indicated by colour changes from blue to red to yellow to white. All images are representative. Scale bar = 100 µm. (b) Cell tracking data was used to calculate cell persistence and cell guidance ratio for both 2D and 3D cell migration. Both cell persistence and cell guidance ratio were significantly increased as pattern dimensions decreased. Cell persistence was significantly greater for 3D migration, while there was no significant difference in cell guidance ratio between 2D and 3D migration. (*p<0.05).
In order to quantitatively analyze the effects of changing the size of PEG-RGDS micropatterns, cell persistence and cell migration ratio were analyzed. The persistence of cells migrating within 3D hydrogels increased from 0.44 for the larger 100 × 100 µm patterns to 0.75 for the smaller 10 × 10 µm patterns. A similar trend was present for cells migrating on 2D hydrogel surfaces, although considerably less pronounced. This trend is expected as the decreasing pattern dimensions influence the cells to progress farther in a given direction, thereby increasing the contribution of their distance travelled to their overall displacement. Interestingly, cell persistence was markedly decreased on patterned 2D hydrogel surfaces as opposed to within 3D hydrogels (Figure 4b).
The cell guidance ratio measures how well cells are guided in the direction of the pattern, regardless of ultimate cell displacement. The cell guidance ratio for cells migrating in both 2D and 3D increased as pattern dimensions were decreased. In this case, however, the discrepancy between 2D and 3D cell migration was no longer present as the cell guidance ratio between 2D and 3D cell migration was statistically identical for all of the pattern sizes. The combination of the persistence and migration ratio results indicate that while cells in 2D and 3D are both increasingly guided by the micropatterns of smaller dimensions, cells on 2D surfaces have a higher tendency to reverse their migration direction than those in 3D. After reversing direction, 2D cells continue to migrate parallel to the pattern but have a decreased displacement, resulting in decreased persistence, while at the same time maintaining a high cell guidance ratio.
Variation of PEG-RGDS Concentration to Manipulate Cell Migration
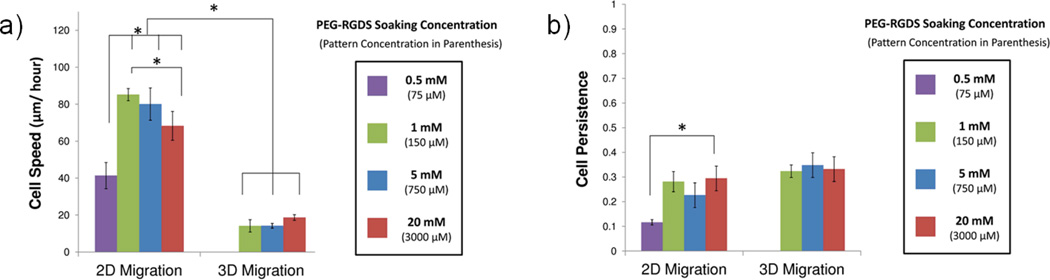
The use of TP-LSL and PEG-based hydrogels allows for the independent modulation of PEG-RGDS concentration to manipulate cell migration. The effect of various PEG-RGDS concentrations on cells migrating on hydrogel surfaces (2D) and within enzymatically degradable hydrogels (3D) was examined. Hydrogels (for both 2D and 3D) were patterned using various soaking concentrations to achieve a range of functional PEG-RGDS concentrations. Specifically, hydrogels were patterned with 0.5 mM, 1 mM, 5 mM, and 20 mM soaking concentrations of PEG-RGDS resulting in functional PEG-RGDS concentrations of 75 µM, 150 µM, 750 µM, and 3000 µM, respectively. Cell migration was then observed via time-lapse microscopy, and each cell was analyzed for cell speed and cell persistence. HT1080s migrating on 2D surfaces were observed to be significantly affected by changing PEG-RGDS concentration. Cell speed varied biphasically, with the maximum cell speed occurring at intermediate PEG-RGDS concentrations (Figure 5a). This observation is in agreement with previous studies of 2D cell migration that report a biphasic dependence of cell speed on adhesive molecule concentration due to the balance of forces between leading cell edge attachment and the trailing cell edge detachment6,17. Also in agreement with previous results, migration speed was significantly lower for 3D cell migration than 2D cell migration at every tested PEG-RGDS concentration. The cell speed of HT1080s migrating in 3D was not greatly influenced by PEG-RGDS as it was in 2D. When examining cell persistence (Figure 5b), cells on 2D surfaces exhibited a lower persistence at the lowest concentration of PEG-RGDS, but a persistence trend was not detected above this threshold level. The persistence of HT1080s migrating in 3D was also not significantly related to changes in PEG-RGDS concentration.
Fig. 5.
To evaluate the effect of PEG-RGDS concentration, HT1080 cells were tracked while migrating on both PEG-DA surfaces with 100 µm wide patterns of PEG-RGDS (2D) and within PEG-PQ hydrogels with 100 × 100 µm areas of the same adhesive ligand (3D). The initial soaking concentrations of PEG-RGDS are displayed with the functional concentrations of patterned PEG-RGDS below in parenthesis. (a) Cell speed in 2D had biphasic dependence on ligand concentration, while in 3D it did not vary with PEG-RGDS concentration. Notably, cell speed was higher in 2D than in 3D for every PEG-RGDS concentration. (b) Cell persistence increased in 2D between low and high PEG-RGDS concentrations but this effect was not evident in 3D. Cell persistence did, however, trend higher in 3D than in 2D. (* p <0.05).
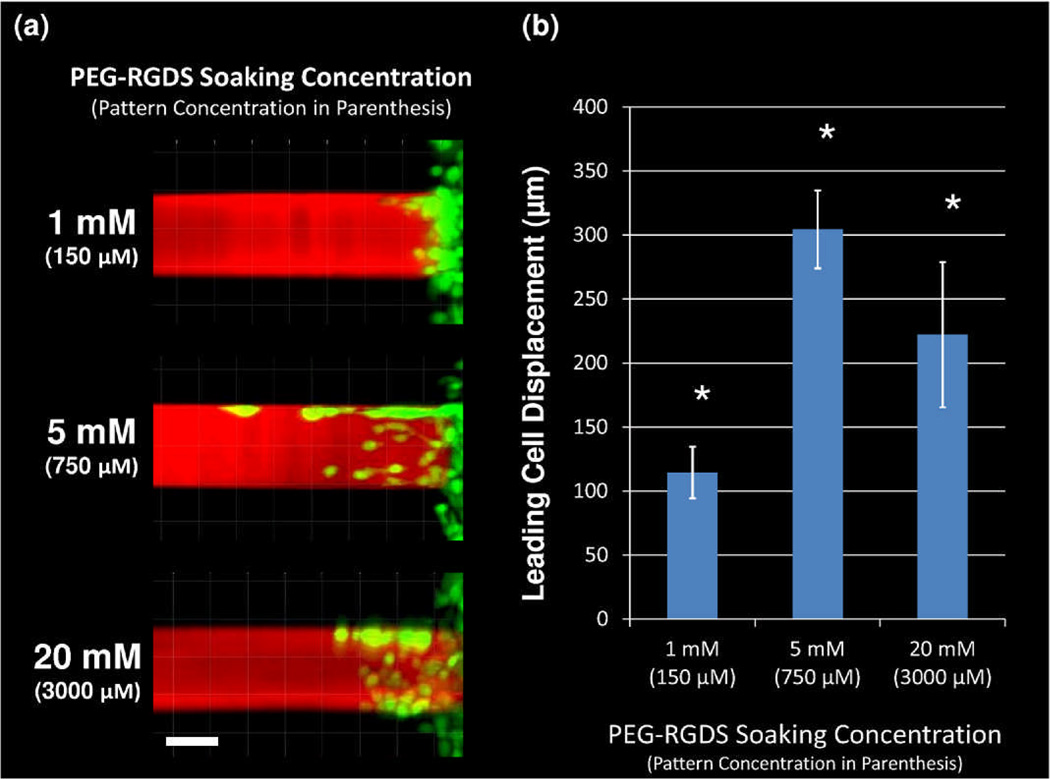
While variation of PEG-RGDS concentration did not affect 3D cell migration in terms of speed or persistence, a significant dependence was observed in terms of total displacement of the leading cell into the PEG-RGDS pattern. Specifically, migrating HT1080s were imaged for each different PEG-RGDS concentration exactly 12 h after cell encapsulation next to the 3D micropatterns (Figure 6a). The leading cell (the cell which had migrated farthest into the pattern) invaded a significantly greater distance into the pattern at an intermediate concentration of PEG-RGDS (750 µM functional concentration) than at either a high or low PEG-RGDS concentration. Upon quantification (Figure 6b), the displacement of leading cell migration in an intermediate PEG-RGDS micropattern was determined to be over 300 µm. Contrastingly, the leading cell in the high PEG-RGDS concentration migrated 220 µm into the pattern, while the leading cell in the low PEG-RGDS concentration migrated only 115 µm. This biphasic dependence of maximal leading cell displacement on PEG-RGDS concentration may reflect that leading cells are better able to degrade and migrate through the PEG-PQ hydrogel when ligated by an intermediate concentration of adhesive ligand.
Fig. 6.
(a) After 1 day, HT1080s (green) have migrated into 3D micropatterns of varying concentrations of PEG-RGDS (red) within PEG-PQ hydrogels. The soaking concentration of PEG-RGDS is indicated with the functional PEG-RGDS concentration of each pattern below in parenthesis. All images are representative. Scale bar = 50 µm. (b) The total displacement of the leading cell along each 3D micropattern has a significant (*p<0.05) biphasic dependence on PEG-RGDS concentration.
Cell Speed and Persistence within 3D Hydrogels Increase over Time
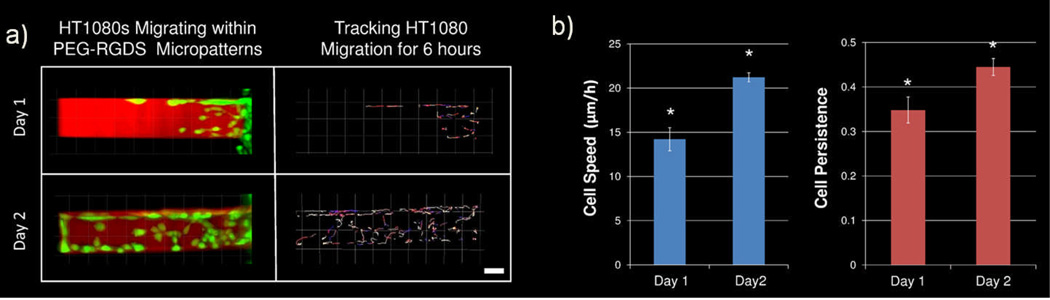
HT1080 cell migration within 3D PEG-PQ hydrogels was studied at early and late time points in order to determine the effects of the passage of time within the system (Figure 7). After 12 h (day 1) cells had moved into the micropattern and were taken to a confocal microscope and monitored for migration via time-lapse microscopy. HT1080s were also monitored for migration 36 h (day 2) after cell encapsulation. Representative images of HT1080s migrating through PEG-RGDS patterns at day 1 and day 2 are shown in Figure 7a. An increased number of cells were present at day 2 and cells had migrated all the way to the end of the 500 µm long pattern. Cells were tracked on both day 1 and day 2 with the passage of time indicated by the changing colour of the tracking lines with early times indicated by blues and reds and later times indicated by yellows and whites. Both cell speed and cell persistence were seen to be significantly increased on day 2 as opposed to day 1 (Figure 7b). Specifically, cell speed was increased from an average of 14 µm/h on day 1 to 22 µm/h on day 2. Cell persistence was also increased from 0.34 to 0.45, indicating that a larger percentage of cell movement contributed to overall displacement from the cell starting position.
Fig. 7.
(a) HT1080s (green) are shown migrating within PEG-RGDS micropatterns (red) both 1 day and 2 days after cell seeding. HT1080 cells were then tracked for 6 h on both days, with the passage of time indicated by colour changes from blue to red to yellow to white. All images are representative. Scale bar = 50 µm. (b) Cell tracking data was used to calculate cell speed and cell persistence on both day 1 and day 2 with both significantly increased (*p<0.05) over this time period.
Hydrogel Composition Affects Leading Cell Displacement into Hydrogel
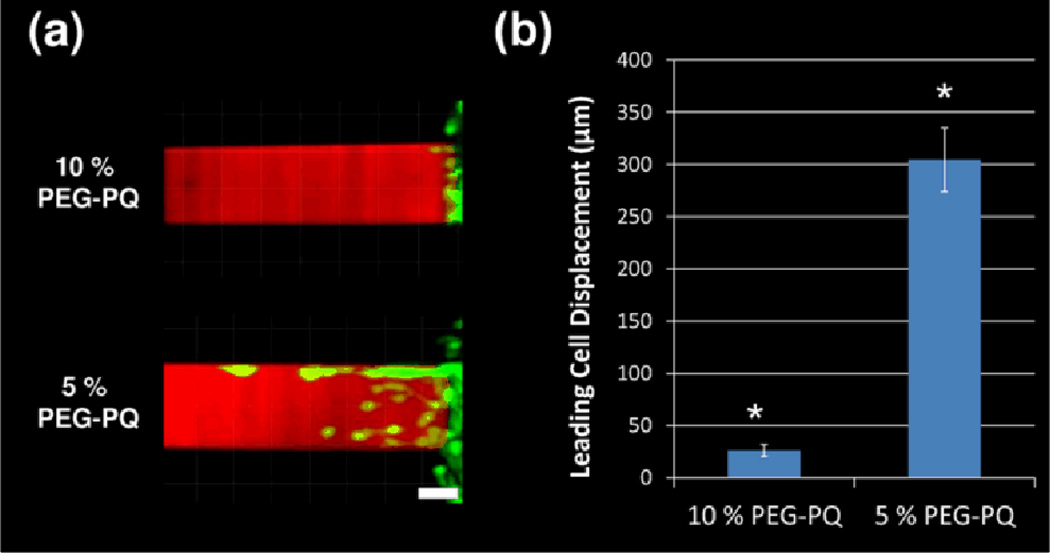
The PEG-based hydrogel system allows for the straightforward variation of the hydrogel composition while keeping the concentration of adhesive ligand constant. PEG-PQ hydrogels of 2 different polymer concentrations (5% and 10%), and thus stiffnesses, were micropatterned with PEG-RGDS channels and 3D cell migration was again accessed via time-lapse confocal microscopy. Representative images of HT1080 migration into the patterned hydrogels 12 h after cell encapsulation are shown in Figure 8a. Very few cells were observed to migrate into the 10% PEG-PQ hydrogels (elastic modulus of 42 kPa) while migration into the 5% PEG-PQ hydrogels (elastic modulus of 21.5kPa) was robust. Due to a lack of cells migrating in 10% hydrogels, cell speed and persistence were not quantified. Rather, to quantify differences in cell migration, the total displacement of the leading cell into the PEG-RGDS pattern for each of the hydrogels was measured. Leading cell displacement was approximately 12 times greater for 5% PEG-PQ hydrogels as compared to 10% PEG-PQ hydrogels (300 µm vs. 25 µm) (Figure 8b).
Fig. 8.
(a) After 1 day, HT1080s (green) have migrated into 3D micropatterns of PEG-RGDS (red) within PEG-PQ hydrogels of 2 different polymer weight percents. All images are representative. Scale bar = 50 µm. (b) The total displacement of the leading cell along each 3D micropattern is significantly (*p<0.05) greater in 5 % PEG-PQ hydrogels as compared to 10% PEG-PQ hydrogels.
Discussion
New technologies to investigate complex biological phenomena have become increasingly important across the bioengineering discipline. More specifically, a detailed understanding of cell motility will be critical to both the advancement of tissue engineering as well as the discovery of new therapeutic strategies to combat disease1. In this work, TP-LSL has been applied to systematically study cell migration in 2 and 3 dimensions through the micropatterning of PEG-based hydrogels. Throughout the studies presented here, fibrosarcoma-derived HT1080 cells were utilized as a model cell line for several important reasons. First, HT1080s migrate rapidly and release large amounts of proteases which allows for significant 3D migration through PEG-PQ hydrogels within a relatively short time period. HT1080s are also a highly invasive cancer cell line that can be employed to study the unique migratory behaviours of metastatic tumour cells18. Additionally, HT1080s are commonly used to study cell motility in 3D19,18,20, and the results of the current work may be directly compared to both previous and future results.
In this work, HT1080s were observed to migrate at faster speeds and with lower persistence on 2D PEG-DA hydrogel surfaces when compared to 3D migration within PEG-PQ hydrogels. Further, cells migrating in 3D were smaller and more elongated than those in 2D. These observations are due to the steric hindrances placed on cells migrating within a 3D matrix. In 2D, cells are free to spread out and move around on a flat surface without having to degrade a matrix and then navigate over and around 3D obstacles. Without these hindrances, cells in 2D move faster and reverse direction more rapidly, leading to decreased persistence. Cells migrating in 3D however must secrete enzymes to degrade the hydrogel before they can migrate. Even after local matrix degradation, cells must deform through irregular 3D pathways, leading to a slower speed, a more elongated shape, and more difficulty reversing direction.
The use of TP-LSL in combination with PEG-based hydrogels to investigate the differences between 2D and 3D cell migration offered several advantages over traditional approaches. In traditional studies comparing 2D and 3D cell migration, confounding factors make it difficult to draw direct conclusions or trends. For example, when comparing 2D tissue culture plastic to 3D naturally derived matrix, cells experience differences in substrate stiffness, pore size, proteolytic susceptibility, adhesive ligand concentration, and biochemical growth factors1. With this many variables, it is difficult to parse out the cause of observed differences in cell migration. In the PEG-based hydrogel system presented here, identical concentrations of PEG-RGDS are presented in both 2D and 3D. Additionally, the non-adhesive nature of PEG ensures that no other biochemical factors influence the results. Finally, the 5% PEG-DA (6000 Da) and 5% PEG-PQ (~7800 Da) hydrogels used in this study have similar mechanical properties (elastic modulus ~ 20–25 kPa) and pore sizes (on the order of nanometres). Therefore, the only true differences between 2D and 3D cell migration in this study are the effects of the biomaterial matrix on all sides of the cells (3D) rather than only below them (2D). The TP-LSL system also has the advantage of increasingly precise guidance of cells in both 2D and 3D. The dimensions of PEG-RGDS patterns were narrowed from 100 µm to 10 µm so that cell persistence and cell migration ratio were increased and the cell direction was precisely guided. This ability to precisely guide cells in 3D will be important for a variety of purposes as research continues. For example, the ability to dictate exactly where cells migrate will be critical for testing how cells respond in controlled biomaterial microenvironments such as engineered cancer models or biomaterial tissue constructs.
This work also investigated the effects of varying the concentration of PEG-RGDS on both 2D and 3D cell migration. On patterned 2D hydrogel surfaces, cell speed was seen to have a biphasic dependence on PEG-RGDS concentration. These results have been well documented on a variety of surfaces6,17, and are a result of the balance of forces between leading cell edge attachment and the trailing cell edge detachment during cell migration. Cell persistence on 2D surfaces was not strongly correlated with PEG-RGDS concentration, a result also in agreement with previous studies1,21. For cells migrating within enzymatically degradable PEG-PQ hydrogels, PEG-RGDS concentration did not significantly affect cell speed or persistence. This result initially appears to be in conflict with previous studies that have reported a biphasic dependence of adhesive ligand concentration on cell speed and persistence for cells migrating in 3D 4,22. However, PEG-RGDS concentration did have a biphasic effect on the displacement of the leading cell migrating into the PEG-RGDS micropatterns, with the maximum cell displacement occurring at intermediate PEG-RGDS concentrations. A similar result was reported by Gobin et al. who utilized a modified Boyden chamber assay to demonstrate that a larger number human dermal fibroblasts migrated the required distance when an intermediate concentration of PEG-RGDS was utilized in PEG hydrogels. The leading cell displacement results in the current study as well as the results reported by Gobin et al. focus only on the farthest cells migrating at an end point of the assay. In each of these cases, leader cells degrade a pathway within the matrix for later migrating cells to follow. Functionally, this means that leading HT1080 cells follow the reported biphasic model4,22 as they are better able to degrade and move through the PEG-PQ hydrogel when ligated by an intermediate concentration of adhesive ligand. HT1080s that migrate into the hydrogel after the leading cells were observed to follow behind leading cells where the matrix has already been degraded. These cells may migrate via a different mechanism in which cell speed is not dependent on PEG-RGDS concentration. In fact, HT1080s have previously been reported to possess the ability to migrate via an amoeboid-like mechanism that was only weakly dependent on cell adhesion18.
3D cell migration was also investigated both 1 day and 2 days after initial encapsulation of cells. Cells migrating within micropatterns of a fixed level of PEG-RGDS demonstrated increases in cell speed and cell persistence on day 2 as opposed to day 1: a result that is explained by the increased degradation of the hydrogel within the PEG-RGDS pattern as cells continue to invade and migrate over an additional 24 h. Hydrogel degradation leads to increased pore size and reduces the need for cells to secrete enzymes to disperse the matrix before moving. This result is in agreement with previous work that has demonstrated increased MMP activity increases cell persistence in 3D collagen matrices23.
Finally, the displacement of leading cells was investigated in PEG-PQ hydrogels of different weight percents. Cell displacement into the PEG-RGDS micropattern was significantly increased in 5% PEG-PQ hydrogels as opposed to 10% hydrogels after 12 h. This effect is due to the increased number of PEG chains that must be degraded within higher percentage PEG-PQ hydrogels before cells are able to move. Additionally, the increased mechanical stiffness of a 10% hydrogel compared to a 5% hydrogel also plays a role in decreased cell displacement as high substrate stiffness has previously been shown to decrease cell speed in 3D4.
The experiments and analysis presented in this work have been specifically selected to shed light on some of the core questions regarding cell motility. However, the studies shown here are by no means an exhaustive study of cell migration. In fact, much research remains in order to understand the complexities of how cells move within 3D microenvirenments. For example, analysis of cell extensions, cell contractability, and integrin interactions must all be investigated as cells migrate through TP-LSL fabricated micropatterns within biomaterials in order to more completely comprehend cell migration processes. Fortunately, the TP-LSL patterning technique presented here utilizes a conventional two-photon microscope commonly available in most research facilities, and therefore, future investigation into cell migration should be readily translated into biological research laboratories across the world. Overall, the research presented here has provided a valuable new technical innovation that opens the door for a wide range of biomimetic experiments that will soon lead to a more complete understanding of cell motility.
Conclusion
In this work, TP-LSL and PEG-based hydrogels have been utilized to guide cell migration under controlled conditions in order to quantitatively study cell motility. PEG-RGDS channels were micropatterned within PEG-DA and PEG-PQ hydrogels and the migration of HT1080 cells were tracked in both 2 and 3 dimensions. HT1080s were shown to migrate faster and with lower persistence on 2D surfaces, while cells migrating in 3D were smaller and more elongated. Further, cell migration was shown to have a biphasic dependence on PEG-RGDS concentration and cells moving within PEG-RGDS micropatterns were seen to move faster and with more persistence over time. Finally, markedly less cell displacement into PEG-RGDS micropatterns was seen for PEG-PQ hydrogels with higher weight percents. These results begin to parse out many of the traditional confounding factors that affect cell migration and establish TP-LSL as a strong tool to probe cell behaviour within 3D engineered biomaterials.
Supplementary Material
Acknowledgments
This work was supported by NIH ROI grants R01 EB005173 and R01 HL097520.
Footnotes
Electronic Supplementary Information (ESI) available: [Supplementart Figures]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Kim H-D, Peyton SR. Integrative Biology. 2012;4:37. doi: 10.1039/c1ib00069a. [DOI] [PubMed] [Google Scholar]
- 2.Mierke CT, Rösel D, Fabry B, Brábek J. European journal of cell biology. 2008;87:669–676. doi: 10.1016/j.ejcb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 4.Zaman MH, Trapani LM, Siemeski A, MacKellar D, Gong HY, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle AD, Wang FW, Matsumoto K, Yamada KM. Journal of Cell Biology. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gobin AS, West JL. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2002;16:751–753. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 7.Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519–2524. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 8.Saik JE, Gould DJ, Watkins EM, Dickinson ME, West JL. Acta Biomater. 2010 doi: 10.1016/j.actbio.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon JJ, Lee SH, West JL. Biomacromolecules. 2007;8:42–49. doi: 10.1021/bm060452p. [DOI] [PubMed] [Google Scholar]
- 10.Mann BK, West JL. Journal of biomedical materials research. 2002;60:86–93. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann JC, West JL. Soft Matter. 2010;6:5056–5063. [Google Scholar]
- 12.Lee S-H, Moon JJ, West JL. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culver JC, Hoffmann JC, Poche RA, Slater JH, West JL, Dickinson ME. Adv. Mater. 2012;24:2344–2348. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn MS, Miller JS, West JL. Adv. Mater. 2006;18:2679–2684. [Google Scholar]
- 15.Leslie-Barbick JE, Moon JJ, West JL. J. Biomater. Sci., Polym. Ed. 2009;20:1763–1779. doi: 10.1163/156856208X386381. [DOI] [PubMed] [Google Scholar]
- 16.Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. FASEB J. 2007;21:790–801. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 17.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz MP, Fairbanks BD, Rogers RE, Rangarajan R, Zaman MH, Anseth KS. Integrative biology: quantitative biosciences from nano to macro. 2010;2:32–40. doi: 10.1039/b912438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tayalia P, Mendonca CR, Baldacchini T, Mooney DJ, Mazur E. Advanced Materials. 2008;20:4494–4498. [Google Scholar]
- 20.Wolf K, Mazo I, Leung H, Engelke K, Andrian UHvon, Deryugina EI, Strongin AY, Bröcker E-B, Friedl P. The Journal of cell biology. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wacker BK, Alford SK, Scott EA, Thakur MDas, Longmore GD, Elbert DL. Biophysical journal. 2008;94:273–285. doi: 10.1529/biophysj.107.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaman MH, Matsudaira P, Lauffenburger DA. Annals of Biomedical Engineering. 2007;35:91–100. doi: 10.1007/s10439-006-9205-6. [DOI] [PubMed] [Google Scholar]
- 23.Kim H-D, Guo TW, Wu AP, Wells A, Gertler FB, Lauffenburger DA. Molecular biology of the cell. 2008;19:4249–4259. doi: 10.1091/mbc.E08-05-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.